Abstract
Purpose
Dedifferentiated liposarcoma (DDLPS) is an aggressive malignancy that can recur locally or disseminate even after multidisciplinary care. Genetically amplified and expressed MDM2, often referred to as a “hallmark” of DDLPS, mostly sustains a wild-type p53 genotype, substantiating the p53-MDM2 axis as a potential therapeutic target for DDLPS. Here we report on the preclinical effects of SAR405838, a novel and highly selective MDM2 small-molecule inhibitor, in both in vitro and in vivo DDLPS models.
Experimental Design
The therapeutic effectiveness of SAR405838 was compared to the known MDM2 antagonists Nutlin-3a and MI-219. The effects of MDM2 inhibition were assessed in both in vitro and in vivo. In vitro and in vivo microarray analyses were performed to assess differentially expressed genes induced by SAR405838, as well as the pathways that these modulated genes enriched.
Results
SAR405838 effectively stabilized p53 and activated the p53 pathway, resulting in abrogated cellular proliferation, cell cycle arrest, and apoptosis. Similar results were observed with Nutlin-3a and MI-219; however, significantly higher concentrations were required. In vitro effectiveness of SAR405838 activity was recapitulated in DDLPS xenograft models where significant decreases in tumorigenicity were observed. Microarray analyses revealed genes enriching the p53 signaling pathway as well as genomic stability and DNA damage following SAR405838 treatment.
Conclusion
SAR405838 is currently in early phase clinical trials for a number of malignancies, including sarcoma, and our in vitro and in vivo results support its use as a potential therapeutic strategy for the treatment of DDLPS.
Keywords: Dedifferentiated liposarcoma, MDM2, p53, SAR405838, small-molecule inhibitors
Introduction
Dedifferentiated liposarcoma (DDLPS) is a rare malignancy (1/330,000 persons/year) originating from an adipogenic precursor cell (1); it is a high-grade biphasic tumor often found juxtaposed to a well-differentiated (WDLPS) lesion (2). Unlike WDLPS, which retains lipotomatous features, DDLPS is a non-lipotomatous, highly cellular malignancy with a much more aggressive phenotype (3). DDLPS frequently presents as a large (>15 cm), painless, retroperitoneal mass (4). Few new treatment options have emerged, and surgery remains the mainstay of care. However, a high recurrence rate (80%) often necessitates serial resections, which are associated with significant post-surgical morbidity (5,6). In addition to local recurrence, DDLPS metastasizes to distant sites resulting in a five-year disease specific survival rate of <5% for advanced disease (4). Molecular genetics reveals supernumerary rings and/or giant rod chromosomes in the 12q13~15 region of DDLPS. Within this amplicon there are over 150 amplified genes; high levels of MDM2 amplification has been found in nearly 100% of DDLPS (7–9). MDM2 is a well characterized oncogene, supporting tumorigenesis and progression in many malignancies, primarily due to its negative regulation of the p53 tumor suppressor (10).
Although p53 is inactivated in approximately 50% of human cancers as a result of mutations or loss of heterozygosity, p53 is wild-type (WT) in the remainder, suggesting functional impairment by other mechanisms (11,12). These alternatives include overexpression of MDM2, the primary negative regulator of p53 (13). Functioning as an E3 ubiquitin ligase, MDM2 binds p53, thereby regulating the protein levels of p53 and its transcriptional activity. Interestingly, p53 is a transcription factor for MDM2, which results in an autoregulatory feedback loop (14). Accordingly, restoration of p53 activity through the use of small-molecule inhibitors targeting the hydrophobic protein-protein interaction site between MDM2 and p53 has become a feasible targeted therapeutic strategy for various cancers (15,16).
To date, multiple MDM2 antagonists have been investigated (17). Nutlin-3a was the first specific small-molecule inhibitor discovered to target the p53-MDM2 complex (18), displacing the p53 protein from MDM2 through its cis-imidazoline core structure. Shortly thereafter, improved MDM2:p53 inhibitors have been developed to enhance specificity and efficacy (19).
Here we report on the effects of a novel and selective small-molecule MDM2 inhibitor, SAR405838. This highly optimized spiro-oxindole compound was developed by Sanofi-Aventis as a derivative of the MDM2-inhibitor-219 (MI-219). SAR405838 binds to the MDM2 protein with remarkable specificity and selectivity, resulting in p53 activation at low nanomolar concentrations; thus effectively inducing cell cycle arrest and apoptosis in a p53-dependent manner (20).
The presence of MDM2 amplification coinciding with a WT p53 genotype suggests DDLPS as an ideal model system in which to examine the antagonistic effects of the MDM2 inhibitor SAR405838. Biological insights presented here may have preclinical relevance for DDLPS patients, most of whom succumb to their disease in the absence of effective systemic therapies.
Materials and Methods
Cell culture and reagents
Human LPS cell lines Lipo224, Lipo246, Lipo863, Lipo815 and PLS1 were established in our lab as previously reported (30). SW872 was obtained from the American Type Culture Collection (ATCC). LiSa2 and LS2B were kindly given to us from Dr. Kiki Broccoli (The Curtis and Elizabeth Anderson Cancer Institute, Savannah, GA, USA). Dr. Jonathan Fletcher (Brigham and Women’s Hospital, Boston, MA) generously provided us with LPS141 cells. All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) and supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 U/mL streptomycin. Human mesenchymal stem cells (hMSC-AT) and pre-adipocytes (PreAdip) derived from visceral adipose tissue were obtained from PromoCell and were cultured in mesenchymal stem cell medium supplemented with the manufacturer’s supplemental mix. These cells were cultured under normal conditions in a humidified chamber delivering 5% CO2 at 37°C.
Chemical compounds and biologic reagents
Nutlin-3a was purchased from Cayman Biochemicals (Ann Arbor, MI). MI-219 and SAR405838 were provided by Sanofi-Aventis (Sanofi-Aventis, Paris, France). Serial dilutions were then made to obtain final dilutions for cellular assays with a final concentration of DMSO not exceeding 0.1%.
Fluorescence in situ hybridization (FISH)
FISH was performed on fixed cell cultured cells specimens as previously described (30). BAC labeled probe cocktails were purchased from the Children’s Hospital Oakland Research Institute, Oakland CA, USA, specific for the 12q15 region and for the centromeric region of chromosome 12 (Abbott Molecular, DesPlaines, IL, USA). MDM2 amplification was determined at the ratio of MDM2/CEP-12 ≥ 2.1 per 100 cells.
gDNA and mRNA extraction and quantitative real-time PCR (qRT-PCR)
gDNA was extracted from DDLPS cell lines using the QIAamp DNA Mini Kit (Qiagen) as per the manufacturer’s instructions. mRNA was extracted from cell pellets using the Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA, USA) and mRNA was quantified using a NanoDrop 2000 instrument (manufacturer specifics). Bio-Rad’s iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) was used to perform reverse transcription and a Light Cycler 480 SYBR Green 1 Master Mix (Roche) was used for quantitative detection of transcripts. Primer sequences are detailed in Supplementary Table 1.
In PD studies, tumor tissues were thinly sliced and frozen in RNAlater (Qiagen) until analysis. They were then homogenized with RLT buffer (Qiagen) in a tissue lyser (Qiagen) with stainless steel beads. Total RNA was extracted with the kit RNeasy kit (Qiagen) following the internal standard operating procedure. cDNA was synthesized from 2μg of total RNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with Oligo dt (Eurogentech). Next, TaqMan gene expression assays were performed by using MDM2 (Hs99999008-m1), p21 (CDKN1A) (Hs00355782-m1) and PUMA (BBC3) (Hs00248075) gene-specific primer/probe sets (Applied Biosystems) for real-time PCR amplification in an Applied Biosystems 7900 thermocycler. RPL37a was used for normalisation using probes and primers from Applied Biosystems. Relative quantification of mRNA was calculated by comparative cycle threshold (Ct) method.
Protein expression analysis
Western blot analysis was performed by standard methods as previously described (31). Relevant commercially available antibodies were used as indicated per experiment: p53, p21, GADD45a (Santa Cruz); MDM2, Nup107 (Abcam). Horseradish peroxidase (HRP)-conjugated secondary antibodies (Santa Cruz) were detected by ECL (PerkinElmer).
In PD studies, tumor tissues were frozen until analysis. They were then lysed after mechanical dissociation using a bead beating Precellys homogenizer 24 (Ozyme) with a Cryolys cooling system in ice-cold lysis buffer: 10 mM Tris (pH 7.5), 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton, 1 mM NaF, 20 mM Na4P2O, 1 mM activated Na3VO4, 10% glycerol supplemented with protease inhibitor tablets. Lysates were incubated for 2 hours at 4°C, clarified by centrifugation and supernatants were collected, aliquoted and stored at −80°C until analysis. Total protein concentrations were determined with the BCA protein assay kit (Thermo scientific).
For Mesoscale Discovery assays, protein analysis was performed after appropriate dilutions with following kits: MDM2 (K152FID), p21 (N45ZA-1), p53, Cleaved Caspase 3 and cleaved PARP (K15102D-1) with a detection on Sector Imager 2400. Results were normalized with total protein concentration.
p53 mutational analysis
p53 mutational analysis was conducted on all cell strains/lines established in our lab as previously described (32). In brief, QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA) was used to extract genomic DNA as per manufacturer’s instructions. PCR amplification was performed on genomic DNA for the entire p53 gene and the sequence analysis from the PCR product was performed with a Sequence Scanner (version 1.0, Applied Biosystems).
Proliferation assays
DDLPS cells were seeded in 96-well plates and incubated with DMSO control or increasing doses of inhibitor for up to 96 h. Cell proliferation was determined using the CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay kit (Promega), per manufacturer’s instructions. Absorbance was read at 490 nm (Beckman Coulter).
Cell cycle and apoptosis assays
DDLPS cells were incubated with MDM2 inhibitors or with a DMSO control for 48 or 96 hours as designated. For cell cycle analysis, cells were stained with propidium iodide (PI) containing 10 μg/mL prior to DNA content analysis by flow cytometry. Apoptosis was measured using the Apoptosis Detection kit 1 (BD Pharmagen, San Diego, CA, USA). Cells were stained with Annexin V/FITC (BD Pharmagen, San Diego, CA, USA) and propidium iodide (Sigma Aldrich Co.); FACScan (Becton Dickinson) was used to analyze samples by emitting excitation light at 488 nm. CellQuest software (Becton Dickinson) was used to analyze the data.
Microarray analysis of gene expression
Total RNA extracted from DDLPS cell lines and xenograft tissue was converted into cRNA (Illumina TotalPrep Amplification Kit, Ambion) for gene expression prolifiling using the Illumina Human Ref-12 Expression BeadChip (Illumina Inc., San Diego, CA).
For in vitro gene expression analysis, two independent biological replicates of different DDLPS cell lines (Lipo224, Lipo815, Lipo246 and Lipo863) were treated with 0.003% DMSO (control), 1 μM, and 3 μM of SAR405838 for 24 hours. Differential gene expression was based on t-statistic, and false discovery rate (FDR) was used to adjust the P-value for multiple testing. In these studies, significance was established with an FDR 0.05 and fold change >1.4 for any three of the four cell lines. For in vivo studies, Fold change >2 or <0.5 and p < 0.01 was used to determine significance.
The function of the differentially expressed genes was mapped to biologic pathways using the Ingenuity Pathway Analysis software (IPA, http://www.ingenuity.com), revealing SAR405838-affected pathways in DDLPS cells and DDLPS xenograft models. Only significantly up- and down-expressed genes compared to controls (p < 0.05; Fold change 2.0) were imported into the software for analysis.
siRNA knockdown
Small-interfering RNA (siRNA) targeting p53 (SMARTpool: ON-TARGETplus p53 siRNA, # L-003329-00-0005), and a non-targeting control (ON-TARGETplus Non-targeting siRNA #1, #D-001810-01-20) were purchased from Dharmacon. siRNA was resuspended in 5X siRNA Buffer (Dharmacon). siRNA was transfected into DDLPS cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Each transfection sample was prepared as followed: 20 pmol siRNA oligomer was diluted in 50 μL of Opti-MEM Reduced Serum Medium without serum (Opti) (Life Technologies) along with 1 μL of Lipofectamine 2000.
SAR405838 pharmacokinetic and therapeutic efficacy in DDLPS xenograft models
For PK studies, groups of 6-week-old female balb/c nude mice were given 100 or 200 mg/kg of SAR405838 orally into Lipo863 tumor bearing xenograft model mice. Blood samples were taken at 6, 24, 48 and 72 hours post treatment by intraocular puncture and transferred into heparinized tubes. The plasma was collected (BD Pharmagen) after centrifugation (3 min, 7600g). Plasma was organically extracted with 300 μL ACN, and supplemented with 2H5-SAR405838 as an internal standard. Liquid chromatography-mass spectrometry was performed for pharmacokinetics analysis.
DDLPS xenograft cells were subcutaneously injected (2–3.5×106 cells) into female balb/c nude SCID mice (Charles River Laboratories International, Inc.) and tumor size was monitored 2–3 times a week until the study was terminated; 5–10 mice per group were used per study. Once palpable tumors formed after approximately two weeks, mice were randomized into vehicle control or SAR405838 treatment arms for single agent efficacy studies, SAR405838 was administered orally (P.O.) (concentrations were designated per experiment, and ranged from 50–200 mg/kg). Upon termination, tumors were resected, weighed, and either snap-frozen in liquid nitrogen or paraffin embedded for sectioning and IHC staining. Mice were housed in pathogen free conditions as per The University of Texas MD Anderson Cancer Center and Institutional Animal Care and Use Committee (IACUC) regulations. Animals received humane care as per the Animal Welfare Act and the NIH “Guide for the Care and Use of Laboratory Animals”.
Statistical analysis
All experiments are represented as averages ± S.E.M. Student t tests were performed to determine statistically significant differences using GraphPad Prism (version 6.0). Probability values of <0.05 were considered to be statistically significant. For in vivo experiments, the average volume (mm3) and SEM for all group variables per study were recorded and calculated. A two-sample t-test was used to assess the differences of an outcome at a single time point. Xenograft growth was assessed using a two-tailed Student’s t-test. Significance was set at p ≤ 0.05.
Results
Characterization of dedifferentiated liposarcoma cell lines
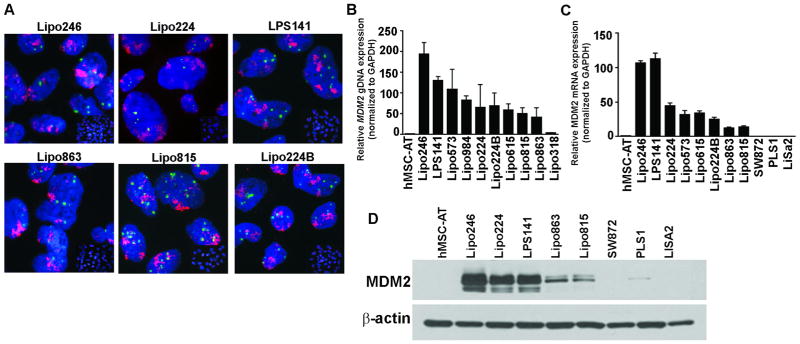
DDLPS cell lines were established in our laboratory due to the lack of commercially available bioresources that accurately recapitulate DDLPS biology. Pleomorphic liposarcoma (PLS) cell lines (SW872, PLS1, and LiSa2) were used as controls due to their variable MDM2 and p53 expression. All liposarcoma (LPS) cells lines were examined for MDM2 amplification and expression. Fluorescence in situ hybridization (FISH) analyses showed MDM2 amplification in DDLPS cells (Fig. 1A); PLS cell lines do not contain the 12q13~15 amplicon, and as a result do not overexpress MDM2. qRT-PCR assays confirmed this amplification of genomic MDM2 (Fig. 1B) and relative MDM2 mRNA expression levels (Fig. 1C). Western blot analysis (Fig. 1D) further demonstrated the high levels of MDM2 protein expression in DDLPS cell lines as compared to PLS cell lines. TP53 sequencing was performed to eliminate the possibility that the amplification of MDM2 may have been due to TP53 mutations in DDLPS cells. P53 mutational status and the translational amplification of MDM2 in DDLPS cell lines are summarized in Supplementary Table S2.
Figure 1. MDM2 amplification and expression in DDLPS cell panel.
(A) Representative images of fluorescence in situ hybridization of the 12q13~15 amplicon in a cell line panel (MDM2: red signal and CEP12: green signal); DDLPS: Lipo246, Lipo224, Lipo863, Lipo815, Lipo224B, LPS141; Pleomorphic LPS: SW872, PLS1, LiSa2. (B) q-PCR was performed for MDM2 genomic amplification on a large DDLPS cell line panel, along with hMSC-AT as a normal control (C) q-PCR analyzed MDM2 mRNA expression levels in DDLPS cell lines, hMSC-AT, and in two pleomorphic LPS cell lines, PLS1 and LiSa2. (D) Western blot analysis of MDM2 protein expression in DDLPS cell lines versus control cell lines.
SAR405838 stabilizes p53 and induces the expression of p53 transcriptional targets in DDLPS
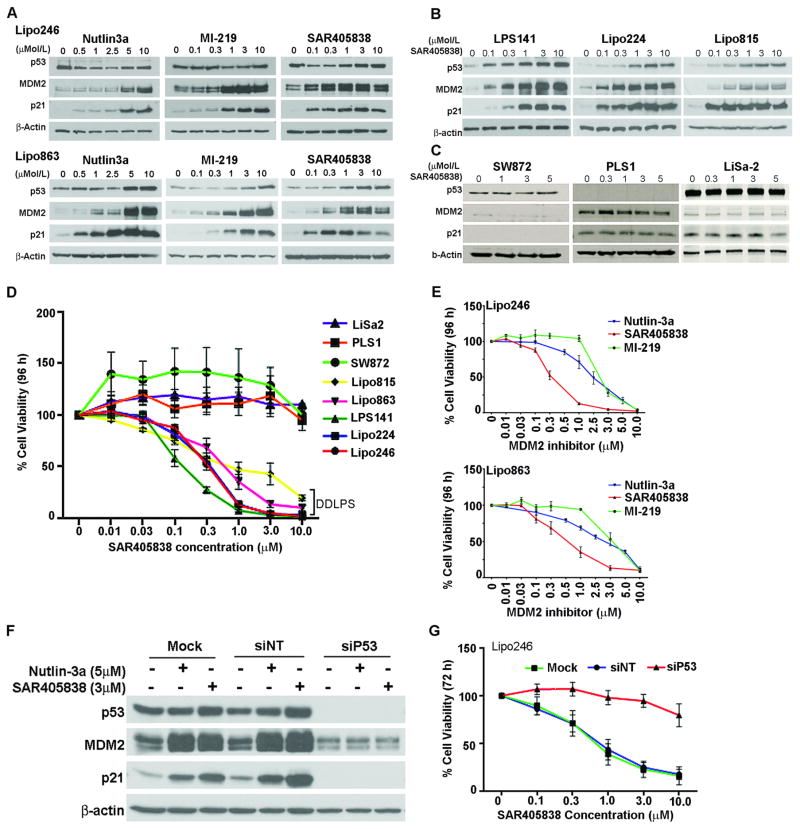
Two previously reported MDM2 inhibitors, Nutlin-3a and MI-219, were used to confirm the expected cellular signaling response of SAR405838 in DDLPS cells. Mechanistically, MDM2 antagonism relieves p53 from MDM2 ubiquitination and degradation, resulting in p53 stabilization and an increased p53-mediated transcriptional activity. We show that when DDLPS cells were incubated with increasing concentrations of Nutlin-3a, MI-219, and SAR405838 (0–10 μM; 24 h), a dose-dependent accumulation of p53 resulted together with an enhancement in MDM2 and p21 p53 transcriptional targets. This effect was seen in all DDLPS cells (Fig. 2A, B); however there was no increase in p53, MDM2, or p21 in PLS control cells (Fig. 2C).
Figure 2. Activation of the p53 pathway in DDLPS cell lines.
(A) Lipo246 (upper panel) and Lipo863 (lower panel) DDLPS cells were incubated with increasing concentrations of Nutlin-3a, MI-219, and SAR405838 for 24 h; p53, MDM2 and p21 proteins were analyzed via Western blot. (B) As in (A), an additional panel of DDLPS cell lines was evaluated via western blot for their cellular response to SAR405838 treatment. (C) SAR405838 had no effect on p53-dependent signaling in control cell lines; SW872, PLS1 and LiSa-2. (D) MTS assays measured dose-dependent effects on cell viability following exposure to SAR405838 for 96 h on exponentially growing DDLPS cells. (E) MTS assays measured cell viability of Lipo246 (left panel) and Lipo863 after exposure to increasing concentrations of Nutlin-3a, SAR405838, and MI-219 after 96h. (F) Western blot analysis assessed the effects of transient p53 knockdown in Lipo246 cells (72 h) using mock treated cells, siNT, and siP53 when treated with either Nutlin-3a or SAR405838. (H) siP53 knockdown reduces the effects of SAR405838 on cell viability. Data represent the mean ± SEM from ≥ 3 independent experiments.
SAR405838 inhibits cell proliferation in DDLPS cells
Activation of the p53 pathway has been shown to result in abrogated cell viability and growth rates (21,22). As anticipated, exposure of DDLPS cells to increasing doses of SAR405838 for 96 hours resulted in a dose-dependent decrease in cell viability, whereas PLS cell lines remained unaffected (Fig. 2D). The half-maximal growth-effective concentration (EC50) was established for each cell line (Fig. 2E, Supplemental Table S2). SAR405838 resulted in a substantially lower EC50 value (0.31 μM) compared to Nutlin-3a (2.90 μM) and MI-219 (2.34 μM) treated Lipo246 cells. Similarly, in the Lipo863 cell line, we found that the EC50 value using SAR405838 was 0.49 μM, 2.75 μM for Nutlin-3a, and 3.03 μM for MI-219.
To confirm that the anti-DDLPS effects of SAR405838 relies on the presence of functional p53, Lipo246 cells were transiently transfected with control (mock), a pool of non-targeting (siNT) small interfering RNAs (siRNAs), or with siRNAs targeting p53 (siP53). Lipo246 siP53 cells were subsequently treated with a DMSO control, Nutlin-3a, or with SAR405838. The transient knockdown of p53 abrogated the expression of p53 transcriptional target genes following MDM2 inhibition (Fig. 2F). Additionally, cell viability was unperturbed by SAR405838 following transient p53 knockdown compared to mock and non-targeting controls (Fig. 2G).
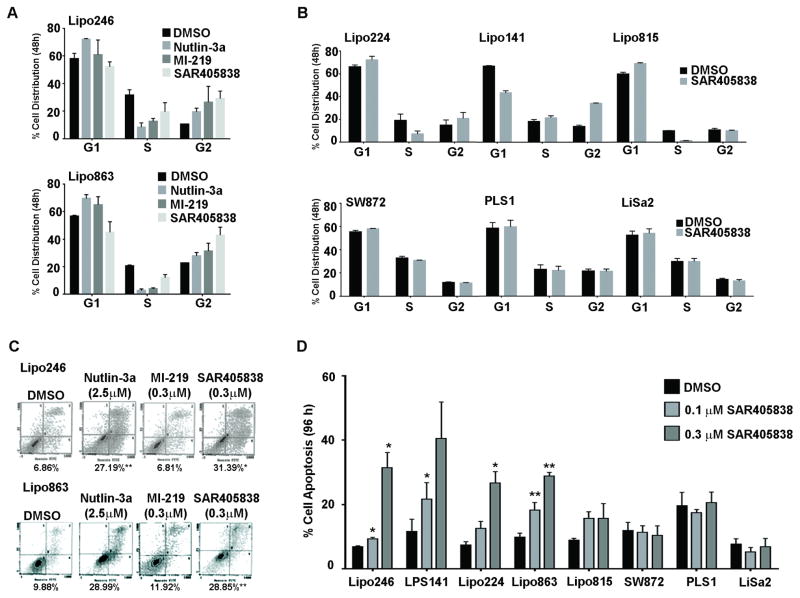
SAR405838 induces cell cycle arrest and apoptosis in DDLPS
p53 is a crucial component in cell cycle regulation, and the reactivation of p53 via MDM2 inhibition induces cell cycle arrest and/or apoptosis in WT p53 tumor cells (23). In agreement with the increased expression of the p21cell cycle marker in Figure 2A, we observed changes in cell cycle profiles for DDLPS cell lines following 48 hour of SAR405838 treatment (Fig. 3A). Lipo246 and Lipo863 demonstrated significant cell cycle arrest. Importantly, SAR405838-induced cell cycle arrest was dependent on p53 status as depicted in Figure 3B.
Figure 3. SAR405838 effectively induces p53 pathway activation by inhibiting cell cycle and survival.
(A) Representative graphs of Lipo246 (upper panel) and Lipo863 (lower panel) were incubated for 48 hours with Nutlin-3a (5 μM), MI-219 (3 μM), or SAR405838 (3 μM) and their cell cycle profiles were analyzed via FACS. (B) LPS cell lines cell-cycle distributions following 48h treatment with SAR405838 (1 μM, 3 μM) or DMSO are shown graphically; SAR405838 effectively arrested DDLPS cells in G1 and G2 phases but had no effect on cells lines with lacking WT p53 or amplified MDM2 (SW872 and PLS1, and LiSa2; respectively). (C) Lipo246 (upper panel) and Lipo863 (lower panel) were incubated with MDM2 inhibitors (96 h); the degree of apoptosis was analyzed by Annexin V/FITC staining (representative images). (D) Apoptotic responses were measured in a larger panel of cell lines by FACS analyses following treatment with DMSO or 0.1 μM, or 0.3 μM of SAR405838 for 96 hours. Data shown represents the average percent of apoptosis induced by the indicated inhibitor; n=3 experiments ± SEM; t-test: *=p < 0.05.
This cell cycle arrest was followed by the induction of apoptosis, since SAR405838 induced early- and late-stage apoptosis in a dose-dependent manner in DDLPS cell lines. It is noteworthy that Nutlin-3a required a much higher concentration to achieve effects similar to SAR405838; however, there was no change in apoptosis when DDLPS cells were treated with MI-219 at the same concentrations as SAR405838 (Fig. 3C). Coinciding with our MTS assay EC50 values, SAR405838 induced apoptosis to a much higher degree than Nutlin-3a and MI-219 in DDLPS cells, with no significant increase in apoptosis observed in control PLS cells (Fig. 3D).
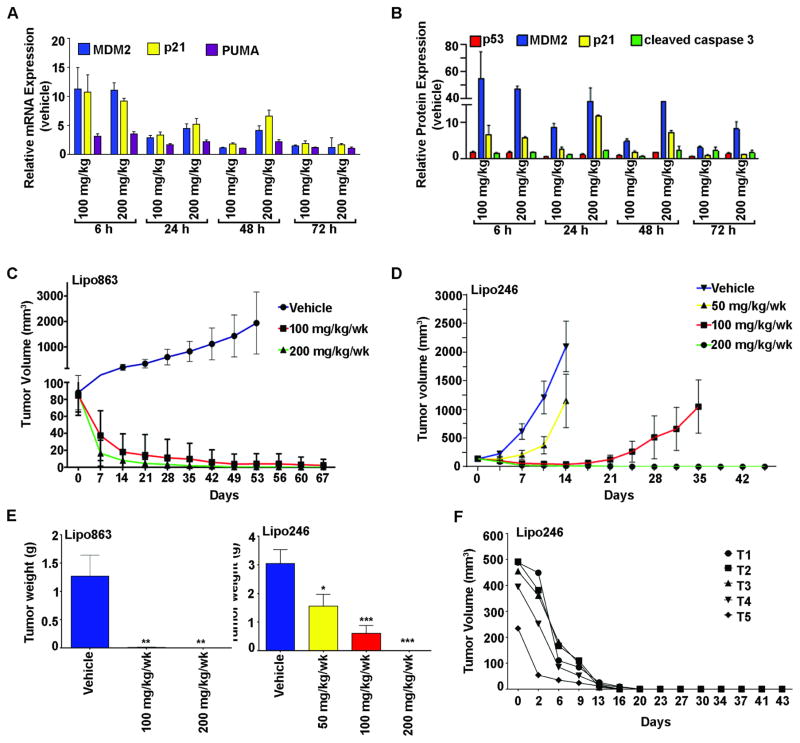
SAR405838 elicits strong anti-tumorigenic responses in DDLPS xenograft models
DDLPS xenografts were utilized to study the tumor penetration and stability of SAR405838. Pharmacokinetic studies revealed that plasma drug concentrations reached the concentration maximum value (Cmax) following 6 hour administration of SAR405838 for both 100 and 200 mg/kg doses (8200 ng/mL and 1630 ng/mL, respectively) (Supp. Fig. 1). All data were below the lower limit of quantitation (5 ng/mL) following 72 hours of drug administration. The data indicates that drug accumulation was dose-dependent; the area under the curve (AUC0-72) increased in a dose-dependent manner and was approximately twice as high with the 200 mg/kg dose versus the 100 mg/kg dose. These results suggested that the Cmax and AUClast increased proportionally with the dose of SAR405838 administered.
To demonstrate that the p53-induced in vitro effects were recapitulated in vivo, Lipo863 xenograft mice were treated with a single oral dose of vehicle, 100 or 200 mg/kg and tumors were harvested at 6, 24, 48 and 72 hours post treatment. Multiple p53 transcriptionally-regulated gene (Fig. 4A) and protein (Fig. 4B) expression levels were assessed in response to treatment, suggesting that the concentration-effect relationship increased in a dose- and time-dependent manner. Taken together, the effects of SAR405838 led to p53 pathway activation, as suggested by increased p53 transcriptional targets.
Figure 4. Antitumor effects of SAR405838 in DDLPS xenograft models.
A) mRNA expression levels from DDLPS xenografts following SAR405838 treatment. B) Protein expression levels of selected genes in response to time (6–72 h) and a single dose (100 and 200 mg/kg, p.o.) of SAR405838 or vehicle. Data represents mean ± SEM, n=3. Oral administration of SAR405838 (50, 100 or 200 mg/kg) or vehicle control in nude mice (n=7–8 per group) bearing s.c. Lipo863 (C) and Lipo246 (D) xenograft tumors, which significantly decreased tumor volumes (± SEM) and (E) tumor weights (mean tumor weight at termination for each group of mice was recorded ± SEM; t-test: *=P<0.05, **=P<0.01, ***=P<0.001). (F) 200 mg/kg/wk was orally administered to Lipo246 xenograft bearing mice (n=5); robust in vivo antitumor activity was seen, and after two treatments tumors were completely eradicated in mice.
Experiments were performed with two DDLPS cell lines (Lipo246 and Lipo863) to evaluate DDLPS tumorigenicity in response to SAR405838 treatment in vivo. Lipo863 tumor bearing mice were separated into three groups and received vehicle or 100 mg/kg/wk, or 200 mg/kg/wk of SAR405838. Following seven weeks of treatment, the control group was terminated due to a large tumor burden; mice receiving 100 or 200 mg/kg/wk were treated for an additional two weeks before termination due to the diminished tumor volumes in each group. As indicated in Figure 4C, SAR405838 effectively reduced tumor volume and growth of Lipo863 tumors compared to the controls. Upon termination, the mean tumor volume of the control group was 1932.84mm3, whereas the mean tumor volumes of the 100 mg/kg/wk and 200 mg/kg/wk treatment groups were 2.55mm3 and 0.57mm3, respectively.
Similarly, Lipo246 tumor bearing mice were separated into four treatment groups, and received vehicle or 50 mg/kg/wk, 100 mg/kg/wk, or 200 mg/kg/wk of SAR405838. Mice from both the vehicle and 50 mg/kg group were terminated on day 15. A modest decrease in tumor volume was observed in mice receiving 50 mg/kg dose compared to vehicle (vehicle: 2722.35mm3, 50 mg/kg: 1821.65mm3). Mice receiving 100 and 200 mg/kg/wk continued receiving treatment for an additional two weeks since they were responding well to treatment. On the last day of measurement, 5/7 mice receiving 100 mg/kg/wk had an average tumor volume of 1047.80mm3; however, even more impressively, 0/8 mice receiving 200 mg/kg/wk had measureable tumors (Fig. 4D). The significantly reduced tumor weight in both xenograft studies is depicted in Figure 4E. In both models, SAR405838 was well tolerated and produced only a minimal (<5%) change in body weight following treatment (data not shown). Taken together, our data suggests that SAR405838 may be an effective therapy for the treatment of DDLPS.
Patients with DDLPS often have very large, deep-seated, and invasive tumors that make complete resection difficult. Therefore, to recapitulate such large patient tumor burdens, Lipo246 tumor bearing mice (n=5, balb/c nude SCID mice) were treated with SAR405838 (200 mg/kg/wk p.o.) once their tumor burden reached 412.49 ± 47.90mm3. The results of this study exemplified the anti-DDLPS effects of SAR405838 treatment in vivo, as SAR405838 treatment significantly abrogated tumor growth in Lipo246 tumor bearing mice; with complete tumor regression after only two treatments (0.00mm3 ± 0.00 after 14 days and termination on day 43) (Fig. 4F).
Molecular changes induced by SAR405838 in DDLPS
In an effort to identify novel biomarkers for therapeutic effectiveness, we performed two Illumina microarray analyses, where global gene expression was compared. The first array investigated differentially expressed genes following SAR405838 treatment in four DDLPS cell lines in vitro; the second array investigated the genes that were modulated in vivo. To obtain an accurate profile of genes altered in response to SAR405838 treatment, the two arrays were overlapped for a final analysis.
Following SAR405838 treatment in vitro, a total of 381 up-regulated and 477 down-regulated genes were significantly different when compared to DMSO controls in at least three out of the four DDLPS cell lines evaluated (p<0.05, FC ±1.4). This stringent analysis revealed that DDLPS gene expression intensities when treated with 1 μM versus 3 μM SAR405838 were highly concordant in a dose-dependent manner (Fig 5A). Ingenuity Pathway Analysis (IPA) software was used to examine the changes in gene interaction profiles that were significantly enriched (Supplemental Fig. S2A). Down regulation of cell cycle, kinetochore, and mitotic regulation are all processes related to cell division. These significantly enriched core pathways that were perturbed due to down regulation of cell division genes implies that cell proliferation processes were decreased following SAR405838 treatment; consistent with our in vivo and in vitro data.
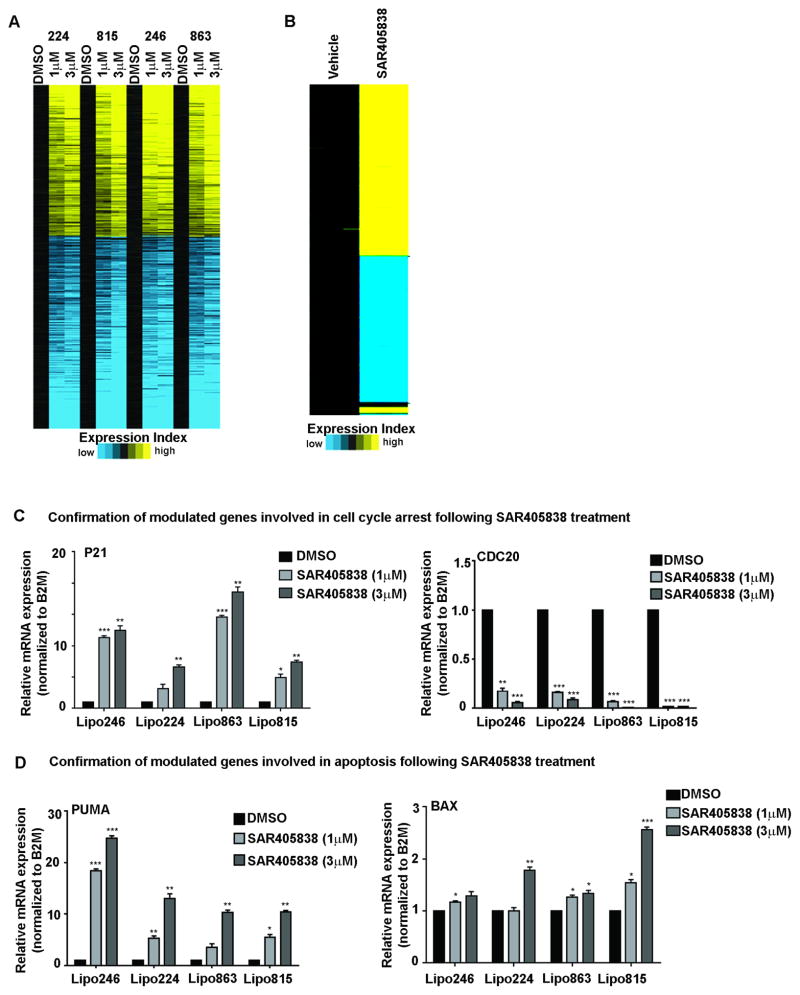
Figure 5. Heatmap of differentially expressed genes in DDLPS in response to SAR405838.
(A) The heatmap of DMSO-treated and SAR405838-treated DDLPS cells that demonstrate 858 most significantly differentially expressed genes in for three of the four DDLPS cells line profiles (1 μM + 3 μM). Within each cell line the gene values are centered on corresponding control profiles (black). Yellow indicates significantly up-regulated genes, and blue indicates down-regulated genes (p < 0.05; FDR 0.05; fold change >1.4). (B) The heatmap represents the 1074 most significant differentially expressed genes in Lipo246 xenografts treated with either the vehicle control or SAR495838 in vivo. Yellow indicates significantly up-regulated genes, and blue indicates down-regulated genes, which were selected at a FDR 0.05 and a fold change >2.0 or <0.5 and a p < 0.01. Confirmation of select p53-modulated genes involved in cell cycle (C) and apoptosis (D). Data shown represents the mean ± SEM from ≥ 3 independent experiments; *=P<0.05, **=P<0.01, ***=P<0.001.
In the second microarray, genes modulated following a single oral dose of SAR405838 (200 mg/kg) in DDLPS xenografts were investigated. Gene expression analyses of tumors identified 586 up- and 488 down-regulated genes that were statistically significantly changed (p<0.01; FC 2.0) in control versus SAR405838-treated mice. Overall IPA revealed that SAR405838 induced gene expression pathways that are highly associated with the p53 pathway, including an overall decrease in cell survival, growth and proliferation, cell cycle, and DNA replication. This suggested that the p53 pathway is active, and that the anti-DDLPS therapeutic response of SAR405838 is p53-dependent in vivo (Supplemental Fig. S2B).
To determine which enriched pathways had significant biological relevance, we crossed and evaluated the common genes between our two independent microarray datasets. We found that 237 genes were significantly differentially regulated: 115 were downregulated and 122 were upregulated. Following IPA analysis of this intersecting gene list, 171 interaction networks were identified (Supplemental Fig. S3A). Of these networks, the p53 signaling pathway contained the highest gene involvement; networks responding to SAR405838 treatment included pathways important in genomic stability such as cell cycle and checkpoint regulation (G2/M and G1/S checkpoint regulation), and response to DNA damage (role of BRCA1 in DNA damage response, and ATM signaling). Since the therapeutic efficacy of SAR405838 depends on p53 activation, we confirmed the expression of select p53-regulated genes that were identified via IPA as enriched network nodes using q-PCR (Fig. 5C, D; Supplemental Fig. 3B). We found that CDC20 decreased significantly in a dose-dependent manner, whereas p21, PUMA, and BAX expression was enhanced with increasing concentrations of SAR405838.
Taken together, these results demonstrate that SAR405838 effectively restores p53 downstream signaling through pharmacological MDM2 inhibition in DDLPS. SAR405838 potently induced anti-tumorigenic responses in DDLPS cell lines expressing WT p53 and overexpressing MDM2.
Discussion
Collectively, our study objectives were two-fold: (1) to investigate the therapeutic efficacy of SAR405838 in DDLPS; and (2) to examine the MDM2-p53 axis role in DDLPS. Genomic characterization of other human cancers has revealed that there is a mutually exclusive relationship between MDM2 amplification and p53 mutations (21), supporting the contention that direct and specific activation of the p53 pathway is a biologically relevant approach for DDLPS as well as many other malignancies. We found that treatment with SAR405838 stabilizes p53 and induces the expression of p53 transcriptional target genes in DDLPS. Although mechanistically similar, SAR405838 more potently induces p53-driven responses compared to older generation inhibitors, such as Nutlin-3a and MI-219. These results substantiate and correlate with results recently reported by Wang et al. (2014) when SAR405838 was used to treat other cancer histologies. In all DDLPS cell lines, SAR405838 induced significant apoptosis as a single agent. At a concentration of 0.3 μM SAR405838, apoptosis was induced by more than 30% and EC50 values were found to occur as low as 0.13 nM. In this study, we evaluated if MDM2 inhibition would function similarly in p53 mutated or p53 negative pleomorphic liposarcoma cases as well, where it was evident that wild-type p53 was necessary for SAR405838 effects. Knockdown of p53 highlighted the difference in findings between MDM2 amplified and WT p53 DDLPS when compared with p53 mutated (SW872) or null (PLS1) pleomorphic liposarcoma cell lines. These sets of experiments highlighted that SAR405838 was not capable of eliciting any p53-driven responses in the aforementioned cell lines. Importantly, functional outcomes of SAR405838 were not only dependent on wild-type status of TP53 but also on the amplification of MDM2, since no effect was observed in LiSa2, a WT p53 cell line that does not overexpress MDM2. This knowledge may inform successful disease treatment selections, as well as explain how MDM2 inhibitors induce apoptosis in cancer cells but not in normal cells where MDM2 inhibition leads to cell cycle arrest and not death (Supplemental Fig. 4).
There are conflicting reports suggesting that the degree of MDM2 amplification is a determinant of MDM2 inhibitor treatment response (17,24–26). To determine if MDM2 status is a biomarker for SAR405838 sensitivity, our DDLPS cell line panel was divided by relative MDM2 expression (MDM2Hi: Lipo224, Lipo246, LPS141; or MDM2LO: Lipo815, Lipo863). The p53 pathway was activated in all DDLPS cell lines following SAR405838 treatment; however, a lower EC50 (0.25 μM vs. 0.43 μM; Supplemental Table. 2), as well as a more robust apoptotic response (36.38% vs. 24.19%; Fig. 3C) was observed in MDM2HI cell lines, suggesting that cells displaying more amplified MDM2 incur increased durable responses. Although plausible, the lack of a universal MDM2 amplification threshold may elicit biased interpretations of results.
SAR405838 demonstrated promising oral pharmacokinetics in mice, achieving dose-dependent and sustained p53 activation in extracted tumor tissues, perhaps the underlying reason for its impressive antitumorigenic capability. Further, when given orally to mice, SAR405838 successfully penetrated human xenografts and elicited significant anti-tumor responses in a dose-dependent manner. Although previously reported (27,28), MDM2 inhibitors have demonstrated therapeutic potential; however, none have established complete tumor regression. SAR405838 generated 100% tumor regression after only two oral doses (200 mg/kg/wk) in our large tumor burden study. Remarkably, no tumor regrowth was observed when treatment was withheld and mice were observed for an additional four weeks. The role of MDM2 in sarcoma is of special interest due to the amplification of MDM2 in nearly one-third of human sarcomas (12). Taken together, the degree of MDM2 amplification may help clinicians predict the therapeutic response of MDM2 inhibition as a single intervention in select DDLPS patients.
SAR405838 is a specific activator of the p53 pathway, inducing up-regulation of p53 in DDLPS cells and, consequently, activating p53-regulated processes (i.e., cell cycle arrest and inducing apoptosis), leading to inhibited cell proliferation, and ultimately cell death. As a result, we questioned which processes controlled by p53 are critical for DDLPS tumor suppression. To address this issue, multiple high-throughput experiments were performed to analyze the responses evoked by p53 reactivation, potentially serving as biomarkers for therapeutic response or as targets in a combinational drug setting.
We identified expression profiles associated with p53 pathway signaling, cell cycle deregulation, and apoptosis (Supplemental Figure S2). The tumor suppressor GADD45a interacts with other intracellular signaling molecules such as p21, which have roles in replication and cell cycle progression; decreased GADD45a expression implies aberrant replication and cell proliferation in DDLPS. Furthermore, the increases in specific p53-induced apoptotic genes suggest that MDM2 targeted therapy is indeed activating the p53 pathway, which was likely due to increased p53 transcription. Moreover, apoptosis associated genes were upregulated following SAR405838 treatment, including PUMA and BAX, thereby demonstrating that apoptosis induced by SAR405838 is triggered by p53.
Gene expression analyses are more objective and quantitative than cellular-based assays; therefore, their results may be more reliably translated to clinical use. Recently, a clinically applied gene expression array was used to predict the recurrence of tamoxifen-treated, node negative breast cancer (29). Such studies may be even more crucial for rare cancers, such as DDLPS, with limited therapeutic options. Towards that end, we are currently investigating gene expression microarray data to further identify genomic subnetworks in the DDLPS tumors. Preliminarily, we have seen a great deal of heterogeneity among MDM2 high and MDM2 low tumors which we believe may correlate to differentially activated pathways that are associated with treatment outcome.
In summary, our data highlights the significant contribution of the deregulated MDM2:p53 axis in DDLPS tumorigenesis. Therefore, by selectively targeting MDM2 with SAR405838, the p53 pathway became activated and elicited anti-DDLPS effects in vitro and in vivo. We investigated pathway enrichment following MDM2 inhibition, identifying possible targetable therapeutic nodes as well as known biomarkers associated with p53 activation. Lastly, our data demonstrates that MDM2 is a critical component of DDLPS tumorigenesis, and that SAR405838 holds remarkable promise as a highly effective candidate therapeutic for patients suffering from this disease.
Supplementary Material
Translational Relevance.
In the 50% of human malignancies that retain wild-type (WT) p53, alternative signaling pathways are used to disable the function of p53, most commonly, overexpression or amplification of MDM2, a negative regulator of p53 proapoptotic function. A unique molecular characteristic of DDLPS is that all of these tumors amplify MDM2 and the majority express WT p53, suggesting that inhibition of MDM2 is a feasible approach for targeting DDLPS. SAR405838, a recently developed spiro-oxindole MDM2 antagonist, binds tightly to the p53 pocket of the MDM2 molecule, thereby inhibiting the binding of MDM2 to p53. Consequently, p53 is stabilized and is activated, leading to decreased cell survival. Therefore, the potential anti-DDLPS therapeutic utility of this approach is suggested by the in vitro and in vivo studies reported here.
Acknowledgments
Financial Support: This manuscript was supported in part by The Amschwand Sarcoma Cancer Foundation (D. Lev) and a grant from the National Cancer Institute of the National Institutes of Health SARC sarcoma SPORE: U54CA168512 (R.E. Pollock). SAR405838 and MI-219 were provided by Sanofi-Aventis.
The authors thank the Lobo and Margolis families for their philanthropic support of our liposarcoma studies, as well as the support of the Amschwand Foundation (to D. Lev). Drugs were generously provided by Sanofi-Aventis, and we thank Dr. Meaux (Sanofi) for her extensive help in data analysis.
Abbreviations
- DDLPS
dedifferentiated liposarcoma
- FISH
fluorescence in situ hybridization
- IPA
Ingenuity Pathway Analysis
- LPS
liposarcoma
- MDM2
mouse double minute 2
- p53
tumor suppressor protein 53
- PLS
pleomorphic liposarcoma
- WDLPS
well-differentiated liposarcoma
- WT
wild-type
Footnotes
Conflicts of Interest: I. Meaux, C. Barriere, and L. Debussche are research investigators at Sanofi-Aventis at the time of work. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Fletcher CDM, Bridge J, Hogendoorn P, Unni KK, Mertens F. In: World Health Organization Classification of Tumours Pathology and Genetics of Tumours of Soft Tissue and Bone. 4. Fletcher CDM, Unni KK, Mertens F, editors. Lyon, France: IARC Press; 2013. [Google Scholar]
- 2.Evans HL. Liposarcoma a study of 55 cases with a reassessment of its classification. Am J Surg Pathol. 1979;3 doi: 10.1097/00000478-197912000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Evans HL, Khurana KK, Kemp BL, Ayala AG. Heterologous Elements in the Dedifferentiated Components of Dedifferentiated Liposarcoma. Am J Clin Pathol. 1994;18:1077–182. doi: 10.1097/00000478-199411000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Ghadimi MP, Al-Zaid T, Madewell J, Peng T, Colombo C, Hoffman A, et al. Diagnosis, management, and outcome of patients with dedifferentiated liposarcoma systemic metastasis. Ann Surg Oncol. 2011;18:3762–70. doi: 10.1245/s10434-011-1794-0. [DOI] [PubMed] [Google Scholar]
- 5.Keung EZ, Hornick JL, Bertagnolli MM, Baldini EH, Raut CP. Predictors of outcomes in patients with primary retroperitoneal dedifferentiated liposarcoma undergoing surgery. J Am Coll Surg. 2014;218:206–17. doi: 10.1016/j.jamcollsurg.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Crago AM, Singer S. Clinical and molecular approaches to well differentiated and dedifferentiated liposarcoma. Curr Opin Oncol. 2011;23:373–8. doi: 10.1097/CCO.0b013e32834796e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meis-Kindblom J, Sjögren H, Kindblom L-G, Peydró-Mellquist A, Röijer E, Åman P, et al. Cytogenetic and molecular genetic analyses of liposarcoma and its soft tissue simulators: recognition of new variants and differential diagnosis. Virchows Arch. 2001;439:141–51. doi: 10.1007/s004280100423. [DOI] [PubMed] [Google Scholar]
- 8.Pedeutour F, Forus A, Coindre JM, Berner JM, Nicolo G, Michiels JF, et al. Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes Cancer. 1999;24:30–41. [PubMed] [Google Scholar]
- 9.Ware PL, Snow AN, Gvalani M, Pettenati MJ, Qasem Sa. MDM2 Copy Numbers in Well-Differentiated and Dedifferentiated Liposarcoma: Characterizing Progression to High-Grade Tumors. Am J Clin Pathol. 2014;141:334–41. doi: 10.1309/AJCPLYU89XHSNHQO. [DOI] [PubMed] [Google Scholar]
- 10.Iwakuma T, Lozano G. MDM2, an introduction. Mol Cancer Res. 2003;1:993–1000. [PubMed] [Google Scholar]
- 11.Ohnstad HO, Castro R, Sun J, Heintz K-M, Vassilev LT, Bjerkehagen B, et al. Correlation of TP53 and MDM2 genotypes with response to therapy in sarcoma. Cancer. 2013;119:1013–22. doi: 10.1002/cncr.27837. [DOI] [PubMed] [Google Scholar]
- 12.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453–9. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 14.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–32. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 15.Vassilev LT. MDM2 inhibitors for cancer therapy. Trends Mol Med. 2007;13:23–31. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Carvajal D, Tovar C, Yang H, Vu BT, Heimbrook DC, Vassilev LT. Activation of p53 by MDM2 antagonists can protect proliferating cells from mitotic inhibitors. Cancer Res. 2005;65:1918–24. doi: 10.1158/0008-5472.CAN-04-3576. [DOI] [PubMed] [Google Scholar]
- 17.Yu S, Qin D, Shangary S, Chen J, Wang G, Ding K, et al. Potent and orally active small-molecule inhibitors of the MDM2-p53 interaction. J Med Chem. 2009;52:7970–3. doi: 10.1021/jm901400z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science (80- ) 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Hu Y. Small Molecule Agents Targeting the p53-MDM2 Pathway for CancerTherapy. Med Res Rev. 2012;32:1159–96. doi: 10.1002/med.20236. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Sun W, Zhao Y, McEachern D, Meaux I, Barriere C, et al. SAR405838: An optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression. Cancer Res. 2014;74:5855–65. doi: 10.1158/0008-5472.CAN-14-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu L, Zhu N, Findley HW, Zhou M. MDM2 antagonist nutlin-3 is a potent inducer of apoptosis in pediatric acute lymphoblastic leukemia cells with wild-type p53 and overexpression of MDM2. Leukemia. 2008;22:730–9. doi: 10.1038/leu.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyachi M, Kakazu N, Yagyu S, Katsumi Y, Tsubai-Shimizu S, Kikuchi K, et al. Restoration of p53 pathway by nutlin-3 induces cell cycle arrest and apoptosis in human rhabdomyosarcoma cells. Clin Cancer Res. 2009;15:4077–84. doi: 10.1158/1078-0432.CCR-08-2955. [DOI] [PubMed] [Google Scholar]
- 23.Conyers R, Young S, Thomas DM. Liposarcoma: molecular genetics and therapeutics. Sarcoma. 2011;483154 doi: 10.1155/2011/483154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haupt Y, Barak Y, Oren M. Cell type-specific inhibition of p53-mediated apoptosis by mdm2. EMBO J. 1996;15:1596–606. [PMC free article] [PubMed] [Google Scholar]
- 25.Constantinidou A, Pollack SM, Jones RL. MDM2 inhibition in liposarcoma: a step in the right direction. Lancet Oncol Elsevier Ltd. 2012;13:1070–1. doi: 10.1016/S1470-2045(12)70457-6. [DOI] [PubMed] [Google Scholar]
- 26.Ray-Coquard I, Blay J-Y, Italiano A, Le Cesne A, Penel N, Zhi J, et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol. 2012;13:1133–40. doi: 10.1016/S1470-2045(12)70474-6. [DOI] [PubMed] [Google Scholar]
- 27.Ding Q, Zhang Z, Liu J-J, Jiang N, Zhang J, Ross TM, et al. Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J Med Chem. 2013;56:5979–83. doi: 10.1021/jm400487c. [DOI] [PubMed] [Google Scholar]
- 28.Tovar C, Graves B, Packman K, Filipovic Z, Higgins B, Xia M, et al. MDM2 small-molecule antagonist RG7112 activates p53 signaling and regresses human tumors in preclinical cancer models. Cancer Res. 2013;73:2587–897. doi: 10.1158/0008-5472.CAN-12-2807. [DOI] [PubMed] [Google Scholar]
- 29.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 30.Peng T, Zhang P, Liu J, Nguyen T, Bolshakov S, Belousov R, et al. An experimental model for the study of well-differentiated and dedifferentiated liposarcoma; deregulation of targetable tyrosine kinase receptors. Lab Invest. 2011;91:392–403. doi: 10.1038/labinvest.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Q-S, Rosenblatt K, Huang K-L, Lahat G, Brobey R, Bolshakov S, et al. Vimentin is a novel AKT1 target mediating motility and invasion. Oncogene. 2011;30:457–70. doi: 10.1038/onc.2010.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolshakov S, Walker CM, Strom SS, Selvan MS, Clayman GL, El-Naggar A, et al. p53 Mutations in Human Aggressive and Nonaggressive Basal and Squamous Cell Carcinomas. Clin Cancer Res. 2003;9:228–34. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.