Abstract
Oncogenic signaling reprograms cancer cell metabolism to augment the production of glycolytic metabolites in favor of tumor growth. The ability of cancer cells to evade immunosurveillance and the role of metabolic regulators in T cell functions suggest that oncogene-induced metabolic reprogramming may be linked to immune escape. Epidermal growth factor (EGF) signaling, frequently dysregulated in triple-negative breast cancer (TNBC), is also associated with increased glycolysis. Here, we demonstrated in TNBC cells that EGF signaling activates the first step in glycolysis, but impedes the last step, leading to an accumulation of metabolic intermediates in this pathway. Furthermore, we showed that one of these intermediates, fructose 1,6 bisphosphate (F1,6BP), directly binds to and enhances the activity of the EGF receptor (EGFR), thereby increasing lactate excretion which leads to inhibition of local cytotoxic T cell activity. Notably, combining the glycolysis inhibitor 2-deoxy-D-glucose (2-DG) with the EGFR inhibitor gefitinib effectively suppressed TNBC cell proliferation and tumor growth. Our results illustrate how jointly targeting the EGFR/F1,6BP signaling axis may offer an immediately applicable therapeutic strategy to treat TNBC.
Keywords: EGFR; TNBC; fructose 1,6 bisphosphate; immune suppression; glycolysis
Introduction
Accelerated glycolysis is a common feature of rapidly proliferating cancer cells. Unlike normal differentiated cells, most cancer cells produce large amounts of lactate regardless of oxygen levels. This metabolic property is often referred to as “aerobic glycolysis” (1,2), a well-known metabolic reprogramming of cancer cells to sustain cell proliferation and a hallmark of cancer (3). In cancer cells, oncogenic receptor tyrosine kinase (RTK) signaling participates in metabolic reprogramming and stimulates the accumulation of cellular metabolites, which are then used as building blocks for cell growth (4). Thus, understanding the cross regulation between RTK signaling and metabolites and/or metabolic enzymes of glycolysis may shed light on new regulation of cancer cell metabolism.
Hexokinase (HK) and pyruvate kinase (PK) are two key enzymes regulating two irreversible steps in glycolysis. HK isozymes 1–4 catalyze the phosphorylation of glucose, which is the first of the two irreversible steps in glycolysis. Although HK2 is associated with cancer promotion and has been proposed as a potential therapeutic target, the detailed molecular mechanisms are not fully understood (5,6). PK catalyzes the dephosphorylation of phosphoenolpyruvate to pyruvate, which is the last irreversible step of glycolysis, and the expression of PK is frequently altered during tumorigenesis (7). The M1 isoform of PK (PKM1) is expressed in most adult tissues whereas M2 (PKM2) is exclusively expressed during embryonic development (8,9). Notably, most tumor cells re-express PKM2, suggesting that the switch from PKM1 to PKM2 leads to aerobic glycolysis, which provides a selective growth advantage in vivo (7,10).
Cancer cells have the ability to disengage immune response by inactivating cytotoxic T cell function via secretion of cytokine or immune checkpoint proteins (11,12). Interestingly, metabolic regulation has been reported to play an important role in T cell differentiation and functions (13). For instance, Myc and HIF1α, which are well-known regulators of metabolism, stimulate T cell receptor activation (14). Moreover, several glycolytic and TCA cycle metabolites, e.g., glucose, acetyl-CoA, and lactate, also regulate T cell proliferation and functions (15,16). Nonetheless, the link connecting oncogenic signaling, metabolism, and immune escape in cancer cells has not been well established.
The epidermal growth factor receptor (EGFR) is one of the major regulators of cell proliferation, cell survival, and metabolism (17). In triple-negative breast cancer (TNBC) patients, EGFR overexpression is frequently observed and associated with poor clinical outcome (18,19). TNBC, which accounts for approximately 15–20% of breast cancers in the United States, lacks the expression of estrogen receptor (ER) and progesterone receptor (PR) as well as amplification of HER2/neu and is associated with poorer outcome compared with other breast cancer subtypes (20–22). Unlike ER-positive, PR-positive, or HER2-overexpressing tumors, the lack of well-defined molecular targets and the heterogeneity of the disease pose a challenge in TNBC treatment (20,22). Clinical outcomes for anti-EGFR targeted therapy in breast cancer have been disappointing compared with those in lung, colon, and head and neck cancers (23–26), suggesting that cancer-specific mechanisms or biological functions of EGFR have yet to be discovered in TNBC.
EGF is known to accelerate glucose consumption and lactate production in cancer cells, including breast cancer (27,28). In addition, EGF-stimulated nuclear translocation of PKM2 promotes tumorigenesis and cell proliferation of glioma cells (29,30). While it has been known for two decades that EGF stimulation leads to a high rate of glycolysis in cells, how this is directly linked to EGFR is not clear yet. Here, we report an EGF/EGFR/fructose-1,6-bisphosphate (F1,6BP) signaling axis in TNBC cells that increases lactate production, which promotes immune evasion. Our findings provide a rationale for combining EGFR tyrosine kinsase inhibitor, gefitinib, with glycolysis inhibitor, 2-DG, as a potential therapeutic strategy for TNBC.
Materials and Methods
Cell culture and treatment
Breast cancer cell lines MDA-MB-468, BT-549, HS578T, BT20, MDA-MB-231, MDA-MB-436, HBL100, AU565, SkBr3, MCF7, T47D, ZR75-1, and human embryonic kidney cell line HEK 293T cells were obtained from American Type Culture Collection. Cell lines were validated by short tandem repeat DNA fingerprinting using the AmpFlSTR Identifiler PCR Amplification Kit (Applied Biosystems catalogue no. 4322288; Life Technologies) according to the manufacturer's instructions. Cells were grown in DMEM supplemented with 10% fetal bovine serum. EGF (Sigma-Aldrich) was prepared according to the manufacturers’ instructions. Cells were treated with 25 ng/ml EGF. Gefitinib (5 µM) was used to inhibit EGFR kinase activity.
Western blot analysis, immunocytochemistry, immunoprecipitation, and immunohistochemical staining
Western blot analysis, immunoprecipitation, and immunocytochemistry were performed as described previously (31). Antibody information is described in the Supplementary Table 3. Image acquisition and quantitation of band intensity were performed using Odyssey® infrared imaging system (LI-COR Biosciences). Immunohistochemical staining (IHC) was performed as previously described (32). To validate the specificity of phospho-Y148-PKM2 antibody in IHC, we performed peptide competition assay by staining human breast tumor sample with phospho-Y148-PKM2 antibody blocked with mock or phospho-Y148-PKM2-peptide or nonphospho-Y148-PKM2-peptide. Duolink® II fluorescence assay was performed as described by the manufacturer (Olink Bioscience, Sweden). In vitro kinase assays were performed as described in Supplementary Information.
Generation of stable cells using lentiviral infection
Human PKM2 ORF clone was obtained from the shRNA/ORF Core Facility (MD Anderson Cancer Center) and cloned into pCDH lentiviral expression vector to establish Flag-PKM2 expression cell lines. The lentiviral-based shRNA (pGIPZ plasmids) used to knockdown expression of PKM2 was purchased from the shRNA/ORF Core Facility (MD Anderson Cancer Center). pGIPZ-shPKM2/Flag-PKM2 dual expression construct to knock down endogenous PKM2 and to reconstitute Flag-PKM2 (by creating a silent mutant which resist to shPKM2) was performed as described in Supplementary Information.
Orthotopic xenograft breast cancer model and treatment
All animal procedures were conducted under the guidelines approved by the Institutional Animal Care and Use Committee (IACUC) at MD Anderson Cancer Center. Female Nu/Nu nude and BALB/c mice were used as hosts for tumor xenografts and 4T1 syngeneic model, respectively. Briefly, orthotopic breast cancer mouse model was established as previously described (33). For MDA-MB-468 PKM2WT and PKM2Y148F cells (1 × 106 cells in 50 µL medium mixed with 50 µL Matrixgel™ (BD Biosciences) were injected into the left and right mammary fat pad, respectively. For 4T1 syngeneic model, PKM2WT- and PKM2Y148F-expressing mouse 4T1-luc cells (5 × 104 cells in 50 µL medium mixed with 50 µL Matrixgel™) were injected to the mammary fat pad. Tumor was measured weekly with a caliper, and tumor volume was calculated by the formula: π/6 × length × width2. For drug treatment, mice were treated with daily oral doses 500 mg/kg 2-DG and 10 mg/kg gefitinib for two weeks (5 days per week). Tumor infiltration lymphocyte profiles in excised tumors were analyzed as described in Supplementary Information.
T cell-mediated tumor cell killing assay
T cell-mediated tumor cell killing assay was performed according to the manufacturer’s protocol (Essen Bioscience). To analyze the killing of tumor cells by T cell inactivation, nuclear restricted RFP expressing tumor cells were co-cultured with activated primary human T cells (Stemcell Technologies) in the presence of caspase 3/7 substrate (Essen Bioscience). T cells were activated by incubation with anti-CD3 antibody (100 ng/ml) and IL-2 (10 ng/ml). After 96 h, RFP and green fluorescent (NucView™ 488 Caspase 3/7 substrate) signals were measured. Green-fluorescent cells were counted as dead cells. Co-culture of Jurkat T cells and tumor cells was performed as described previously (34). Secreted IL-2 level in medium was measured according to the manufacturer’s protocol (Human IL-2 ELISA Kits, Thermo Scientific).
qRT–PCR assays
qRT–PCR assays were performed to measure the expression of mRNA and mature miRNAs as previously described (35). To measure the expression of mRNA or miRNAs, cDNA was synthesized from 1 µg purified total RNA by SuperScript III cDNA synthesis system using random hexamers (Invitrogen) according to the manufacturer’s instructions. For detection of mature miRNAs, TaqMan MicroRNA assay kits for hsa-miR-143, hsa-miR-125a, and hsa-miR-125b (Applied Biosystems) were used following the manufacturer’s protocol. All data analysis was performed using the comparative CT method. Results were normalized to the internal control β-actin mRNA or U6 snRNA.
ECAR/OCR, lactate, pyruvate kinase activity, and glucose uptake measurement
Extracellular acidification (ECAR) and oxygen consumption rate (OCR) were measured using extracellular flux analyzer (XF96) analyzer (Seahorse Bioscience) as described by the manufacturer. More details can be found in Supplementary Information. Lactate production (Lactate Assay kit) and pyruvate kinase activity (Pyruvate Activity Assay kit) were measured as described by the manufacturer (BioVision). To measure glucose uptake, cells were incubated with a fluorescent D-glucose derivative, 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxy-D-glucose (2-NBDG) (Invitrogen) for 10 min after EGF and/or gefitinib treatment. The fluorescence intensity of 2-NBDG was measured using a microplate reader Synergy H4 (BioTeK) and normalized to that of 2-NBDG by total protein amount.
Mass spectrometry
Mass spectrometric analysis for protein identification was performed as previously described (35). To identify PKM2 phosphorylation sites, we purified FLAG-PKM2 with M2 resin (Sigma) that co-expressed with EGFR in 293T cells and analyzed the results by SDS-PAGE and Western blot. The protein band corresponding to PKM2 was excised and subjected to in-gel digestion with trypsin. Samples were analyzed by nanoelectrospray mass spectrometry, using an Ultimate capillary LC system (LC Packings) coupled to a QSTARXL quadrupole time-of-flight mass spectrometer (Applied Biosystem/MDS Sciex).
LC-MS/MS metabolomic analysis
LC-MS/MS metabolomic analysis was performed from dried samples reconstituted in 100 µL 5 mM ammonium acetate in 95% water/5% acetonitrile + 0.5% acetic acid prior to LC-MS analysis. More details can be found in Supplementary Information.
Statistical analysis
Data in bar graphs represents mean fold change relative to untreated or control groups with standard deviation of three independent experiments. Statistical analyses were performed using SPSS (Ver. 20, SPSS Inc. Chicago, IL). The correlation between protein expression and TNBC subset was analyzed using Spearman’s correlation and Mann-Whitney test. Student’s t test was performed for experimental data. A P value < 0.05 was considered statistically significant. Asterisk indicates statistically significant by Student’s t test. All error bars are expressed as ±SD of 3 independent experiments.
Results
EGF signaling stimulates glycolysis in TNBC cells
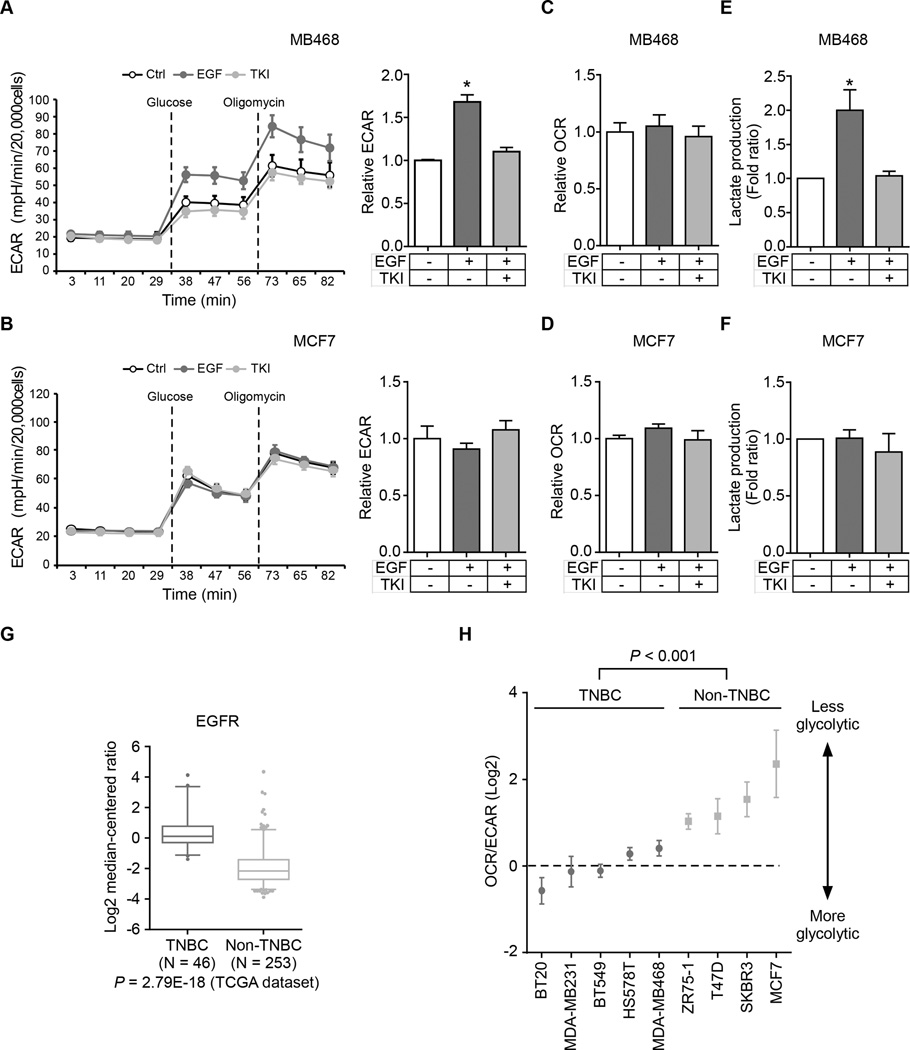
To test whether EGF affects glycolysis or mitochondrial respiration through EGFR, we measured the metabolic profile of an EGFR-overexpressing breast cancer cell line MDA-MB-468. Under EGF stimulation, these cells showed higher glycolytic activities, as indicated by the increased extracellular acidification rate (ECAR), which was attenuated by co-treatment with EGFR-tyrosine kinase inhibitor (TKI) gefitinib (Fig. 1A). In contrast, EGF had no effect on glycolysis in low EGFR-expressing MCF7 breast cancer cells (Fig. 1B). We also measured the O2 consumption rate (OCR) and lactate production in MDA-MB-468 and MCF7 cells. We did not observe any differences in OCR (Fig. 1C and D); however, lactate production (Fig. 1E and 1F) and glucose uptake (Supplementary Fig. S1A) increased only in MDA-MB-468 cells, and the addition of gefitinib eliminated the differences. Both TCGA database (patient samples; Fig. 1G and Supplementary Fig. S1B) and Western blot (cell lines; Supplementary Fig. S1C) analyses indicated higher EGFR expression in TNBC than in non-TNBC, and therefore we further analyzed the changes in glycolysis in TNBC and non-TNBC cell lines under EGF stimulation. Similar to the results observed in MDA-MB-468 and MCF7 cells, five TNBC and one non-TNBC cell line expressing EGFR also had increased glycolysis and lactate production but not OCR under EGF treatment (Supplementary Fig. S1D). To measure the relative contribution of glycolysis and mitochondrial respiration in energy production, we compared the OCR to ECAR ratio in nine breast cancer cell lines and found that TNBC cell lines exhibited more glycolytic phenotype compared with non-TNBC cell lines (Fig. 1H), indicating that EGF signaling enhances glycolysis but not oxidative phosphorylation in TNBC cells. In addition, other EGFR-expressing cancer cells, such as A431 and SW48, also showed enhanced aerobic glycolysis by EGF treatment (Supplementary Fig. S1E). Together, these results implied that EGF signaling specifically stimulates aerobic glycolysis in EGFR-expressing cancer cells. EGFR is highly expressed in TNBC for which no effective clinical treatment is currently available (18,19). Therefore, all subsequent investigations focused on EGFR-expressing TNBC cells although the outcome may apply to other EGFR-expressing cancer cells.
Figure 1.
EGF signaling stimulates glycolysis in TNBC cells. (A and B) ECAR, an indicator of glycolysis, was measured in MDA-MB-468 (A) or MCF7 (B) cells following the addition of glucose (10 mM) or oligomycin (1 µM) in the presence of EGF and/or TKI. Right, relative ECAR of EGFR before and after glucose injection. Ctrl, non-treated control cells; EGF, EGF treated cells; TKI, EGF and TKI co-treated cells. (C and D) OCR was measured in MDA-MB-468 (C) or MCF7 (D) following the addition of oligomycin (1 µM) or FCCP (0.5 µM) in the presence of EGF and/or TKI. (E and F) Lactate production was measured in the medium of MDA-MB-468 (E) or MCF7 (F) cells treated with EGF and/or TKI. (G) A Box-and-Whisker plot of EGFR mRNA expression in TNBC and non-TNBC tumors from the TCGA dataset using Oncomine™. (H) OCR/ECAR values in breast cancer cell lines following the addition of oligomycin (1 µM).
EGFR binds to and phosphorylates PKM2 to inhibit its activity
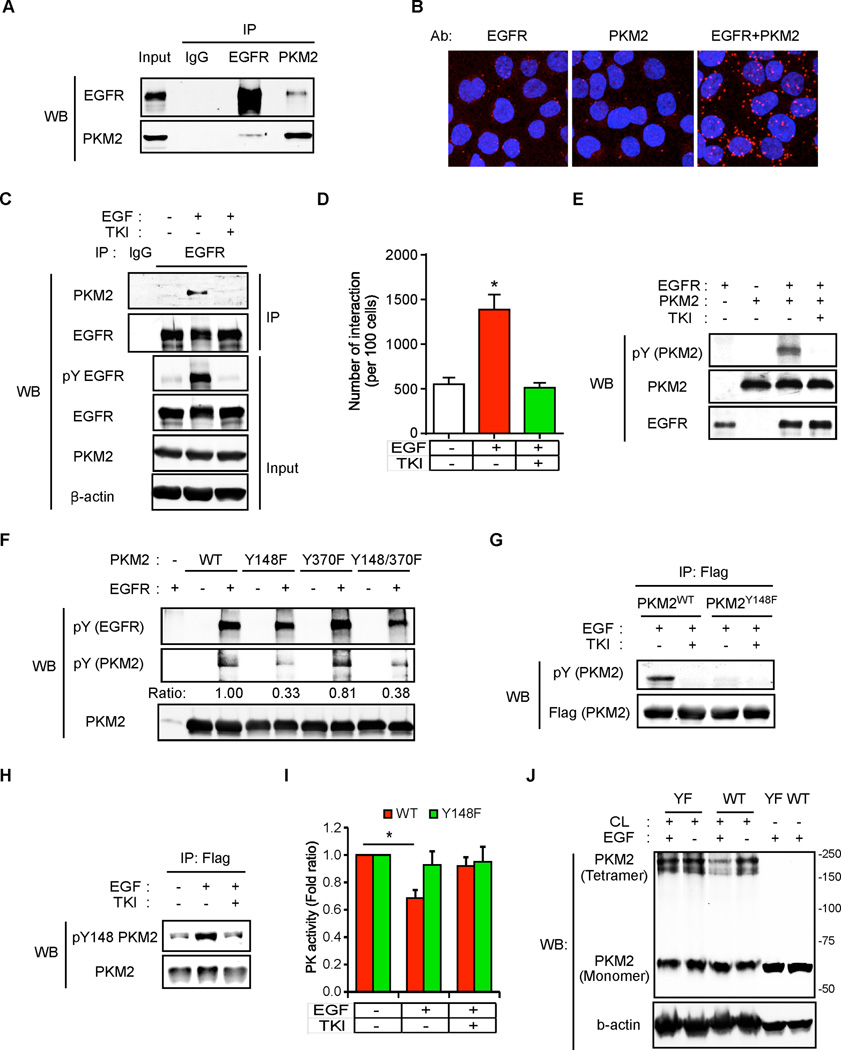
To understand how EGFR regulates glycolysis, we analyzed the EGFR-binding proteins that we previously reported (36) using Ingenuity® Pathway Analysis (IPA) and identified four glycolytic proteins: HK1, phosphofructokinase, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and PKM2. We then validated their interaction with EGFR by endogenous co-immunoprecipitation followed by Western blot and demonstrated that HK1 and PKM2 interacted specifically with EGFR (Supplementary Fig. S2A). However, only the interaction between PKM2 and EGFR was increased by EGF and inhibited by gefitinib as shown by co-immunoprecipitation (Fig. 2A and C) and Duolink® assay (Fig. 2B and D, and Supplementary Fig. S2B). These results indicate that PKM2 is a novel binding partner of EGFR.
Figure 2.
EGFR binds to and phosphorylates PKM2 to inhibit its activity. (A) Lysates from EGF-treated MDA-MB-468 cells were subjected to co-immunoprecipitation using EGFR and PKM2 antibodies followed by Western blotting. IgG, negative control. (B) EGF-treated MDA-MB-468 cells were immunostained with EGFR and/or PKM2 antibodies and assessed using Duolink® II assay as indicated. Red foci indicate interactions between endogenous EGFR and PKM2 proteins. EGFR or PKM2 antibody staining alone served as negative control. (C) Lysates from EGF and/or TKI treated MDA-MB-468 cells were subjected to immunoprecipitation using EGFR antibody followed by Western blotting. IgG, negative control. (D) EGF and/or TKI treated MDA-MB-468 cells were immunostained with EGFR and PKM2 antibodies and then subjected to Duolink® II assay. The number of red foci as described in (B) was calculated based on three randomly selected fields and normalized by nuclear number (per 100 cells). (E) In vitro kinase assay was performed using purified full-length recombinant His-PKM2 and commercially available purified EGFR. PKM2 phosphorylation was detected by phospho-Tyr antibody. (F) In vitro kinase reaction products of mutant PKM2 Y148F and/or Y370F detected by phospho-Tyr antibody. (G) EGF and/or TKI treated Flag-PKM2WT- or PKM2Y148F-expressing MDA-MB-468 cells were immunoprecipitated with Flag M2 affinity resin. Flag-PKM2 proteins were eluted by Flag peptide followed by Western blotting. (H) Flag-PKM2WT-expressing MDA-MB-468 cells were treated with EGF and/or TKI and were immunoprecipitated with Flag M2 affinity resin. Flag-PKM2 proteins were eluted by Flag peptide followed by Western blotting using phospho-Y148 PKM2 antibody. (I) PK activities of Flag-PKM2WT- or PKM2Y148F-expressing MDA-MB-468 cells treated with EGF and/or TKI. (J) EGF-treated Flag-PKM2WT- or PKM2Y148F-expressing MDA-MB-468 cells were cross-linked by glutaraldehyde followed by Western blotting. CL, cross-linker glutaraldehyde.
The interaction between EGFR and PKM2 prompted us to ask whether PKM2 is a phosphorylation substrate of EGFR. As shown in Figure 2E, PKM2 can be phosphorylated by commercially available purified EGFR protein, and this phosphorylation was abolished by gefitinib treatment. To determine the phosphorylation site(s), a tandem mass spectrometric analysis was performed which identified two PKM2 Tyr phosphorylation sites (Y148 and Y370) in EGF-treated cells (Supplementary Fig. S2C). To further characterize these two sites, we generated three PKM2 mutants, PKM2 Y148F, PKM2 Y370F, and PKM2 Y148F/Y370F. Interestingly, phosphorylation of PKM2 Y148F but not Y370F mutant was significantly decreased compared to PKM2 wild type (WT) by an in vitro EGFR kinase assay (Fig. 2F). To measure PKM2 Y148 phosphorylation in vivo, we first established PKM2 WT (PKM2WT) or Y148F mutant (PKM2Y148F) in PKM2-knockdown MDA-MB-468 cells by using a dual-expression construct to knockdown endogenous PKM2 by shRNA and then re-express exogenous Flag-PKM2 (Supplementary Fig. S2D). As expected, phosphorylation of PKM2 was readily detectable in PKM2WT-expressing cells and blocked by gefitinib, but not in PKM2Y148F-expressing cells (Fig. 2G). A phospho-Y148 PKM2 antibody that we generated preferentially recognized phospho-PKM2WT but not PKM2Y148F (Fig. 2H and Supplementary Fig. S2E–S2G), indicating that Y148 is a major EGFR phosphorylation site. Interestingly, EGF treatment decreased the PK activity of PKM2 and of its tetramer, an active form of PKM2 in PKM2WT-expressing but not in PKM2Y148F-expressing PKM2-knockdown MDA-MB-468 cells (Fig. 2I and J and Supplementary S2H). Together these data suggest that EGFR phosphorylation of PKM2 at Y148 reduces active PKM2 tetramer and inhibits its PK activity.
EGF signaling reprograms cancer cell metabolism by PKM2 phosphorylation
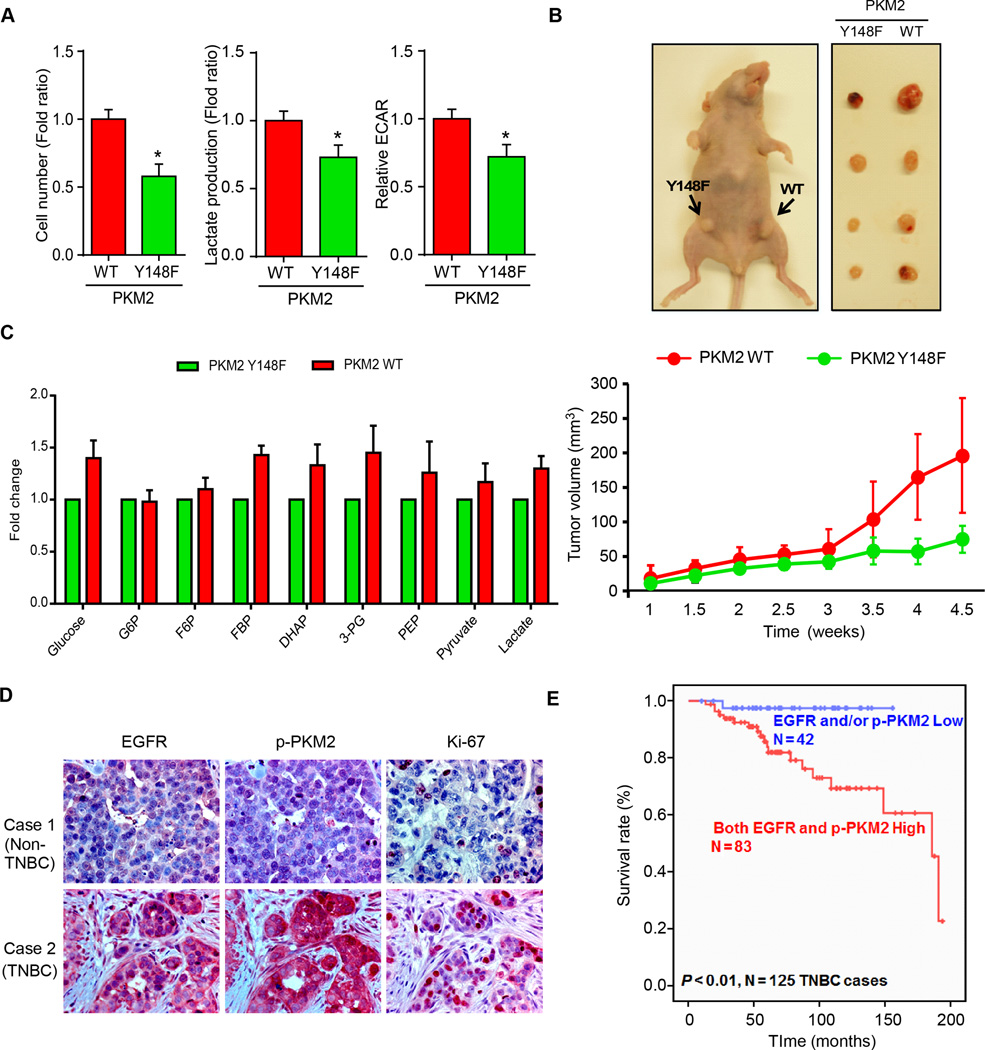
Next, we investigated whether EGFR-mediated phospho-Y148 PKM2 affects biological activities, such as cell proliferation, in vitro and in vivo. Cell proliferation, glycolysis, and lactate production were higher in PKM2WT than in PKM2Y148F cells (Fig. 3A). Furthermore, tumor volumes derived from PKM2WT-expressing BT549 or MDA-MB-468 cells were significantly larger compared with those expressing PKM2Y148F (Fig. 3B and Supplementary Fig. S3A). Since PKM2 regulates the last step of glycolysis, and decreased PKM2 activity leads to an accumulation of glycolytic intermediates and promotes tumor growth (37,38), we also profiled the intracellular glycolysis-related metabolites in EGF-treated PKM2WT and PKM2Y148F cells. Indeed, the levels of most glycolytic intermediates were higher in PKM2WT-expressing MDA-MB-468 cells compared to those expressing PKM2Y148F (Fig. 3C). To further determine the clinical relevance, we analyzed the relationship between phosho-Y148 PKM2, EGFR, and proliferation marker Ki-67 in human breast cancer tissues by immunohistochemistry staining (IHC). A positive correlation was identified between PKM2 Y148 phosphorylation, EGFR expression, and Ki67 in human TNBC but not in non-TNBC tissues (Fig. 3D and Supplementary Tables 1 and 2). High levels of phospho-PKM2 and EGFR also correlated with poorer survival in TNBC but not in non-TNBC patients (Fig. 3E and Supplementary Fig. S3B). Together, these results support that EGFR-induced phosphorylation at PKM2 Y148 confers a proliferation advantage in TNBC cells.
Figure 3.
EGFR reprograms cancer cell metabolism by PKM2 phosphorylation in TNBC cells. (A) Cell proliferation, glycolysis (ECAR), and lactate production were measured in EGF-treated PKM2WT- or PKM2Y148F-expressing MDA-MB-468 cells. (B) Top, nude mice were injected with PKM2WT- and PKM2Y148F-expressing BT549 cells in mammary glands as shown. Tumors were measured and dissected at the endpoint. Bottom, tumors were measured at the indicated time points. N = 8 per group. (C) Fold changes of glycolytic intermediates in EGF-treated MDA-MB-468 cells expressing PKM2WT or PKM2Y148F. G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; DHAP, dihydroxyacetone phosphate; 3-PG, 3-phosphoglyceric acid; PEP, phosphoenolpyruvate. (D) Representative images of IHC staining of EGFR, phospho-Y148 PKM2, and Ki-67 in non-TNBC (case 1) and TNBC (case 2) tumor tissues. (E) Kaplan-Meier survival curve of EGFR and phospho-PKM2 in TNBC tissues (N = 125, P < 0.01).
Upregulation of HK2 expression by EGF signaling also enhances aerobic glycolysis
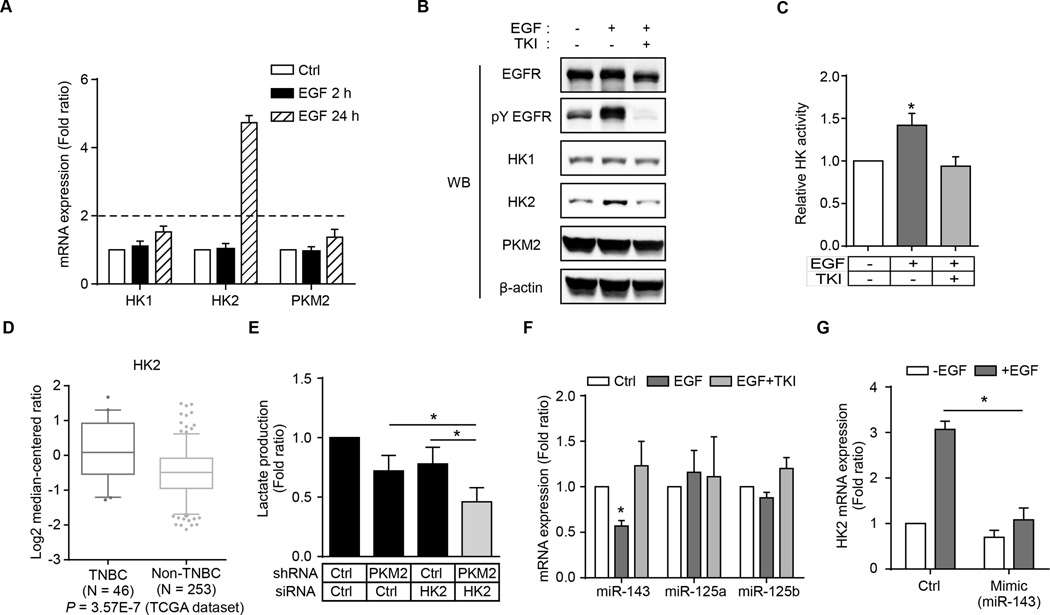
Knockdown of PKM2 abolished EGF-enhanced lactate production under short (2 h) EGF treatment (Supplementary Fig. S4A). Unexpectedly, however, knocking down PKM2 did not abolish EGF-enhanced lactate production under longer (24 h) EGF treatment (Supplementary Fig. S4A). Furthermore, the metabolic intermediates, such as amino acids and nucleotide precursors, were significantly increased in cells exposed to longer EGF treatment (Supplementary Fig. S4B and S4C). These results imply that other mechanisms at a later stage may also be involved in EGF signaling-induced aerobic glycolysis in addition to the early stage PKM2 phosphorylation. To identify these later-stage mechanisms, we asked whether the expression pattern of glycolytic enzymes correlated with EGFR expression in human breast cancer specimens. Analysis of TCGA database using Oncomine™ revealed that several glycolytic enzymes were significantly increased in TNBC tissues and positively correlated with EGFR expression (Supplementary Fig. S4D). To further validate these results, we analyzed mRNA expression of a number of glycolytic enzymes in EGF-treated TNBC and non-TNBC cells. Interestingly, the levels of mRNA and protein expression of HK2, but not HK1, were significantly increased by EGF treatment (24 h) in both MDA-MB-468 and BT549 TNBC cells (Fig. 4A and B and Supplementary Fig. S4E and F). In addition, EGF stimulation also enhanced HK activity (Fig. 4C). From the TCGA dataset, HK2 expression, but not HK1, correlated with TNBC or EGFR expression (Fig. 4D and Supplementary Fig. S4G and S4H). Knocking down both PKM2 and HK2 had more reduction in EGF-induced lactate production than knocking down PKM2 or HK2 alone (Fig. 4E). These data suggested that both EGF-induced upregulation of HK2 and inhibition of PKM2 activity by phosphorylation enhance aerobic glycolysis.
Figure 4.
Upregulation of HK2 expression by EGF signaling contributes to aerobic glycolysis in TNBC cells. (A) The mRNA expression of HK1, HK2, and PKM2 in EGF-treated MDA-MB-468 cells. Ctrl, without EGF treatment. (B) Protein expression of HK1, HK2, and PKM2 in EGF-treated MDA-MB-468 cells. (C) Hexokinase activity was measured in MDA-MB-468 cells after 24-h EGF treatment. (D) A Box-and-Whisker plot of HK2 gene expression in TNBC and non-TNBC tumor tissues. (E) Lactate production of PKM2 and/or HK2 knockdown MDA-MB-468 cells after 24-h EGF treatment. (F) RT-qPCR analysis of miR-143, miR-125a, and miR-125b expression in EGF- and/or TKI-treated MDA-MB-468 cells. Ctrl, without EGF and/or TKI treatment. (G) RT-qPCR analysis of HK2 mRNA expression in MDA-MB-468 cells transfected with miR-143 mimic or microRNA mimic negative control (ctrl) and treated with or without EGF for 24 h.
EGFR has recently been shown to regulate microRNA (miRNA) expression via Ago2 protein (35). Therefore, we asked whether EGF-induced increased in HK2 might be regulated by miRNA. We analyzed the 3’-UTR of HK2 mRNA using TargetScan and identified three miRNAs, miR-143, miR-125a, and miR-125b, predicted to regulate HK2 mRNA. Among these, only expression of miR-143 was significantly decreased in EGF-treated MDA-MB-468 cells, which was restored by gefitinib treatment (Fig. 4F). Furthermore, addition of miR-143 mimic attenuated EGF-induced HK2 mRNA expression (Fig. 4G) but did not affect the level of pY EGFR expression (Supplementary Fig. S4I). Thus, EGF may upregulate HK2 expression by downregulating miR-143.
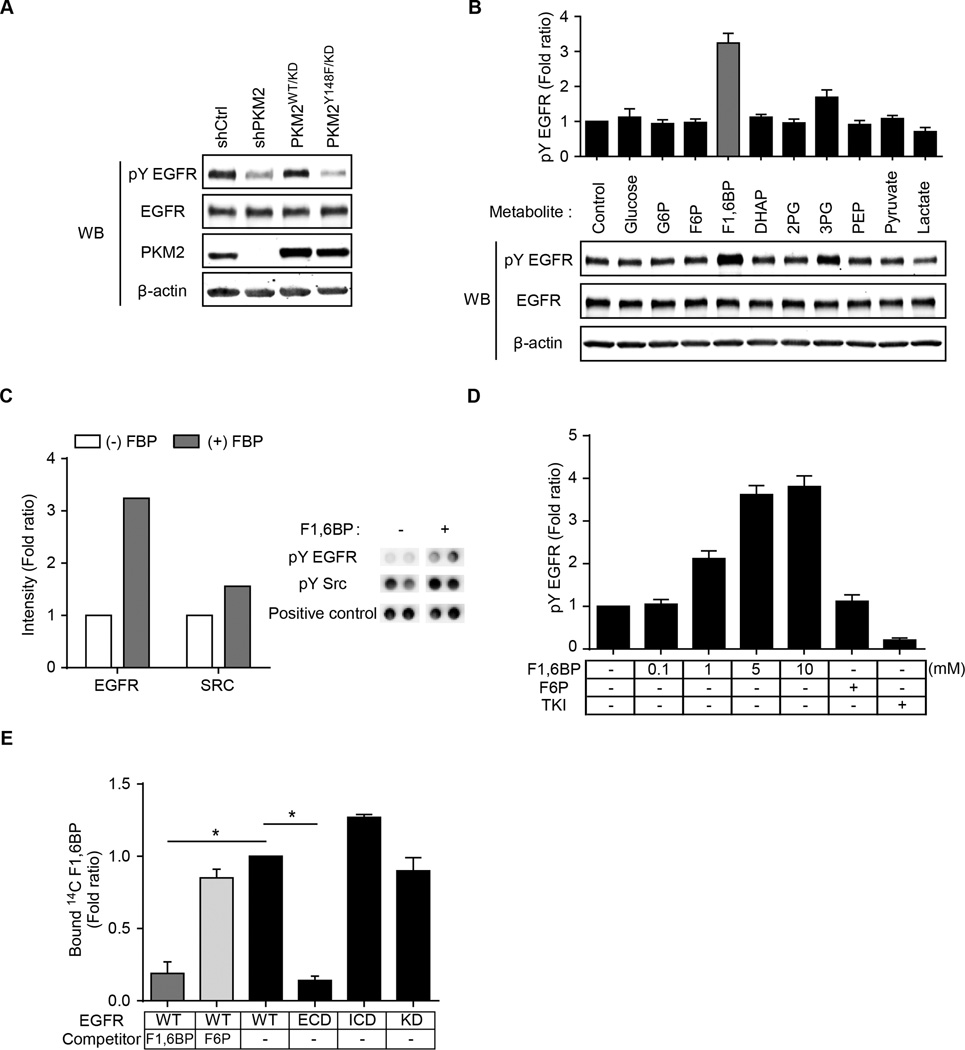
Glycolytic metabolite F1,6BP binds directly to and enhances the activity of EGFR
The level of EGFR tyrosine phosphorylation was substantially reduced in PKM2Y148F-expressing and PKM2-knockdown (shPKM2) MDA-MB-468 cells compared with those expressing PKM2WT (Fig. 5A). However, there was no difference in EGFR phosphorylation when EGFR was incubated with PKM2 WT or PKM2 Y148F, suggesting that PKM2 did not directly phosphorylate EGFR (Supplementary Fig. S5A). We also noticed that the levels of several glycolysis metabolites were increased by EGF treatment in PKM2WT but not PKM2Y148F cells (Fig. 3C). Therefore, we asked whether enhanced EGFR phosphorylation was a result of the increase in the levels of these metabolites. Among them, only the addition of F1,6BP to the culture medium increased EGFR tyrosine phosphorylation by more than 3-fold (Fig. 5B). Consistently, in vitro EGFR kinase assay and RTK array analysis also demonstrated enhanced EGFR tyrosine phosphorylation in the presence of F1,6BP (Fig. 5C and 5D and Supplementary Fig. S5B and S5C). Unlike F1,6BP, fructose-6-phosphate (F6P), which is structurally similar to F1,6BP, did not affect EGFR tyrosine phosphorylation (Fig. 5B and D). In addition, 14C-labeled F1,6BP bound to the intracellular domain (ICD) of EGFR protein, which was blocked by non-labeled F1,6BP (Fig. 5E and Supplementary Fig. S5D). These results suggested that F1,6BP binds directly to EGFR to enhance its phosphorylation.
Figure 5.
F1,6BP enhances EGFR activity through a direct binding with EGFR. (A) Protein expression of tyrosine-phosphorylated (pY) EGFR in MDA-MB-468 cells expressing control shRNA (shCtrl), sh-PKM2, PKM2WT/KD, or PKM2Y148F/KD. (B) Protein expression of pY EGFR in MDA-MB-468 cells treated with various metabolites. Top, quantification of pY EGFR. (C) In vitro kinase assay on RTK array with or without 500 µM F1,6BP. Left, quantification of pY EGFR and pY SRC. Right, representative RTK array images of pY EGFR, pY SRC, and positive control. (D) Quantitation of pY EGFR Western blotting from in vitro EGFR kinase assay on RTK array with the indicated treatments. (E) Quantification of 14C-F1,6BP bound EGFR from in vitro binding assay. WT, wild type; ECD, extracellular domain; ICD, intracellular domain; KD, kinase dead mutant.
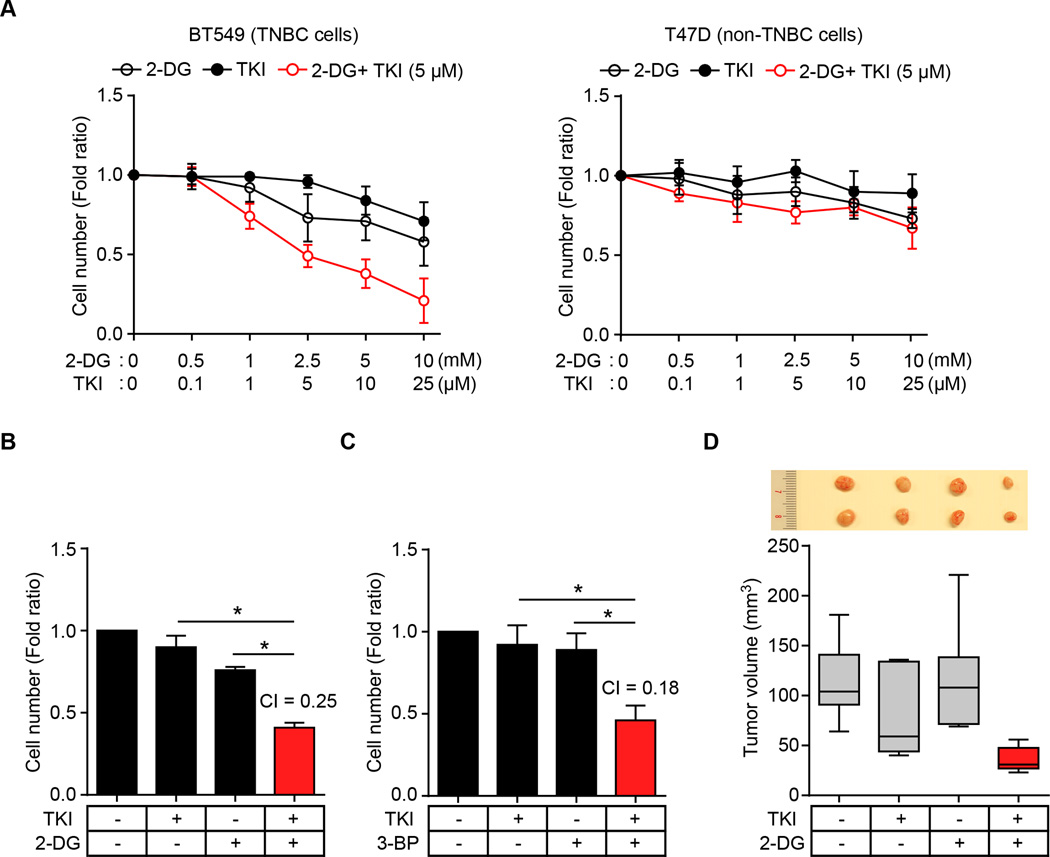
Combined inhibition of EGFR and glycolysis synergistically suppresses TNBC cell proliferation
The crosstalk of EGF signaling and glycolysis metabolite prompted us to determine whether blocking of both EGF signaling and glycolysis may be effective in inhibiting cancer cell proliferation. Although 2-DG is one of the most effective inhibitors of glycolysis (39,40), 2-DG alone is not effective in cancer cell killing and has an unacceptable toxicity at high doses (39–41), and gefitinib alone is also not effective in killing breast cancer cells (Fig. 6A). However, we found that gefitinib sensitized TNBC cells to relatively low concentrations of 2-DG (2.5 mM) compared with 2-DG alone (Fig. 6A). The gefitinib-2-DG combination synergistically suppressed cellular proliferation in TNBC cells (MDA-MB-468 and BT549) but not in non-TNBC cells (T47D) compared with each inhibitor alone (Fig. 6A and B and Supplementary Fig. S6A). The combination of gefitinib and another glycolysis inhibitor, 3-bromo-pyruvate (3-BP), also synergistically suppressed TNBC cell proliferation (Fig. 6C and Supplementary Fig. S6B). Remarkably, gefitinib plus 2-DG also significantly reduced tumor volume in an MDA-MB-468 xenograft tumor model in mice (Fig. 6D), indicating that inhibition of both EGFR and glycolysis inhibition has potential as a combination therapeutic approach to treat TNBC.
Figure 6.
The combination of EGFR and glycolysis inhibitors synergistically suppresses TNBC cell proliferation. (A) Proliferation of BT549 (left) and T47D (right) cells treated with glycolysis inhibitor 2-DG and/or TKI as determined by cell counting assay. (B and C) Proliferation of BT549 cells treated with TKI and/or glycolysis inhibitor 2-DG (B) or 3-BP (C) as measured by cell counting assay. CI, combination index value. CI < 0.8 indicates synergistic effect. (D) Nude mice were injected with BT549 cells in the mammary glands and treated with 2-DG and/or gefitinib. Tumors were measured and dissected at the endpoint, and tumor size (mm3) shown in a Box-and-Whisker plot.
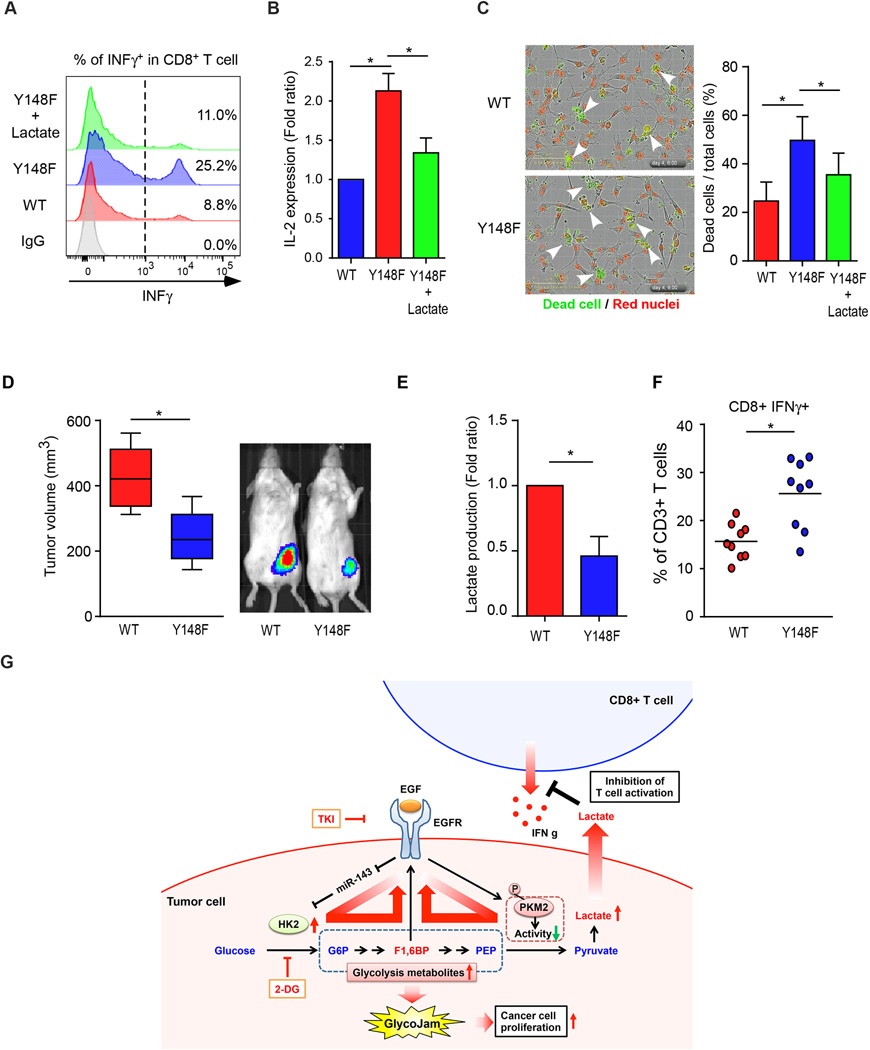
EGF signaling-induced production of lactate inhibits cytotoxic T cell activity
On the basis of our findings the EGF/EGFR signaling induces glycolytic jam, which leads to the accumulation of glycolytic metabolites in tumor cells and increased extracellular level of lactate (Fig. 1) and a previous report demonstrating that extracellular lactate inhibits cytotoxic T cell activity (42), we next asked whether extracellular lactate modulates cytotoxic T cell-related functions in TNBC cells. To this end, we analyzed cytotoxic T-cell activity with or without the addition of lactate by using activated primary human T-cells. Indeed, the population of interferon gamma (INFγ)-positive cytotoxic T cells (CD8+) decreased in culture medium supplemented with lactate (Supplementary Fig. S7). Furthermore, co-culture of T cells and PKM2WT-expressing MDA-MB-468 cells, which excreted elevated levels of lactate (Fig. 1), had less activated T-cell population accompanied by a decrease in INFγ and IL-2 expression compared with those expressing PKM2Y148F (Fig. 7A and B). Consistently, PKM2WT-expressing cells had less T cell-mediated apoptosis than those expressing PKM2Y148F in a T cell-mediated tumor cell-killing assay (Fig. 7C). The addition of lactate also reduced number of apoptotic PKM2Y148F-expressing cells (Fig. 7C). To further investigate whether this phenomenon occurs in vivo, we examined tumor growth, lactate production, and the population of tumor infiltrating lymphocytes (TIL) in PKM2 WT- or PKM2Y148F-expressing 4T1 syngeneic BALB/c mouse model because 4T1 cells are commonly used to investigate TNBC in mice. In line with the results from in vitro experiments, PKM2WT-expressing 4T1 tumors were bigger in mass, and had higher levels of lactate and less activation of cytotoxic T cells compared with those expressing PKM2Y148F (Fig. 7D–F). Together the results suggest that increased levels of extracellular lactate via EGF signaling inhibits cytotoxic T cell activity, which may contribute to the invasive nature of TNBC cells (Fig. 7G, proposed model).
Figure 7.
EGF signaling-induced lactate inhibits cytotoxic T cell activity. (A) Flow cytometric analysis of activated cytotoxic T-cell (INFγ+ and CD8+) population in co-culture of primary T cells and PKM2WT- or PKM2Y148F-expressing MDA-MB-468 cells with or without 10 mM lactate treatment. (B) Levels of soluble IL-2 in co-culture of Jurkat T cells and PKM2WT- or PKM2Y148F-expressing MDA-MB-468 cells. (C) Representative images from phase contrast microscopy showing red fluorescence (nuclear restricted RFP) and/or green fluorescence (NucView™ 488 Caspase 3/7 substrate) from merged images (20×) of PKM2WT- or PKM2Y148F-expressing MDA-MB-468 cells and activated T cell co-cultured in the presence of caspase 3/7 substrate for 96 h. Scale bar, 10 µm. Green fluorescent cells were counted as dead cells. Right, the percentage of dead cells relative to the total number of cells counted. (D) Left, tumors were measured and dissected at the endpoint, and tumor size (mm3) shown in a Box-and-Whisker plot. N = 9 mice per group. Right, representative images of tumor growth of PKM2WT- or PKM2Y148F-expressing mouse 4T1-luc cells in BALB/c mice by IVIS bioluminescence imaging (IVIS100). (E) Lactate production of PKM2WT-or PKM2Y148F-expressing 4T1 tumors. (F) Intracellular cytokine staining of IFNγ in CD8+ CD3+ T cell populations. P < 0.05, two-way ANOVA. N = 9 mice per group. (G) Proposed model of EGF-mediated glycolytic metabolite accumulation and immune escape.
Discussion
Tumor cells are known to reprogram their metabolic pathways to fuel cell proliferation, and many oncogenic signaling pathways such as those activated by RTK contribute to this process. While EGF signaling has been reported to accelerate glucose metabolism in cancer cells (27,28), it is not clear whether this is facilitated directly by EGFR. Our current study identifies a mechanism by which EGF signaling mediates aerobic glycolysis through upregulation of HK2 expression, which speeds up the first step of glycolysis, and downregulation of PKM2 activity, which slows down the last step of glycolysis. As a result, this creates a ‘glycolytic jam’ that leads to an accumulation of metabolic intermediates.
In aerobic glycolysis, the switch from PKM1 to PKM2 is a well-recognized metabolic reprogramming event (7,9,43). Over the past few years, there has been a substantial increase in our understanding of the molecular mechanism by which PKM2 modulates metabolic rearrangement during cancer progression. In tumorigenesis, cell growth signals stimulate the switch from glycolytically active to inactive PKM2, consequently remodeling the glycolytic pathway to channel the carbon source from glucose for biosynthesis (7,9). Posttranslational modifications of PKM2, such as phosho-Y105 and acetyl-K305, which decrease PKM2 activity, have been shown to contribute to tumor cell growth (37,38,44). In addition, PKM2 activator promotes a constitutively active state of PKM2 and suppresses tumor cell proliferation (45). However, a later study demonstrated that knockdown of PKM2 by siRNA suppresses cell proliferation and tumor growth (46). These findings point to a contradictory role of PKM2. Although PKM2 is relatively a well-characterized glycolytic enzyme in tumor cells, ligand-stimulated tumorigenic function of PKM2 by protein-protein interaction or posttranslational modification in specific tumor type, such as TNBC, has not been reported. Here, we demonstrated that PKM2 phosphorylation plays an important role in EGF signaling-mediated aerobic glycolysis in TNBC cells.
Until now, metabolites that accumulated via aerobic glycolysis were considered as a building blocks or fuel source for cancer cell proliferation. The loss of fructose-1,6-bisphosphatase (FBP1), which catalyzes the dephosphorylation of F1,6BP to F6P in gluconeogenesis, was recently determined to be a critical oncogenic event in breast cancer and renal cell carcinoma progression (47,48). These finding suggests that F1,6BP, in addition to serving as a source for cancer cell proliferation, may be important to the regulation of glucose metabolism in cancer. Our findings further support the critical role of F1,6BP in cancer by modulating oncogenic signaling.
As with glycolytic metabolites, lactate was thought to be just a byproduct of glycolysis. However, it was recently shown that lactate functions as an important regulator of cancer development and metastasis through cell-to-cell interactions between cancer, stromal, and endothelial cells. Further studies also demonstrated that extracellular lactate regulates T cell functions (16,42). Here, we provided evidence to support a link between EGF-induced extracellular lactate and cancer cell immune escape through inhibition of cytotoxic T cell activity. These findings implied that oncogenic signaling of cancer cells regulates not only intracellular metabolic pathway but also extracellular immune escape function.
On the basis of our findings, we propose a model (Fig. 7G) illustrating the effect of oncogenic signaling of EGF/EGFR on aerobic glycolysis and immune escape in TNBC cells. Inhibition of PKM2, the last irreversible step of glycolysis, and upregulation of HK2, the first irreversible step of glycolysis, forms a ‘glycolytic jam’ that results in an accumulation of glycolytic intermediates. Accumulated F1,6BP via this glycolytic jam together with enhanced levels of extracellular lactate contribute to oncogenic signaling, tumor growth, and immune escape. The combined inhibition of EGFR to block oncogenic signaling and of glycolysis to reduce the levels of metabolites may represent a new therapeutic strategy to treat TNBC and possibly other EGFR-expressing cancer cells.
Supplementary Material
Acknowledgments
This study was funded in part by National Institutes of Health (CA109311, CA099031, and CCSG CA16672); Susan G. Komen Foundation (SAC100016); Breast Cancer Research Foundation; Patel Memorial Breast Cancer Research Fund; The University of Texas MD Anderson-China Medical University and Hospital Sister Institution Fund (to M.-C.H.); Ministry of Science and Technology, International Research-intensive Centers of Excellence in Taiwan (I-RiCE; MOST 104-2911-I-002-302); Ministry of Health and Welfare, China Medical University Hospital Cancer Research Center of Excellence (MOHW104-TDU-B-212-124-002); Center for Biological Pathways; Susan G. Komen for the Cure Postdoctoral Fellowship (PDF12231298 to S.-O.L.); Basic Science Research Program through the National Research Foundation of Korea funded by the Korea government (MSIP) (NRF-2011-357-C00140 to S.-O.L.); University of Washington and Washington Research Foundation (to D.R.).
Footnotes
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dang CV. Links between metabolism and cancer. Genes & development. 2012;26(9):877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang R, Xiao T, Fang Z, Sun Y, Li F, Gao Y, et al. MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. The Journal of biological chemistry. 2012;287(27):23227–23235. doi: 10.1074/jbc.M112.373084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathupala SP, Ko YH, Pedersen PL. Hexokinase II: cancer's double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25(34):4777–4786. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 8.Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44(27):9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 9.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Seminars in cancer biology. 2005;15(4):300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. The international journal of biochemistry & cell biology. 2011;43(7):969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348(6230):74–80. doi: 10.1126/science.aaa6204. [DOI] [PubMed] [Google Scholar]
- 12.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nature reviews Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollizzi KN, Powell JD. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nature reviews Immunology. 2014;14(7):435–446. doi: 10.1038/nri3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35(6):871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342(6155):1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. The Journal of clinical investigation. 2013;123(9):3685–3692. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nature reviews Clinical oncology. 2010;7(12):683–692. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- 19.Gazinska P, Grigoriadis A, Brown JP, Millis RR, Mera A, Gillett CE, et al. Comparison of basal-like triple-negative breast cancer defined by morphology, immunohistochemistry and transcriptional profiles. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2013 doi: 10.1038/modpathol.2012.244. [DOI] [PubMed] [Google Scholar]
- 20.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. The Journal of clinical investigation. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26(15):2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 22.Metzger-Filho O, Tutt A, de Azambuja E, Saini KS, Viale G, Loi S, et al. Dissecting the heterogeneity of triple-negative breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30(15):1879–1887. doi: 10.1200/JCO.2011.38.2010. [DOI] [PubMed] [Google Scholar]
- 23.Dickler MN, Cobleigh MA, Miller KD, Klein PM, Winer EP. Efficacy and safety of erlotinib in patients with locally advanced or metastatic breast cancer. Breast cancer research and treatment. 2009;115(1):115–121. doi: 10.1007/s10549-008-0055-9. [DOI] [PubMed] [Google Scholar]
- 24.Mueller KL, Yang ZQ, Haddad R, Ethier SP, Boerner JL. EGFR/Met association regulates EGFR TKI resistance in breast cancer. J Mol Signal. 2010;5:8. doi: 10.1186/1750-2187-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, et al. TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30(21):2615–2623. doi: 10.1200/JCO.2010.34.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12(18):5268–5272. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 27.Kaplan O, Jaroszewski JW, Faustino PJ, Zugmaier G, Ennis BW, Lippman M, et al. Toxicity and effects of epidermal growth factor on glucose metabolism of MDA-468 human breast cancer cells. The Journal of biological chemistry. 1990;265(23):13641–13649. [PubMed] [Google Scholar]
- 28.Baulida J, Onetti R, Bassols A. Effects of epidermal growth factor on glycolysis in A431 cells. Biochemical and biophysical research communications. 1992;183(3):1216–1223. doi: 10.1016/s0006-291x(05)80320-1. [DOI] [PubMed] [Google Scholar]
- 29.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480(7375):118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, et al. PKM2 Phosphorylates Histone H3 and Promotes Gene Transcription and Tumorigenesis. Cell. 2012;150(4):685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim SO, Gu JM, Kim MS, Kim HS, Park YN, Park CK, et al. Epigenetic changes induced by reactive oxygen species in hepatocellular carcinoma: methylation of the E-cadherin promoter. Gastroenterology. 2008;135(6):2128–2140. 40 e1–40 e8. doi: 10.1053/j.gastro.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 32.Xia W, Wei Y, Du Y, Liu J, Chang B, Yu YL, et al. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Molecular carcinogenesis. 2009;48(7):610–617. doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang JY, Hsu JL, Meric-Bernstam F, Chang CJ, Wang Q, Bao Y, et al. BikDD eliminates breast cancer initiating cells and synergizes with lapatinib for breast cancer treatment. Cancer cell. 2011;20(3):341–356. doi: 10.1016/j.ccr.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheppard KA, Fitz LJ, Lee JM, Benander C, George JA, Wooters J, et al. PD-1 inhibits T-cell receptor induced phosphorylation of the ZAP70/CD3zeta signalosome and downstream signaling to PKCtheta. FEBS letters. 2004;574(1–3):37–41. doi: 10.1016/j.febslet.2004.07.083. [DOI] [PubMed] [Google Scholar]
- 35.Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497(7449):383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huo L, Wang YN, Xia W, Hsu SC, Lai CC, Li LY, et al. RNA helicase A is a DNA-binding partner for EGFR-mediated transcriptional activation in the nucleus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(37):16125–16130. doi: 10.1073/pnas.1000743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334(6060):1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Science signaling. 2009;2(97):ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aghaee F, Pirayesh Islamian J, Baradaran B. Enhanced radiosensitivity and chemosensitivity of breast cancer cells by 2-deoxy-d-glucose in combination therapy. Journal of breast cancer. 2012;15(2):141–147. doi: 10.4048/jbc.2012.15.2.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dwarakanath BS, Singh D, Banerji AK, Sarin R, Venkataramana NK, Jalali R, et al. Clinical studies for improving radiotherapy with 2-deoxy-D-glucose: present status and future prospects. Journal of cancer research and therapeutics. 2009;5(Suppl 1):S21–S26. doi: 10.4103/0973-1482.55136. [DOI] [PubMed] [Google Scholar]
- 41.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nature reviews Drug discovery. 2011;10(9):671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 42.Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood. 2007;109(9):3812–3819. doi: 10.1182/blood-2006-07-035972. [DOI] [PubMed] [Google Scholar]
- 43.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463(7279):364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Molecular cell. 2011;42(6):719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nature chemical biology. 2012 doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldberg MS, Sharp PA. Pyruvate kinase M2-specific siRNA induces apoptosis and tumor regression. The Journal of experimental medicine. 2012;209(2):217–224. doi: 10.1084/jem.20111487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer cell. 2013;23(3):316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B, Qiu B, Lee DS, Walton ZE, Ochocki JD, Mathew LK, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513(7517):251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.