Abstract
The t(8;21) rearrangement, which creates the AML1-ETO fusion protein, represents the most common chromosomal translocation in acute myeloid leukemia (AML). Clinical data suggest that CBL mutations are a frequent event in t(8;21) AML, but the role of CBL in AML1-ETO-induced leukemia has not been investigated. In this study, we demonstrate that CBL mutations collaborate with AML1-ETO to expand human CD34+ cells both in vitro and in a xenograft model. CBL depletion by shRNA also promotes the growth of AML1-ETO cells, demonstrating the inhibitory function of endogenous CBL in t(8;21) AML. Mechanistically, loss of CBL function confers hyper-responsiveness to thrombopoietin and enhances STAT5/AKT/ERK/Src signaling in AML1-ETO cells. Interestingly, we found the protein tyrosine phosphatase UBASH3B/Sts-1, which is known to inhibit CBL function, is upregulated by AML1-ETO through transcriptional and miR-9-mediated regulation. UBASH3B/Sts-1 depletion induces an aberrant pattern of CBL phosphorylation and impairs proliferation in AML1-ETO cells. The growth-inhibition caused by UBASH3B/Sts-1 depletion can be rescued by ectopic expression of CBL mutants, suggesting that UBASH3B/Sts-1 supports the growth of AML1-ETO cells partly through modulation of CBL function. Our study reveals a role of CBL in restricting myeloid proliferation of human AML1-ETO-induced leukemia, and identifies UBASH3B/Sts-1 as a potential target for pharmaceutical intervention.
Keywords: AML1-ETO, CBL, UBASH3B/Sts-1, miR-9, acute myeloid leukemia
Introduction
Core binding factor (CBF) acute myeloid leukemia (AML) is the most common cytogenetic subtype in AML, defined by the presence of t(8;21) or inv(16)/t(16;16). CBF is a heterodimeric transcription factor complex composed of RUNX1 and CBFB, and plays essential roles in hematopoiesis. The chromosomal aberrations create AML1-ETO (also called RUNX1-RUNX1T1) and CBFB-MYH11 fusion proteins that disrupt the functions of CBF. Numerous laboratory and clinical discoveries have revealed the molecular basis of CBF-AML (reviewed in1, 2). Nevertheless, molecular therapies for CBF leukemia have been difficult to develop, and only recently has some success been seen in targeting the CBF complex itself3, 4. It is therefore important to identify druggable pathways and proteins that are involved in the leukemogenic program in CBF leukemia.
Several recent studies have identified CBL mutations in 5 –10 % of CBF-AML5–9. CBL is also frequently mutated in myelodysplastic/myeloproliferative neoplasms, but rarely mutated in other types of de novo AML10–18. CBL is an E3 ubiquitin ligase and promotes ubiquitination-directed degradation of target proteins, such as EGFR, FLT3, KIT, MPL and Src family kinases19–23. CBL mutations are frequently found in exons 8–9, encoding the linker region and the RING finger domain, which are essential for the E3 ligase activity. Loss of the E3 ligase activity together with additional gain-of-functions induced by these mutations promote malignant transformation24. Multiple CBL interacting proteins have been identified to modulate CBL function25, and deregulation of the CBL regulators are also implicated in the development of malignant diseases26. Among these, the protein tyrosine phosphatase UBASH3B/Sts-1 (also called TULA-2) has been shown to inhibit CBL function to regulate EGFR activity and promote invasion/metastasis of breast cancer27, 28.
The physiologic roles of CBL in hematopoiesis and leukemogenesis have been studied using mouse genetic models. Hematopoietic stem cells (HSCs) of Cbl-deficient mice exhibit hypersensitivity to a variety of cytokines and enhanced long-term repopulating capacity12, 29. Furthermore, Cbl-deficiency accelerates the development of blast crisis in BCR-ABL transgenic mice12, and myeloid leukemia in knockin mice with a Cbl mutation30. Thus, these mouse models revealed a role for Cbl as a negative regulator of HSCs and myeloid leukemogenesis. However, murine hematopoietic cells may differ in their regulation from their human counterparts. Furthermore, the role of CBL in CBF leukemia has not been investigated.
We have established a culture system to model CBF-AML using human cord blood (CB) CD34+ cells31–33. We have also developed a xenograft model for human leukemia using immunodeficient mice with transgenic expression of human SCF, GM-CSF, and IL-3 (three poorly cross-reacting cytokines) in the NOD/SCID/IL2RG−/− background (NOD/LtSz-scid/IL2RG-SGM3, NSGS). The NSGS mice provide optimal conditions for engraftment and expansion of human AML cells in vivo34. Using these human cell-based assays, we demonstrate that CBL mutations collaborate with AML1-ETO to expand human CB cells. Conversely, endogenous CBL inhibits the growth of AML1-ETO cells. Interestingly, we also found that AML1-ETO induces UBASH3B/Sts-1 expression to attenuate the growth-inhibitory function of CBL. Thus, the UBASH3B/Sts-1-CBL axis delicately controls myeloid proliferation in human AML1-ETO leukemia, providing potential therapeutic targets.
Materials and Methods
Human cell culture
Human umbilical CB cells were obtained from Translational Trials Development and Support Laboratory at Cincinnati Children’s Hospital Medical Center according to an institutional review board-approved protocol. Informed consent was obtained in accordance with the Declaration of Helsinki. CD34+ cells were separated using EasySep CD34 selection kit (StemCell Technologies, Vancouver, BC, Canada). We engineered human AML-ETO cells by transducing AML1-ETO into CB cells using retrovirus, as described previously31, 32. Cells were cultured in IMDM media containing 20% BIT9500 (StemCell Technologies) and 10 ng/mL human SCF, TPO, FLT3L, IL-3, and IL-6, as described previously35, 36.
Vectors and viral transduction
Haemagglutinin (HA)-tagged cDNAs of wild-type and mutant CBL (Q369P, Y371S) in a MSCV-based retroviral vector pGCDNsam-IRES-GFP were provided by Dr. S. Ogawa, M. Sanada and Dr. M. Onodera. A deletion mutant of CBL (ΔE8/9) was provided by Dr. K. Spiekermann, and we cloned it into a retroviral vector pMYs-IRES-GFP. HA-tagged AML1-ETO in a pMSCV-IRES-Thy1.1 retroviral vector was used for AML1-ETO expression. Lentiviral vector MISSION pLKO.1-shRNA-puro constructs targeting human CBL [TRCN0000010727 (shCBL)] or UBASH3B/Sts-1 (TRCN0000073150 (shUBASH-1) and TRCN0000073151(shUBASH-2)] were obtained from Sigma-Aldrich. Venus marker was excised from the pLKO.1-Venus construct with BamH1 and Kpn1 and was subcloned into the corresponding sites on the CBL shRNAs to replace the puromycin resistant gene. MSCVpig-miR-9 was described previously37. Viral production was performed by transfecting viral plasmids along with gag, pol, env-expressing plasmids into 293T cells, as described previously33, 38.
Flow Cytometry
Cells were analyzed on a FACSCanto and were sorted with a FACSAria (BD). Antibodies were all purchased from BD Biosciences (San Jose, California) (Table S1). Cells were stained with fluorochrome-conjugated antibodies incubated for 30 min at 4°C and were washed with 2%FBS in PBS prior to analysis. Cell-cycle analysis (Vybrant® DyeCycle™ Violet stain, Invitrogen) and apoptosis analysis (AnnexinV-APC kit, BD Biosciences) were performed according to the manufacturer’s recommendations.
Immunoprecipitation and western blotting
Western blotting was performed as described previously33. Signals were detected with SuperSignalWest Pico or Femto Chemiluminescent Substrate (Pierce), or with LI-COR Odyssey Infrared Imaging System. Band intensity was measured using LabWorks Version 4.5 software (UVP, LLC). For immunoprecipitation, cell lysates were incubated with the anti-CBL monoclonal antibody (D4E10) overnight at 4 °C. Then, the samples were incubated with Dynabeads Protein G (Life Technologies) for 30 min at 4 °C. The precipitates were washed three times with the cell lysis buffer (Cell Signaling Technology, #9803) containing 1mM PMSF (Cell Signaling Technology, #8553), subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and analyzed by western blotting. Antibodies used are listed in Table S1.
WST-1 assays
Human AML1-ETO cells expressing vector, wild-type or mutant CBL, and CB CD34+ cells were plated with the indicated cytokines (10 ng/ml), or titrating doses of drugs (Dasatinib; 0 – 30 μM, INCB018424; 0 – 10 μM, TG101209; 0 – 10 μM), in triplicate. After 48 hours, 10 μL WST-1 cell proliferation assay premix (MK 400; Takara Bio Inc) was added to each well. Plates were read at 450–560 nm to measure optical density (O.D.).
Xenograft assay
NOD/SCID/IL2rg−/−SGM3 (NSGS) mice were generated by crossing NOD/SCID/IL2rg−/− mice (Jackson Laboratory, Bar Harbor, Maine) with NOD/SCID mice with transgenic expression of hSCF, hGM-CSF and hIL-3 (a kind gift from Dr. C. Eaves), and were used as recipients34. Human CB CD34+ cells were transduced with Vector, wild-type or mutant CBL (coexpressing GFP) together with AML1-ETO (coexpressing Thy1.1), and were injected by intrafemoral injection into sublethally irradiated (250cGy from a cesium source) 6- to 8-week-old male or female mice. The recipient mice were euthanized 12 – 16 weeks after transplantation for analyses. We used human cord blood cells derived from a single donor for each experiment, and performed three independent experiments using 6 recipient mice in total for each group (three mice for cord blood 1, one mouse for cord blood 2, and two mice for cord blood 3). Randomization and blinding were not performed in this study.
Gene and miRNA expression analyses and ChIP-Seq analysis
Generation of microarray data was described (GSE8023)39. ChIP-Seq and RNA-Seq analyses in Kasumi-1 cells were performed as described previously40, 41. miRNA expression was examined as described previously37. We also used the following public databases: Hemaexplorer (http://servers.binf.ku.dk/hemaexplorer/)42, cBioPortal (http://www.cbioportal.org/public-portal/)43, 44, starBase v2.0 (http://starbase.sysu.edu.cn/)45, 46, and TargetScan (http://www.targetscan.org/)47.
Statistics
Unpaired and two-tail t-test was used to evaluate differences between groups in WST-1 assay (Figure 4A). Error bars representing S.D. indicate variation for each group and variance was found to be similar between compared groups. Unpaired t-test with Welch’s correction was used in Figure 2C and Figure 5B. The one sample t-test was used in Figure 4A and 4B.
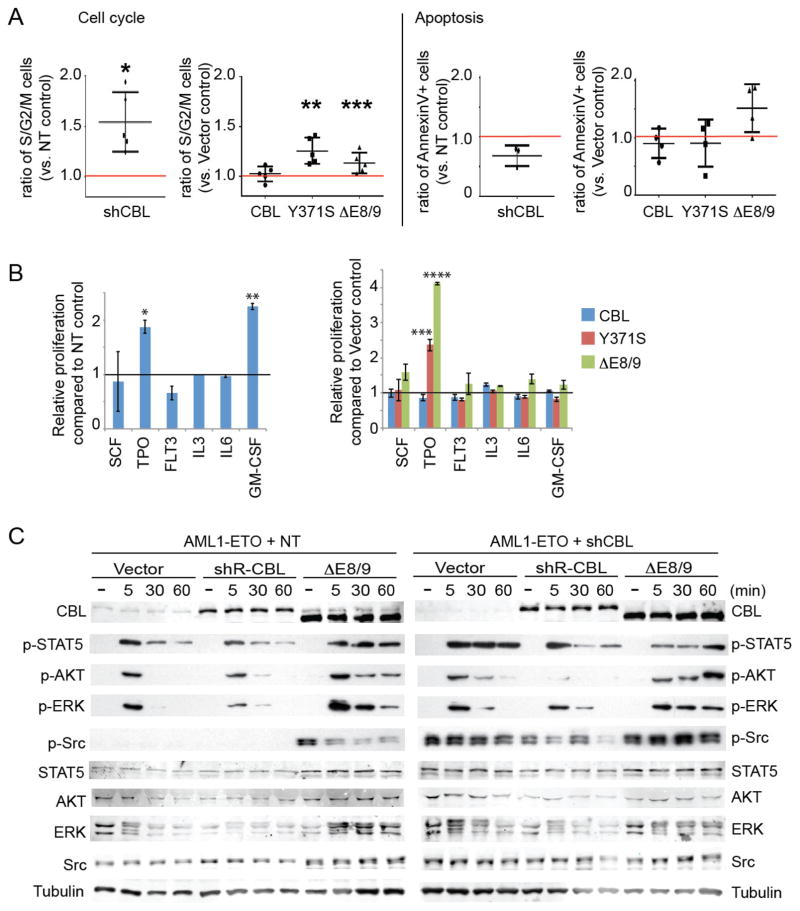
Figure 4. CBL mutations/depletion modulates STAT5, AKT, ERK, and Src pathways.
A. Cell cycle status and apoptosis were assessed on days 5 through 7 of culture. The frequency of S/G2/M phase cells or AnnexinV+ cells was normalized to that of NT or Vector control. At least three independent experiments were performed for each analysis, and data are shown as the mean ± SD. See also Figure S7A, B. B. The growth of AML1-ETO cells transduced with NT/shCBL (left) or various CBL constructs (right) was measured by WST-1 assay 48 hours after the indicated cytokine stimulation. CBL-depleted cells showed hyperresponsiveness to TPO and GM-CSF (left). Mutant CBL, but not wild-type CBL, conferred hyperresponsiveness to TPO (right). Data are shown as Mean+/−SD (N=3). *p=0.014, **p<0.001. ***p=0.039, ****p<0.001. C. AML1-ETO-expressing CB cells were first transduced with NT or shCBL, and were then transduced with shRNA-resistant versions of CBL constructs. The cells were left unstimulated or stimulated with 6 cytokines (SCF, TPO, FLT3L, IL-3, IL-6, GM-CSF, 10 ng/ml each) for the indicated times. Total cell lysates were analyzed by western blotting using antibodies to STAT5, AKT, ERK, Src and their phosphorylated forms. Note the sustained signal of phosphorylated STAT5 and Src in CBL-depleted cells and ΔE8/9-expressing cells following cytokine stimulation. ΔE8/9 mutant also increased AKT and ERK phosphorylation in both NT and shCBL-transduced cells. See also Figure S8.
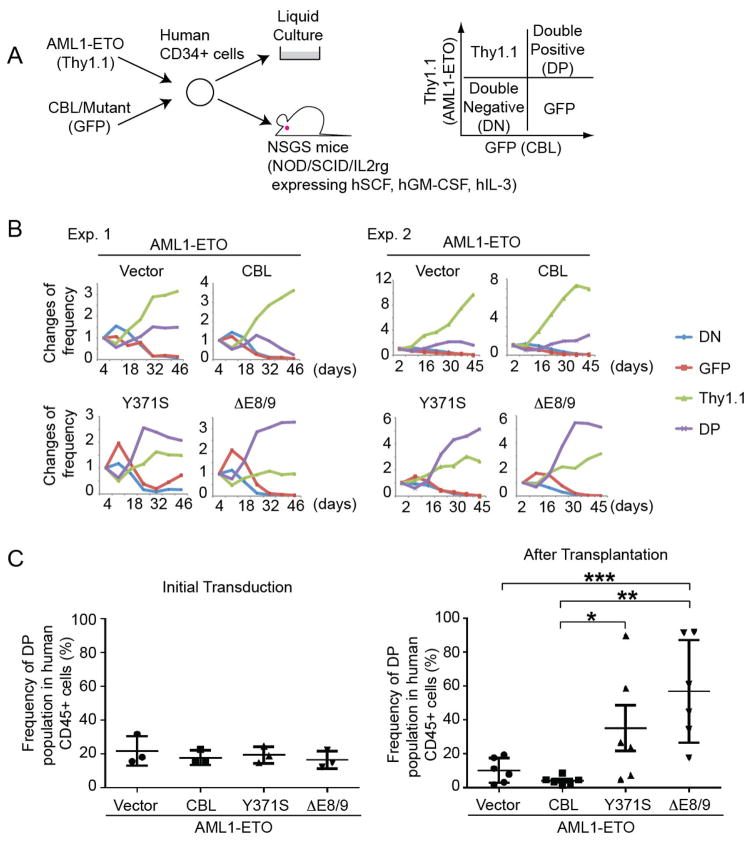
Figure 2. Mutant CBL promotes the growth of human AML1-ETO cells.
A. Experimental scheme used in Figures 2B, C. Human CB CD34+ cells were cotransduced with various CBL constructs (marked with GFP) together with AML1-ETO (marked with Thy1.1), and were cultured with cytokines, or were injected (5x105 cells) into the left femur of NSGS mice. Bone marrow cells from the injected bone were analyzed 12–16 weeks after injection. B. Changes of GFP/Thy1.1 expression in culture were observed for 46 days. Results are normalized to the frequency of each population at day 3, set to 1. GFP/Thy1.1-double positive (DP) cells became dominant when the CD34+ cells were cotransduced with AML1-ETO and CBL mutants, suggesting their cooperativity. See also Figure S3A. C. Frequency of GFP/Thy1.1-DP cells before and after transplantation. Three independent experiments were performed, and data are shown as the mean ± SEM. There was a strong trend that mutant CBL increases the frequency of GFP/Thy1.1-DP cells. *p=0.044, **p=0.008, ***p=0.012. See also Figure S4.
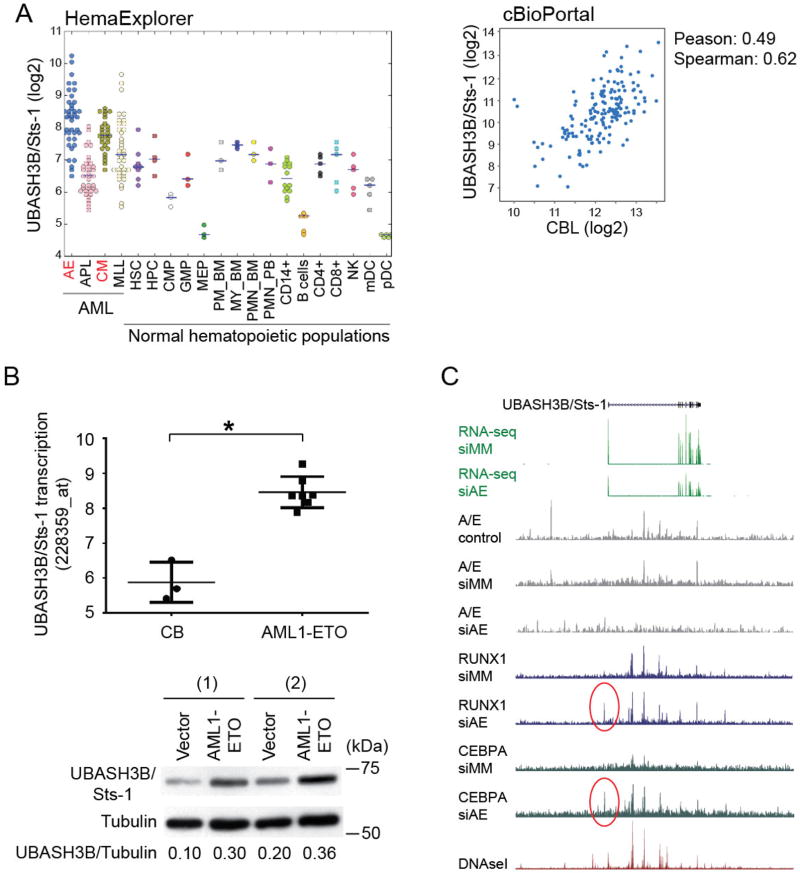
Figure 5. AML1-ETO transcriptionally induces UBASH3B/Sts-1 upregulation.
A. (Left) A plot of UBASH3B/Sts-1 expression in human AML samples and human normal hematopoietic populations was drawn using HemaExplorer. UBASH3B/Sts-1 expression is higher in AML1-ETO (AE) and CBFB-MYH11 (CM) leukemia compared to any normal hematopoietic populations including hematopoietic stem cells (HSC). Meaning of other abbreviations is available in the HemaExplorer website (http://servers.binf.ku.dk/hemaexplorer/). (Right) A scatter plot showing CBL expression against UBASH3B/Sts-1 expression in 187 AML patient samples was drawn using cBioPortal. Pearson’s correlation= 0.49, Spearman’s correlation= 0.62. B. (Upper) UBASH3B/Sts-1 expression detected by a probe (228359_at) in CB CD34+ cells (3 samples) and AML1-ETO-expressing CB cells (7 samples) cultured for 2–3 weeks. Other probes for UBASH3B showed similar results (data not shown). *p=0.006. (Lower) Two independent CB cells were transduced with vector or AML1-ETO, and were subjected to immunoblotting with UBASH3B/Sts-1 and tubulin antibodies 7 days after transduction. Protein expression of UBASH3B/Sts-1 was upregulated in AML1-ETO-transduced cells. Relative densitometry values of UBASH3B/Sts-1 are shown underneath blots as ratios relative to the levels of Tubulin. C. UCSC genome browser screenshot showing UBASH3B/Sts-1 expression, the binding patterns of AML1/ETO (A/E), RUNX1, C/EBPα, and DNase I hypersensitive sites (DHS) at UBASH3B/Sts-1 locus in Kasumi-1 cells. siMM: control siRNA, siAE: AML1/ETO siRNA. Regions with changing RUNX1 and C/EBPα binding sites are highlighted by red circles.
Results
Mutant CBL promotes transient proliferation of human cord blood cells
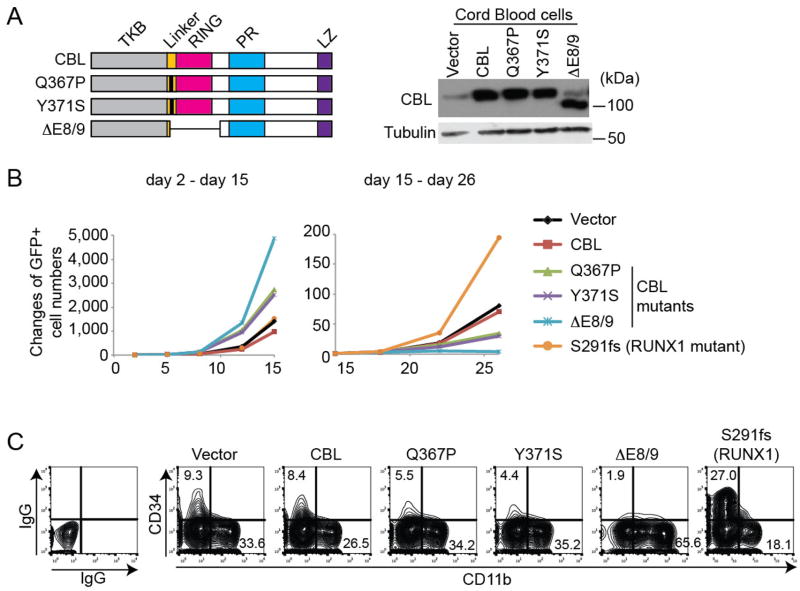
We first examined the effect of enforced expression of wild-type and mutant CBL in human CB CD34+ cells. Q367P and Y371S are CBL mutants in the linker domain, and ΔE8/9 is a splice variant of CBL lacking exons 8 and 912,7 (Figure 1A). These mutations were found in myeloid diseases including CBF-AML7, 12. We transduced these constructs (co-expressing GFP) into CB CD34+ cells, and monitored the changes of GFP frequency in culture. CBL mutants, but not wild-type CBL, showed a growth-promoting effect on CB cells at the beginning of culture (Figure 1B left, Figure S1C). However, all the CBL mutant-expressing CB cells stopped growing earlier than normal CB cells due to rapid terminal differentiation as evidenced by the loss of primitive CD34+ cells and increased expression of CD11b (Figure 1B right, C, Figure S1A, B, D). These properties of CBL mutants contrast sharply with those of a C-terminal truncation mutant of RUNX1 (S291fs), another frequently mutated gene in myeloid neoplasms (Figure 1B, C, Figure S1A, B, D). Thus, in contrast to the RUNX1 mutant that was shown to increase self-renewal of CB cells48, CBL mutations enhance the transient proliferation of CB cells but do not increase their long-term proliferative potential.
Figure 1. Mutant CBL promotes the growth of human cord blood cells.
A. (Left) Schematic presentation and confirmation of expression of wild-type and mutant CBL constructs. Q367P and Y371S have a point mutation (indicated by a bold line) in the Linker domain. ΔE8/9 is a splice variant lacking the sequences of exon 8 and 9. (Right) Expression of the individual proteins was confirmed in CB cells. TKB: Tyrosine Kinase binding domain, Linker: Linker domain, RING: RING finger domain, PR: Proline-Rich domain, LZ: leucine zipper motif. B. Changes of GFP+ (vector/CBL/mutant-expressing) cell numbers in CB cell cultures. CBL mutants showed a growth-promoting effect in CB cells at the beginning of culture (days 2 – 15), while later (days 15 – 26), the CBL-mutant expressing cells stopped growing. In contrast, RUNX1 mutant (S291fs)-expressing CB cells grew better than control cells at the late phase (day 15 – day 26) of culture. Results are normalized to the number of GFP+ cells at day 2 (left) or day 15 (right), set to 1. See also Figure S1C. C. Frequency of CD34+ and CD11b+ cells in CB cells transduced with various CBL constructs or a mutant RUNX1 (S291fs) at 2 weeks of culture. CBL mutants decreased CD34 and increased CD11b expression in CB cells compared to vector control, indicating their effects to promote myeloid maturation. In contrast, a RUNX mutant (S291fs)-expressing CB cells contained more CD34+ cells and less CD11b+ cells, indicating the effect of the RUNX1 mutant to promote self-renewal. See also Figure S1A.
Mutant CBL promotes myeloid proliferation of human AML1-ETO cells
We next transduced the CBL constructs into CB cells expressing AML-ETO, which can grow for over 6 months in culture retaining primitive CD34+ cells31, 32(Figure S2A). All CBL mutants showed a robust growth-promoting effect in AML1-ETO cells (Figure S2B, C), suggesting the functional cooperation between CBL mutants and AML1-ETO to promote proliferation of CB cells. To further analyze the cooperativity between AML1-ETO and mutant CBL in the generation of human AML, we co-transduced wild-type or mutant CBL (co-expressing GFP) together with AML1-ETO (co-expressing Thy1.1) into human CB CD34+ cells, and assessed the expression changes of GFP (i.e. CBL) and Thy1.1 (i.e. AML1-ETO) in culture or in a xenograft model (Figure 2A). In agreement with our previous reports31, 32, Thy1.1+ cells (AML1-ETO expressing cells) became dominant within 6 weeks of culture, reflecting their long-term proliferative capacity. GFP-only cells (CBL expressing cells) were not observed at 6 weeks in any of the cultures, indicating that neither wild-type nor mutant CBL extends the lifespan of CB cells. However, mutant CBL, but not wild-type CBL, collaborated with AML1-ETO to promote myeloid proliferation in vitro, as evidenced by the increased frequency of GFP/Thy1.1-double positive (DP) cells by 6 weeks of culture (Figure 2B, Figure S3A). Strong expression of mutant CBL in long-term cultured cells was confirmed by immunoblotting (Figure S3B). We also directly transplanted human CD34+ cells transduced with wild-type or mutant CBL together with AML1-ETO into NSGS mice34, and analyzed bone marrow cells between 12 and 16 weeks after transplantation. Consistent with the in vitro results, we found a substantial increase of GFP/Thy1.1-DP population in the mutant CBL transduced cells, which was not seen in vector or wild-type CBL transduced cells (Figure 2C, Figure S4). The engrafted human cells expressing mutant CBL and AML1-ETO were myeloid progenitors (CD33+, CD19-, CD13+, CD11b+/−, CD14+/−) in almost all cases, except for one mouse in which lymphoid progenitors (CD19+, CD79a+, CD34+/−, CD33-, MPO-) were expanded (Figure S5A, B). The human GFP+ cells were also detected in the non-injected bones and to a lesser extent in the spleen of mice, suggesting hematogenous spreading (Figure S5C). Moreover, Wright-Giemsa staining showed that the AML1-ETO/CBL-mutant coexpressing cells contained immature cells exhibiting a blast-like morphology, with larger cell size, higher nuclear-to-cytoplasmic ratio, and less condensed chromatin structure (Figure S5A). Both mutant CBL and AML1-ETO proteins were indeed expressed in GFP/Thy1.1 DP cells (Figure S5D). Taken together, AML1-ETO/CBL-mutant co-expressing cells recapitulate several features consistent with progression toward human AML. However, despite significantly increased engraftment of these cells in bone marrow, we did not detect overt leukemia development. Moreover, these cells were not serially transplantable (data not shown).
Endogenous CBL inhibits the proliferation of human AML1-ETO cells
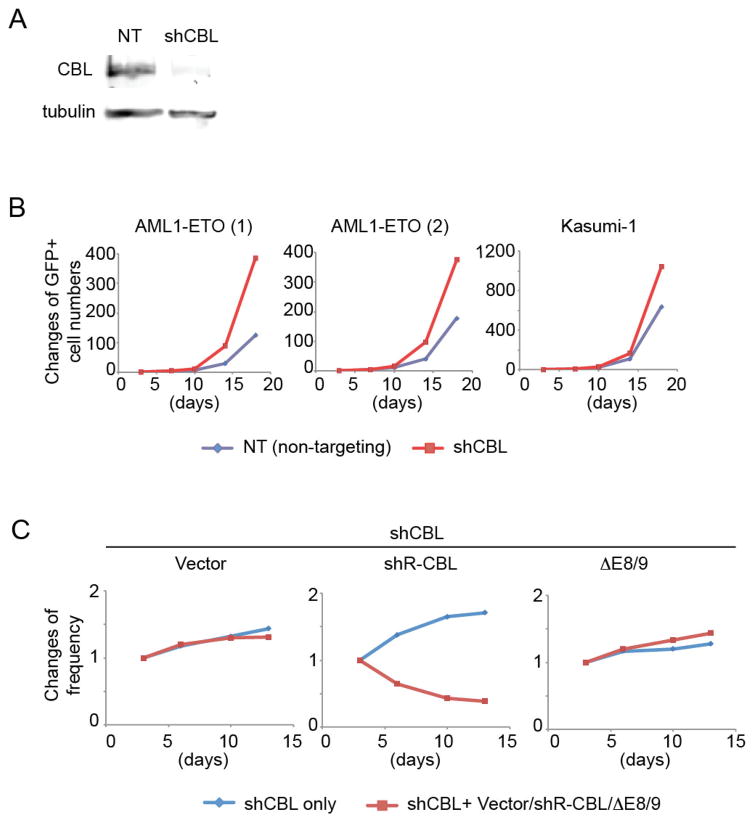
CBL was abundantly expressed in all the hematopoietic/leukemic cells we examined: CB CD34+ cells, the “engineered” AML cells (AML1-ETO-, CBFB-MYH11-, and MLL-AF9-expressing CB cells)31–33, 49, and several myeloid cell lines (THP1, K562, HEL, OCI-AML3, Kasumi-1) (Figure S6A). To assess the role of endogenous CBL, we knocked down CBL expression in AML1-ETO-expressing CB cells and the AML1-ETO harboring Kasumi-1 cell line50, 51 using a CBL-specific shRNA lentivirus (shCBL) that showed efficient knockdown of CBL protein (Figure 3A). CBL depletion promoted the growth of AML-ETO-expressing CB cells and Kasumi-1 cells (Figure 3B). To rule out the possible off-target effects of shRNA, we then examined whether the reintroduction of CBL could reverse the growth-inhibitory effect of shCBL. We constructed an shCBL resistant version of CBL (shR-CBL) by introducing silent mutations. The ΔE8/9 mutant is also resistant to shCBL because it lacks the region targeted by shCBL (Figure S6B, C). We expressed vector control, shR-CBL, or ΔE8/9 together with the shCBL in AML1-ETO cells, and compared the growth of shCBL-transduced cells and shCBL-Vector/shR-CBL/ΔE8/9 co-transduced cells. CBL reintroduction suppressed the enhanced cell growth by shCBL, indicating that the growth-promoting effect of the CBL shRNA is in fact due to CBL downregulation. In contrast, the ΔE8/9 mutant did not reverse the effect of shCBL (Figure 3C, Figure S6D).
Figure 3. CBL depletion promotes the growth of human AML1-ETO cells.
A. shCBL showed efficient CBL knockdown and growth-promoting effect in AML1-ETO cells. B. AML1-ETO cells were transduced with NT or shCBL and were cultured with cytokines. Changes of Venus+ (shRNA-transduced) cell numbers in culture are shown. CBL-depleted AML1-ETO expressing CB cells and Kasumi-1 cells grow faster than cells transduced with a non-targeting shRNA (NT). Results are normalized to the number of Venus+ cells at day 3, set to 1. C. AML1-ETO cells were transduced with shCBL (marked with Venus) in combination with vector, shRNA-resistant version of wild-type CBL (shR-CBL) or ΔE8/9 (marked with GFP). The growth of Venus+ (shRNA only) and GFP+Venus+ (shRNA + vector or shR-CBL or ΔE8/9) cells in each culture were monitored. Results are normalized to the frequency of Venus+ cells or that of GFP+Venus+ cells at day 3, set to 1. The growth-promoting effect of shCBL was abrogated by shR-CBL, but not by the ΔE8/9 mutant. See also Figure S6D.
CBL inactivation promotes cell cycle progression, confers hyper-responsiveness to TPO, and activates STAT/AKT/ERK/Src pathways
To characterize the increased cell growth induced by CBL mutations/depletion, we performed cell cycle and apoptosis analyses. CBL depletion by shCBL as well as forced expression of CBL mutants consistently increased the proportion of S/G2/M phase cells in human AML1-ETO cells. The effect of CBL mutations/depletion on apoptosis varied among experiments (Figure 4A, Figure S7). Thus, it appears that CBL inactivation promotes cell growth mainly through increased cell cycle progression. We next examined the role of CBL in the response of AML1-ETO cells to various cytokines. Both CBL-depleted and mutant-expressing AML1-ETO cells showed an enhanced proliferative response to TPO compared to control cells (Figure 4B). These results, together with the findings in our recent report52, indicate an essential role of TPO in the development of AML1-ETO leukemia.
We then analyzed the effects of CBL depletion on the amplitude and duration of STAT/AKT/ERK signaling induced by cytokine stimulation. CBL depletion resulted in prolonged phosphorylation of STAT5 in AML1-ETO-expressing CB cells, which was reduced by reintroduction of wild-type CBL (Figure 4C, Figure S8, Figure S9). In the Kasumi-1 cell line that harbors AML1-ETO and a c-Kit mutation50, 51, CBL depletion induced enhanced phosphorylation of AKT and ERK (Figure S9). We also observed a prolonged activation of STAT/AKT/ERK pathways induced by the CBL mutant (ΔE8/9) in both control and CBL-depleted AML1-ETO cells (Figure 4C, Figure S8). Stimulation by TPO alone was sufficient to induce prolonged STAT5 phosphorylation caused by the ΔE8/9 mutant (Figure S10). Furthermore, we found the increased Y416 phosphorylation (i.e. activation) of Src, a non-receptor tyrosine kinase that is known to interact with CBL53, in CBL-depleted and mutant-transduced AML1-ETO cells. The Src phosphorylation in CBL-depleted cells was attenuated by wild-type CBL, while ΔE8/9 mutant further increased the level of phosphorylation (Figure 4C, Figure S8). Thus, CBL depletion/mutations result in activation of multiple signaling pathways including STAT, AKT, ERK and Src in AML1-ETO cells.
Expression of AML1-ETO leads to upregulation of UBASH3B/Sts-1
CBL is known to interact with multiple proteins to fine-tune signal transduction. We found one of the CBL-interacting proteins, UBASH3B/Sts-1, is abundantly expressed in CBF-AML. Expression level of UBASH3B/Sts-1 RNA is higher in AML1-ETO and CBFB-MYH11 AML than in other types of AML and normal hematopoietic populations. Furthermore, UBASH3B/Sts-1 expression correlates well with that of CBL in AML patient samples, suggesting the functional interaction between these proteins in AML cells (Figure 5A). Expression of UBASH3A/Sts-2, another member of Sts family, is not upregulated in CBF-AML and does not correlate with that of CBL (Figure S11). Consistent with these data, enforced expression of AML1-ETO in CB cells led to upregulation of UBASH3B/Sts-1 at both mRNA and protein levels (Figure 5B). We then examined ChIP-Seq and RNA-Seq data in Kasumi-1 cells40, 41, and found multiple peaks of AML1-ETO binding to the genomic locus of UBASH3B/Sts-1. The loss of these peaks upon AML1-ETO-knockdown in Kasumi-1 cells coupled with UBASH3B/Sts-1 downregulation indicates a direct role of AML1-ETO in the regulation of UBASH3B/Sts-1 expression. Many of the peaks are within DNaseI hypersensitive sites (DHSs), the marker of transcriptionally active regions of the genome. Interestingly, ChIP data also revealed the binding of RUNX1 in this region, suggesting dynamic competition between AML1-ETO and RUNX1 for UBASH3B/Sts-1 expression. Moreover, the UBASH3B/Sts-1 promoter contains multiple RUNX1 and C/EBPα motifs (data not shown), and AML1-ETO-depleted Kasumi-1 cells showed increased binding of RUNX1 and C/EBPα to the region (Figure 5C). Combined with our recent demonstration that AML1-ETO depletion activates a C/EBPα-driven transcriptional network41 and the fact that C/EBPα is required to repress stem cell specific genes such as SOX454, our data suggest that AML1-ETO binding may induce UBASH3B/Sts-1 upregulation by interfering with the repressive activity of a RUNX1/C/EBPα-complex. In line with this, UBASH3B/Sts-1 is also highly expressed in AMLs with loss-of-function RUNX1 mutations55.
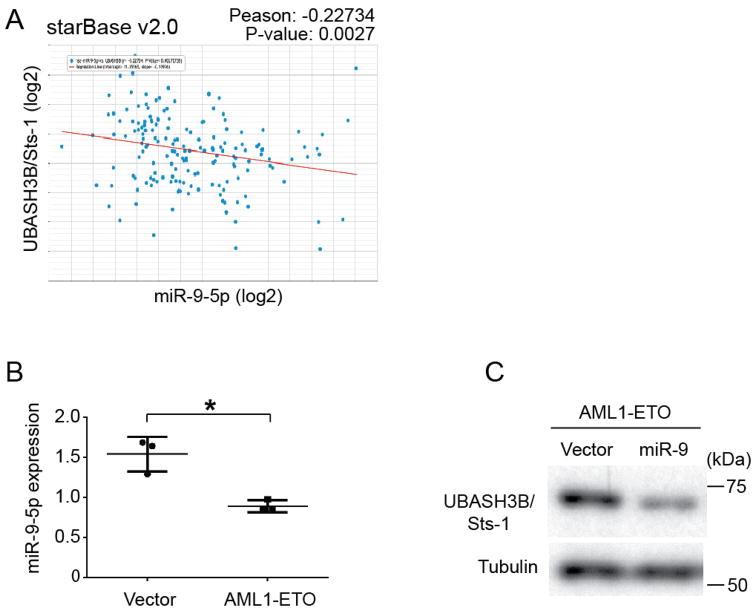
In addition to the transcriptional control, miRNAs may also participate in the regulation of UBASH3B/Sts-1 expression, as was shown in triple negative breast cancer28. Interestingly, miR-9-5p, which was shown to be downregulated in AML1-ETO leukemia56, exhibits a significant inverse correlation to UBASH3B/Sts-1 expression in AML patient samples (Figure 6A). There is an evolutionally conserved miR-9-5p binding site located in the 3′ UTR region of UBASH3B/Sts-1 (Figure S12). miR-9-5q level was downregulated in AML1-ETO-expressing CB cells (Figure 6B), and enforced expression of miR-9 resulted in the reduction of UBASH3B/Sts-1 protein level (Figure 6C). Thus, the low expression of miR-9-5p also contributes to the high expression of UBASH3B/Sts-1 in AML1-ETO cells.
Figure 6. AML1-ETO induces UBASH3B/Sts-1 upregulation through miR-9 downregulation.
A. A scatter plot showing miR-9-5p expression against UBASH3B/Sts-1 expression in 172 AML patient samples was drawn using starBase v2.0 (http://starbase.sysu.edu.cn/). Pearson’s correlation= −0.22734. P-Value= 0.0027. B. miR-9-5p expression in 3 independent vector- or AML1-ETO-expressing CB cells cultured for 2 – 3 weeks. *p=0.025 C. AML1-ETO-expressing CB cells were transduced with vector or miR-9, and were subjected to immunoblotting with UBASH3B/Sts-1 and tubulin antibodies 7 days after transduction.
UBASH3B/Sts-1 modulates CBL phosphorylation and enhances proliferation in AML1-ETO cells
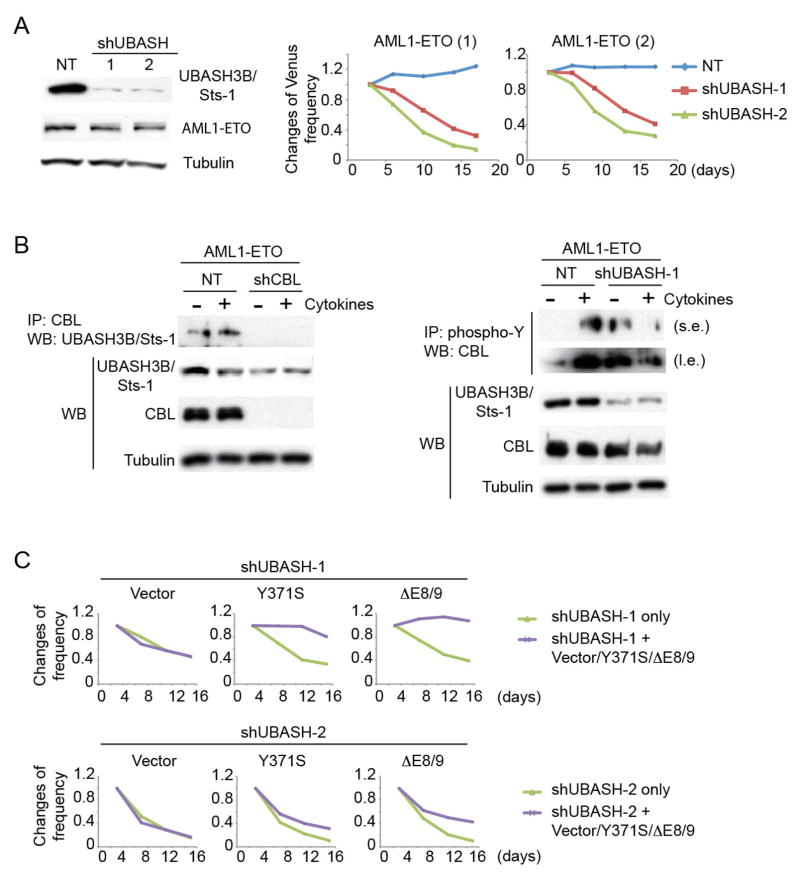
To determine the role of UBASH3B/Sts-1 in AML1-ETO leukemia, we assessed the effect of UBASH3B/Sts-1 depletion in AML1-ETO-expressing CB cells using 2 shRNAs (shUBASH-1 and shUBASH-2). Both shRNAs showed efficient knockdown of UBASH3B/Sts-1 protein and growth-inhibitory effect in AML1-ETO cells (Figure 7A). We next examined physical and functional interactions between UBASH3B/Sts-1 and CBL in AML1-ETO leukemia. We observed constitutive binding of UBASH3B/Sts-1 to CBL irrespective of cytokine stimulation (Figure 7B left). In agreement with previous reports showing the phosphatase activity of UBASH3B/Sts-128, 57, 58, tyrosine phosphorylation of CBL at Y700 and potentially other residues was increased in UBASH3B/Sts-1-depleted cells in the absence of cytokines. However, we observed unexpected downregulation of CBL phosphorylation after cytokine stimulation in UBASH3B/Sts-1-depleted cells (Figure 7B right, Figure S13A). The distinct pattern of phosphorylation was also observed for CBL mutants (Y371S and ΔE8/9) in UBASH3B/Sts-1-depleted AML1-ETO cells (Figure S13B). Thus, UBASH3B/Sts-1-depletion did not simply increase CBL phosphorylation, but changed the pattern of tyrosine phosphorylation in CBL that will affect its activity. We then examined if ectopic expression of CBL mutants can rescue the growth-inhibition caused by UBASH3B/Sts-1 depletion. Both Y371S and ΔE8/9 mutants, but not wild-type CBL, reversed the growth-inhibitory effect of shUBASH-1, and partially reversed that of shUBASH-2 (Figure 7C, Figure S13C). Taken together, these results indicate that UBASH3B/Sts-1 interacts with CBL and controls myeloid proliferation of AML1-ETO cells through modulation of CBL function. It is not clear why shUBASH-2 showed a stronger negative effect than shUBASH-1 despite the similar knockdown efficiency of the two shRNAs. The precise effects of UBASH3B loss in human AML1-ETO cells need to be examined using genome-editing techniques in future studies.
Figure 7. UBASH3B/Sts-1 promotes the growth of AML1-ETO cells through the interaction with CBL.
A. Two shRNAs showed efficient UBASH3B/Sts-1 knockdown and growth-inhibitory effect in AML1-ETO cells. AML1-ETO cells were transduced with NT or shUBASH and were cultured with cytokines. Changes in frequency of Venus+ cells (shRNA-transduced cells) in two independent AML1-ETO cell cultures are shown. Results are normalized to the frequency of Venus+ cells at day 3, set to 1. B. (Left) Interaction between endogenous CBL and UBASH3B/Sts-1 in AML1-ETO cells irrespective of cytokine stimulation. NT- or shCBL-transduced AML1-ETO-expressing CB cells were left unstimulated or stimulated with 6 cytokines (SCF, TPO, FLT3L, IL-3, IL-6, GM-CSF) for 5 minutes. Total cell lysates were immunoprecipitated with anti-CBL antibody, and CBL-bound UBASH3B/Sts-1 was detected by western blotting. (Right) NT- or shUBASH-1-transduced AML1-ETO-expressing CB cells were left unstimulated or stimulated with 6 cytokines for 5 minutes. Total cell lysates were immunoprecipitated with anti-phospho-tyrosine antibody, and tyrosine phosphorylated CBL was detected by western blotting. s.e., short exposure; l.e., long exposure. C. AML1-ETO cells were transduced with shUBASH-1 or shUBASH-2 (marked with Venus) in combination with vector or CBL mutants (Y371S or ΔE8/9, marked with GFP), and the growths of Venus+ (shRNA only) and GFP+Venus+ [shRNA + (vector or Y371S or ΔE8/9)] cells were monitored. Results are normalized to the frequency of Venus+ cells or that of GFP+Venus+ cells at day 2, set to 1. Y371S and ΔE8/9 fully reversed the negative effect of shUBASH-1, and partially reversed that of shUBASH-2. See also Figure S13C.
CBL mutations do not predict sensitivity to JAK/Src inhibition
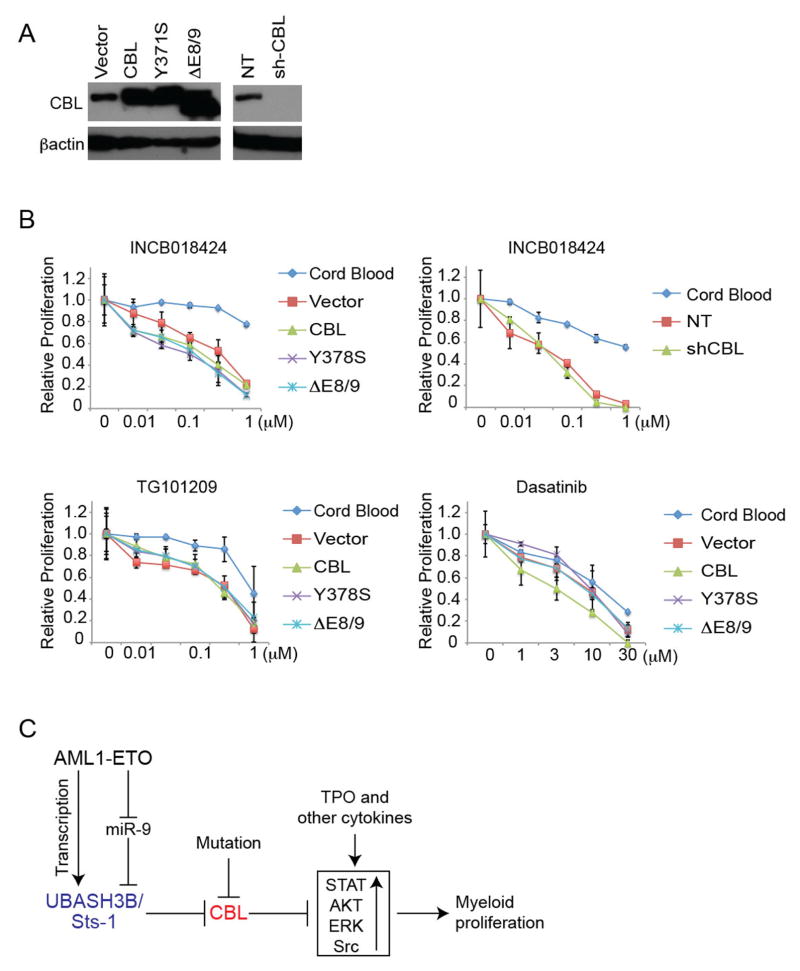
Targeting the signaling pathways activated by CBL mutations is an area of intense investigation24. Using the AML-ETO-expressing CB cells with CBL mutations/depletion (Figure 8A), we first tested the effect of JAK inhibitors that were recently shown to suppress AML1-ETO leukemia in a mouse transplantation model59. As expected, AML1-ETO cells were more sensitive to the growth-inhibition by JAK inhibitors (INCB018424 and TG101209) than normal human CD34+ cells. However, neither the expression of CBL mutants nor CBL depletion increased the sensitivity of AML1-ETO cells to these inhibitors. We then examined the effect of Dasatinib, a dual inhibitor of Src and RTKs. Dasatinib showed equal efficacy on the growth of normal CD34+ cells and AML1-ETO cells with/without CBL mutations (Figure 8B). Thus, the presence of CBL mutations did not increase the sensitivity of AML1-ETO cells to the inhibitors for JAK/Src pathways.
Figure 8. In vitro drug testing using AML1-ETO-expressing CB cells with CBL mutations/depletion.
A. AML1-ETO cells were transduced with various CBL constructs or shRNAs, as indicated. Expression of individual proteins and knockdown of endogenous CBL by shCBL were confirmed by western blotting. B WST-1 cell proliferation assay using the cells described in (A) and CB CD34+ cells, treated with INCB018424 (JAK inhibitor), TG101209 (another JAK inhibitor) or Dasatinib (Src inhibitor) at indicated concentrations for 48 hours in triplicate. AML1-ETO cells were more sensitive to JAK inhibitors, but were equally sensitive to Dasatinib, compared to normal CB CD34+ cells, irrespective of CBL function. Data are normalized to vehicle control (0 μM group), and are shown as Mean+/−SD. C. A regulatory network to control signal transduction and myeloid proliferation in AML1-ETO leukemia.
Discussion
Using the human cell-based system, we demonstrated a critical role for CBL to restrict myeloid proliferation in AML-ETO-induced leukemia. Given the high mutation rate in RAS-associated pathway genes including CBL in AMLs with CBFB-MYH11 rearrangement6, CBL is likely to play a similar growth-inhibitory role in CBFB-MYH11-induced leukemia. Indeed, CBL depletion by shCBL promoted the growth of CBFB-MYH11-expressing CB cells, but not of MLL-AF9-expressing CB cells, indicating the specific role for CBL in CBF leukemia (Figure S14). CBL depletion induced hyper-activation of AKT/ERK pathways even in Kasumi-1 cells that harbor KIT and TP53 mutations in addition to AML1-ETO rearrangement, suggesting that these mutations do not abrogate the suppressive function of CBL. In line with this, CBL mutations are not mutually exclusive to KIT mutations in CBF leukemia8, 60. Interestingly, AML1-ETO induces UBASH3B/Sts-1 upregulation through both transcriptional control and miR-9-mediated regulation. The high expression of UBASH3B/Sts-1 counteracts the suppressive role of CBL to sustain the efficient growth of AML1-ETO cells. Thus, our study reveals a novel network connecting a transcription factor (AML1-ETO), a miRNA (miR-9), a protein tyrosine phosphatase (UBASH3B/Sts-1), and an E3 ubiquitin ligase (CBL) to control signal transduction and leukemic proliferation (summarized in Figure 8C). Given that UBASH3B/Sts-1 is expressed higher in CBF-AML cells compared to normal HSCs (Figure 5A), UBASH3B/Sts-1 could be a target for pharmacological intervention. Development of UBASH3B/Sts-1 inhibitors merits further investigation.
UBASH3B/Sts-1 has been shown to dephosphorylate not only CBL but also several kinases including Src and Syk57, 58. The aberrant pattern of CBL phosphorylation in UBASH3B/Sts-1-depleted cells suggests a complex crosstalk among UBASH3B/Sts-1, CBL, and other kinases to transmit signals in AML1-ETO leukemia. Interestingly, we found relatively high expression of UBASH3B/Sts-1 in CBL-mutated CBF-AMLs compared to their CBL wild-type counterparts (Table S2). Whether UBASH3B/Sts-1 still participates in the regulation of signal transduction in CBL-mutated cells remains to be elucidated (Supplemental Discussion). In addition, CBL-mediated processes of receptor ubiquitination and/or endocytosis in CBF-AML need to be clarified in future studies. Although it has been shown that KIT and MPL, which play important roles in AML1-ETO leukemia52, 61–65, are targets of CBL-mediated ubiquitination and degradation21, 23, we did not observe consistent upregulation of these receptor tyrosine kinases (RTKs) in AML1-ETO cells with CBL depletion/mutation (data not shown). Thus, downregulation of these receptors may not be the major mechanisms of CBL-meditated signal transduction. It is also possible that more sensitive assays are required to precisely track dynamic changes of receptor expression in CBL-depleted/mutated cells.
Point mutants (Q367P, Y371S) and a deletion mutant (ΔE8/9) of CBL may have slightly different functions. In particular, ΔE8/9 mutant has much stronger activity to promote transient proliferation and differentiation than point mutants in CB cells. Clinical data suggest that deletion mutants of CBL are usually heterozygous and are found mainly in core-binding factor leukemia5, 8, while point mutations of CBL, often found in a homozygous form, are most commonly detected in MDS/MPN patients12, 15. Taken together, it appears that deletion-type CBL mutations are functionally more potent and need a cooperating mutation with strong self-renewal ability, such as AML1-ETO, to promote leukemogenesis.
From a clinical point of view, identification of therapeutic vulnerability in CBL-dysregulated leukemia is an important challenge. Suppressing FLT3 signaling was shown to prevent leukemia development in c-Cbl ring finger mutant mice30, 66. However, withdrawal of FLT3L from the culture did not inhibit the growth of AML1-ETO-expressing CB cells over a period of at least several weeks (data not shown). Several reports have shown the efficacy of the Src inhibitor Dasatinib in treating leukemia cell lines with CBL mutations67, 68. However, we did not observe enhanced efficacy of Dasatinib to AML1-ETO cells expressing CBL mutations (Figure 8B). The discrepancy suggests the tumor-specific role of CBL, and large-scale drug screening will be necessary to identify compounds that selectively target CBL-mutated CBF-AML. The AML1-ETO-expressing CB cells with CBL mutations/depletion established in this study will provide efficient tools for this purpose. Of note, consistent with a previous report59, AML1-ETO-expressing human CB cells are highly sensitive to JAK inhibitors irrespective of CBL function. The therapeutic effect of JAK inhibitors on AML1-ETO leukemia should be tested in a clinical trial.
The classic “2-hit” model of AML was proposed based on experimental data in mouse transplantation assays69. Indeed, several reports have shown that the combined expression of a CBF-fusion gene and an activated tyrosine kinase in mouse hematopoietic progenitors is sufficient to produce in vivo leukemia62, 65, 70, 71. However, it appears that human cells are more resistant to oncogene-induced transformation, and more than “2-hits” may be required to generate human AML-ETO leukemia. As shown in this study, ectopic expression of AML1-ETO and CBL mutants enhanced repopulating ability of human CB cells but did not induce overt leukemia. Similarly, expressions of Nras(G12) or KIT together with AML1-ETO were not sufficient to produce human AML in xenograft models63, 72. Interestingly, recent studies have identified frequent mutations in ASXL1 and ASXL2 in AML1-ETO leukemia60, 73. Because these ASXL proteins are thought to have distinct functions from the signal regulators such as CBL and KIT, the ASXL mutations may be the 3rd hit to promote leukemogenic transformation in human CD34+ cells. Alternatively, the host mouse environment may be suboptimal to support the development of human t(8;21) AML. Although we used NSGS mice expressing human SCF, GM-CSF, and IL-3 in the current experiments, the mice still do not express several cytokines that are species-specific and important for AML development. Given the essential role of the TPO/MPL pathway in human AML1-ETO cells52, 64, lack of human TPO in these mice may be a factor in stunting the growth of human AML1-ETO cells. In fact, the common experience in the field has been a resounding lack of success in engrafting primary t(8;21) patient samples into the commercially available immunodeficient mouse strains, questioning whether this readout is accurately representing leukemic transformation for these samples. A recent presentation at the 2014 American Society of Hematology meeting indicated that a hTPO knockin mouse may resolve this problem74. Whether the mice expressing human TPO and other human cytokines75, 76 will reproducibly support the propagation of human AML1-ETO leukemia should also be examined. Such effort will lead to the establishment of disease models for human AML1-ETO leukemia that allow testing of novel therapies in vivo.
Supplementary Material
Acknowledgments
We thank Dr. Seishi Ogawa, Dr. Masashi Sanada, Dr. Masafumi Onodera, Dr. Karsten Spiekermann, for plasmids. We thank the Flow Cytometry Core and the Mouse Core at Cincinnati Children’s Hospital Medical Center for their help. This work was supported by a grant from the CancerFree Kids Foundation for Cancer Research (J.C.M. and S.G.), an Institutional Clinical and Translational Science Award, NIH/NCRR Grant Number 1UL1RR026314-01, Translational Trials Development and Support Laboratory award (U.S.P.H.S. Grant Number MO1 RR 08084), a Center of Excellence in Molecular Hematology P30 award (DK090971), JSPS Postdoctoral Fellowship for Research Abroad (S.G.), CA178454 (J.C.), the Coleman Leukemia Research Foundation (C.D.B), CA180861 (C.D.B), CA101140 (C.D.B), CA140158 (C.D.B), grants from Leukaemia Lymphoma Research (12007, C.B.) and (12055, O.H). A. G. and N.N.N. were supported by a grant from the Leukemia and Lymphoma Society. JCM is a Leukemia and Lymphoma Society Scholar.
Footnotes
Supplementary information is available at Leukemia’s website.
Authorship and Conflict of Interest Statements
S.G. conceived the project, designed and performed the research, analyzed the data, and wrote the paper. J.S. performed the research and analyzed the data. M.S., S.L., and K.A.L. assisted with experiments. A.G. and N.N.N. actively participated in designing and assisting with the experiments regarding UBASH3B/Sts-1. J.C. assisted with the experiments regarding miR-9. S.P.W., C.D.B. and D.N. analyzed expression profiles of CBF-AML patients. S.A., A.P., O.H., C.B. performed ChIP-Seq and RNA-Seq analyses. J.C.M. secured funding, analyzed the data, and participated in writing the paper. Authors declare no conflict of interest.
References
- 1.Goyama S, Mulloy JC. Molecular pathogenesis of core binding factor leukemia: current knowledge and future prospects. International journal of hematology. 2011 Aug;94(2):126–133. doi: 10.1007/s12185-011-0858-z. [DOI] [PubMed] [Google Scholar]
- 2.Link KA, Chou FS, Mulloy JC. Core binding factor at the crossroads: determining the fate of the HSC. Journal of cellular physiology. 2010 Jan;222(1):50–56. doi: 10.1002/jcp.21950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cunningham L, Finckbeiner S, Hyde RK, Southall N, Marugan J, Yedavalli VR, et al. Identification of benzodiazepine Ro5–3335 as an inhibitor of CBF leukemia through quantitative high throughput screen against RUNX1-CBFbeta interaction. Proc Natl Acad Sci U S A. 2012 Sep 4;109(36):14592–14597. doi: 10.1073/pnas.1200037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Illendula A, Pulikkan JA, Zong H, Grembecka J, Xue L, Sen S, et al. Chemical biology. A small-molecule inhibitor of the aberrant transcription factor CBFbeta-SMMHC delays leukemia in mice. Science. 2015 Feb 13;347(6223):779–784. doi: 10.1126/science.aaa0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aranaz P, Migueliz I, Hurtado C, Erquiaga I, Larrayoz MJ, Calasanz MJ, et al. CBL RING finger deletions are common in core-binding factor acute myeloid leukemias. Leuk Lymphoma. 2012 Jul 17; doi: 10.3109/10428194.2012.709629. [DOI] [PubMed] [Google Scholar]
- 6.Haferlach C, Dicker F, Kohlmann A, Schindela S, Weiss T, Kern W, et al. AML with CBFB-MYH11 rearrangement demonstrate RAS pathway alterations in 92% of all cases including a high frequency of NF1 deletions. Leukemia. 2010 May;24(5):1065–1069. doi: 10.1038/leu.2010.22. [DOI] [PubMed] [Google Scholar]
- 7.Reindl C, Quentmeier H, Petropoulos K, Greif PA, Benthaus T, Argiropoulos B, et al. CBL exon 8/9 mutants activate the FLT3 pathway and cluster in core binding factor/11q deletion acute myeloid leukemia/myelodysplastic syndrome subtypes. Clin Cancer Res. 2009 Apr 1;15(7):2238–2247. doi: 10.1158/1078-0432.CCR-08-1325. [DOI] [PubMed] [Google Scholar]
- 8.Abbas S, Rotmans G, Lowenberg B, Valk PJ. Exon 8 splice site mutations in the gene encoding the E3-ligase CBL are associated with core binding factor acute myeloid leukemias. Haematologica. 2008 Oct;93(10):1595–1597. doi: 10.3324/haematol.13187. [DOI] [PubMed] [Google Scholar]
- 9.Becker H, Yoshida K, Blagitko-Dorfs N, Claus R, Pantic M, Abdelkarim M, et al. Tracing the development of acute myeloid leukemia in CBL syndrome. Blood. 2014 Mar 20;123(12):1883–1886. doi: 10.1182/blood-2013-10-533844. [DOI] [PubMed] [Google Scholar]
- 10.Dunbar AJ, Gondek LP, O’Keefe CL, Makishima H, Rataul MS, Szpurka H, et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008 Dec 15;68(24):10349–10357. doi: 10.1158/0008-5472.CAN-08-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grand FH, Hidalgo-Curtis CE, Ernst T, Zoi K, Zoi C, McGuire C, et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood. 2009 Jun 11;113(24):6182–6192. doi: 10.1182/blood-2008-12-194548. [DOI] [PubMed] [Google Scholar]
- 12.Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S, et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009 Aug 13;460(7257):904–908. doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- 13.Muramatsu H, Makishima H, Jankowska AM, Cazzolli H, O’Keefe C, Yoshida N, et al. Mutations of an E3 ubiquitin ligase c-Cbl but not TET2 mutations are pathogenic in juvenile myelomonocytic leukemia. Blood. 2010 Mar 11;115(10):1969–1975. doi: 10.1182/blood-2009-06-226340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loh ML, Sakai DS, Flotho C, Kang M, Fliegauf M, Archambeault S, et al. Mutations in CBL occur frequently in juvenile myelomonocytic leukemia. Blood. 2009 Aug 27;114(9):1859–1863. doi: 10.1182/blood-2009-01-198416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makishima H, Cazzolli H, Szpurka H, Dunbar A, Tiu R, Huh J, et al. Mutations of e3 ubiquitin ligase cbl family members constitute a novel common pathogenic lesion in myeloid malignancies. J Clin Oncol. 2009 Dec 20;27(36):6109–6116. doi: 10.1200/JCO.2009.23.7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohlmann A, Grossmann V, Klein HU, Schindela S, Weiss T, Kazak B, et al. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol. 2010 Aug 20;28(24):3858–3865. doi: 10.1200/JCO.2009.27.1361. [DOI] [PubMed] [Google Scholar]
- 17.Schnittger S, Bacher U, Alpermann T, Reiter A, Ulke M, Dicker F, et al. Use of CBL exon 8 and 9 mutations in diagnosis of myeloproliferative neoplasms and myeloproliferative/myelodysplastic disorders: an analysis of 636 cases. Haematologica. 2012 Jun 24; doi: 10.3324/haematol.2012.065375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niemeyer CM, Kang MW, Shin DH, Furlan I, Erlacher M, Bunin NJ, et al. Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat Genet. 2010 Sep;42(9):794–800. doi: 10.1038/ng.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sargin B, Choudhary C, Crosetto N, Schmidt MH, Grundler R, Rensinghoff M, et al. Flt3-dependent transformation by inactivating c-Cbl mutations in AML. Blood. 2007 Aug 1;110(3):1004–1012. doi: 10.1182/blood-2007-01-066076. [DOI] [PubMed] [Google Scholar]
- 20.Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999 Dec;4(6):1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 21.Saur SJ, Sangkhae V, Geddis AE, Kaushansky K, Hitchcock IS. Ubiquitination and degradation of the thrombopoietin receptor c-Mpl. Blood. 2010 Feb 11;115(6):1254–1263. doi: 10.1182/blood-2009-06-227033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yokouchi M, Kondo T, Sanjay A, Houghton A, Yoshimura A, Komiya S, et al. Src-catalyzed phosphorylation of c-Cbl leads to the interdependent ubiquitination of both proteins. J Biol Chem. 2001 Sep 14;276(37):35185–35193. doi: 10.1074/jbc.M102219200. [DOI] [PubMed] [Google Scholar]
- 23.Zeng S, Xu Z, Lipkowitz S, Longley JB. Regulation of stem cell factor receptor signaling by Cbl family proteins (Cbl-b/c-Cbl) Blood. 2005 Jan 1;105(1):226–232. doi: 10.1182/blood-2004-05-1768. [DOI] [PubMed] [Google Scholar]
- 24.Ogawa S, Shih LY, Suzuki T, Otsu M, Nakauchi H, Koeffler HP, et al. Deregulated intracellular signaling by mutated c-CBL in myeloid neoplasms. Clin Cancer Res. 2010 Aug 1;16(15):3825–3831. doi: 10.1158/1078-0432.CCR-09-2341. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt MH, Dikic I. The Cbl interactome and its functions. Nat Rev Mol Cell Biol. 2005 Dec;6(12):907–918. doi: 10.1038/nrm1762. [DOI] [PubMed] [Google Scholar]
- 26.Ryan PE, Davies GC, Nau MM, Lipkowitz S. Regulating the regulator: negative regulation of Cbl ubiquitin ligases. Trends in biochemical sciences. 2006 Feb;31( 2):79–88. doi: 10.1016/j.tibs.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Kowanetz K, Crosetto N, Haglund K, Schmidt MH, Heldin CH, Dikic I. Suppressors of T-cell receptor signaling Sts-1 and Sts-2 bind to Cbl and inhibit endocytosis of receptor tyrosine kinases. J Biol Chem. 2004 Jul 30;279(31):32786–32795. doi: 10.1074/jbc.M403759200. [DOI] [PubMed] [Google Scholar]
- 28.Lee ST, Feng M, Wei Y, Li Z, Qiao Y, Guan P, et al. Protein tyrosine phosphatase UBASH3B is overexpressed in triple-negative breast cancer and promotes invasion and metastasis. Proc Natl Acad Sci U S A. 2013 Jul 2;110(27):11121–11126. doi: 10.1073/pnas.1300873110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rathinam C, Thien CB, Langdon WY, Gu H, Flavell RA. The E3 ubiquitin ligase c-Cbl restricts development and functions of hematopoietic stem cells. Genes Dev. 2008 Apr 15;22(8):992–997. doi: 10.1101/gad.1651408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rathinam C, Thien CB, Flavell RA, Langdon WY. Myeloid leukemia development in c-Cbl RING finger mutant mice is dependent on FLT3 signaling. Cancer Cell. 2010 Oct 19;18(4):341–352. doi: 10.1016/j.ccr.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Mulloy JC, Cammenga J, MacKenzie KL, Berguido FJ, Moore MA, Nimer SD. The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood. 2002 Jan 1;99(1):15–23. doi: 10.1182/blood.v99.1.15. [DOI] [PubMed] [Google Scholar]
- 32.Mulloy JC, Cammenga J, Berguido FJ, Wu K, Zhou P, Comenzo RL, et al. Maintaining the self-renewal and differentiation potential of human CD34+ hematopoietic cells using a single genetic element. Blood. 2003 Dec 15;102(13):4369–4376. doi: 10.1182/blood-2003-05-1762. [DOI] [PubMed] [Google Scholar]
- 33.Wunderlich M, Krejci O, Wei J, Mulloy JC. Human CD34+ cells expressing the inv(16) fusion protein exhibit a myelomonocytic phenotype with greatly enhanced proliferative ability. Blood. 2006 Sep 1;108(5):1690–1697. doi: 10.1182/blood-2005-12-012773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wunderlich M, Chou FS, Link KA, Mizukawa B, Perry RL, Carroll M, et al. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia. 2010 Oct;24( 10):1785–1788. doi: 10.1038/leu.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wunderlich M, Mulloy JC. Model systems for examining effects of leukemia-associated oncogenes in primary human CD34+ cells via retroviral transduction. Methods Mol Biol. 2009;538:263–285. doi: 10.1007/978-1-59745-418-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulloy JC, Wunderlich M, Zheng Y, Wei J. Transforming human blood stem and progenitor cells: a new way forward in leukemia modeling. Cell Cycle. 2008 Nov 1;7(21):3314–3319. doi: 10.4161/cc.7.21.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen P, Price C, Li Z, Li Y, Cao D, Wiley A, et al. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia. Proc Natl Acad Sci U S A. 2013 Jul 9;110(28):11511–11516. doi: 10.1073/pnas.1310144110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chou FS, Griesinger A, Wunderlich M, Lin S, Link KA, Shrestha M, et al. The THPO/MPL/Bcl-xL pathway is essential for survival and self-renewal in human pre-leukemia induced by AML1-ETO. Blood. 2012 Feb 14; doi: 10.1182/blood-2012-01-403212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krejci O, Wunderlich M, Geiger H, Chou FS, Schleimer D, Jansen M, et al. p53 signaling in response to increased DNA damage sensitizes AML1-ETO cells to stress-induced death. Blood. 2008 Feb 15;111(4):2190–2199. doi: 10.1182/blood-2007-06-093682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ptasinska A, Assi SA, Mannari D, James SR, Williamson D, Dunne J, et al. Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia. 2012 Aug;26(8):1829–1841. doi: 10.1038/leu.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ptasinska A, Assi SA, Martinez-Soria N, Imperato MR, Piper J, Cauchy P, et al. Identification of a Dynamic Core Transcriptional Network in t(8;21) AML that Regulates Differentiation Block and Self-Renewal. Cell reports. 2014 Sep 25;8(6):1974–1988. doi: 10.1016/j.celrep.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bagger FO, Rapin N, Theilgaard-Monch K, Kaczkowski B, Thoren LA, Jendholm J, et al. HemaExplorer: a database of mRNA expression profiles in normal and malignant haematopoiesis. Nucleic acids research. 2013 Jan;41(Database issue):D1034–1039. doi: 10.1093/nar/gks1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013 Apr 2;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012 May;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic acids research. 2011 Jan;39(Database issue):D202–209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic acids research. 2014 Jan;42(Database issue):D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005 Jan 14;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 48.Goyama S, Schibler J, Cunningham L, Zhang Y, Rao Y, Nishimoto N, et al. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. The Journal of clinical investigation. 2013 Sep 3;123(9):3876–3888. doi: 10.1172/JCI68557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei J, Wunderlich M, Fox C, Alvarez S, Cigudosa JC, Wilhelm JS, et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 2008 Jun;13(6):483–495. doi: 10.1016/j.ccr.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asou H, Tashiro S, Hamamoto K, Otsuji A, Kita K, Kamada N. Establishment of a human acute myeloid leukemia cell line (Kasumi-1) with 8;21 chromosome translocation. Blood. 1991 May 1;77(9):2031–2036. [PubMed] [Google Scholar]
- 51.Larizza L, Magnani I, Beghini A. The Kasumi-1 cell line: a t(8;21)-kit mutant model for acute myeloid leukemia. Leuk Lymphoma. 2005 Feb;46(2):247–255. doi: 10.1080/10428190400007565. [DOI] [PubMed] [Google Scholar]
- 52.Chou FS, Griesinger A, Wunderlich M, Lin S, Link KA, Shrestha M, et al. The thrombopoietin/MPL/Bcl-xL pathway is essential for survival and self-renewal in human preleukemia induced by AML1-ETO. Blood. 2012 Jul 26;120(4):709–719. doi: 10.1182/blood-2012-01-403212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanjay A, Houghton A, Neff L, DiDomenico E, Bardelay C, Antoine E, et al. Cbl associates with Pyk2 and Src to regulate Src kinase activity, alpha(v)beta(3) integrin-mediated signaling, cell adhesion, and osteoclast motility. J Cell Biol. 2001 Jan 8;152(1):181–195. doi: 10.1083/jcb.152.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang H, Alberich-Jorda M, Amabile G, Yang H, Staber PB, Di Ruscio A, et al. Sox4 is a key oncogenic target in C/EBPalpha mutant acute myeloid leukemia. Cancer Cell. 2013 Nov 11;24(5):575–588. doi: 10.1016/j.ccr.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mendler JH, Maharry K, Radmacher MD, Mrozek K, Becker H, Metzeler KH, et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J Clin Oncol. 2012 Sep 1;30(25):3109–3118. doi: 10.1200/JCO.2011.40.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emmrich S, Katsman-Kuipers JE, Henke K, Khatib ME, Jammal R, Engeland F, et al. miR-9 is a tumor suppressor in pediatric AML with t(8;21) Leukemia. 2014 May;28(5):1022–1032. doi: 10.1038/leu.2013.357. [DOI] [PubMed] [Google Scholar]
- 57.Mikhailik A, Ford B, Keller J, Chen Y, Nassar N, Carpino N. A phosphatase activity of Sts-1 contributes to the suppression of TCR signaling. Mol Cell. 2007 Aug 3;27( 3):486–497. doi: 10.1016/j.molcel.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas DH, Getz TM, Newman TN, Dangelmaier CA, Carpino N, Kunapuli SP, et al. A novel histidine tyrosine phosphatase, TULA-2, associates with Syk and negatively regulates GPVI signaling in platelets. Blood. 2010 Oct 7;116(14):2570–2578. doi: 10.1182/blood-2010-02-268136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lo MC, Peterson LF, Yan M, Cong X, Hickman JH, Dekelver RC, et al. JAK inhibitors suppress t(8;21) fusion protein-induced leukemia. Leukemia. 2013 Dec;27(12):2272–2279. doi: 10.1038/leu.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krauth MT, Eder C, Alpermann T, Bacher U, Nadarajah N, Kern W, et al. High number of additional genetic lesions in acute myeloid leukemia with t(8;21)/RUNX1-RUNX1T1: frequency and impact on clinical outcome. Leukemia. 2014 Jul;28(7):1449–1458. doi: 10.1038/leu.2014.4. [DOI] [PubMed] [Google Scholar]
- 61.Wang YY, Zhou GB, Yin T, Chen B, Shi JY, Liang WX, et al. AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia: implication in stepwise leukemogenesis and response to Gleevec. Proc Natl Acad Sci U S A. 2005 Jan 25;102(4):1104–1109. doi: 10.1073/pnas.0408831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang YY, Zhao LJ, Wu CF, Liu P, Shi L, Liang Y, et al. C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci U S A. 2011 Feb 8;108(6):2450–2455. doi: 10.1073/pnas.1019625108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wichmann C, Quagliano-Lo Coco I, Yildiz O, Chen-Wichmann L, Weber H, Syzonenko T, et al. Activating c-KIT mutations confer oncogenic cooperativity and rescue RUNX1/ETO-induced DNA damage and apoptosis in human primary CD34+ hematopoietic progenitors. Leukemia. 2014 Jun 4; doi: 10.1038/leu.2014.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pulikkan JA, Madera D, Xue L, Bradley P, Landrette SF, Kuo YH, et al. Thrombopoietin/MPL participates in initiating and maintaining RUNX1-ETO acute myeloid leukemia via PI3K/AKT signaling. Blood. 2012 Jul 26;120(4):868–879. doi: 10.1182/blood-2012-03-414649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nick HJ, Kim HG, Chang CW, Harris KW, Reddy V, Klug CA. Distinct classes of c-Kit-activating mutations differ in their ability to promote RUNX1-ETO-associated acute myeloid leukemia. Blood. 2012 Feb 9;119(6):1522–1531. doi: 10.1182/blood-2011-02-338228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor SJ, Dagger SA, Thien CB, Wikstrom ME, Langdon WY. Flt3 inhibitor AC220 is a potent therapy in a mouse model of myeloproliferative disease driven by enhanced wild-type Flt3 signaling. Blood. 2012 Nov 8;120(19):4049–4057. doi: 10.1182/blood-2012-06-436675. [DOI] [PubMed] [Google Scholar]
- 67.Makishima H, Sugimoto Y, Szpurka H, Clemente MJ, Ng KP, Muramatsu H, et al. CBL mutation-related patterns of phosphorylation and sensitivity to tyrosine kinase inhibitors. Leukemia. 2012 Jul;26(7):1547–1554. doi: 10.1038/leu.2012.7. [DOI] [PubMed] [Google Scholar]
- 68.Bunda S, Kang MW, Sybingco SS, Weng J, Favre H, Shin DH, et al. Inhibition of SRC corrects GM-CSF hypersensitivity that underlies juvenile myelomonocytic leukemia. Cancer Res. 2013 Apr 15;73(8):2540–2550. doi: 10.1158/0008-5472.CAN-12-3425. [DOI] [PubMed] [Google Scholar]
- 69.Gilliland DG, Jordan CT, Felix CA. The molecular basis of leukemia. Hematology Am Soc Hematol Educ Program. 2004:80–97. doi: 10.1182/asheducation-2004.1.80. [DOI] [PubMed] [Google Scholar]
- 70.Schessl C, Rawat VP, Cusan M, Deshpande A, Kohl TM, Rosten PM, et al. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. The Journal of clinical investigation. 2005 Aug;115(8):2159–2168. doi: 10.1172/JCI24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao L, Melenhorst JJ, Alemu L, Kirby M, Anderson S, Kench M, et al. KIT with D816 mutations cooperates with CBFB-MYH11 for leukemogenesis in mice. Blood. 2012 Feb 9;119(6):1511–1521. doi: 10.1182/blood-2011-02-338210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chou FS, Wunderlich M, Griesinger A, Mulloy JC. N-Ras(G12D) induces features of stepwise transformation in preleukemic human umbilical cord blood cultures expressing the AML1-ETO fusion gene. Blood. 2011 Feb 17;117(7):2237–2240. doi: 10.1182/blood-2010-01-264119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Micol JB, Duployez N, Boissel N, Petit A, Geffroy S, Nibourel O, et al. Frequent ASXL2 mutations in acute myeloid leukemia patients with t(8;21)/RUNX1-RUNX1T1 chromosomal translocations. Blood. 2014 Aug 28;124(9):1445–1449. doi: 10.1182/blood-2014-04-571018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ellegast Jana M, Saito Yasuyuki, Flavell Richard A, Manz Markus G. MISTRG Mice Support Good-Risk AML Engraftment. ASH 2014 Annual Meeting Abstracts. 2014:3808. [Google Scholar]
- 75.Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL, et al. Development and function of human innate immune cells in a humanized mouse model. Nature biotechnology. 2014 Apr;32(4):364–372. doi: 10.1038/nbt.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rongvaux A, Willinger T, Takizawa H, Rathinam C, Auerbach W, Murphy AJ, et al. Human thrombopoietin knockin mice efficiently support human hematopoiesis in vivo. Proc Natl Acad Sci U S A. 2011 Feb 8;108(6):2378–2383. doi: 10.1073/pnas.1019524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.