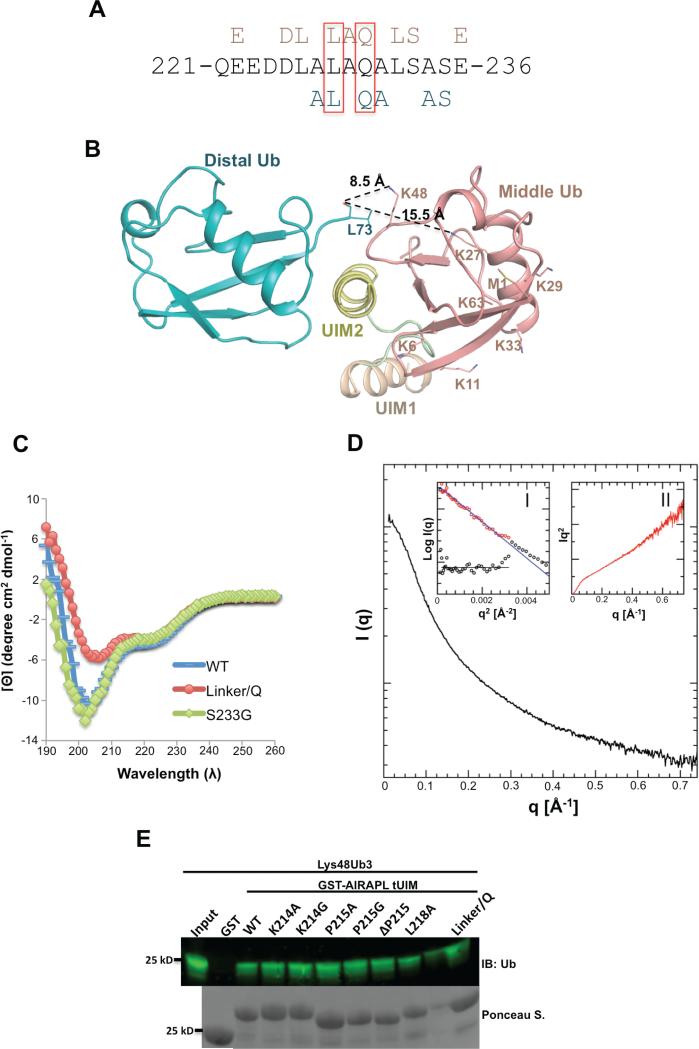
Figure 4. The roles of AIRAPL UIM2 and inter-UIM linker in defining specificity towards Lys48-linked ubiquitin chains.
(A) UIM2 sequence comprising amino acid residues 221-236. Residues from UIM2 that interact with the middle (top, pink) and/or distal (below, blue) ubiquitins are indicated. Leu228 and Gln230 that are involved in the interactions with both distal and middle ubiquitins are marked by red boxes. (See also Fig. S5a) (B) Orientation of the distal and middle ubiquitins exclusively allows Lys48 linkage binding. The last C-terminal residues (74-76) of distal ubiquitins are not visible in the electron density map. In spite of that, Lys48 is the closest lysine residue from middle ubiquitin to the main chain carbonyl group of Leu73 (distance: 8.5 Å). The second closest lysine (Lys27) is 15.5 Å away from Leu73 (See also Fig. S5b-d) (C) Circular Dichroism (CD) measurements of AIRAPL tUIM WT and two mutants (S233G and linker/Q). The data is reported as mean residue ellipticities in the range of λ: 190-260 nm and indicate ~32% helical content for tUIM WT and S233G, and 25% for the linker/Q mutant. In linker/Q mutant all residues of the linter/UIM linker are substituted with glutamine. (D) SAXS intensity of AIRAPL tUIM in solution. Inset I: the Guinier analysis of the data exhibits good linearity within the fit region (red circles) with unbiased residuals (depicted enlarged by a factor of two as purple circles distributed around a straight line in the lower left corner). Inset II: the Kratky representation of the SAXS data monotonically increases in the high q-region and does not show a pronounced peak at lower q indicative of an extended, flexible structure. (E) Binding of GST-tagged tUIM WT and inter-UIM linker mutants to Lys48-linked tri-ubiquitin chains was analyzed by immunoblotting using anti-ubiquitin antibody. Loading of GST-tagged proteins was determined by Ponceau S staining.