Abstract
Gangliosides are sialic acid-containing glycosphingolipids that are most abundant in the nerve tissues. The quantity and expression pattern of gangliosides in brain change drastically throughout development and are mainly regulated through stage-specific expression of glycosyltransferase (ganglioside synthase) genes. We previously demonstrated that acetylation of histones H3 and H4 on the N-acetylgalactosaminyltransferase I (GalNAcT, GA2/GM2/GD2/GT2-synthase) gene promoter resulted in recruitment of trans-activation factors. In addition, we reported that epigenetic activation of the GalNAcT gene was also detected as accompanied by an apparent induction of neuronal differentiation in neural stem cells responding to an exogenous supplement of ganglioside GM1. Here, we present evidence supporting the concept that nuclear GM1 is associated with gene regulation in neuronal cells. We found that nuclear GM1 binds acetylated histones on the promoters of the GalNAcT and NeuroD1 genes in differentiated neurons. Our study demonstrates for the first time that GM1 interacts with chromatin via acetylated histones at the nuclear periphery of neuronal cells.
Keywords: brain development, epigenetic regulation, glycosyltransferase, GM1 ganglioside, histone acetylation, neural stem cell, neural progenitor cell, neuronal differentiation
Introduction
Epigenetic regulation of gene expression involving DNA methylation and histone modifications is an important mechanism governing stage-specific gene expression in developing mammalian brains as well as other tissues [3, 4]. The most common post-translational histone modification is histone acetylation, which occurs as an addition of acetyl groups to lysine residues of the amino-terminal tails of core histones. Acetylation of histone H3 or H4 can alter the interaction between histones and DNA and allow relaxation of chromatin. Thus, changes in histone acetylation status are often associated with transcriptional activation and repression [5, 6]. In neural stem cells (NSCs), induction of histone acetylation promotes neuronal differentiation and inhibits glial differentiation through up-regulation of neurogenic transcription factors [7]. Histone acetylation is controlled by histone acetyl transferases (HATs) and histone deacetylases (HDACs) [8]. Intriguingly, recent studies have suggested that glycans can contribute to epigenetic gene regulation as modulators. For example, a “carbohydrate-like” molecule, inositol tetraphosphate (IP4; inositol 1, 3, 4, 5–tetrakisphosphate) acts as a stimulator to the activity of class I HDACs (HDAC1/2/3) [9-11]. Additionally, O-GlcNAcylation has been reported to regulate localization, activity, and stability of many transcription factors, as well as their interactions with other proteins and DNA [12, 13]. As such, glycosyl modification, including O-GlcNAcylation, are now considered as an important modulatory mechanism to epigenetic control of gene expression [14].
Glycolipid-enriched microdomains (GEMs, also known as lipid rafts) on the cell plasma membrane surface are increasingly recognized as an important site for signaling transmission. Nuclear lipid domains on the nuclear envelope have also recently been suggested to play a similar role [15]. Gangliosides are sialic acid-containing glycosphingolipids expressed primarily, but not exclusively, on the plasma membrane of cells in all vertebrates. Both gangliosides GM1 and GD1a have been detected in the inner and outer nuclear membranes [16]. The outer nuclear membrane is a continuum of the endoplasmic reticulum (ER), whereas the inner nuclear membrane is connected with the nuclear lamina. The composition of nuclear gangliosides in the developing brain reflects their composition in the total brain [17, 18], i.e., GD3 is abundant in the nuclear membranes of the embryonic brain and adult brain type gangliosides (GM1, GD1a, GD1b, GT1b) become more abundant in the nuclei of postnatal brain cells [18]. Interestingly, GT3, a member of the c-series gangliosides, is also plentiful in the nuclei of embryonic brain cells, and its concentration is decreased drastically in postnatal-brain nuclei [18]. It is known that GM1 enhances neurite outgrowth, and up-regulation of GM1 in the nuclear membrane accompanies the process of neurite outgrowth [19, 20]. This observation prompted Ledeen et al. to propose that the elevated level of GM1 has a modulatory effect on Ca2+ homeostasis in the nucleus, which is mediated by a tight association of GM1 with the Na+/Ca2+exchanger [21, 22]. Another ganglioside, GD3, is reported to interact with histone H1 in the nucleus [23]. It has also been shown that nuclear sphingolipids participate in epigenetic regulation of gene expression by controlling histone acetylation [24]. For these reasons, it is reasonable to assume that membrane lipids, including glycolipids, may contribute to building of the nuclear membranes in a stage- and cell-specific manner to modulate gene expression.
The nuclear envelope, including the nuclear lamina and nuclear pore complexes, is a key structure to maintain chromatin architecture and cell-specific gene expression [25]. Stem cells and differentiated cells have distinct nuclear envelope compositions as well as specific epigenetic modifications, which play important roles to determine nuclear spatial localization of individual genes. During cell differentiation, tissue-specific genes can be re-localized in the nuclear space [25, 26]. Interestingly, differential ganglioside expression has been detected in nuclear membranes during neuronal development [18] and hence likely regulates cell-specific gene expression. It is not clear, however, how nuclear ganglioside expression and gene activity are correlated. Here, we investigated the hypothesis that nuclear GM1 is associated with gene regulation in differentiated neuronal cells. We found that nuclear GM1 binds with acetylated histones on the promoters of the GalNAcT as well as on the NeuroD1 genes in differentiated neurons. Our study demonstrated for the first time that GM1 interacts with active chromatin via acetylated histones at the nuclear periphery, resulting in changes in gene expression.
Experimental Procedure
Cell culture and differentiation
NSCs were isolated from the telencephalons of C57BL/6 mouse embryos (E14.5) as previously described [27-30]. Neuronal precursor cells (NPCs) were isolated by a positive selection of polysialic acid-neural cell adhesion molecule (PSA-NCAM)-harboring NSCs, following a negative selection of A2B5 antigen expression, using the Magnetic-Activated Cell Sorting (MACS) method with microbeads pre-labeled with specific antibodies (Meltenyi Biotec, Bergisch Gladbach, Germany). NPCs were cultured in 2 mM of L-glutamine- and B27-supplemented Neurobasal-A medium (Life Technology, Carlsbad, CA, USA) containing 10 ng/ml of basic fibroblast growth factor (FGF2) (Peprotech, Rocky Hill, NJ, USA) on dishes coated with poly-L-ornithine and bovine fibronectin (Sigma-Aldrich, MO, USA) at 37°C in a humidified 5% carbon dioxide atmosphere. Neuron-committed NPCs were induced into neurogenic differentiation with a B27 supplement without FGF2 for 7 days.
Fluorescence staining and imaging
Cells grown on a coverslip were fixed with 4% paraformaldehyde in PBS for 20 min, and then permeablized by exposing briefly to 0.5% Triton X-100 for 5 min at ambient temperature. To immunostain nuclear lamin B1 and nucleoporin, cells were incubated with specific primary antibodies diluted in 1% BSA/PBS overnight at 4°C (Abcam, ab16048, 1:500 dilution; and Abcam, ab24609, 1:1000 dilution, respectively), followed by treatment with Alexa Fluor-488-conjugated anti-rabbit IgG or anti-mouse IgG secondary antibody (Invitrogen), respectively, diluted 1:800 in 1% BSA/PBS for 1 hr at ambient temperature. To visualize GM1 and counterstain the nucleus, Alexa Fluor-594-conjugated cholera toxin B (CtxB, Invitrogen, 1:800 dilution) and Hoechst 33342 (Invitrogen, 1:500 dilution) were included in the staining solution. After staining, the specimen was mounted on a glass slide with anti-fade reagent (Invitrogen S36940). Samples were observed on a Zeiss LSM 510 confocal microscope equipped with a 63×, NA 1.4 Plan Apochromatic objective, and fluorescent images were acquired using the software ZEN 2012 (Carl Zeiss GmbH, Jena, Germany).
Chromatin immunoprecipitation (ChIP) assay
A method for nuclear fractionation was adapted from a previous study [31]. Briefly, to isolate nuclei, cultured cells (NPCs or differentiated neurons) were suspended in a hypotonic lysis buffer [10 mM KCl, 1.5 mM MgCl2, 10 mM HEPES (pH 7.4), 10% glycerol, 0.05% NP-40, 1mM dithiothreitol, and EDTA-free protease inhibitor cocktail (Roche)], and incubated on ice for 20 min. After a 5-sec vortex and centrifugation at 2,000 g for 5 min, the pellet was washed once with the lysis buffer and then spun down as the isolated nuclei. Besides, the supernatants were also pooled to serve as the post-nuclear fraction (cytoplasm). Whole cells, isolated nuclei, and the post-nuclear fraction were incubated in 1% paraformaldehyde for 10 min at ambient temperature to crosslink the interacting partners of cellular components [28, 32]. Cells or cellular parts with the cross-linked complexes were subjected to lysis by sonication with six 20-sec pulses at the power scale 7 controlled by a sonicator (Sonic Dismembrator Model 100, Fisher Scientific). After centrifugation at 12,000 rpm on a tabletop centrifuge for 15 min, the supernatants from the whole cells nuclear and cytoplasmic lysate fractions were collected for the following experiments.
To immunoprecipitate chromatins or the GM1-associated complexes, 1 µg each of anti-acetyl histone H3 (AcH3, Millipore, 06-599), anti-acetyl histone H4 (AcH4, Millipore, 06-866), anti-trimethylated lysine residue 27 of histone H3 (H3K27me3) (Millipore, 07-449), or anti-GM1 antibodies (Abcam, ab23943) that had been subjected to prior absorption with Protein A/G magnetic beads was added to the lysate, and the mixture was incubated overnight at 4°C. In experiments as indicated, the supernatant of anti-GM1 immunoprecipitation was collected for the anti-AcH3 and anti-AcH4 ChIP assay. After washing for 4 times with PBS containing 0.5% NP-40, the precipitated chromatins were released by heating with 0.1 M sodium bicarbonate solution containing 1% SDS. The sample was de-crosslinked for 4-6 hrs at 65°C, followed by purification of the genomic DNA. The amounts of the DNA fragments of interest were analyzed by quantitative PCR using specific primer pairs. The primer sequences are described elsewhere [28, 32-35].
Co-immunoprecipitation of histones with nuclear GM1
The nuclear fraction and the post-nuclear fraction prepared from hypotonic treatment of differentiated neurons prior to formaldehyde cross-linking were used for anti-GM1, anti-AcH3, anti-AcH4, or anti-H3K27me3 immunoprecipitation. Polyvinylidene difluoride (PVDF) membranes on which the precipitated complex was dot-blotted were blocked with 5% BSA, and then probed with HRP-conjugated CtxB. Signals were visualized with Western Lightning Western blot chemiluminescence reagent (Perkin Elmer Life and Analytical Sciences, Boston, MA)
Treatment of inhibitors of cytoskeleton polymerization
The neuronal culture was incubated with 500 nM of nocodazole or cytochalasin D (both from Sigma-Aldrich) for 4-6 hrs at 37°C in a humidified 5% carbon dioxide atmosphere.
In-situ hybridization
For DNA probe preparation, PCR reactions containing digoxigenin-11-dUTP (Roche) were performed to simultaneously amplify and label a 332-bp fragment of the B4galnt1 (GalNAcT) promoter. The sequences of the primer pair are: 5’-TTT GGG GAC GAA GGG ATG TG-3’; 5’-GAC TCC GGG GCT TTG TAG AC-3’. Procedures for in-situ hybridization were as previously described [36]. The genomic DNA targeted by the probe was revealed by an anti-digoxigenin antibody (Roche; clone 1.71.256; mouse IgG1) (1:500 dilution in 1% BSA/PBS) followed by Alexa Fluo-conjugated secondary antibody.
Statistical evaluation
Data are expressed as means ± standard deviation (SD) from three to four independent experiments. Statistical significance was determined using unpaired two-tailed Student’s t-test, and p<0.05 was regarded as significant.
Results
Localization of GM1 at the nuclear periphery
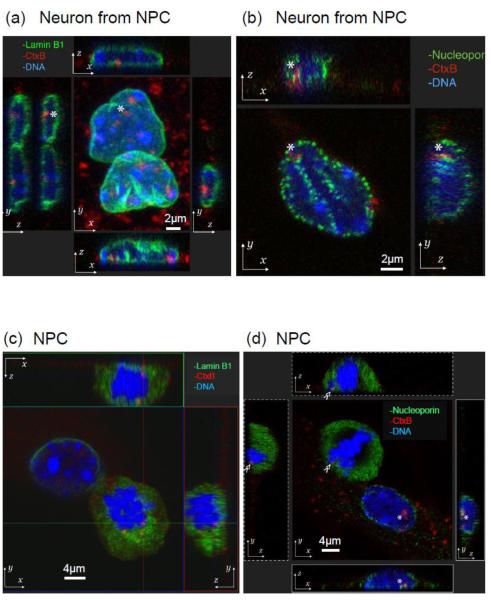
In this study, we utilized mouse NPCs and differentiated neurons derived from NPCs [28]. It is well known that GM1 enhances neurite outgrowth in primary neuronal cultures. More recent findings also revealed that GM1 is present in the nuclear membranes as well as the plasma membranes, and up-regulation of the GM1 level in the nuclear membranes accompanies the process of neurite outgrowth [19, 20]. We confirmed that GM1 is indeed present and colocalized with lamin B1 and nucleoporin at the nuclear periphery of neuronal cells derived from NPCs (Fig. 1). Nuclear lamins are type V intermediate filamentous proteins and are the major components of the nuclear lamina. The nuclear lamina constitutes the molecular interface between the inner nuclear envelope membrane and the peripheral chromatin domains. Lamin B1 is expressed in nearly every cell type and has been shown to play important roles for transcriptional regulation and chromatin organization [37]. We confirmed that GM1 is co-localized with the nuclear lamin of neurons (Fig.1a).
Fig. 1.
Localization of GM1 at the nuclear periphery in neurons. Differentiated neurons (a, b) or NPSs (c, d) were fixed, stained with fluorescent cholera toxin B subunit (CtxB, red fluorescent) to determine the localization of GM1. Cells were co-stained with either lamin B1 (green fluorescence in a, c) or nucleoporin (green fluorescence in b, d). Nuclear DNA was counterstained with Hoechst 33258. Z-projection: top and bottom panels are x-z plane, and side panels are y-z plane.
Nuclear pore complexes are the other structure on nuclear membranes. Nucleoporins are best known as the constituent building blocks of nuclear pore complexes, membrane-embedded channels that mediate nuclear transport across the nuclear envelope. Recent evidence suggests that several nucleoporins have additional roles in gene regulation, such as the activating and silencing of developmental genes [25]. We observed that GM1 is localized in the nuclear periphery and is associated with nucleoporins (Fig. 1b); thus, it is highly likely that it may also participate in gene regulation once it has gained entry into the nucleus.
Z-stack images reveal that GM1 can be localized at the nuclear periphery in neural cells (Fig. 1a-b). Despite the lack of knowledge of how GM1 gains entry into the nucleus in interphase, we observed GM1’s localization during the breakdown of the nuclear envelope at mitosis. In dividing NPCs, the co-existence of GM1 with lamin B1 and nucleoporin was found, some even proximal to chromosomes (Fig.1c-d). It is possible that GM1 may hitchhike into the nucleus with nuclear envelope vesicles or with chromosomes as a passenger preceding the next nuclear formation.
GM1 associated with modified histones binds to promoters
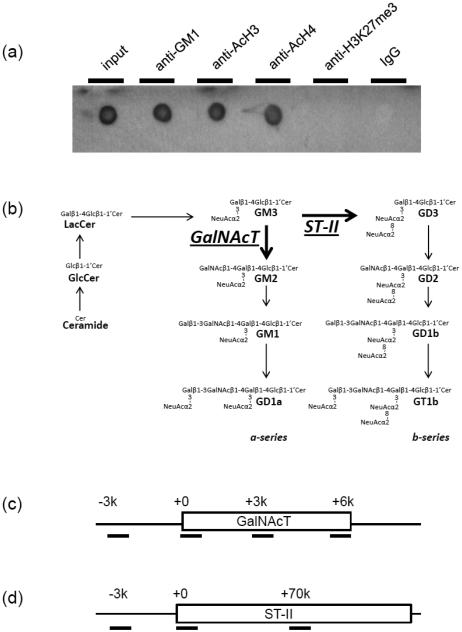
We previously demonstrated that NSCs/NPCs cultured with a supplement of GM1 exhibited a significantly enhanced neurogenic effect [28]. During the enhanced neurogenic process, we found that exogenously added GM1 induced NSCs to transcribe an elevated level of GalNAcT (GM2/GD2S) mRNA with a higher amount of acetylated histones on the GalNAcT gene’s promoter. In the present study, we confirmed the expression of GM1 at the nuclear periphery (Fig. 1). To examine whether endogenous GM1 was indeed associated with the GalNAcT gene in the nucleus, nuclear fractions were analyzed by co-immunoprecipitation experiments. The results revealed that nuclear GM1 genuinely interacted with AcH3 and AcH4, which are both active epigenetic marks, but not with H3K27me3, which is an inactive epigenetic mark (Fig. 2a). However, without prior cross-linking, we did not detect any histone signals in an assay on GM1-containing precipitate (data not shown). This experiment showed that GM1-engaged chromatins are transcriptionally active.
Fig. 2.
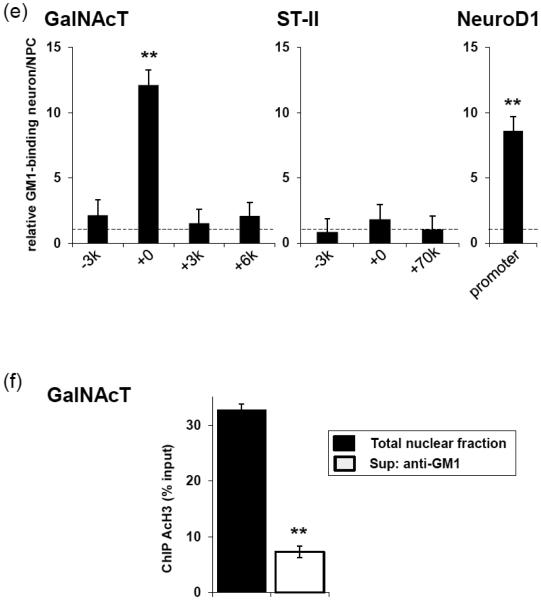
GM1 is accumulated in the activated GalNAcT and NeuroD1 genes. (a) The nuclear fraction of differentiated neurons with prior formaldehyde cross-linking was used for anti-GM1, anti-AcH3, anti-AcH4, or anti-H3K27me3 immunoprecipitation. PVDF membrane on which the precipitated complex was dot-blotted was probed with HRP-conjugated CtxB. (b) Structure and metabolic pathways of a- and b-series gangliosides. LacCer, lactosylceramide; GlcCer, glucosylceramide. (c and d) Regions of GalNAcT and ST-II genes amplified by PCR after ChIP are indicated by horizontal bars. GalNAcT-3000 (−3k) or ST-II-3000 (−3k) represents upstream region of the gene; GalNAcT+0 or ST-II+0, the 5′ promoter region; GalNAcT+3000 (+3k), GalNAcT+6000 (+6k), or ST-II+70000 (+70k), the gene body. (e) The amount of specific DNA fragments co-precipitated with GM1 was analyzed by quantitative real-time PCR. The data indicated the relative GM1 binding ability in neurons. The value of NPC samples was defined as 1.1 and represented with a dashed line. (f) The supernatant of anti-GM1 immunoprecipitation (neurons) was collected for the succeeding anti-AcH3 and ChIP assay. Amounts of the DNA fragments of GalNAcT (+0) were analyzed by quantitative PCR using specific primer pairs. The data, normalized to those of input signals, indicate the histone H3 acetylation levels of the gene. Each bar represents mean ± SD of 3-4 independent experiments (n = 3-4). ** (p<0.01) indicates the level of significance in two-tailed t-tests of differences between differentiated neurons versus NPCs.
GalNAcT and ST-II are two critical enzymes at a branching point of the ganglioside biosynthesis pathways that governs the pathway-shift for the expression of GD3 and N-acetylgalactosaminyl-containing “brain-type” gangliosides, including GM1 (Fig. 2b). We next investigated the interaction between GM1 and putative genetic loci, including the GalNAcT and ST-II genes. ChIP assay was performed using anti-GM1 antibody, and we found that GM1 was increased at the promoter region of the GalNAcT gene. To compare the GM1-associated DNA content before and after neuronal induction of NPCs, quantitative real-time PCR analyses were performed. As shown in Fig. 2e, the 5′ proximal promoter region (+0) had significantly more recruitment of GM1 in differentiated neurons than in the precursor counterpart. Neither the 5’ distal region (−3k) or the downstream gene body (+3K, +6k) showed any change. Such a tendency did not appear for the ST-II gene loci (2e). Interestingly, after neuronal differentiation, more GM1 was accumulated on the promoter of the neurogenic transcription factor NeuroD1 gene, which commits the transition of NSCs to NPCs (Fig. 2e). These results suggest that GM1 is involved in the chromatin complex that is associated with neuronal differentiation.
We next investigated whether GM1-associated genetic loci enriched with AcH3 could represent active chromatin. The supernatant of anti-GM1 immunoprecipitation was collected as the non-GM1-interacting chromatin fraction. The total nuclear fraction and non-GM1-interacting chromatin were compared in an anti-AcH3 ChIPn assay. Our result showed that GalNAcT gene which contains histone acetylation exclusively binds GM1 (Fig. 2f). This result supports that GM1 indeed participates in transcriptional activation of the GalNAcT gene.
Next, we investigated the co-localization of GM1 and GalNAcT gene by in-situ hybridization. We found that GM1 and the promoter region of the GalNAcT gene are in close proximity in the nucleus of a neuron (Fig. 3a). Despite the low expression of nuclear GM1 in NPCs, they did not co-localize therein. This result suggests that the interaction of GM1 and the GalNAcT gene promoter occurs in a developmental stage-specific manner.
Fig. 3.
Co-localization of GM1 and GalNAcT genes. Differentiated neurons (a) or NPSs (b) were fixed, and localization of the GalNAcT genes was determined by in-situ hybridization (green fluorescence). The cells were then stained with fluorescent cholera toxin B subunit (CtxB, red fluorescence) to determine the localization of GM1. DNA was counterstained with Hoechst 33258.
Inhibition of microtubule polymerization reduces GM1 binding to the promotor of the GalNAcT gene
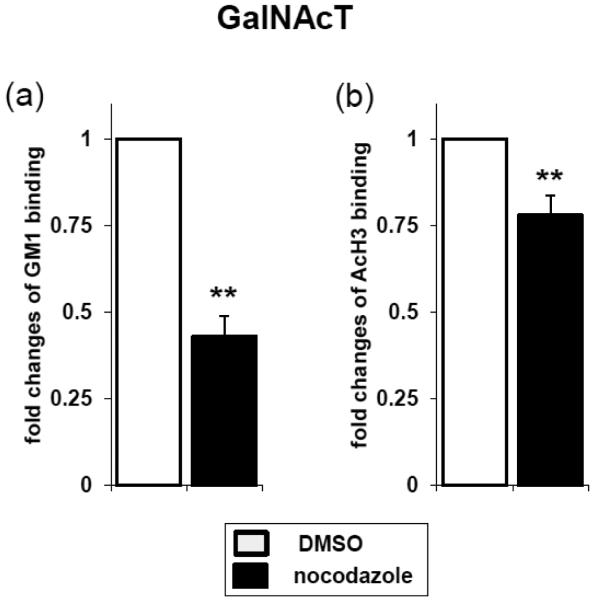
We next investigated the possibility of GM1 interacting with the promotor of the GalNAcT gene. The cytoskeleton is considered to have functional roles in chromatin remodeling and gene transcription [38]. To examine the role of the cytoskeleton in regulating the association between GM1 and GalNAcT gene promoter, we treated neurons with nocodazole or cytochalasin D to specifically depolymerize the microtubules or actin network, respectively. Nocodazole and cytochalasin D are known to induce nuclear deformation [39]. Nocotazole treatment reduced the binding of GM1 and AcH3 to the promoter region of the GalNAcT gene (Fig. 4). On the other hand, cytochalasin D did not have these effects (data not shown). These results suggest that microtubules, but not actin, are involved in the interaction between GM1 and the GalNAcT gene promoter. The molecular mechanisms of interaction between GM1 and gene promotors need to be defined in future studies.
Fig. 4.
Perturbing the nuclear envelope structure resulted in the detachment of the GalNAcT gene from the GM1 domains. The neuronal culture was incubated with 500 nM of nocodazole for 4-6 hrs. ChIP assay of GalNAcT (+0) gene in differentiated neurons was performed. Regions of the GalNAcT (+0) gene amplified by PCR after ChIP are indicated as horizontal bars. Genomic DNAs and histones (i.e., chromatin) were sonicated and precipitated by anti-GM1 (a) or anti-AcH3 (b) antibodies. GM1-associated chromatin (a) or acetylated chromatin were detected by quantitative real-time PCR. The data indicate fold changes between control (DMSO) and nocodazole-treated cells. The values of control samples were defined as 1.0. Each bar represents mean ± SD of 3 independent experiments (n = 3). ** (p<0.01) indicates the level of significance in two-tailed t-tests of differences between nocodazole treated versus control samples.
Discussion
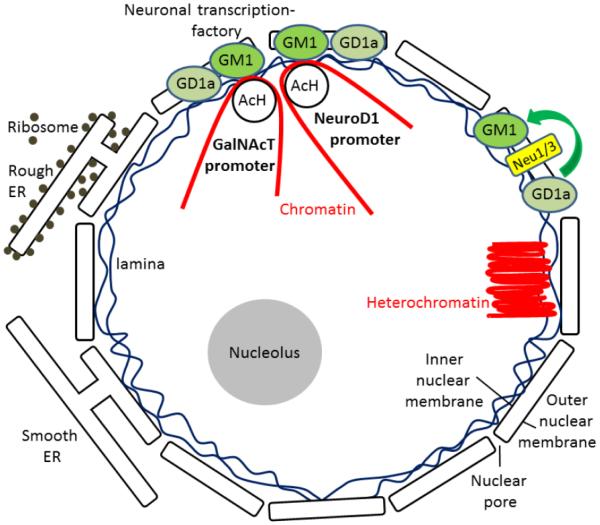
We investigated the hypothesis that nuclear GM1 is associated with gene regulation in neuronal cells. We found that GM1 is co-localized with lamin B1 at the nuclear lamin and nucleoporin at the nuclear pore complex in the neuronal nuclear envelope. Co- immunoprecipitation experiments revealed that nuclear GM1 interacts with acetylated histones of active epigenetic marks. On the other hand, nuclear GM1 did not bind with H3K27me3, which is an inactive epigenetic mark. Rather, it bound to acetylated histones on the promoters of the GalNAcT and NeuroD1 genes in differentiated neurons, but not in undifferentiated NPCs. However, little or no GM1 interacted with the ST-II gene promoter, consistent with our previous finding that only GalNAcT was responsible for the pathway shifts of ganglioside biosynthesis pathways during brain development [32, 40]. The binding of GM1 and AcH3 with the GalNAcT gene promoter is mediated by microtubules as shown by the nocodazole experiment. Thus, the present study provides the first direct evidence that GM1 interacts with active chromatin via acetylated histones at the nuclear periphery in neuronal cells (Fig. 5).
Fig. 5.
Proposed model of epigenetic regulation of the glycosyltransferase GalNAcT gene by nuclear GM1.
Our study agrees with several previous reports documenting the presence of GM1 in the nucleus [18-22]. In our previous study, we demonstrated that NSCs in culture supplemented with GM1 exhibited a significantly enhanced neurogenic effect [28]. It is possible that in this neurogenesis enhancement process, exogenous GM1 induces NSCs to transcribe more GalNAcT mRNA with a higher level of acetylated histones on the GalNAcT gene promoter region where more transcription factors are recruited. On the other hand, the ST-II gene did not show any significant changes. This result might represent a potential mechanism accounting for the correlations between the ganglioside pattern shift and epigenetic modifications of ganglioside synthase expression during neuronal differentiation and neural development. In this regard, GM1 might have a role in modulating the “pathway switch” in ganglioside expression in the developing brain. We suggest that GM1 may generate a positive feedback loop for NSCs to enhance neuronal differentiation and thereby to produce more GM1 and other “brain-type” gangliosides, such as GD1a, GD1b and GT1b by increasing the GalNAcT message level. Since the content of GM1 in the nuclear membrane is increased during neuronal differentiation [20], it is possible that the nuclear GM1-lipid domains may serve as a docking site at the nuclear periphery for specific active region of chromatin. Our scheme for the role of nuclear gangliosides epigenetically modulating the gene expression in neuronal cells is shown schematically in Fig. 5.
An important question concerns the source of GM1 residing in the nucleus. It has been established that GM1 and GD1a are present in the nuclear envelope [21, 41]. Sialidase activity has also been identified in the nuclear membrane of rodent brains [42, 43]. Further, it has been reported that neuraminidases (sialidases) Neu3 and Neu1 are present in the inner and outer nuclear membranes, respectively [16]. Since these neuraminidases can convert GD1a to GM1, GD1a may serve as a nuclear storage reserve precursor of GM1. Fig. 1c-d show the other potential sources of nuclear GM1. GM1 is found to be associated with lamin B1, nucleoporin, and chromosomes during breakdown of the nuclear envelope at mitosis. Our observations suggest that GM1 may hitchhike into the nucleus with nuclear envelope vesicles or with chromosomes as a passenger.
In conclusion, our results suggest that nuclear GM1 is capable of modulating transcriptional activity of neurogenic genes, such as GalNAcT and NeuroD1. We envision that signaling for differentiation and proliferation could be modulated by nuclear gangliosides via a novel epigenetic gene modification mechanism during neural differentiation. Since ganglioside expression profiles are associated not only with neural development, but also with pathogenic mechanisms of diseases in the central nervous system, future studies on epigenetic regulation of cell surface glycosphingolipid expression should provide clues as to the disease mechanisms, which should be useful in providing novel strategies for disease intervention and neural repair.
Acknowledgments
We thank Dr. Wei-Hua Wu for helpful comments on this manuscript. This work was supported in part by a VA Merit Award (1 IO1BX001388 to RKY) and NIH grants (RO1 NS26994 and RO1 NS11853 to RKY).
Abbreviations
- AcH3
acetylated histone H3
- AcH4
acetylated histone H4
- ChIP
chromatin immunoprecipitation
- GalNAcT
N-acetylgalactosaminyltransferase I (GA2/GM2/GD2/GT2-synthase)
- H3K27me3
histone H3 with trimethylation on lysine 27
- NPC
neuronal precursor cell
- NSC
neural stem cell
- ST-II
sialyltransferase II (GD3-synthase). Gangliosides are abbreviated using the nomenclatural rules of IUPAC-IUB [1] and according to Svennerholm [2]
References
- 1.The nomenclature of lipids. Recommendations (1976) IUPAC-IUB Commission on Biochemical Nomenclature. Lipids. 1977;12:455–468. [PubMed] [Google Scholar]
- 2.Svennerholm L. Chromatographic Separation of Human Brain Gangliosides. J Neurochem. 1963;10:613–623. doi: 10.1111/j.1471-4159.1963.tb08933.x. [DOI] [PubMed] [Google Scholar]
- 3.Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci. 2010;11:377–388. doi: 10.1038/nrn2810. [DOI] [PubMed] [Google Scholar]
- 4.Jobe EM, McQuate AL, Zhao X. Crosstalk among Epigenetic Pathways Regulates Neurogenesis. Front Neurosci. 2012;6:59. doi: 10.3389/fnins.2012.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsieh J, Gage FH. Epigenetic control of neural stem cell fate. Current opinion in genetics & development. 2004;14:461–469. doi: 10.1016/j.gde.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Mehler MF. Epigenetics and the nervous system. Annals of neurology. 2008;64:602–617. doi: 10.1002/ana.21595. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition- mediated neuronal differentiation of multipotent adult neural progenitor cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 9.Jamaladdin S, Kelly RD, O'Regan L, Dovey OM, Hodson GE, Millard CJ, Portolano N, Fry AM, Schwabe JW, Cowley SM. Histone deacetylase (HDAC) 1 and 2 are essential for accurate cell division and the pluripotency of embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9840–9845. doi: 10.1073/pnas.1321330111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson PJ, Fairall L, Santos GM, Schwabe JW. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millard CJ, Watson PJ, Celardo I, Gordiyenko Y, Cowley SM, Robinson CV, Fairall L, Schwabe JW. Class I HDACs share a common mechanism of regulation by inositol phosphates. Molecular cell. 2013;51:57–67. doi: 10.1016/j.molcel.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozcan S, Andrali SS, Cantrell JE. Modulation of transcription factor function by O-GlcNAc modification. Biochimica et biophysica acta. 2010;1799:353–364. doi: 10.1016/j.bbagrm.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardiville S, Hart GW. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell metabolism. 2014;20:208–213. doi: 10.1016/j.cmet.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis BA, Hanover JA. O-GlcNAc and the epigenetic regulation of gene expression. The Journal of biological chemistry. 2014;289:34440–34448. doi: 10.1074/jbc.R114.595439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucki NC, Sewer MB. Nuclear sphingolipid metabolism. Annual review of physiology. 2012;74:131–151. doi: 10.1146/annurev-physiol-020911-153321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Wu G, Miyagi T, Lu ZH, Ledeen RW. Sialidase occurs in both membranes of the nuclear envelope and hydrolyzes endogenous GD1a. J Neurochem. 2009;111:547–554. doi: 10.1111/j.1471-4159.2009.06339.x. [DOI] [PubMed] [Google Scholar]
- 17.Yu RK, Itokazu Y. Glycolipid and glycoprotein expression during neural development. Advances in neurobiology. 2014;9:185–222. doi: 10.1007/978-1-4939-1154-7_9. [DOI] [PubMed] [Google Scholar]
- 18.Saito M, Sugiyama K. Characterization of nuclear gangliosides in rat brain: concentration, composition, and developmental changes. Archives of biochemistry and biophysics. 2002;398:153–159. doi: 10.1006/abbi.2001.2725. [DOI] [PubMed] [Google Scholar]
- 19.Wu G, Lu ZH, Ledeen RW. GM1 ganglioside in the nuclear membrane modulates nuclear calcium homeostasis during neurite outgrowth. J Neurochem. 1995;65:1419–1422. doi: 10.1046/j.1471-4159.1995.65031419.x. [DOI] [PubMed] [Google Scholar]
- 20.Wu G, Lu ZH, Ledeen RW. Induced and spontaneous neuritogenesis are associated with enhanced expression of ganglioside GM1 in the nuclear membrane. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1995;15:3739–3746. doi: 10.1523/JNEUROSCI.15-05-03739.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ledeen RW, Wu G. The multi-tasked life of GM1 ganglioside, a true factotum of nature. Trends in biochemical sciences. 2015;40:407–418. doi: 10.1016/j.tibs.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Xie X, Wu G, Lu ZH, Ledeen RW. Potentiation of a sodium-calcium exchanger in the nuclear envelope by nuclear GM1 ganglioside. J Neurochem. 2002;81:1185–1195. doi: 10.1046/j.1471-4159.2002.00917.x. [DOI] [PubMed] [Google Scholar]
- 23.Tempera I, Buchetti B, Lococo E, Gradini R, Mastronardi A, Mascellino MT, Sale P, Mosca L, d'Erme M, Lenti L. GD3 nuclear localization after apoptosis induction in HUT-78 cells. Biochemical and biophysical research communications. 2008;368:495–500. doi: 10.1016/j.bbrc.2007.12.196. [DOI] [PubMed] [Google Scholar]
- 24.Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, Spiegel S. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325:1254–1257. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talamas JA, Capelson M. Nuclear envelope and genome interactions in cell fate. Frontiers in genetics. 2015;6:95. doi: 10.3389/fgene.2015.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demmerle J, Koch AJ, Holaska JM. Emerin and histone deacetylase 3 (HDAC3) cooperatively regulate expression and nuclear positions of MyoD, Myf5, and Pax7 genes during myogenesis. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2013;21:765–779. doi: 10.1007/s10577-013-9381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Yu RK. Interaction of ganglioside GD3 with an EGF receptor sustains the self- renewal ability of mouse neural stem cells in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:19137–19142. doi: 10.1073/pnas.1307224110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai YT, Yu RK. Epigenetic activation of mouse ganglioside synthase genes: implications for neurogenesis. J Neurochem. 2014;128:101–110. doi: 10.1111/jnc.12456. [DOI] [PubMed] [Google Scholar]
- 29.Nakatani Y, Yanagisawa M, Suzuki Y, Yu RK. Characterization of GD3 ganglioside as a novel biomarker of mouse neural stem cells. Glycobiology. 2010;20:78–86. doi: 10.1093/glycob/cwp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itokazu Y, Yu RK. Amyloid beta-Peptide 1-42 Modulates the Proliferation of Mouse Neural Stem Cells: Upregulation of Fucosyltransferase IX and Notch Signaling. Mol Neurobio. 2014 doi: 10.1007/s12035-014-8634-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wysocka J, Reilly PT, Herr W. Loss of HCF-1-chromatin association precedes temperature- induced growth arrest of tsBN67 cells. Molecular and cellular biology. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki Y, Yanagisawa M, Ariga T, Yu RK. Histone acetylation-mediated glycosyltransferase gene regulation in mouse brain during development. J Neurochem. 2011;116:874–880. doi: 10.1111/j.1471-4159.2010.07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu RK, Lee SH. In vitro biosynthesis of sialosylgalactosylceramide (G7) by mouse brain microsomes. The Journal of biological chemistry. 1976;251:198–203. [PubMed] [Google Scholar]
- 34.Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. The EMBO journal. 2007;26:5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kizuka Y, Kitazume S, Yoshida M, Taniguchi N. Brain-specific expression of N- acetylglucosaminyltransferase IX (GnT-IX) is regulated by epigenetic histone modifications. The Journal of biological chemistry. 2011;286:31875–31884. doi: 10.1074/jbc.M111.251173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai YT, Lin CI, Chen HK, Lee KM, Hsu CY, Yang SJ, Yeh NH. Chromatin tethering effects of hNopp140 are involved in the spatial organization of nucleolus and the rRNA gene transcription. Journal of biomedical science. 2008;15:471–486. doi: 10.1007/s11373-007-9226-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng X, Kim Y, Zheng Y. Identification of lamin B-regulated chromatin regions based on chromatin landscapes. Molecular biology of the cell. 2015;26:2685–2697. doi: 10.1091/mbc.E15-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osmanagic-Myers S, Dechat T, Foisner R. Lamins at the crossroads of mechanosignaling. Genes & development. 2015;29:225–237. doi: 10.1101/gad.255968.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tremblay D, Andrzejewski L, Leclerc A, Pelling AE. Actin and microtubules play distinct roles in governing the anisotropic deformation of cell nuclei in response to substrate strain. Cytoskeleton. 2013;70:837–848. doi: 10.1002/cm.21148. [DOI] [PubMed] [Google Scholar]
- 40.Ngamukote S, Yanagisawa M, Ariga T, Ando S, Yu RK. Developmental changes of glycosphingolipids and expression of glycogenes in mouse brains. J Neurochem. 2007;103:2327–2341. doi: 10.1111/j.1471-4159.2007.04910.x. [DOI] [PubMed] [Google Scholar]
- 41.Kotzerke J, Stibane C, Dralle H, Wiese H, Burchert W. Screening for pheochromocytoma in the MEN 2 syndrome. Henry Ford Hospital medical journal. 1989;37:129–131. [PubMed] [Google Scholar]
- 42.Saito M, Hagita H, Ito M, Ando S, Yu RK. Age-dependent reduction in sialidase activity of nuclear membranes from mouse brain. Experimental gerontology. 2002;37:937–941. doi: 10.1016/s0531-5565(02)00021-9. [DOI] [PubMed] [Google Scholar]
- 43.Saito M, Fronda CL, Yu RK. Sialidase activity in nuclear membranes of rat brain. J Neurochem. 1996;66:2205–2208. doi: 10.1046/j.1471-4159.1996.66052205.x. [DOI] [PubMed] [Google Scholar]