Abstract
Linker histones H1 are ubiquitous chromatin proteins that play important roles in chromatin compaction, transcription regulation, nucleosome spacing and chromosome spacing. H1 function in DNA and chromatin structure stabilization is well studied and established. The current paradigm of linker histone mode of function considers all other cellular roles of linker histones to be a consequence from H1 chromatin compaction and repression. Here we review the multiple processes regulated by linker histones and the emerging importance of protein interactions in H1 functioning. We propose a new paradigm which explains the multi functionality of linker histones through linker histones protein interactions as a way to directly regulate recruitment of proteins to chromatin.
Introduction
In eukaryotes, genomic DNA is packaged into a complex nucleoprotein structure called chromatin, composed of repetitive arrays of nucleosomes. The nucleosome consists of 145-147 base pairs (bp) of DNA wrapped around an octamer of two core histone H2A-H2B dimers and one core histone H3-H4 tetramer. DNA that connects two adjacent nucleosomes is called linker DNA and usually is 10–80 bp in length [1]. In contrast to core histones, the H1 linker histones are not components of the nucleosome core [2,3], but instead bind to the linker DNA at the nucleosome entry and exit sites, stabilizing the entire complex [4,5]. Linker histones are abundant, with a stoichiometry of ~0.8 total linker histones per nucleosome in most tissues and closer to one in highly condensed heterochromatin [1]. The linker histones are multifunctional, with roles in chromatin compaction [6–8], regulation of gene expression [9–12], and other key aspects of genome biology [13–15]. This review focuses on the emerging role of protein-protein interactions in linker histone action.
Linker histones have a tripartite organization, consisting of an unstructured N-terminal domain (NTD, 13–40 amino acids in length), a central folded globular domain (GD, ~80 amino acids) and a C-terminal unstructured domain (CTD, ~100 amino acids) [16]. The globular domain is a winged helix DNA binding domain that interacts with nucleosomes. The CTD interacts with linker DNA and is essential for stabilizing higher-order chromatin structure [17]. The CTD and NTD of H1 are highly basic and enriched in disorder promoting amino acids, such as lysine, alanine and proline [8]. These regions are predicted to be intrinsically disordered, i.e. they do not have a well-defined secondary structure. The flexibility of intrinsically disordered proteins (IDP) allows them to accommodate different structures when interacting with proteins and/or nucleic acids, adapt to their ligands and acquire some interaction-dependent secondary structure [8]. Accordingly, the disordered H1 NTD and CTD acquire secondary structure when bound to DNA [18,19], nucleosomes [17,20] and, possibly other proteins.
Linker histones are the most diverse class of histone proteins. There are 11 different histone isoforms in mammalian cells. Five of them are somatic variants specific to the S phase of mammalian cells and are expressed in a replication dependent manner (H1.1, H1.2, H1.3, H1.4 and H1.5). Two more are somatic variants encoded by solitary genes expressed in a replication independent mode (H1.0 and H1x) and more prevalent in differentiated cells [21]. Three other variants are testis specific (H1t, H1T2 and HILS1) and one is specific to oocytes (H1oo). Somatic linker histone variants differ mostly in their CTD primary sequences, but have very similar amino acid composition [22,23]. A triple knock-out of three of the somatic isoforms accompanied by 50% reduction in total H1 content results in embryonic lethality in mice [24,25]. The H1.0, H1.1, H1.2, and H1.3 isoforms were found to be more prevalent in euchromatin regions, while histones H1.4 and H1.5 are more abundant in heterochromatin regions of the nucleus [26]. Various isoforms were reported to have different affinities for the nucleosome [26,27]. H1.1 is associated with more actively transcribed regions of chromatin, while H1.2, H1.4 and H1.5 are associated with less active to inactive regions [25,28–31]. As will be discussed further below, the H1 isoforms show both redundancy and specificity in protein-protein interactions. As is discussed in detail in chapters six and seven of this special issue and has recently been reviewed [32,33], all of the isoforms have numerous sites of post-translational modifications. In particular, the NTD is acetylated and phosphorylated throughout its entire sequence. The CTD is acetylated, methylated, and phosphorylated, primarily in the central region of the domain. The GD is acetylated, ubuiquitinated, and formylated. As will be discussed throughout the article, H1 post-translational modifications likely serve as functional switches for modulating chromatin architecture and protein-protein interactions.
The prevailing linker histone functional paradigm
H1 first and foremost is thought of as a nucleosome and DNA binding chromatin architectural protein [6,10,34,35]. H1 stabilizes nucleosomes by binding to the dyad region between the entry and exit sites of DNA. The H1 GD is sufficient for binding to the nucleosome. Binding of the GD protects an additional 10bp from endonuclease digestion at both sides of nucleosome [4]. The role of H1 in the formation and stabilization of higher order chromatin structures has long been known [36–38]. In vitro, H1 facilitates both local nucleosome-nucleosome interactions that cause folding into compact 30nm fibers [7,17,39] and the intermolecular nucleosome-nucleosome interactions that bring chromatin fibers together to form highly condensed chromatin oligomers [7,17]. The CTD binding to linker DNA is essential for H1-dependent stabilization of folded and oligomeric chromatin [17,38,40]. Interestingly, CTD amino acid composition, and not its primary sequence, is the primary determinant of CTD function during chromatin compaction [23], consistent with its intrinsically disordered structure. When H1 is depleted in vivo nuclei become enlarged [41,42], consistent decreased chromatin condensation in the knockout cells. Collectively, there is considerable evidence spanning nearly four decades indicating that H1 promotes stabilization of higher order chromatin structures [6,7,17,37,38].
H1 is also known to be a mediator of transcription. Based on the in vitro studies, H1 was long presumed to be a general repressor of transcription [10,35] by promoting a more condensed chromatin architecture. Indeed, the binding of H1 to DNA and chromatin in vitro impedes the access of RNA polymerases and represses transcription [43,44]. However, it is now clear that in vivo H1 functions both as a repressor and activator of specific gene transcription [11,12,14,45]. In an H1 knockout strain of Tetrahymena thermophila the total level of transcribed mRNA does not change, ruling out the role of H1 as a general repressor of transcription [11]. The authors showed that H1 was acting as either repressor or activator of transcription depending on the specific gene [11,12,45]. Likewise, in the H1 triple knockout mouse system, the expression of certain specific genes was increased, while other genes were decreased [24]. Moreover, it was demonstrated that the histone H1 immunosignal was detected at higher levels at transcriptionally active genes than repressed genes [46], indicating that H1 is still bound to the chromatin template during transcription by RNA polymerase. H1 posttranslational modifications likely have an important role in transcription regulation. For example, H1 partial phosphorylation was reported to decrease the affinity of H1 for chromatin, resulting in less compacted chromatin and increased transcription [47–50]. Likewise, as will be discussed below, H1 post-translational modifications may modulate specific protein-protein interactions. There is an increasing evidence of H1 roles in other cellular processes. H1 is involved in global temporal regulation of DNA replication [15]. H1 represses homologous recombination [51] and DNA damage repair [52] and is essential for chromosome segregation [13]. Ultimately, it is difficult to explain the effects of linker histones on transcription and other genomic functions purely in terms of modulation of chromatin architecture.
In this regard, there is increasing evidence that H1-protein interactions are implicated in transcription regulation. The repression of at least subset of genes, namely p53 target genes, is mediated by the interaction of H1 with CHD8 protein [53]. The complex that H1.2 forms with other proteins was shown to be repressive for p53-dependent Bax gene transcription by repressing p300-mediated chromatin acetylation [54]. Recently, another study demonstrated interaction of prothymosin alpha (ProTα) with the H1.2 CTD domain resulting in a decrease of p53-dependent transcription repression [55]. The remainder of this article focuses on H1-dependent protein-protein interactions in the nucleus and nucleolus
Incorporating protein-protein interactions into the H1 paradigm
A well established property of intrinsically disordered proteins is their ability to bind to multiple macromolecular partners, and many bind both protein and DNA [8]. Although H1 is a well characterized DNA binding protein, the extensive disorder present in the NTD and CTD raises the possibility that H1 also interacts specifically with other proteins. An emerging body of structural data supports this view.
There are many different ways to identify protein-protein interactions. One of the first studies demonstrating H1-protein interactions was performed using immunoprecipitations with exogenous Flag-tagged Msx1 and anti-Flag antibodies. They identified H1.5 as a major interacting partner of Msx1, and that the reported interaction was associated with repressed chromatin [9]. Another study used antibodies which specifically recognized either the NTD or CTD of D. melanogaster H1 and pull-downs with nuclear extract from D. melanogaster Kc cells, followed by mass spectrometry to identify interaction partners [12]. The authors found that H1 co-immunoprecipitates with the 40S and 60S ribosomal components (L6e, L7, L7a, L8, L9, L10, L11, L13, L15, L18, L19, L22, L23, S3, S2, S3a, S4, S6, S8) and two heterogeneous nuclear ribonucleoproteins (hnRNP48 and hnRNP36) [12]. Sucrose gradient co-purification showed that H1 is in the same complex as tagged ribosomal protein L22. The authors suggested this complex is chromatin-specific and involved in transcription repression of certain genes since both overexpression of tagged H1 and H1 knockdown affected RNA transcription and both H1 and L22 were present on chromatin of most of affected genes as revealed by ChIP [12].
In another approach An and co-workers overexpressed FLAG- and HA-tagged H1.2 in HeLa-S cells [54]. They prepared nuclear extract and purified from it an H1.2-containing complex using cation-exchange chromatography followed by affinity chromatography with immobilized anti-FLAG antibodies. The authors showed that H1.2 co-purified as a single multiprotein complex, and identified its components using mass spectrometry. They detected four ribosomal proteins (L13a, L7a, L22, and S3), four co-repressor proteins (YB1, FIR, PARP1, and PURα), hnRNPK and several additional factors (ASXL1, nucleolin, β-catenin, TGase7, CAPERα, Importin7/90, DNA-PK, PP1 and WDR5) [54]. They also showed that the identified complex had an inhibitory effect on the transcription of in vitro assembled chromatin templates [54].
Several articles have addressed whether the described H1 interactions are direct. Widlak et al [56] used recombinant proteins to show that all somatic H1 isoforms activate DFF40/CAD nuclease activity acting through the H1 CTD. Recombinant proteins and immunoblotting also were used to confirm that some of the components of the H1 complex discussed above directly interacted with GST-tagged H1.2 [54]. Direct H1.2 interaction partners included ASXL1, PARP1, FIR, CAPERα, YB1, PURα and WDR5. Our laboratory has used a fluorescence quenching assay to measure the KD for recombinant H1.0 binding to recombinant SRSF1, U2AF65 and FACT, and showed that the interactions occurred with moderate (μM) to strong (nM) affinity [57].
Recently, proteomics methods were used to investigate the H1 binding proteins present in human nuclear and nucleolar extracts. H1.0 was covalently immobilized to sepharose using the HaloTag system and mixed with nuclear extracts from four different human cell lines (HeLa, CEM, RPE-1 and U2OS). All proteins that bound to HaloTag-fused H1.0 were subjected to mass spectrometry [57]. This approach identified 107 proteins that interacted with H1.0 either directly or indirectly. In this type of affinity binding experiment one has to be concerned with specificity. In particular, it is possible that the highly basic and disordered H1.0 protein (calculated pI of 10.9) simply binds non-specifically to many acidic proteins in the extracts. However, the calculated pIs of the nuclear H1 binding proteins ranged from 3.8 to 11.9 with nearly 50% of the identified proteins having a pI ≥ 9.0 (Fig. 1). This indicates that the H1.0–protein interactions did not result solely from non-specific electrostatic binding [57]. Moreover, the KDs of H1.0 for three of the proteins identified in the extract experiments (SRSF1, U2AF65, FACT) were in the micromolar to nanomolar range, well below the millimolar affinities that would be expected for a non-specific interaction. Most of the interactions persisted with RNAse treatment, suggesting that the H1-protein interactions generally were not mediated by RNA. At this point, the available evidence favors the interpretation that the majority of the H1-protein interactions identified in nuclear extracts are direct and specific. However, to fully answer this question it will be necessary to perform physicochemical studies of many more H1-protein interactions in purified systems. In preliminary experiments we observed that about 75% of the proteins in nuclear extracts that bound immobilized H1.0 also bound immobilized H1.3 (unpublished). On the other hand, Skoultchi and co-workers have demonstrated that H1 binding to DNMT1 and DNMT3B is isoform-specific (see Transcription section below). Thus, as is the case with many other aspects of H1 function [23,24,26,58], for protein-protein interactions some isoform specificity appears to be overlaid on top of general isoform redundancy. Because so many of the nuclear H1 binding proteins had nucleolar functions, the same proteomics approach was used to survey H1.0-dependent protein-protein interactions in human nucleolar extracts. We identified 175 nucleolar H1.0 binding proteins [59]. About 25% of the nucleolar H1.0 binding proteins overlapped with the nuclear H1.0 binding proteins, and the nucleolar H1.0 binding proteins had the same broad distribution of calculated pIs. Together, our biochemical studies demonstrated that the scope of H1-protein interactions in the nucleus is much greater than had been previously appreciated.
Figure 1.
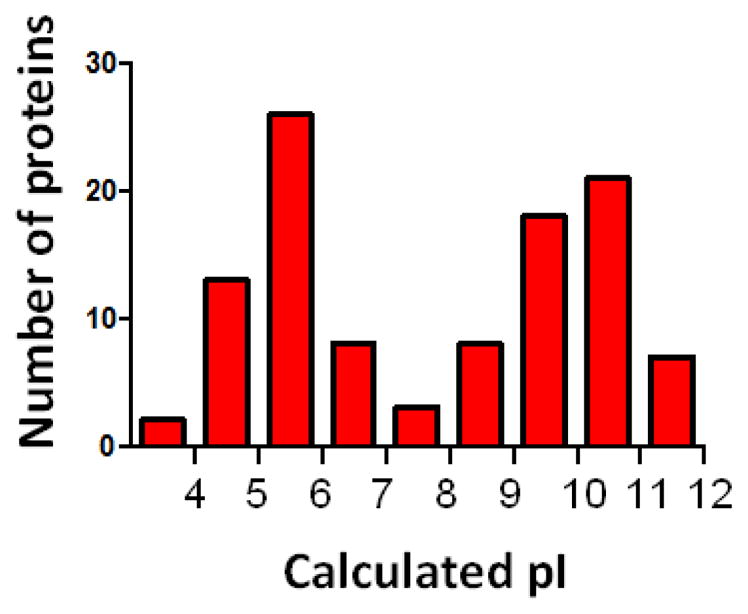
Distribution of calculated pIs for nuclear H1-binding proteins.
Categories of H1-protein interactions
The number of H1 binding proteins identified to date is large and encompasses a range of different functional categories. Discussed below are classes of H1 binding proteins that are relevant to genome biologists.
Pre-mRNA splicing
The single largest category of H1.0 binding proteins identified by the proteomics experiments is pre-mRNA splicing. Splicing is carried out by the spliceosome complex [60]. Proteomic analyses of spliceosomes assembled onto pre-mRNAs in HeLa nuclear extracts identified several hundred human spliceosome-associated proteins [61,62]. Thirty-three spliceosomal proteins in nuclear extracts bound to immobilized H1.0; together these constitute nearly a third of the H1.0 interaction partners identified. Similarly, of the 175 H1.0 binding proteins identified in human nucleolar extracts, one third are components of the spliceosome [59]. Pre-mRNA splicing is initiated when U1 snRNP particle binds to the 5′ end of the intron, while the 3′ end is recognized by U2AF35-U2AF65 heterodimer. The serine/arginine-rich splicing factors (SR proteins) facilitate these steps. H1.0 interacts with U2AF35, U2AF65 and two SR proteins (SRSF1, SRSF2). Moreover, the interactions of H1.0 with U2AF65 and SRSF1 are direct, implying that H1.0 may play a role in spliceosome recruitment to genomic locations. hnRNP proteins repress splice site recognition by competing for the same pre-RNA binding sites as the SR proteins [63]. H1.0 pulled down numerous hnRNPs (A2/B1, C1/C2, D, F, G, H, M, U, UL1). This raises the possibility that linker histones can both activate and repress mRNA splicing, and hence specific gene expression [24], via regulatory interactions with SRs, hnRNPs, U2AF, and U1 snRNP proteins. Future studies will be necessary to directly test the functional role of H1-splicing protein interactions. For example, analysis of RNA sequencing data obtained from wild type and triple-H1 knockout mouse embryonic stem cells [24] should indicate whether splice site recognition or other steps in pre-mRNA splicing is both positively and negatively regulated at specific genes by H1 in vivo.
Core histone chaperones
Three core histone binding chaperone proteins present in human nuclear extracts (FACT, Nap1L1, ANP32E) formed complexes with immobilized H1.0 in our affinity binding experiments [57]. FACT consists of the Spt16 and SSRP1 subunits [64], and both subunits were identified by mass spectrometry as interacting with H1.0. FACT binds H2A/H2B dimers with nanomolar affinity [65], and can mediate both disassembly and reassembly of nucleosomal substrates in vitro [66]. The finding that FACT binds strongly and directly to H1.0 (KD ~32 nM) suggests that FACT also may be a linker histone chaperone that can deposit and remove H1 on chromatin fibers. NAP1L1 is a structurally unrelated histone chaperone that mediate nucleosome formation and disassembly, and play roles in transport and deposition of histone H2A-H2B dimer [67]. In nucleolar extracts, immobilized H1.0 bound to FACT, nucleolin (NCL), nucleophosmin (NPM), SET, DEK, NAP1L1, NAP1L4, Spt6, HIRAIP3, and Brd4 [59], all core histone chaperones. Thus, in almost every case core histone chaperones also appear to be linker histone binding proteins. The functional role(s) that H1-histone chaperone interactions play in storage, removal and deposition of H1 in chromatin in vivo remains to be determined.
Transcription
H1.0 binds to a number of transcription activators and facilitators, such as transcription elongation factor FACT, RNA binding protein 39, TATA-binding protein-associated factor 2N, and casein kinase II subunits, YBX1, ILF2, nucleolin and HDAC2 [57]. Interestingly, a previous study found that H1.2 is a component of a stable multi-protein complex that includes YB1, PURα, and nucleolin [54]. Recently it was demonstrated that H1 plays an important role in epigenetic silencing [68]. Skoultchi and co-workers studied triple-H1 knockout (H1ΔTKO) mouse embryonic stem cells with reduced levels of H1.2, H1.3 and H1.4. They detected a significant reduction in the level of DNA methylation of genes. They also demonstrated that these changes in methylation are due to reduced recruitment of two specific DNA methyltransferases, DNMT1 and DNMT3B. DNMT1 and DNMT3b are recruited to chromatin through direct interaction with H1.3 and the CTD is essential for this interaction. H1.1, H1.4 and H1.5 also interacted with DNMT1 and DNMT3b, but H1.2 did not show any binding to DNMT1 and DNMT3b or recruitment of these DNA methyltransferases to genes. H1.3 also inhibited binding of lysine methyltransferase SET7/9 to chromatin and methylation of lysine 4 on histone H3 tail. These studies have documented that protein-protein interactions involving H1 play a role in establishing epigenetically silent chromatin. H1.3 and several other isoforms of H1 may function in epigenetic silencing of chromatin through direct interaction with epigenetic modifiers. Taken together, these results suggest a second molecular mechanism (in addition to regulation of pre-mRNA splicing) through which H1 can positively or negatively influence specific RNA polymerase II-dependent transcription, in this case by mediating direct interactions with positive or negative regulatory transcription proteins.
DNA damage response
One of the most prominent H1.0 interaction partners in our proteomics experiments was valosin-containing protein (VCP). In neural cells with abnormal protein accumulation VCP translocates to the nucleus, leading to core histone deacetylation and transcription suppression [69]. VCP has also been shown to play a role in DNA damage repair [70]. VCP acts as a “screw” on the sites of DNA damage repair, pulling away the proteins and making DNA more accessible for DNA repair machinery. [70]. H1.0 also interacts with other proteins involved in DNA damage response, such as Ku86 (XRCC5) and Ku70 (XRCC6) [57]. Following DNA damage, global chromatin compaction stabilized by linker histone H1.0 occurs [71]. This compaction could play a role in cell protection from further damage [71]. H1.0 interaction with DNA damage repair factors may be crucial for its deposition on chromatin and/or its epigenetic modifications following DNA damage.
Translation
In early studies it was demonstrated that Drosophila H1 co-immunoprecipitates with 40S and 60S ribosomal components [12], and that overexpressed H1.2 co-purified as a single complex with several ribosomal proteins [54]. In our H1.0 affinity binding experiments we identified six 40S and sixteen 60S ribosomal proteins, five eukaryotic translation initiation factor 3 (eIF3) subunits, and three signal recognition particle subunits in nuclear extracts [57]. Of note, only a few ribosomal proteins and one eIF3 subunit were identified in nucleolar extracts [59], suggesting a role for H1 in ribosome function in the nucleoplasm more so than ribosome pre-assembly in the nucleolus. Nuclear translation, while controversial, has been reported by several groups [72–74]. Also of potential interest, a “translasome” complex composed in part of ribosomes, eIF3, 26S proteosome subunits and actin has been identified in the cytoplasm and nucleus of S. pombe [75]. All of these proteins are present in human nuclear extracts and interact with H1.0 ([59], and unpublished data).
H1-dependent protein-protein interactions and the nucleolus
When we scrutinized the list of H1.0 binding proteins identified in nuclear extracts, we noticed most of the proteins were reported to function in part in the nucleolus. The nucleolus is a non-membranous multifunctional nuclear organelle [76]. Besides functioning in ribosome biogenesis, which consists of the coupled processes of rRNA synthesis, rRNA processing, and pre-ribosome assembly, the nucleolus is involved in cell cycle control, DNA damage repair, apoptosis and development [76]. To directly investigate the role of H1-protein interactions in the nucleolus we performed affinity binding experiments with immobilized H1.0 and extracts prepared from purified Jurkat cell nucleoli, as well as proteomics profiling of nucleoli isolated from wild type and H1ΔTKO mouse embryonic stem cells.
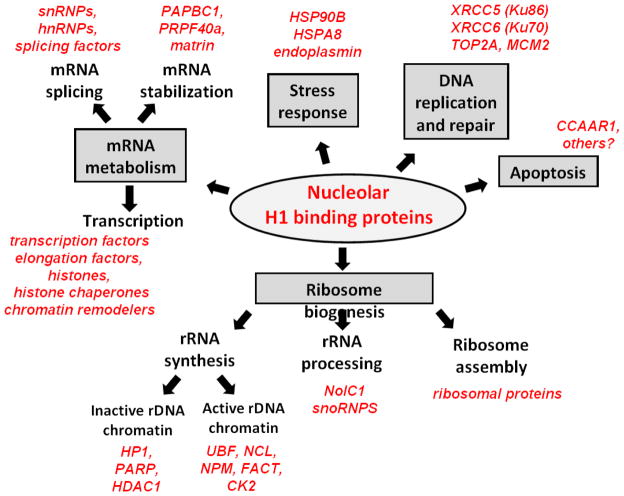
The affinity binding experiments identified 175 proteins in nucleolar extracts that bound to immobilized H1.0 [59], encompassing most aspects of nucleolar function (Fig. 2). H1.0 bound to HP1, PARP, and HDAC1, which work together to repress rRNA genes [77–79], and UBF, NCL, NPM, TOPI, FACT and CK2, which all have a role in active rRNA transcription [80–85]. These results suggest that H1 may regulate rRNA transcription, consistent with the finding that phosphorylated nucleolar H1.2 and H1.4 are associated with increased RNA polymerase I activity and rRNA synthesis [50]. Mechanistically, we speculate that H1 acts through chromatin-coupled protein-protein interactions [59] rather than by modulating the higher order structure of rDNA chromatin as proposed by others [80]. NolC1 and snoRNPS are H1.0 binding proteins that function in rRNA processing [86,87]. The affinity binding studies also identified many nucleolar H1.0 binding proteins involved in Pol II transcription (transcription factors, elongation factors, histone chaperones, chromatin remodelers), pre-mRNA splicing (snRNPS, hnRNPS, many splicing factors), and mRNA processing (PAPBC1, matrin). Non-traditional nucleolar functions were represented as well, including stress response (HSP90B, HSPA8), DNA replication and repair (XRCC5/Ku86, XRCC6/Ku70, TOP2A, MCM2), and apoptosis (CCAAR1). Based on the wide range of nucleolar proteins detected in our affinity binding experiments, we hypothesized that H1 is a key hub protein [59] necessary for the proper structure and function of the nucleolus. Consistent with this notion, H1 forms stable complexes within the nucleolus. Using size-exclusion chromatography we detected endogenous H1 in nucleolar fractions corresponding to ~378 to >670 kDa [59]. Nine proteins identified in the affinity binding experiments were observed to co-elute with endogenous H1 by Western Blot. Interestingly, the peaks for concentration of different proteins were non-overlapping, suggesting that H1 is a part of a number of large macromolecular complexes in the nucleolus.
Figure 2.
A synopsis of H1-protein interactions in the nucleolus. Adapted from [59].
A direct prediction of the hypothesis that H1 is a nucleolar hub protein is that cellular depletion of H1 would disrupt the H1 protein-protein interaction network and influence the structure and function of the nucleolus. To address this hypothesis, nucleoli were purified from wild type and H1ΔTKO mouse embryonic stem cells and subjected to mass spectrometry to determine the nucleolar protein composition [59]. Of the 613 different proteins identified in the wild type nucleoli, 239 proteins were absent from the H1ΔTKO nucleoli. Concomitantly, the H1ΔTKO nucleoli were reduced in size by 20% and RNA content by 2.5-fold [59]. Spectral counting analysis quantitated 135 proteins that were significantly affected by H1 depletion. The ontological functional groups of the affected proteins showed a strong overlap with groups identified by the H1 affinity binding experiments, and included gene silencing, nucleosome organization, transport proteins, rRNA processing, and mRNA splicing. Taken together, our affinity binding and proteomics profiling analyses support a model in which H1 is extensively involved in nucleolar protein-protein interactions. However, while the proteomics profiling data add physiological relevance to the biochemical results, a direct functional role for H1 in the nucleolus has yet to be demonstrated.
Concluding remarks
The current linker histone paradigm holds that H1 is a ubiquitous chromatin architectural protein with important roles in maintaining nuclear chromatin structure [36], [4,5]. However, the data summarized in this review support a new paradigm in which the multifunctionality of H1 originates at least in part from interactions with numerous nuclear and nucleolar proteins. It should be noted that the evidence in support of H1-protein interactions is almost exclusively biochemical. As such, there is very little data to indicate that these protein-protein interactions are functionally relevant, and direct functional studies of H1-protein interactions in vivo will be an important area of future research.
What is the mechanistic basis for H1-protein interactions? In particular, what are the contributions of the NTD, GD, and CTD? Our proteomics studies indicate that ~25% of the H1.0-protein interactions detected in the nuclear extracts are completely abolished by removal of the CTD [57]. Thus, the CTD, NTD and GD all make important contributions depending on the specific H1-protein interaction. For example, FACT binds to full-length H1.0 with a KD of ~32 nM [57], whereas the KD for FACT binding to the NTD-GD fragment is ~75 nM (unpublished). In this particular case, the NTD-GD interaction provides the bulk of the binding energy, although the CTD participates as well. For most of the identified interactions the relative contributions of the three H1 domains responsible for these interactions have yet to be identified. While it is possible that in some cases H1 functions as a modular protein with different domains responsible for different interactions, we suspect that the concerted contribution of all three domains underlies the ability of H1 to specifically interact with most proteins. In the case of the CTD and NTD, the structural malleability inherent to these intrinsically disordered domains is likely to facilitate a wide range of specific protein-protein interactions [57,59]. Another important factor is likely to be H1 post-translational modifications. Different specific post-translational modifications may modulate protein-protein interactions by affecting the affinity of the interaction, leading to downstream functional effects For example, H1.0 was found to bind to both positive (SRSFs) and negative (hnRNPs) splice site recognition proteins [57], raising the question of how H1 can regulate the direction of splice site recognition at a specific gene. One explanation is that post-translational modifications may control whether H1 binds to SRSFs or hnRNPs, allowing the modifications to mediate the functionality of the H1-protein interaction. In another example, Lys26-methylation of histone H1.4 promotes specific binding to HP1, while simultaneous Ser27 phosphorylation negates H1-HP1 interactions [88].
H1 is bound throughout most eukaryotic genomes at nearly one H1 protein per nucleosome [1], implying critical chromatin-based functions. For many years attention has been focused on H1-dependent modulation of chromatin architecture. In view of the evidence discussed in this article, we speculate that another key function of H1 may be to physically couple nuclear processes mediated by protein-protein interactions to the chromatin environment in which they occur. For example, in the studies of Skoultchi and co-workers showed that decreased methylation of specific upregulated genes resulted from reduced recruitment of two specific DNA methyltransferases, DNMT1 and DNMT3B to chromatin through direct interaction with H1.3 [68]. H1.3 also inhibited binding of lysine methyltransferase SET7/9 to chromatin and methylation of lysine 4 on histone H3 tail [68]. Thus, H1-dependent protein-protein interactions involving H1 play a role in establishing epigenetically silent chromatin. In the future it will be interesting to determine the extent to which H1 can make ternary complexes with nucleosomes and its protein binding partners. Linker histones are highly mobile in the nucleus [89,90], suggesting that there is a pool of free H1 transiently available to interact with other proteins in nuclear chromatin and the nucleolus. Moreover, H1 in principle should participate in the same interactions in both the nucleoplasm and the nucleolus. The nucleolus is a steady-state structure whose protein constituents are in equilibrium with the nucleoplasm [91,92]. By linking events that take place in nuclear chromatin to those that occur in the nucleolus, we speculate that H1 functions as a master regulator of nuclear function acting through chromatin-coupled protein-protein interactions.
Highlights for Review.
A new paradigm is discussed focusing on the role of protein-protein interactions in linker histone H1 action.
The involvement of nuclear H1-protein interactions is reviewed.
The involvement of H1-protein interactions in the nucleolus is highlighted.
Future directions are discussed.
Acknowledgments
THIS WORK WAS SUPPORTED BY AMERICAN HEART ASSOCIATION GRANT #15POST22770011 (KALASHNIKOVA), INTERNATIONAL RETT SYNDROME FOUNDATION AWARD # 2822 (KALASHNIKOVA) AND NIH GRANT GM45916 (HANSEN)
Footnotes
Statement of Conflicts of Interests
The authors declare no conflicts of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosom Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Zhou YB, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 1998;395:402–405. doi: 10.1038/26521. [DOI] [PubMed] [Google Scholar]
- 4.Syed SH, Goutte-Gattat D, Becker N, Meyer S, Shukla M, Hayes J, et al. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc Natl Acad Sci U S A. 2010;107:9620–5. doi: 10.1073/pnas.1000309107. 10.1073/pnas.1000309107/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1000309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer S, Becker NB, Syed SH, Goutte-Gattat D, Shukla MS, Hayes JJ, et al. From crystal and NMR structures, footprints and cryo-electron-micrographs to large and soft structures: Nanoscale modeling of the nucleosomal stem. Nucleic Acids Res. 2011;39:9139–9154. doi: 10.1093/nar/gkr573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bednar J, Horowitz Ra, Grigoryev Sa, Carruthers LM, Hansen JC, Koster aJ, et al. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci U S A. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carruthers LM, Bednar J, Woodcock CL, Hansen JC. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: Mechanistic ramifications for higher-order chromatin folding. Biochemistry. 1998;37:14776–14787. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- 8.Hansen JC, Lu X, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J Biol Chem. 2006;281:1853–1856. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]
- 9.Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- 10.Zlatanova J. Histone H1 and the regulation of transcription of eukaryotic genes. Trends Biochem Sci. 1990;15:273–276. doi: 10.1016/0968-0004(90)90053-E. [DOI] [PubMed] [Google Scholar]
- 11.Shen X, Gorovsky Ma. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475–83. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 12.Ni JQ, Liu LP, Hess D, Rietdorf J, Sun FL. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 2006;20:1959–1973. doi: 10.1101/gad.390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maresca TJ, Freedman BS, Heald R. Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J Cell Biol. 2005;169:859–869. doi: 10.1083/jcb.200503031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinbach OC, Wolffe aP, Rupp Ra. Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature. 1997;389:395–399. doi: 10.1038/38755. [DOI] [PubMed] [Google Scholar]
- 15.Thiriet C, Hayes JJ. Linker histone phosphorylation regulates global timing of replication origin firing. J Biol Chem. 2009;284:2823–2829. doi: 10.1074/jbc.M805617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allan AFJ, Hartman PG, Crane-Robinson C. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–9. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 17.Lu X, Hansen JC. Identification of Specific Functional Subdomains within the Linker Histone H10 C-terminal Domain. J Biol Chem. 2004;279:8701–8707. doi: 10.1074/jbc.M311348200. [DOI] [PubMed] [Google Scholar]
- 18.Vila R, Ponte I, Collado M, Arrondo JLR, Jiménez MA, Rico M, et al. DNA-induced α-Helical Structure in the NH2-terminal Domain of Histone H1. J Biol Chem. 2001;276:46429–46435. doi: 10.1074/jbc.M106952200. [DOI] [PubMed] [Google Scholar]
- 19.Roque A, Iloro I, Ponte I, Arrondo JLR, Suau P. DNA-induced secondary structure of the carboxyl-terminal domain of histone H1. J Biol Chem. 2005;280:32141–32147. doi: 10.1074/jbc.M505636200. [DOI] [PubMed] [Google Scholar]
- 20.Fang H, Clark DJ, Hayes JJ. DNA and nucleosomes direct distinct folding of a linker histone H1 C-terminal domain. Nucleic Acids Res. 2012;40:1475–1484. doi: 10.1093/nar/gkr866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terme JM, Sesé B, Millán-Ariño L, Mayor R, Belmonte JCI, Barrero MJ, et al. Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency. J Biol Chem. 2011;286:35347–35357. doi: 10.1074/jbc.M111.281923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendzel MJ, Lever Ma, Crawford E, Th’Ng JPH. The C-terminal Domain Is the Primary Determinant of Histone H1 Binding to Chromatin in Vivo. J Biol Chem. 2004;279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- 23.Lu X, Hamkalo B, Parseghian MH, Hansen JC. NIH Public Access. Biochemistry. 2009;48:164–172. doi: 10.1021/bi801636y.Chromatin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 25.Alami R, Fan Y, Pack S, Sonbuchner TM, Besse A, Lin Q, et al. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc Natl Acad Sci U S A. 2003;100:5920–5925. doi: 10.1073/pnas.0736105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Th’ng JPH, Sung R, Ye M, Hendzel MJ. H1 family histones in the nucleus: Control of binding and localization by the C-terminal domain. J Biol Chem. 2005;280:27809–27814. doi: 10.1074/jbc.M501627200. [DOI] [PubMed] [Google Scholar]
- 27.Orrego M, Ponte I, Roque A, Buschati N, Mora X, Suau P. Differential affinity of mammalian histone H1 somatic subtypes for DNA and chromatin. BMC Biol. 2007;5:22. doi: 10.1186/1741-7007-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millán-Ariño L, Islam ABMMK, Izquierdo-Bouldstridge A, Mayor R, Terme JM, Luque N, et al. Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Res. 2014;42:4474–4493. doi: 10.1093/nar/gku079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li JY, Patterson M, Mikkola HKa, Lowry WE, Kurdistani SK. Dynamic Distribution of Linker Histone H1.5 in Cellular Differentiation. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parseghian MH, Newcomb RL, Hamkalo Ba. Distribution of somatic H1 subtypes is non-random on active vs. inactive chromatin II: Distribution in human adult fibroblasts. J Cell Biochem. 2001;83:643–659. doi: 10.1002/jcb.1224. [DOI] [PubMed] [Google Scholar]
- 31.Parseghian M, Newcomb S, Winokur Robert L, Hamkalo Ba. The distribution of somatic H1 subtypes is non-random on active vs. inactive chromatin: distribution in human fetal fibroblasts. Chromosom Res. 2000;8:405–24. doi: 10.1023/a:1009262819961. [DOI] [PubMed] [Google Scholar]
- 32.Harshman SW, Young NL, Parthun MR, Freitas Ma. H1 histones: Current perspectives and challenges. Nucleic Acids Res. 2013;41:9593–9609. doi: 10.1093/nar/gkt700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Godde JS, Ura K. Cracking the enigmatic linker histone code. J Biochem. 2008;143:287–293. doi: 10.1093/jb/mvn013. [DOI] [PubMed] [Google Scholar]
- 34.Nalabothula N, McVicker G, Maiorano J, Martin R, Pritchard JK, Fondufe-Mittendorf YN. The chromatin architectural proteins HMGD1 and H1 bind reciprocally and have opposite effects on chromatin structure and gene regulation. BMC Genomics. 2014;15:92. doi: 10.1186/1471-2164-15-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Croston GE, Kerrigan LA, Lira LM, Marshak DR, Kadonaga JT. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991;251:643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 36.Vignali M, Workman JL. Location and function of linker histones. Nat Struct Biol. 1998;5:1025–1028. doi: 10.1038/4133. [DOI] [PubMed] [Google Scholar]
- 37.Thoma Koller F, Klug TA. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- 39.Robinson PJ, Rhodes D. Structure of the “30 nm” chromatin fibre: A key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Thomas J. Histone H1 : location and role. Curr Opin Cell Biol. 1999;11:312–317. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 41.Lu X, Wontakal SN, Emelyanov AV, Morcillo P, Konev AY, Fyodorov DV, et al. Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev. 2009;23:452–465. doi: 10.1101/gad.1749309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen X, Yu L, Weir JW, Gorovsky Ma. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 1995;82:47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 43.Shimamura A, Sapp M, Rodriguez-Campos A, Worcel A. Histone H1 represses transcription from minichromosomes assembled in vitro. Mol Cell Biol. 1989;9:5573–5584. doi: 10.1128/mcb.9.12.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laybourn PJ, Kadonaga JT. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA Polymerase II. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- 45.Lin Q, Inselman A, Han X, Xu H, Zhang W, Handel MA, et al. Reductions in linker histone levels are tolerated in developing spermatocytes but cause changes in specific gene expression. J Biol Chem. 2004;279:23525–23535. doi: 10.1074/jbc.M400925200. [DOI] [PubMed] [Google Scholar]
- 46.Ericsson C, Grossbach U, Bjorkroth B, Daneholt B. Presence of histone H1 on an active Balbiani ring gene. Cell. 1990;60:73–83. doi: 10.1016/0092-8674(90)90717-S. [DOI] [PubMed] [Google Scholar]
- 47.Herrera RE, Chen F, Weinberg Ra. Increased histone H1 phosphorylation and relaxed chromatin structure in Rb-deficient fibroblasts. Proc Natl Acad Sci U S A. 1996;93:11510–11515. doi: 10.1073/pnas.93.21.11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roque A, Ponte I, Arrondo JLR, Suau P. Phosphorylation of the carboxy-terminal domain of histone H1: Effects on secondary structure and DNA condensation. Nucleic Acids Res. 2008;36:4719–4726. doi: 10.1093/nar/gkn440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee H, Archer TK. Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J. 1998;17:1454–1466. doi: 10.1093/emboj/17.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng Y, John S, Pesavento JJ, Schultz-Norton JR, Schiltz RL, Baek S, et al. Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J Cell Biol. 2010;189:407–415. doi: 10.1083/jcb.201001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li C, Mueller JE, Elfline M, Bryk M. Linker histone H1 represses recombination at the ribosomal DNA locus in the budding yeast Saccharomyces cerevisiae. Mol Microbiol. 2008;67:906–919. doi: 10.1111/j.1365-2958.2007.06101.x. [DOI] [PubMed] [Google Scholar]
- 52.Menoni H, Shukla MS, Gerson V, Dimitrov S, Angelov D. Base excision repair of 8-oxoG in dinucleosomes. Nucleic Acids Res. 2012;40:692–700. doi: 10.1093/nar/gkr761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishiyama M, Oshikawa K, Tsukada Y, Nakagawa T, Iemura S, Natsume T, et al. CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nat Cell Biol. 2009;11:172–182. doi: 10.1038/ncb1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim K, Choi J, Heo K, Kim H, Levens D, Kohno K, et al. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J Biol Chem. 2008;283:9113–9126. doi: 10.1074/jbc.M708205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zakharova NI, Sokolov VV, Suvorova aa, Shiau A-L, Wu C-L, Evstafieva aG. Prothymosin alpha interacts with C-terminal domain of histone H1 and dissociates p53-histone H1 complex. Mol Biol. 2011;45:624–633. doi: 10.1134/S0026893311040157. [DOI] [PubMed] [Google Scholar]
- 56.Widlak P, Kalinowska M, Parseghian MH, Lu X, Hansen JC, Garrard WT. The histone H1 C-terminal domain binds to the apoptotic nuclease, DNA fragmentation factor (DFF40/CAD) and stimulates DNA cleavage. Biochemistry. 2005;44:7871–7878. doi: 10.1021/bi050100n. [DOI] [PubMed] [Google Scholar]
- 57.Kalashnikova AA, Winkler DD, McBryant SJ, Henderson RK, Herman JA, Deluca JG, et al. Linker histone H1.0 interacts with an extensive network of proteins found in the nucleolus. Nucleic Acids Res. 2013;41:4026–4035. doi: 10.1093/nar/gkt104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Happel N, Doenecke D. Histone H1 and its isoforms: Contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 59.Szerlong HJ, Herman JA, Krause CM, Deluca G, Skoultchi A, Winger QA, et al. Proteomic characterization of the nucleolar linker histone H1 interaction network. J Mol Biol. 2015;427:2056–2071. doi: 10.1016/j.jmb.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2003;13:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 63.Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. 2012;3:1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winkler DD, Luger K. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J Biol Chem. 2011;286:18369–74. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winkler DD, Muthurajan UM, Hieb AR, Luger K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem. 2011;286:41883–92. doi: 10.1074/jbc.M111.301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–16. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 67.Okuwaki M, Kato K, Nagata K. Functional characterization of human nucleosome assembly protein 1-like proteins as histone chaperones. Genes to Cells. 2010;15:13–27. doi: 10.1111/j.1365-2443.2009.01361.x. [DOI] [PubMed] [Google Scholar]
- 68.Yang S, Kim BJ, Norwood Toro L, Skoultchi AI. H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc Natl Acad Sci U S A. 2013;110:1708–13. doi: 10.1073/pnas.1213266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koike M, Fukushi J, Ichinohe Y, Higashimae N, Fujishiro M, Sasaki C, et al. Valosin-containing protein (VCP) in novel feedback machinery between abnormal protein accumulation and transcriptional suppression. J Biol Chem. 2010;285:21736–21749. doi: 10.1074/jbc.M109.099283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meerang M, Ritz D, Paliwal S, Garajova Z, Bosshard M, Mailand N, et al. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat Cell Biol. 2011;13:1376–1382. doi: 10.1038/ncb2367. [DOI] [PubMed] [Google Scholar]
- 71.Hamilton C, Hayward RL, Gilbert N. Global chromatin fibre compaction in response to DNA damage. Biochem Biophys Res Commun. 2011;414:820–825. doi: 10.1016/j.bbrc.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iborra FJ, Jackson Da, Cook PR. Coupled transcription and translation within nuclei of mammalian cells. Science. 2001;293:1139–1142. doi: 10.1126/science.1061216. [DOI] [PubMed] [Google Scholar]
- 73.David A, Dolan BP, Hickman HD, Knowlton JJ, Clavarino G, Pierre P, et al. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol. 2012;197:45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilkinson MF, Shyu aB. Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays. 2001;23:775–787. doi: 10.1002/bies.1113. [DOI] [PubMed] [Google Scholar]
- 75.Sha Z, Brill LM, Cabrera R, Kleifeld O, Scheliga JS, Glickman H, et al. The eIF3 interactome reveals the translasome, a supercomplex linking protein synthesis and degradation machineries. Mol Cell. 2010;36:141–152. doi: 10.1016/j.molcel.2009.09.026.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pederson T. The Nucleolus. Cold Spring Harb Perspect Biol. 2015;3 doi: 10.1038/153687a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lund aH, Lohuizen VMb. Epigenetics and cancer. Genes Dev. 2004;18:2315–2335. doi: 10.1101/gad.1232504.Per. [DOI] [PubMed] [Google Scholar]
- 78.Guetg C, Scheifele F, Rosenthal F, Hottiger MO, Santoro R. Inheritance of Silent rDNA Chromatin Is Mediated by PARP1 via Noncoding RNA. Mol Cell. 2012;45:790–800. doi: 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 79.Grummt I, Längst G. Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim Biophys Acta - Gene Regul Mech. 2013;1829:393–404. doi: 10.1016/j.bbagrm.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 80.Sanij E, Poortinga G, Sharkey K, Hung S, Holloway TP, Quin J, et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J Cell Biol. 2008;183:1259–1274. doi: 10.1083/jcb.200805146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rickards B, Flint SJ, Cole MD, LeRoy G. Nucleolin is required for RNA polymerase I transcription in vivo. Mol Cell Biol. 2007;27:937–948. doi: 10.1128/MCB.01584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meder VS, Boeglin M, de Murcia G, Schreiber V. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J Cell Sci. 2005;118:211–222. doi: 10.1242/jcs.01606. [DOI] [PubMed] [Google Scholar]
- 83.Grierson PM, Acharya S, Groden J. Collaborating functions of BLM and DNA topoisomerase I in regulating human rDNA transcription. Mutat Res - Fundam Mol Mech Mutagen. 2013;743–744:89–96. doi: 10.1016/j.mrfmmm.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Birch JL, Tan BCM, Panov KI, Panova TB, Andersen JS, Owen-Hughes TA, et al. FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 2009;28:854–865. doi: 10.1038/emboj.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Panova TB, Panov KI, Russell J, Zomerdijk JCBM. Casein kinase 2 associates with initiation-competent RNA polymerase I and has multiple roles in ribosomal DNA transcription. Mol Cell Biol. 2006;26:5957–5968. doi: 10.1128/MCB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gao X, Wang Q, Li W, Yang B, Song H, Ju W, et al. Identification of nucleolar and coiled-body phosphoprotein 1 (NOLC1) minimal promoter regulated by NF-κB and CREB. BMB Rep. 2011;44:70–75. doi: 10.5483/BMBRep.2011.44.1.70. [DOI] [PubMed] [Google Scholar]
- 87.Terns M, Terns R. Noncoding RNAs of the H/ACA family. Cold Spring Harb Symp Quant Biol. 2006;71:395–405. doi: 10.1101/sqb.2006.71.034. [DOI] [PubMed] [Google Scholar]
- 88.Hale TK, Contreras A, Morrison AJ, Herrera RE. Phosphorylation of the Linker Histone H1 by CDK Regulates its binding to HP1alpha. Mol Cell. 2006;22:693–699. doi: 10.1016/j.molcel.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 89.Misteli T, Gunjan a, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–81. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 90.Lever Ma, Th’ng JP, Sun X, Hendzel MJ. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–6. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- 91.Andersen JS, Lam YW, Leung AKL, Ong SE, Lyon CE, Lamond AI, et al. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03180.1. [DOI] [PubMed] [Google Scholar]
- 92.Hernandez-Verdun D. Nucleolus: From structure to dynamics. Histochem Cell Biol. 2006;125:127–137. doi: 10.1007/s00418-005-0046-4. [DOI] [PubMed] [Google Scholar]