Abstract
Primates tend to be long-lived for their size with humans being the longest lived of all primates. There are compelling reasons to understand the underlying age-related processes that shape human lifespan. But the very fact of our long lifespan that makes it so compelling, also makes it especially difficult to study. Thus, in studies of aging, researchers have turned to non-human primate models, including chimpanzees, baboons, and rhesus macaques. More recently, the common marmoset, Callithrix jacchus, has been recognized as a particularly valuable model in studies of aging, given its small size, ease of housing in captivity, and relatively short lifespan. However, little is known about the physiological changes that occur as marmosets age. To begin to fill in this gap, we utilized high sensitivity metabolomics to define the longitudinal biochemical changes associated with age in the common marmoset. We measured 2104 metabolites from blood plasma at three separate time points over a 17-month period, and we completed both a cross-sectional and longitudinal analysis of the metabolome. We discovered hundreds of metabolites associated with age and body weight in both male and female animals. Our longitudinal analysis identified age-associated metabolic pathways that were not found in our cross-sectional analysis. Pathways enriched for age-associated metabolites included tryptophan, nucleotide, and xenobiotic metabolism, suggesting these biochemical pathways might play an important role in the basic mechanisms of aging in primates. Moreover, we found that many metabolic pathways associated with age were sex specific. Our work illustrates the power of longitudinal approaches, even in a short time frame, to discover novel biochemical changes that occur with age.
Keywords: metabolomics, aging, marmoset, longitudinal, body weight, metabolic pathways
1. Introduction
Many studies have attempted to determine biomarkers of age and age-related survivorship of individuals (reviewed in Johnson 2006; Mather et al. 2011). However, most have been unsuccessful. There are numerous factors that might account for our relative lack of success in finding biomarkers of aging, one of which is that most studies have looked at cross-sectional, rather than longitudinal, data. Cross-sectional studies compare changes that occur among all individuals as they age, but fail to track changes within individuals. There are molecular changes that occur throughout an individual's life that may have profound effects on aging and age-related disease, yet are not revealed by traditional cross-sectional aging studies. The most meaningful biomarkers might be those whose long-term trajectories, rather than their static values at one time point, predict pathology or death. To identify these biomarkers, we need to turn to longitudinal data analysis. In fact, many longitudinal studies in humans have identified genetic and environmental correlates of longevity (e.g. Colditz and Hankinson 2005; Ferrucci 2008). However, these studies take many decades to complete, and at considerable expense. With this in mind, we turned to the common marmoset, Callithrix jacchus, in which we could use longitudinal approaches to search for systems biology predictors of healthspan and lifespan.
The marmoset, a small, relatively short-lived non-human primate, offers us a powerful, translational model to understand the causes and correlates of aging. Though most studies have utilized large primates, such as the rhesus macaque, recent studies have pointed to the common marmoset, Callithrix jacchus, as an ideal non-human primate model of aging (Fischer and Austad 2011; Tardif et al. 2011). Marmosets have age-associated pathologies also seen in humans, and they have a relatively short lifespan (average 8-12 years and maximum 16.5-21.5) compared to other well-studied primates (Tardif et al. 2011; Nishijima et al. 2012).
We believe that by studying the biochemical changes that occur throughout the life of the marmoset, we may be able to better detect specific, accurate biomarkers of aging. One way to do this is through the use of high-resolution metabolomics (Jones et al. 2012), the study of small molecules in an organism. This can provide us with a snapshot of metabolic changes that occur over time. Cross-sectional approaches in non-human primates have been completed to identify metabolites correlated with age (Muehlenbein et al. 2003; Kuehnel et al. 2012). However, longitudinal studies of metabolites in primates have been confined to earlier developmental stages (Higley et al. 1992; Beckstrom et al. 2012). Research in model organisms show that the analysis of metabolites has the potential to identify metabolic pathways that might be implicated in aging (Fuchs et al. 2010; Houtkooper et al. 2011; Hoffman et al. 2014).
Metabolomic studies have rarely been used in the analysis of natural aging in non-human primates, with previous studies focusing on biomarkers of specific diseases, rather than the effects of natural aging (e.g. Patterson et al. 2011; Liu et al. 2013). Moreover, metabolomic studies that do look at metabolites associated with age in the marmoset have relied on cross-sectional analyses (Kuehnel et al. 2012; Roede et al. 2013), which as described earlier, may be missing important changes within individuals. This leaves a potentially important gap in our understanding of how individual metabolites and metabolic pathways change over the life of an animal.
We take advantage of the marmoset's short lifespan to gain insight into the longitudinal changes in the metabolome, and use this system as a potential model for human aging metabolomics. While the common marmoset has been proposed as a new non-human primate model of aging, very little is known about how its biochemical makeup changes with age. Here, we present the first longitudinal study of age related changes in the metabolome of a large colony of marmosets.
2. Methods
2.1 Marmosets and sample collection
Marmosets were housed at the New England Primate Research Center and were maintained as described in Soltow et al. (2013). Briefly, most marmosets were pair housed in cages and fed commercial marmoset chow that was supplemented with various fruits, vegetables, seeds, and mealworms. Animals were given water ad libitum, and water was changed daily. Animal cages were cleaned three times a week. All rooms containing animals were temperature and humidity controlled, and the animals were given various life enrichments including toys, food treats, and music.
Blood plasma samples were collected at three different time points over a 17-month period (June 2012, October 2012, and November 2013) during routine physical exams of the animals under sedation with 0.2mL of ketamine as described in Roede et al. (2013). Previous reproductive history of the animals was unknown; however, no females pregnant at the time of sampling were included in the population. This 17-month period represents at least 12% of the mean lifespan in this species (Tardif et al. 2011).
2.2 Metabolomic analysis
Metabolites were analyzed by high-resolution mass spectrometry (MS; LTQ-Velos Orbitrap, Thermo Fisher) coupled to liquid chromatography (LC) using a reverse-phase C18 column (Soltow et al. 2013). Briefly, 50 μL of plasma was added to 100 μL of acetonitrile along with a 2.5 μL aliquot containing stable isotope standards. Samples were mixed, incubated at 4° C for 30 min, and centrifuged to remove protein. Supernatants were analyzed in triplicate by LC-MS (Soltow et al. 2013; Go et al. 2014). Data were extracted using apLCMS (Yu et al. 2009) with xMSanalyzer (Uppal et al. 2013) as m/z features, where an m/z feature is defined by m/z (mass-to-charge ratio), retention time, and ion intensity (Johnson et al. 2010).
2.3 Data analysis
Data analyses were carried out using the statistics package R, version 3.0.2 (R Core Team 2013) unless otherwise stated. For each time point, the data structure originally consisted of over 20,000 metabolites measured for each individual. As a first quality control step, all metabolites were normalized using a log transformation. We then measured the repeatability of technical replicates for each metabolite, measured as the signal-to-noise ratio (SNR, ), where is the mean intensity of metabolite i across all samples, and is the mean of the standard deviation of technical replicates within each biological replicate for metabolite i, averaged across all biological replicates. Following Hoffman et al. (2014), only those metabolites with SNR ≥ 15 were kept for further analysis. These quality control measures were executed separately for each time point.
Next, to ensure we combined the same metabolites across the three different time points, we used the R package xMSanalyzer (Uppal et al., 2013). This program identifies the same metabolite from different datasets by analyzing both the mass to charge (m/z) ratio and column retention time. Metabolites were considered identical across the time points if they had m/z ratios within 10 ppm of each other and their retention times varied by less than 10 seconds.
We first wanted to discover specific metabolites that were associated with age and body weight within each time point individually (i.e., cross-sectional analysis). We ran a linear model with body weight and age as fixed effects predicting metabolite intensity. The sexes were analyzed separately. Metabolites were considered significantly associated with either factor if they passed a false discovery rate (FDR) of α = 0.1 (Benjamini and Hochberg 1995).
We then looked at metabolite associations with weight change. For those animals that had data from the first and last time points, we calculated the change in weight over the 17-month period. We then ran a linear model looking at associations of weight change and metabolite values in the first time point. Thus we were determining if metabolite values were correlated with future weight changes.
To determine if the changes observed across ages within a time point continued across time and individuals as well as to discover novel metabolites associated with aging in the marmoset, we sought metabolites with significant longitudinal changes in intensity across all individuals. The sexes were again analyzed separately. Our longitudinal analysis was carried out by implementing a random effects model in the nlme package (Pinherio et al. 2012), with age and weight as fixed effects and individual as a random effect. Thus, we wanted to determine if the correlations between metabolite intensity and age and body weight in our cross-sectional analysis were recapitulated across time within individuals in our longitudinal analysis. The linear mixed effects approach enables us to include all individuals, including those for which we have only a single time point (Bernal-Rusiel et al. 2013). We were also interested in discovering if specific metabolites were associated with age or body weight in one statistical approach but not the other. This would also give us insight into whether a cross-sectional or longitudinal approach is more sensitive to changes in metabolite concentration with age. There are batch effects in the data such that the mean metabolite concentration across all samples can vary between time points, and we were interested in the relative metabolite intensity among individuals. Accordingly, for this longitudinal analysis, we centered all metabolites within a marmoset to a mean of zero. We again used an FDR with α = 0.1 to correct for multiple comparisons.
For those metabolites that were discovered to be associated either with age or weight in the cross sectional or longitudinal analysis, we looked for specific metabolic pathways that were overrepresented using the pathway enrichment program mummichog (Li et al. 2013). Mummichog annotates m/z features and determines specific metabolic pathway enrichment using the MetaCyc database (Caspi et al. 2014). In particular, we set mummichog to query the “human_mfn” reference metabolic pathway, as humans are the most closely related organism to the marmoset for which metabolite profiles and pathways have been well defined. For this analysis, we considered pathways significantly enriched for a factor if the mummichog-adjusted P value was less than 0.05. The adjusted P-value is calculated from resampling the reference input file using a gamma distribution which penalizes pathways with fewer reference hits, thus giving more significance to larger pathways.
3. Results
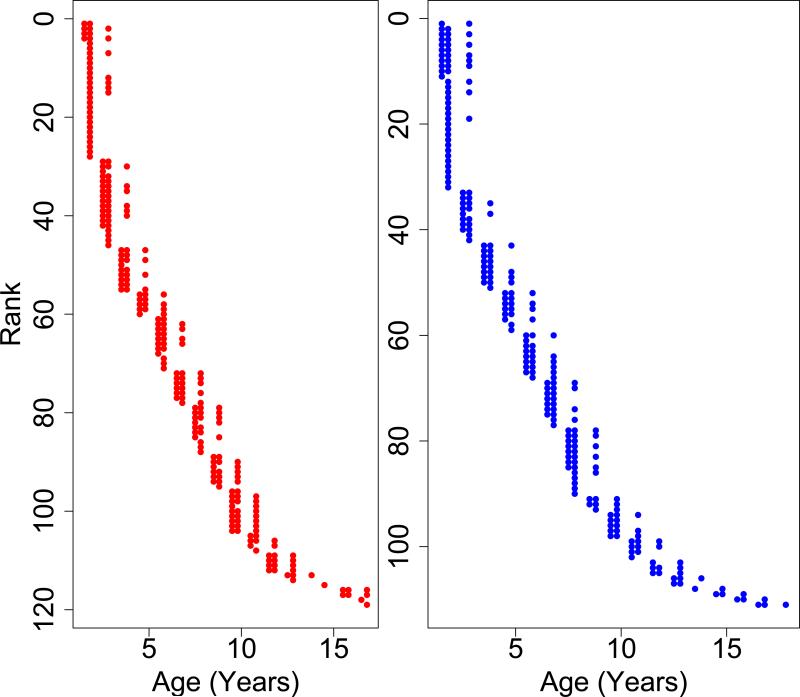
Our final dataset consisted of information from 230 individual marmosets across a period of 17-months. Of the 230 animals, 77 were sampled twice, and 84 were sampled three times (Figure 1). The animals ranged in age from 1-17 years with an average age of 6.3 years. Age distributions in each time point are shown in Figure 1. Females were slightly but not significantly larger than males on average across all time points combined, (407.4 g (+/− 62.8 SD) and 403.6 g (+/− 49.3 SD), respectively). Age and weight means and ranges for each time point individually are shown in Table 1. We also found significant differences in weight changes over the 17 months between young and old animals. In both sexes, young animals (those under 4 years at the first time point) lost weight on average, while old animals (those over 10 years at the first time point) gained weight on average. The distributions of weight changes over the course of the experiment were significantly different between old and young monkeys (t-test: P<0.01 for both sexes).
Figure 1. Plot of all ages of each individual maramoset.
Marmosets are ranked by age. Each point represents a blood sample that was taken from an individual marmoset. The y-axis values represents a single marmoset while the x-axis values represents the age at which it was sampled. Animals were sampled either 1, 2, or 3 times. Females are in red on the left; males on the right in blue.
Table 1. Numbers of metabolites associated with either age or body weight.
Results from the cross sectional and longitudinal analysis are shown. Mean age and weight along with ranges are also shown.
| Factor | June 2012 | October 2012 | November 2013 | Longitudinal | ||||
|---|---|---|---|---|---|---|---|---|
| Females | Males | Females | Males | Females | Males | Females | Males | |
| N | 80 | 76 | 90 | 85 | 75 | 69 | 119 | 111 |
| Mean age in years (range) | 6.4 (1.4-16.4) | 6 (1.4-16.4) | 5.8 (1.8-16.8) | 5.8 (1.8-16.8) | 7 (1.8-16.8) | 6.8 (1.8-17.8) | 6.4 (1.4-16.8) | 6.2 (1.4-17.8) |
| Mean weight in grams (range) | 408.6 (306-627) | 403.3 (304-551) | 410.6 (308-620) | 405.3 (322-522) | 402.2 (309-608) | 401.6 (309-569 | 407.4 (306-627) | 403.6 (304-569) |
| Increase with age | 268 | 366 | 985 | 706 | 224 | 100 | 272 | 196 |
| Decrease with age | 362 | 452 | 78 | 67 | 40 | 49 | 163 | 199 |
| Increase with body weight | 84 | 0 | 93 | 37 | 51 | 13 | 125 | 55 |
| Decrease with body weight | 13 | 0 | 0 | 0 | 0 | 5 | 142 | 2 |
Our final metabolomics dataset consisted of 2104 metabolites that were present in all three time points analyzed. Of these metabolites, we were able to putatively annotate 250 (11.9%) using the program mummichog.
We analyzed our data in two steps. First, we ran a cross-sectional analysis of each time point individually to identify metabolites associated with age and body weight among individuals within a time-specific cohort. We were able to discover hundreds of metabolites that increased (up to 46% of metabolites analyzed) or decreased (up to 21% of metabolites analyzed) with age, as well as dozen of metabolites that were positively correlated with body mass (Table 1). Relatively few metabolites were negatively correlated with body weight.
Metabolite enrichment analysis using mummichog pointed to many metabolic pathways associated with age within the individual time points. Among metabolites that increased with age, we found significant enrichment for purine and pyrimidine metabolism in both sexes (Table 2). In contrast, we did not find any pathways that were clearly enriched for metabolites that decreased with age (Table 3).
Table 2. Pathways associated with an increase with age.
Metabolic pathways enriched for individual time points and the longitudinal analysis. Values reported are adjusted p-values from the metabolic pathway enrichment program mummichog. Pathways were considered significant if their adjusted p-values were less than 0.05. Tables of actual metabolites detected increasing with age and in the entire annotated metabolome shown in Table S1.
| Increase with age | ||||||||
|---|---|---|---|---|---|---|---|---|
| Pathway | June 2012 | October 2012 | November 2013 | Longitudinal | ||||
| Females | Males | Females | Males | Females | Males | Females | Males | |
| 3-Chloroacrylic acid degradation | 0.0033 | |||||||
| Androgen and estrogen biosynthesis and metabolism | 0.0023 | 0.0049 | 0.0301 | |||||
| Arginine and Proline Metabolism | 0.0016 | |||||||
| Ascorbate (Vitamin C) and Aldarate Metabolism | 0.0093 | |||||||
| Aspartate and asparagine metabolism | 0.0018 | |||||||
| Beta-Alanine metabolism | 0.0451 | |||||||
| C21-steroid hormone biosynthesis and metabolism | 0.0291 | |||||||
| Caffeine metabolism | 0.0089 | 0.0048 | ||||||
| Carnitine shuttle | 0.0183 | |||||||
| D4&E4-neuroprostanes formation | 0.0086 | |||||||
| Drug metabolism - cytochrome P450 | 0.0162 | 0.0421 | ||||||
| Fatty acid activation | 0.0053 | |||||||
| Fructose and mannose metabolism | 0.0301 | |||||||
| Glycolysis and Gluconeogenesis | 0.0282 | |||||||
| Glycerophospholipid metabolism | 0.0035 | |||||||
| Glycosphingolipid metabolism | 0.0049 | |||||||
| Histidine metabolism | 0.0451 | |||||||
| Hyaluronan Metabolism | 0.0300 | |||||||
| Linoleate metabolism | 0.0098 | |||||||
| Methionine and cysteine metabolism | 0.0035 | 0.0042 | ||||||
| N-Glycan biosynthesis | 0.0063 | |||||||
| Pentose phosphate pathway | 0.0248 | |||||||
| Phosphatidylinositol phosphate metabolism | 0.0145 | |||||||
| Phytanic acid peroxisomal oxidation | 0.0163 | |||||||
| Porphyrin metabolism | 0.0046 | 0.0029 | 0.0057 | 0.0035 | ||||
| Purine metabolism | 0.0240 | 0.0263 | 0.0135 | 0.0029 | 0.0012 | 0.0030 | 0.0005 | |
| Pyrimidine metabolism | 0.0183 | 0.0084 | ||||||
| Squalene and cholesterol biosynthesis | 0.0041 | 0.0475 | 0.0209 | |||||
| TCA cycle | 0.0191 | 0.0040 | 0.0079 | |||||
| Tryptophan metabolism | 0.0242 | |||||||
| Tyrosine metabolism | 0.0131 | |||||||
| Vitamin B3 (nicotinate and nicotinamide) metabolism | 0.0021 | 0.0025 | ||||||
| Vitamin B9 (folate) metabolism | 0.0163 | 0.0204 | ||||||
| Vitamin E metabolism | 0.0020 | |||||||
| Xenobiotics metabolism | 0.0320 | 0.0034 | 0.0476 | |||||
Table 3. Pathways associated with a decrease with age.
Metabolic pathways enriched for individual time points and the longitudinal analysis. Values reported are adjusted p-values from the metabolic pathway enrichment program mummichog. Pathways were considered significant if their adjusted p-values were less than 0.05. Tables of actual metabolites detected increasing with age and in the entire annotated metabolome shown in Table S2.
| Decrease with age | ||||||||
|---|---|---|---|---|---|---|---|---|
| Pathway | June 2012 | October 2012 | November 2013 | Longitudinal | ||||
| Females | Males | Females | Males | Females | Males | Females | Males | |
| Biopterin metabolism | 0.0014 | 0.0025 | ||||||
| Chondroitin sulfate degradation | 0.0025 | |||||||
| Dynorphin metabolism | 0.0004 | 0.0014 | 0.0025 | |||||
| Fatty acid activation | 0.0218 | |||||||
| Glycerophospholipid metabolism | 0.0319 | |||||||
| Glycine, serine, alanine and threonine metabolism | 0.0131 | 0.0118 | ||||||
| Glycosphingolipid biosynthesis - ganglioseries | 0.0112 | 0.0192 | ||||||
| Heparan sulfate degradation | 0.0025 | |||||||
| Histidine metabolism | 0.0235 | |||||||
| Leukotriene metabolism | 0.0035 | 0.0122 | ||||||
| Methionine and cysteine metabolism | 0.0215 | |||||||
| Prostaglandin formation from arachidonate | 0.0123 | |||||||
| Prostaglandin formation from dihomo gama-linoleic acid | 0.0025 | 0.0042 | ||||||
| Purine metabolism | 0.0118 | |||||||
| Pyrimidine metabolism | 0.0072 | |||||||
| Sialic acid metabolism | 0.0389 | |||||||
| Starch and Sucrose Metabolism | 0.0394 | 0.0126 | ||||||
| TCA cycle | 0.0093 | 0.0389 | ||||||
| Tryptophan metabolism | 0.0111 | 0.0032 | 0.0036 | 0.0287 | ||||
| Tyrosine metabolism | 0.0332 | 0.0218 | 0.0287 | |||||
| Valine, leucine and isoleucine degradation | 0.0160 | |||||||
| Vitamin A (retinol) metabolism | 0.0319 | |||||||
| Vitamin E metabolism | 0.0046 | 0.0389 | ||||||
We then looked at the associations with metabolite values and changes in weight. Our regression analysis found no metabolites in females that in the first time point (June 2012) were associated with future change in body weight. However, in males we discovered 127 metabolites that were correlated with future changes in weight. The majority of these metabolites were found to be associated with a future increase in weight (115), yet as with the metabolites associated with body weight before, we did not find enrichment for any metabolic pathways.
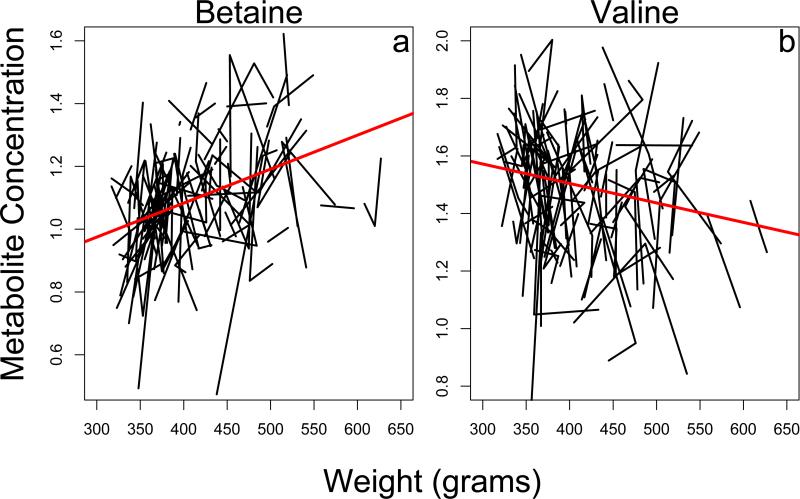
After carrying out the cross-sectional analysis, we used the random effects model for longitudinal data analysis, with the goal of identifying those metabolites whose trajectory changed consistently with age and/or body weight within individuals throughout the population. Unlike our cross-sectional analysis, in females, we were able to find many metabolites both positively correlated (Table 1, Figure 2a) and negatively correlated (Table 1, Figure 2b) with body weight. We found many fewer metabolites either positively or negatively associated with body weight in males (Table 1). In neither sex did we identify enrichment in any specific metabolic pathways for body weight.
Figure 2. Plot of individual metabolites that change with body weight.
a) metabolite increasing with body weight. b) metabolite decreasing with body weight. Each line represents an individual marmoset. Data are only shown for female animals.
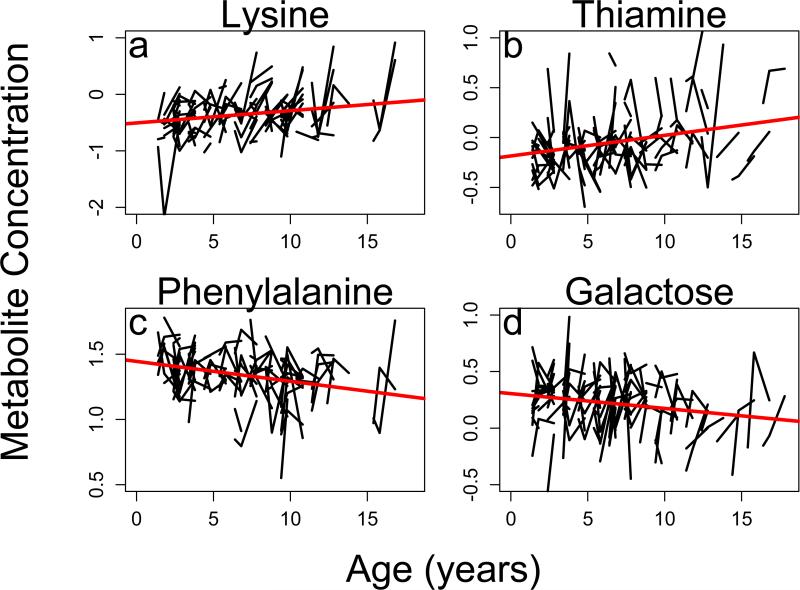
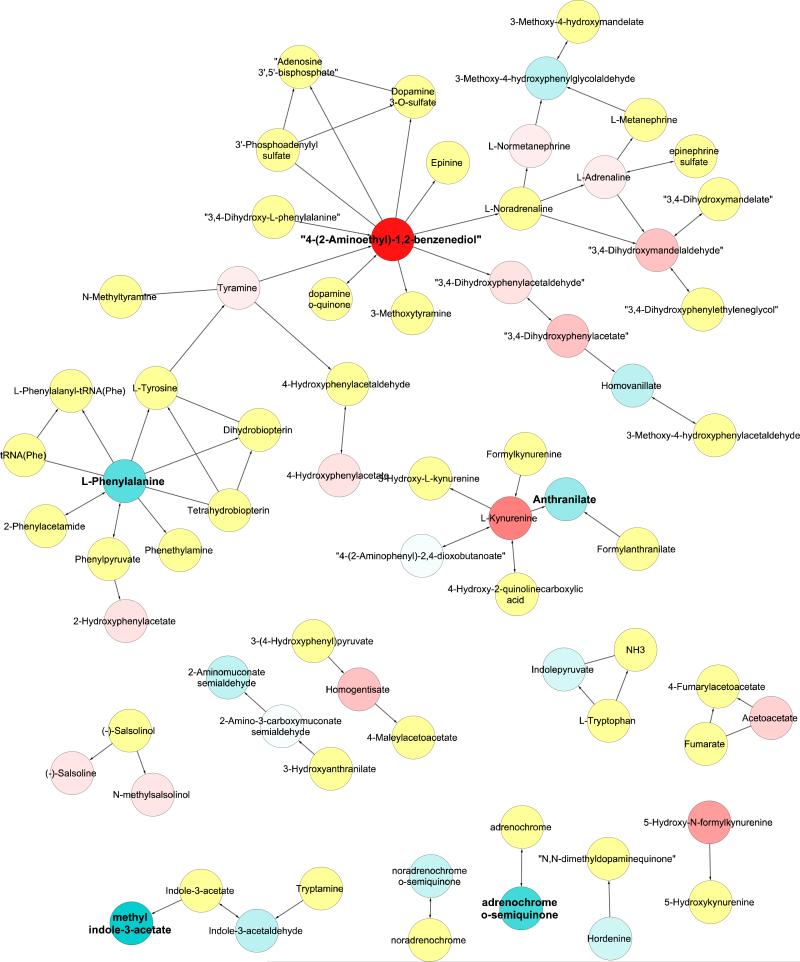
In our longitudinal analysis we identified hundreds of specific metabolites that both increased (Table 1, Figure 3a and 3b) and decreased with age (Table 1, Figure 3c and 3d) across individuals. Our mummichog pathway analysis was then able to determine 27 specific metabolic pathways that were significantly associated with age-related change in at least one sex (Table 2 and 3). Numbers of actual metabolites found in each metabolic pathway are shown in Tables S1 and S2. Over 30% (10) of the metabolic pathways associated with age in our longitudinal analysis were shared between the sexes. Moreover, ten metabolic pathways associated with age in the longitudinal analysis were not discovered in any of the cross-sectional analyses. Several of the metabolic pathways found to be associated with age are associated with tryptophan and tyrosine, nucleotide, and xenobiotic metabolism. The changes seen in tryptophan and tyrosine metabolism metabolic pathway are shown in Figure 4.
Figure 3. Plot of individual metabolites associated with age.
a) female increase with age. b) male increase with age. c) female decrease with age. d) male decrease with age. Each line represents an individual marmoset.
Figure 4. Metabolites in Tryptophan and Tyrosine metabolism pathway.
Each circle represents a metabolite in the metabolic pathway, and lines with arrows are known metabolic interactions. Blue circles decrease with age while red circles increase with age. The degree of saturation indicates the strengths of the response. Metabolites in the metabolic pathway but not found in our dataset are colored yellow. Bold names indicate metabolites with significant changes with age (FDR<0.1). Results shown are for males only.
4. Discussion
Here, we have presented the largest longitudinal metabolomics study to date, and we show the power longitudinal studies have to identify metabolites associated with age that are not found in cross sectional analyses. Cross sectional studies provide great insight into how metabolites are associated with age; however, our findings suggest that longitudinal studies, even over relatively short periods of time, may better identify metabolic correlates of aging.
We were able to discover hundreds of metabolites significantly associated with age in the marmoset, and our 17-month longitudinal analysis was able to cover between twelve and eighteen percent of the average lifespan of the marmoset (depending on reported population averages). This is comparable to nine to fourteen years of human lifespan, yet previous longitudinal metabolomics studies in humans have covered a maximum of seven years (Yousri et al. 2014),. The results of this study show the power of the common marmoset to discover new potential biomarkers of aging in a much shorter time frame than studies on humans (and other non-human primates).
We have shown that many metabolic pathways associated with age in the animals are only found in one sex. While many studies have looked at sex differences in metabolomic profiles and age (e.g. Slupsky et al. 2007; Psihogios et al. 2008), few have attempted to determine how metabolomic profiles change with age between the sexes (e.g. Lawton et al. 2008; Yu et al. 2012). Our results suggest future metabolomics and aging studies should look at the sexes independently as the metabolic causes and consequences of aging may be sex specific, similar to results previously shown in human metabolomic profiles (Yu et al. 2012). Moreover, the sex-specific trajectories observed here point to the possibility that sex-specific metabolomic analysis might point to mechanisms that underlie sex-differences in aging and aging-related disease.
Among metabolites whose concentrations increased or decreased significantly with age, we found substantial enrichment for tryptophan and tyrosine metabolism, nucleotide metabolism, and xenobiotic metabolism. Here we discuss each of these metabolic pathways in turn.
Our results suggest that in the marmoset there is significant enrichment across time points for changes in tryptophan and tyrosine metabolism with age (Figure 4). Previous studies have implicated tryptophan metabolism in aging in a variety of model organisms (reviewed in van der Goot and Nollen 2013), especially in its breakdown to kynurenine (e.g. Coburn and Gems 2013). Tyrosine and tryptophan metabolism can involve the production of monoamine neurotransmitters, which have recently been associated with aging in flies (Hoffman et al. 2014). Both tryptophan and tyrosine metabolism have been associated with inflammation in elderly human subjects (Capuron et al. 2011). Our results combined with previous research suggest there is the potential that changes in tryptophan metabolism may be a conserved metabolic pathway that is associated the age and longevity across distantly related organisms. Future studies are needed to determine if the age-related changes that we see in the tryptophan metabolic pathway affect patterns of aging, or rather are a secondary consequence of aging.
We also found evidence that pathways associated with xenobiotic metabolism are enriched for metabolites that increase with age. Xenobiotic metabolism involves the breakdown of foreign metabolites that are taken in from the environment. The ability to break down xenobiotics may decrease in older individuals, leading to a buildup of these metabolites. Previous research in mice has shown that longer-lived strains exhibit increased expression of genes linked to xenobiotic metabolism (Amador-Noguez et al. 2007; Steinbaugh et al. 2012). These results, combined with our own, suggest that xenobiotic metabolism might be an important factor associated with aging.
We also found strong evidence that the concentration of metabolites associated with nucleotide metabolite, especially purine metabolism, increased with age. The majority of metabolites associated with purine metabolism were found to be increased with age, and it is conserved across both sexes and multiple time points (Table 2). While DNA damage is often cited as a potential causative agent of aging and longevity (e.g. Gensler and Bernstein 1981; Schumacher et al. 2008; Hoeijmakers 2009), few aging studies have shown changes in the metabolites that make up nucleotide metabolism pathways. Previous metabolomic work in yeast has shown that decreases in nucleotide concentrations are significantly associated with extended longevity (Yoshida et al. 2010), and nucleotide metabolism has been shown to influence tumor growth (Aird and Zhang 2015). These results in other species, coupled with our significant changes in nucleotide metabolism in the marmoset suggest changes in nucleotide metabolism may have a significant impact on the aging phenotype. Future studies are needed to determine the specific effects nucleotides have on natural aging and longevity within different organisms.
While not the focus of this study, we were able to discover dozens of metabolites in both sexes that were significantly associated with body weight longitudinally. Interestingly, while we failed to find many metabolites that decline with increases in body weight in our cross sectional analysis, we found over 100 metabolites in the longitudinal study. This again points to the power of longitudinal studies, even over a relatively short time frame, to detect potential biomarkers of phenotypes of interest. We were also able to discover that some metabolites appear to be associated with future changes in body weight, but in males only. Previous work has shown the metabolome to be associated with changes in body weight; however the sexes were analyzed together in this analysis (Wahl et al. 2015).
Although we were able to discover many metabolites associated with body weight and change in body weight (both positively and negatively), we were not able to discern any specific pathways in either sex that were significantly enriched for these metabolites. This suggests that while metabolomic profiles are associated with body weight, these associations might occur throughout the metabolome, rather than focused on specific pathways. Interestingly, metabolites were more likely to be associated with body weight in females, but more likely to be associated with changes in body weight in males. This suggests there may be sex differences not only in metabolite correlates with body weight, but also in how metabolites respond to changes in weight. Our results are consistent with previous work showing sex-specificity of metabolite-body weight associations (Szymanska et al. 2012). Future studies are necessary to more fully understand the effects of body size on metabolomic profiles as an animal ages.
Although the results of this study represent the largest metabolomic longitudinal analysis to date, the time period investigated is overall quite short. As described above the three time points only represent about 15% or so of the average lifespan of the animal, yet by looking at metabolites across time points we were able to discover many metabolic pathways that might have been missed in a cross-sectional analysis. Eleven metabolic pathways were found to be enriched solely in the longitudinal analysis, and 28 pathways were found in only one individual time point. This suggests that if we had only sampled the animals as one point, there are many metabolic pathways associated with age that we would have missed.
While we were able to analyze thousands of metabolites and find hundreds that significantly change with age and body weight across the sexes, there were several limitations to this study. First, metabolomic profiles are known to change on a daily basis and even cyclically within the day (Queiroz 1974). However, we were not able to draw marmoset blood samples on the same day let alone the same time of day. These daily changes could explain, in part, the large variances seen in metabolite concentrations even within animals of the same age, which might have limited our ability to detect metabolites that change with age. Second, blood samples were drawn while the primates were anesthetized with ketamine, which could potentially disrupt normal metabolomic profiles. While the effects of ketamine on blood metabolomics are unknown, previous research in macaques has shown that ketamine does not change blood hormones levels and has less pronounced effects than other forms of anesthesia (Zaidi et al. 1982). Both of these factors are expected to add noise to the data; however, it does suggest that those patterns we see in the data are reliable correlations, making these results all the more impressive.
The nature of this dataset points to two additional caveats to these findings. First, the parameters we used to combine metabolites across time points were conservative (especially the retention time). Each time point had over 10,000 metabolites left after quality control, but only 2104 could be conservatively said to be the same across all three time points, which suggests that we might have discarded information on some metabolites that were actually present in the data from all three time points. Our study was also limited by the lack of metabolite annotation matches available. Very little is known about specific marmoset metabolites, so we used the human metabolome as the reference. However, mummichog was only able to annotate a small proportion of the metabolites used in this study (~12%). While some of these undefined metabolites might have simply been adducts of metabolites that we did identify, others might have been novel, unknown chemicals.
5. Conclusion
Here we have presented the first large-scale longitudinal metabolomics study in a non-human primate, and we were able to discover many metabolic pathways that show consistent changes with age. We believe longitudinal studies are underutilized as a method for determining changes in metabolites and metabolic pathways that are involved in the aging process, and future metabolomics studies should try to incorporate multiple measurements of the same individuals. The marmosets in this colony will continue to be followed, and as the animals age and die, we hope to identify long-term changes in metabolomic profiles that are predictive of risk of morbidity and mortality.
Supplementary Material
Highlights.
Longitudinal studies give insights into aging not seen in cross sectional analyses
The common marmoset provides an ideal model to study aging metabolomics
The metabolome is highly associated with age of an individual
Tryptophan, nucleotide, and xenobiotic metabolism are associated with age
Acknowledgements
This work was funded in part by NIH grant AG038746 to DPJ and DELP. JMH was supported in part by NIH training grant T32 GM007103. We thank the two anonymous reviewers for their helpful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aird KM, Zhang R. Nucleotide metabolism, oncogene-induced senescence and cancer. Cancer Letters. 2015;356:204–210. doi: 10.1016/j.canlet.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador-Noguez D, Dean A, Huang WD, Setchell K, Moore D, Darlington G. Alterations in xenobiotic metabolism in the long-lived Little mice. Aging Cell. 2007;6:453–470. doi: 10.1111/j.1474-9726.2007.00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstrom AC, Tanya P, Humston EM, Snyder LR, Synovec RE, Juul SE. The perinatal transition of the circulating metabolome in a nonhuman primate. Pediatric Research. 2012;71:338–344. doi: 10.1038/pr.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Bernal-Rusiel JL, Greve DN, Reuter M, Fischl B, Sabuncu MR, Alzheimer's Disease Neuroimaging I. Statistical analysis of longitudinal neuroimage data with Linear Mixed Effects models. NeuroImage. 2013;66:249–260. doi: 10.1016/j.neuroimage.2012.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuron L, Schroecksnadel S, Feart C, Aubert A, Higueret D, Barberger-Gateau P, Laye S, Fuchs D. Chronic low-grade inflammation in elderly persons is associated with altered tryptophan and tyrosine metabolism: role in neuropsychiatric symptoms. Biol Psychiatry. 2011;70:175–182. doi: 10.1016/j.biopsych.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Weerasinghe D, Zhang PF, Karp PD. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014;42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chignell CF. Structure Activity Relationships in the Free-Radical Metabolism of Xenobiotics. Environ Health Persp. 1985;61:133–137. doi: 10.1289/ehp.8561133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn C, Gems D. The mysterious case of the C. elegans gut granule: death fluorescence, anthranilic acid and the kynurenine pathway. Frontiers in Genetics. 2013;4:151. doi: 10.3389/fgene.2013.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colditz GA, Hankinson SE. The Nurses' Health Study: Lifestyle and health among women. Nat Rev Cancer. 2005;5:388–396. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- Ferrucci L. The Baltimore Longitudinal Study of Aging (BLSA): A 50-Year-Long Journey and Plans for the Future. The Journals of Gerontology. Series A, Biological Sciences and Medical sciences. 2008;63:1416–1419. doi: 10.1093/gerona/63.12.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer KE, Austad SN. The Development of Small Primate Models for Aging Research. Ilar J. 2011;52:78–88. doi: 10.1093/ilar.52.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S, Bundy JG, Davies SK, Viney JM, Swire JS, Leroi AM. A metabolic signature of long life in Caenorhabditis elegans. BMC Biol. 2010;8 doi: 10.1186/1741-7007-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensler HL, Bernstein H. DNA damage as the primary cause of aging. The Quarterly Review of Biology. 1981;56:279–303. doi: 10.1086/412317. [DOI] [PubMed] [Google Scholar]
- Go Y-M, Uppal K, Walker DI, Dury L, Strobel FH, Baudichon-Cortay H, Roede JR, Jones DP. Raftery D, editor. Mitochondrial Metabolomics Using High- Resolution Fourier-Transform Mass Spectrometry, in Mass Spectrometry in Metabolomics Methods and Protocols. Methods in Molecular Biology. 2014 doi: 10.1007/978-1-4939-1258-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higley JD, Suomi SJ, Linnoila M. A Longitudinal Assessment of Csf Monoamine Metabolite and Plasma-Cortisol Concentrations in Young Rhesus- Monkeys. Biol Psychiat. 1992;32:127–145. doi: 10.1016/0006-3223(92)90016-s. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. DNA damage, aging, and cancer. The New England Journal of Medicine. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Hoffman JM, Soltow QA, Li SZ, Sidik A, Jones DP, Promislow DEL. Effects of age, sex, and genotype on high-sensitivity metabolomic profiles in the fruit fly, Drosophila melanogaster. Aging Cell. 2014;13:596–604. doi: 10.1111/acel.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Argmann C, Houten SM, Canto C, Jeninga EH, Andreux PA, Thomas C, Doenlen R, Schoonjans K, Auwerx J. The metabolic footprint of aging in mice. Scientific Reports. 2011;1 doi: 10.1038/srep00134. Article 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Yu TW, Strobel FH, Jones DP. A practical approach to detect unique metabolic patterns for personalized medicine. Analyst. 2010;135:2864–2870. doi: 10.1039/c0an00333f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TE. Recent results: biomarkers of aging. Exp Gerontol. 2006;41:1243–1246. doi: 10.1016/j.exger.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Jones DP, Park Y, Ziegler TR. Nutritional metabolomics: progress in addressing complexity in diet and health. Annual Review of Nutrition. 2012;32:183–202. doi: 10.1146/annurev-nutr-072610-145159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehnel F, Grohmann J, Buchwald U, Koeller G, Teupser D, Einspanier A. Parameters of haematology, clinical chemistry and lipid metabolism in the common marmoset and alterations under stress conditions. J Med Primatol. 2012;41:241–250. doi: 10.1111/j.1600-0684.2012.00550.x. [DOI] [PubMed] [Google Scholar]
- Lawton KA, Berger A, Mitchell M, Milgram KE, Evans AM, Guo L, Hanson RW, Kalhan SC, Ryals JA, Milburn MV. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008;9:383–397. doi: 10.2217/14622416.9.4.383. [DOI] [PubMed] [Google Scholar]
- Li SZ, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, Jones DP, Pulendran B. Predicting network activity from high throughput metabolomics. Plos Comput Biol. 2013;9:e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Wang D, Chen YN, Sun HJ, He SR, Wang CS, Yang G, Shi MM, Zhang J, Ren Y, Wang L, Lu YR, Cheng JQ. H-1 NMR-based metabonomic analysis of serum and urine in a nonhuman primate model of diabetic nephropathy. Mol Biosyst. 2013;9:2645–2652. doi: 10.1039/c3mb70212j. [DOI] [PubMed] [Google Scholar]
- Mather KA, Jorm AF, Parslow RA, Christensen H. Is telomere length a biomarker of aging? A review. The Journals of Gerontology. Series A, Biological Sciences and Medical sciences. 2011;66:202–213. doi: 10.1093/gerona/glq180. [DOI] [PubMed] [Google Scholar]
- Muehlenbein MP, Campbell BC, Richards RJ, Svec F, Phillippi- Falkenstein KM, Murchison MA, Myers L. Dehydroepiandrosterone sulfate as a biomarker of senescence in male non-human primates. Exp Gerontol. 2003;38:1077–1085. doi: 10.1016/j.exger.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Nishijima K, Saitoh R, Tanaka S, Ohsato-Suzuki M, Ohno T, Kitajima S. Life span of common marmoset (Callithrix jacchus) at CLEA Japan breeding colony. Biogerontology. 2012;13:439–443. doi: 10.1007/s10522-012-9388-1. [DOI] [PubMed] [Google Scholar]
- Patterson AD, Bonzo JA, Li F, Krausz KW, Eichler GS, Aslam S, Tigno X, Weinstein JN, Hansen BC, Idle JR, Gonzalez FJ. Metabolomics Reveals Attenuation of the SLC6A20 Kidney Transporter in Nonhuman Primate and Mouse Models of Type 2 Diabetes Mellitus. J Biol Chem. 2011;286:19511–19522. doi: 10.1074/jbc.M111.221739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinherio J, Bates D, DebRoy S, Sarkar D, Team RC. nlme: Linear and Nonlinear Mixed Effect Models. R package version 3. 2012:1–118. [Google Scholar]
- Psihogios NG, Gazi IF, Elisaf MS, Seferiadis KI, Bairaktari ET. Gender-related and age-related urinalysis of healthy subjects by NMR-based metabonomics. NMR in Biomedicine. 2008;21:195–207. doi: 10.1002/nbm.1176. [DOI] [PubMed] [Google Scholar]
- Queiroz O. Circadian-Rhythms and Metabolic Patterns. Annu Rev Plant Phys. 1974;25:115–134. [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- Roede JR, Uppar K, Yongliang L, Promislow DEL, Wachtman LM, Jones DP. Characterization of plasma thiol redox potential in a common marmoset model of aging. Redox Biology. 2013;1:387–393. doi: 10.1016/j.redox.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, Garinis GA, Hoeijmakers JH. Age to survive: DNA damage and aging. Trends in Genetics : TIG. 2008;24:77–85. doi: 10.1016/j.tig.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Slupsky CM, Rankin KN, Wagner J, Fu H, Chang D, Weljie AM, Saude EJ, Lix B, Adamko DJ, Shah S, Greiner R, Sykes BD, Marrie TJ. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Analytical Chemistry. 2007;79:6995–7004. doi: 10.1021/ac0708588. [DOI] [PubMed] [Google Scholar]
- Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High-performance metabolic profiling with dual chromatography-Fourier- transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics. 2013;9:S132–S143. doi: 10.1007/s11306-011-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbaugh MJ, Sun LY, Bartke A, Miller RA. Activation of genes involved in xenobiotic metabolism is a shared signature of mouse models with extended lifespan. Am J Physiol-Endoc M. 2012;303:E488–E495. doi: 10.1152/ajpendo.00110.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanska E, Bouwman J, Strassburg K, Vervoort J, Kangas AJ, Soininen P, Ala-Korpela M, Westerhuis J, van Duynhoven JP, Mela DJ, Macdonald IA, Vreeken RJ, Smilde AK, Jacobs DM. Gender-dependent associations of metabolite profiles and body fat distribution in a healthy population with central obesity: towards metabolomics diagnostics. Omics : A Journal of Integrative Biology. 2012;16:652–667. doi: 10.1089/omi.2012.0062. [DOI] [PubMed] [Google Scholar]
- Tardif SD, Mansfield KG, Ratnam R, Ross CN, Ziegler TE. The Marmoset as a Model of Aging and Age-Related Diseases. Ilar J. 2011;52:54–65. doi: 10.1093/ilar.52.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K, Soltow QA, Strobel FH, Pittard WS, Gernert KM, Yu TW, Jones DP. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics. 2013;14 doi: 10.1186/1471-2105-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goot AT, Nollen EA. Tryptophan metabolism: entering the field of aging and age-related pathologies. Trends in Molecular Medicine. 2013;19:336–344. doi: 10.1016/j.molmed.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Wahl S, Vogt S, Stuckler F, Krumsiek J, Bartel J, Kacprowski T, Schramm K, Carstensen M, Rathmann W, Roden M, Jourdan C, Kangas AJ, Soininen P, Ala-Korpela M, Nothlings U, Boeing H, Theis FJ, Meisinger C, Waldenberger M, Suhre K, Homuth G, Gieger C, Kastenmuller G, Illig T, Linseisen J, Peters A, Prokisch H, Herder C, Thorand B, Grallert H. Multi-omic signature of body weight change: results from a population-based cohort study. BMC Medicine. 2015;13:48. doi: 10.1186/s12916-015-0282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Tamura T, Takaoka C, Harada K, Kobayashi A, Mukai Y, Fukusaki E. Metabolomics-based systematic prediction of yeast lifespan and its application for semi-rational screening of ageing-related mutants. Aging Cell. 2010;9:616–625. doi: 10.1111/j.1474-9726.2010.00590.x. [DOI] [PubMed] [Google Scholar]
- Yousri NA, Kastenmuller G, Gieger C, Shin SY, Erte I, Menni C, Peters A, Meisinger C, Mohney RP, Illig T, Adamski J, Soranzo N, Spector TD, Suhre K. Long term conservation of human metabolic phenotypes and link to heritability. Metabolomics. 2014;10:1005–1017. doi: 10.1007/s11306-014-0629-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Park Y, Johnson JM, Jones DP. apLCMS-adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25:1930–1936. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZH, Zhai GJ, Singmann P, He Y, Xu T, Prehn C, Roemisch-Margl W, Lattka E, Gieger C, Soranzo N, Heinrich J, Standl M, Thiering E, Mittelstrass K, Wichmann HE, Peters A, Suhre K, Li YX, Adamski J, Spector TD, Illig T, Wang-Sattler R. Human serum metabolic profiles are age dependent. Aging Cell. 2012;11:960–967. doi: 10.1111/j.1474-9726.2012.00865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi P, Wickings EJ, Nieschlag E. The Effects of Ketamine Hcl and Barbiturate Anesthesia on the Metabolic-Clearance and Production-Rates of Testosterone in the Male Rhesus-Monkey, Macaca-Mulatta. J Steroid Biochem. 1982;16:463–466. doi: 10.1016/0022-4731(82)90061-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.