Abstract
KRAS is one of the most frequently mutated proto-oncogenes in human cancers. The dominant oncogenic mutations of KRAS are single amino acid substitutions at codon 12, in particular G12D and G12V present in 60–70% of pancreatic cancers and 20–30% of colorectal cancers. The consistency, frequency, and tumor specificity of these “neo-antigens” make them attractive therapeutic targets. Recent data associates T cells that target mutated antigens with clinical immunotherapy responses in patients with metastatic melanoma, lung cancer, or cholangiocarcinoma. Using HLA-peptide prediction algorithms, we noted that HLA-A*11:01 could potentially present mutated KRAS variants. By immunizing HLA-A*11:01 transgenic mice, we generated murine T cells and subsequently isolated T-cell receptors (TCRs) highly reactive to the mutated KRAS variants G12V and G12D. Peripheral blood lymphocytes (PBLs) transduced with these TCRs could recognize multiple HLA-A*11:01+ tumor lines bearing the appropriate KRAS mutations. In a xenograft model of large established tumor, adoptive transfer of these transduced PBLs reactive with an HLA-A*11:01, G12D-mutated pancreatic cell line could significantly reduce its growth in NSG mice (P = 0.002). The success of adoptive transfer of TCR-engineered T cells against melanoma and other cancers support clinical trials with these T cells that recognize mutated KRAS in patients with a variety of common cancer types.
Keywords: KRAS mutation, T cell receptor, MHC restriction, immunotherapy, adoptive cell transfer
Introduction
KRAS is a proto-oncogene that plays a key role in numerous human cancers. It is a master activator of multiple cell pathways essential to cell division and metabolism, especially the MAP kinase pathway (1). The RAS family of proteins all contain a GTPase activity critical to turning off their function, and they interact with GTPase-activating proteins (GAPs) that facilitate this hydrolysis to their GDP-bound inactive state. Activating mutations in KRAS typically impair this GTPase activity, often by disrupting the interaction with GAP, and are difficult to therapeutically modulate with small molecule kinase inhibitors (2,3). With the exception of non-small cell lung cancer, the most frequent mutations found are in codon 12 with either aspartic acid or valine substituted for the native glycine (4). These two mutations alone are found in approximately 60–70% of all pancreatic adenocarcinomas (5,6) and 20–30% of all colorectal cancers (7,8). Because of their completely tumor-specific nature, their ubiquity, and the limited diversity of their mutated variants, we sought to target mutated KRAS variants immunologically by developing T-cell receptors specific for codon 12 mutations in KRAS.
The immune recognition of the products of tumor-specific mutations is recognized as a significant component of the endogenous host response to cancer. Although the presence of such immunological activity in patients with cancer has been recognized for 20 years (9), recent data has shown that this activity is associated with clinical responses to checkpoint blockade with antibodies to CTLA4 and PD1 in patients with melanoma and non-small cell lung cancer (10,11). Tumor infiltrating lymphocytes (TIL) have been commonly found in patients with melanoma and adoptive transfer of these TIL can achieve durable complete regressions of widespread metastatic disease (12). In one reported case in a patient with metastatic cholangiocarcinoma, isolation and adoptive transfer of a nearly clonal population of CD4+ T cells, reactive to a mutated epitope in ERB2-interacting protein, induced a durable and ongoing regression of metastatic disease (13). Other studies have shown that autologous patient lymphocytes can be made reactive with tumor-associated antigens through engineered TCRs or chimeric antigen receptors, and induce tumor regression upon adoptive transfer (14–16). TCRs cloned from both patients and vaccinated mice (transgenic for human HLA molecules) have been utilized in this manner to induce objective responses in patients with metastatic diseases (14,17,18). TCRs of HLA-transgenic murine origin have the advantage of not pairing with human TCR chains, thus avoiding the danger of “mispaired’ heterodimers generating unpredictable reactivity (19). The major limitation of this gene-engineered T-cell transfer approach has been the paucity of safe and active tumor antigen targets. “Neo-epitopes” generated by tumor-specific mutations are in many ways the ideal tumor antigens. They are completely tumor specific and they are “non-self” so they are not subjected to negative thymic selection and can be highly immunogenic. There are major drawbacks to targeting such antigens. First, they require completely personalized therapies, as most mutations will differ from patient to patient, even when the same gene is involved. Second, many mutations are non-driver mutations and may be heterogeneously expressed or lost without consequence. Finally, many mutated epitopes will not be naturally processed by the proteasome and presented by an HLA allele. Targeting the most common mutated forms of KRAS would address the first and second problems, as long as we can identify those naturally processed mutated epitopes that can be presented by a known HLA allele. T cells from cancer patients can recognize peptides derived from mutated KRAS variants (20–22), indicating the existence of immune reactivity to mutated KRAS. This study attempts to identify naturally processed epitopes from mutated KRAS that contain the most common mutated variants present in common human cancers, clone mutated KRAS-reactive TCRs, and test treatment efficacy of these TCRs in a xenograft mouse model. Ultimately these mutated KRAS-reactive TCRs will be applied clinically to treat cancer patients who carry these mutations.
Materials and Methods
Mice and tumor lines
HLA-A*11:01 transgenic mice (Taconic Bioscience), and NOD/scid/gamma (NSG) immunodeficient mice (Jackson Laboratory) were maintained in the NIH animal facility. NSG mice lack mature T cells, B cells and nature killer cells and deficient in cytokine signaling pathways. Expression of HLA-A*11:01 in transgenic mice was confirmed by staining with anti-HLA-A*11 antibody (One Lambda). All mouse studies were approved by the National Cancer Institute Animal Care and Use Committee.
Human pancreatic tumor lines were purchased from ATCC or gifts from Dr. Rudloff in 2014, and maintained in RPMI 1640 (Life Technologies), DMEM (Life Technologies), or IMDM (Lonza) with 10% fetal bovine serum (FBS; Life Technologies). The cell lines were authenticated by HLA genotyping and KRAS mutation status (Supplementary Table S1), and retrovirally transduced with retrovirus encoding HLA-A*11:01 if they were not HLA-A*11:01 positive. The cell lines were maintained in the cell culture only when they are needed in the experiments and usually kept in culture for approximately a month.
Antibodies and peptides
Monoclonal antibodies (mAbs) including fluorescein isothiocyanate (FITC)-labeled anti-human CD3 (clone SK7), phycoerythrin (PE)-labeled anti-human CD8 (clone SK-1), allophycocyanin (APC)-labeled anti-mouse TCRβ (clone H57-597) were purchased from BD Pharmingen.
All KRAS related peptides used in the study were custom synthesized by peptide 2.0, with purity greater than 90% by mass spectroscopy. Hepatitis B virus core peptide (HBVc128-140; TPPAYRPPNAPIL) was purchased from GenScript with purity >95%.
KRAS mutation-specific qRT-PCR
RNA was isolated from pancreatic tumor lines using RNeasy Mini kit (Qiagen) and cDNA was synthesized by reverse transcription (Life Technologies). Allele-specific primers, reference primers, and probes were custom synthesized according to a previous study(23). CFX96 Touch System (Biorad) was used for real-time PCR analysis, and results were presented relative to βactin (ACTB) expression.
Immunization of HLA-A*11:01 transgenic mice, in vitro stimulation of murine T cells, and reactivity of murine anti-KRAS G12D or G12V T cells
HLA-A*11:01 transgenic mice were injected subcutaneously at the base of the tail and footpads with KRAS G12V7-16 or KRAS G12D7-16, and helper peptide HBVc128-140 emulsified in incomplete Freund’s adjuvant (Sigma). Mice were immunized twice with KRAS G12V7-16, or three times with KRAS G12D7-16, with at least a 2-week interval between immunizations. Seven days after the final immunization, splenic and lymph node lymphocytes were harvested, pulsed with corresponding peptides at concentrations of 1 μM, 0.1 μM, or 0.01 μM, and then cultured in a 24-well plate at concentration of 3 × 106/ml in 2 ml of mouse T-cell medium including RPMI1640 plus 10% FBS, non-essential amino acid (Life Technologies), serum pyruvate (Life Technologies), β-mercaptoethanol (β-ME; Life Technologies) and recombinant human interleukin 2 (rhIL2; 30 IU/ml). Cell growth was monitored daily, and cultures split or replenish with fresh mouse T cell medium and rhIL2 when necessary. Seven days after in vitro stimulation, effector T cells (1 × 105) were cocultured with appropriate target cells (5 × 104) overnight, and the supernatant was harvested for IFNγ measurement by ELISA.
Clonotypic analysis of KRAS G12V or G12D-reactive murine T cells
For each KRAS G12V or G12D-reactive murine T-cell population, total RNA was isolated using RNeasy mini kits (Qiagen). TCR α and β chains were then identified using 5′-rapid amplification of cDNA ends (RACE)-PCR. 5′ RACE reaction was performed by SMARTer RACE cDNA amplification kit (Clontech) following the manufacturer’s instructions. The RACE cDNAs (~600bp) were obtained with primers complementary to the constant regions of TCR alpha or beta chains and then inserted into the pCR2.1 vector by TA cloning (Life Technologies). Primers for the TCR alpha or beta chain were synthesized by IDT, and their sequences were 5′-gttgctccaggcaatggccccattgctc or 5′-ggtccgtgctgaccccactgtggacctc, respectively. After TA cloning, 48 colonies were picked from each 5′ RACE product of both TCR alpha and beta chains and their variable regions and complementarity determining region 3 (CDR3) were sequenced.
Retroviral production, transduction of anti-CD3 stimulated PBL, and reactivity of transduced cells
cDNAs encoding selected full-length TCR α and β chains (Genbank accession number KU254560 to KU254565) were cloned into the pMSGV1 plasmid, which has been described in previous publications with some modification (24). Briefly, full-length TCR α and β chain cDNAs were amplified by PCR using the pairs appropriate to corresponding sequences of each TCR α and β chain with a P2A sequence used as the spacer in between.
To produce retrovirus, 293gp cells were transfected with 9 μg of pMSGV1-TCR and 4.5μg of plasmid RD114 using Lipofectamine 2000 (Life Technologies; 60 μl). Two days later, the supernatants were harvested and used to transduce anti-CD3–stimulated PBLs. Allogeneic donor PBLs were stimulated with soluble OKT-3 (50 ng/ml) and rhIL2 (300 IU/ml) for 2 days before transduction was performed. The stimulated cells were added to 24-well plates initially coated with RetroNectin (Takara) and subsequently precoated with retrovirus by spinoculation (2000xg, 32°C, 2 hrs) at 5 × 105/ml. The plates were then centrifuged at 1000 × g for 10 min, and incubated overnight at 37°C in a 5% CO2 incubator. This procedure was repeated the next day and cells were split as necessary to maintain cell density between 0.5 and 1 × 106/ml. Transduction efficiency was determined by analyzing mouse TCRβ expression of retrovirally-transduced cells. CD8 and CD4 enrichment was performed using CD8 and CD4 T cell isolation kits (Miltenyi Biotec). In some experiments, pancreatic tumor lines were pre-treated with IFNγ (10 ng/ml) for 48 hrs, and washed three times before coculture with T cells. Immune recognition was assessed by measuring IFNγ in the supernatant after 24 hours of coculture.
In vitro proliferation and CD107a degranulation assays
To test the in vitro proliferation potential of TCR-transduced T cells, carboxyfluorescein succinimidyl ester (CFSE) cell proliferation assay was performed (ThermoFisher Scientific). Briefly, CFSE-labeled anti-mutated KRAS TCR-transduced T cells (1 × 105) were cocultured with various target cells (5 × 104), including COS7 stably transfected with KRAS variants and pancreatic cell lines. Three days after incubation, cells were further labeled with anti-human CD3 PE-cy7 and anti-murine TCR beta APC Abs followed by flow analysis.
CD107a degranulation assay was performed to test the cytolytic potential of the anti-mutated KRAS TCRs. Similarly, 1×105 TCR-transduced T cells were co-cultured with various targets (4 × 105) in the presence of FITC conjugated anti-CD107a Ab (BD Biosciences) for 2 to 4 hr., and then labeled with anti-human CD3 PE-cy7, CD8 PE and anti-murine TCR beta APC. Flow analysis was performed thereafter.
Tumor challenge and adoptive cell transfer
NSG mice were injected subcutaneously with 1 × 106 FA6-2/A11 pancreatic tumor cells. Ten days after inoculation, mice received 107 intravenous human T cells retrovirally transduced with KRAS G12D-specific TCR or controls, followed by intraperitoneal administration of 200,000 IU of rhIL2 per day for 3 days. Treatment group included 10 mice, whereas control groups had five. All groups were randomized and tumor measurements were performed by a blinded impartial observer. For analyzing MHC class I expression and persistence of transferred T cells, spleen and tumors were processed to obtain single-cell suspension, and then labeled with antibodies to HLA-A11 or to human CD3, CD8 and mTCRβ.
Statistical Analysis
Wilcoxon rank-sum test was used to compare tumor slopes between each treatment groups, and log-rank test was used to analyze survival.
Results
Generation of Mutated KRAS-reactive T cells with HLA-A*11:01 transgenic mice
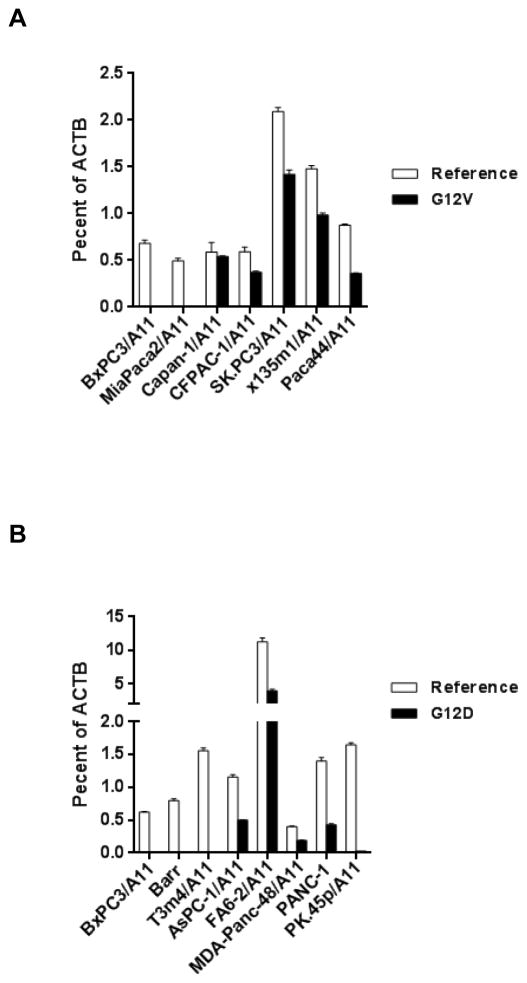
Using algorithms that predict HLA-peptide binding (25), we identified HLA-A*11:01 as one of the MHC molecules that may bind peptides that harbor mutations of KRAS amino acid 12, such as KRAS G12D7-16 (VVVGADGVGK), KRAS G12D8-16 (VVGADGVGK), G12V7-16 (VVVGAVGVGK), and G12V8-16 (VVGAVGVGK) (Supplementary Table S2). High affinity murine TCRs reactive to human antigens, such as P53, MAGE-A3 and CEA, have been generated using HLA-A*02:01 transgenic mice (17, 26, 27), so we applied a similar approach to generate HLA-A*11:01–restricted murine T cells to mutated KRAS G12V or G12D. HLA-A*11:01 transgenic mice were immunized with KRAS G12V7-16 or G12D7-16, and their spleens and lymph nodes harvested and stimulated in vitro with corresponding cognate peptides. KRAS G12V or G12D-reactive T cells were identified by testing against a panel of target cells expressing KRAS G12V or G12D, including pancreatic tumor lines carrying corresponding KRAS mutations. To assess KRAS mutation status in pancreatic tumor lines, we first confirmed KRAS mutations by direct sequencing of genomic DNA (Supplementary Table S1), and then analyzed expression of KRAS mutation by allele-specific quantitative RT-PCR (23) (Fig. 1). Primers specific for all KRAS genes irrespective of mutation status (referred to as “Reference”) showed copy numbers that were between 0.4% and 2% of β-actin copy numbers in the pancreatic tumor lines (Fig. 1A and B). In pancreatic tumor lines that harbor KRAS G12V mutation, expression of the mutated gene was approximately 0.3% to 1.4% of β-actin, whereas control cell lines without such mutations, such as BxPC3/A11 and MiaPaca2/A11, showed no expression of KRAS G12V (Fig. 1A). Expression of KRAS G12D, however, varied significantly among five KRAS G12D-positive pancreatic tumor lines. FA6-2/A11 had the highest expression (~4%), whereas PK.45p/A11 showed the lowest expression (~0.02%) with the other lines varying from 0.2% to 0.4% of β-actin. No expression was detected in KRAS G12D-negative tumor lines (Fig. 1B).
Figure 1.
Quantitative RT-PCR analysis of expression of mutated KRAS mRNA in pancreatic tumor lines. (A) cDNAs from five KRAS G12V-positive tumor lines and two KRAS G12V-negative tumor lines were synthesized for expression analysis. Mutation status was verified by sequencing of genomic DNA shown in table S1. Primers specific for KRAS gene regardless of mutation status (designated as “Reference”), or for mutated KRAS G12V (designated as “G12V”) were used in the analysis. Results are presented relative to ACTB mRNA (encoding β-actin). (B) Similar analysis of KRAS expression of five KRAS G12D-positive tumor lines and three KRAS G12D-negative tumor lines.
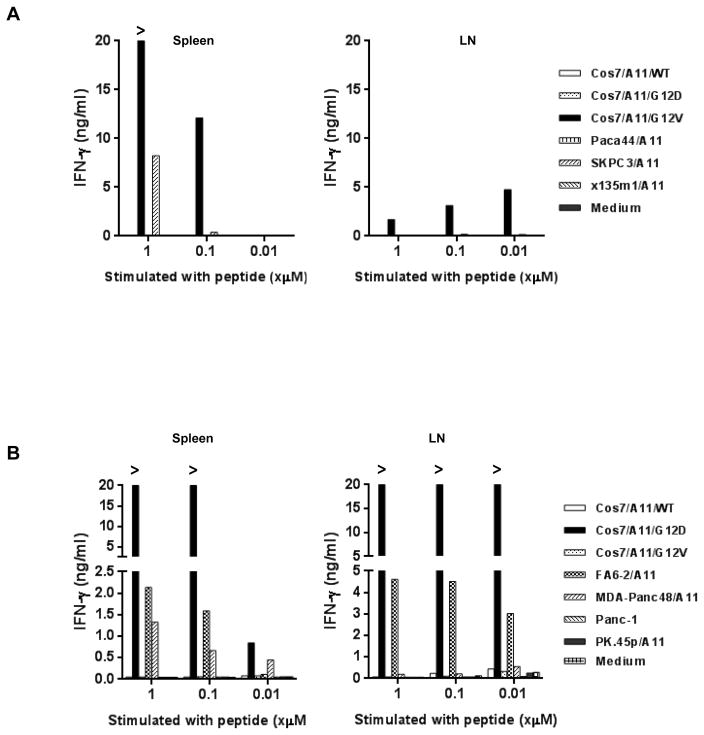
After in vitro stimulation with different concentrations of the appropriate cognate peptide, anti-KRAS G12V-specfic T cells were found in both spleen and LN; these T cells secreted IFNγ only when cocultured with COS7 stably transduced with HLA-A*11:01 and a KRAS G12V minigene encoding aa 1–23 (COS7/A11/G12V), but not with other KRAS minigenes, such as wild-type (COS7/A11/WT) and G12D (COS7/A11/G12D) (Fig. 2A). Among these cultures, splenocytes stimulated with 1 μM of G12V7-16 in vitro appeared to be most reactive: they not only recognized COS7/A11/G12V, but were also highly reactive to a KRAS G12V-positive pancreatic tumor line, SK.PC3, transduced with HLA-A*11:01 (SK.PC3/A11). Similarly, KRAS G12D-specific T cells could also be detected in both spleens and LNs, and splenocytes stimulated with 1 μM of G12D7-16 had the highest reactivity against HLA-A*11:01-positive KRAS G12D-positive pancreatic tumor lines (Fig. 2B). This is the first evidence of T cell immunity against naturally processed and presented epitopes from mutated KRAS, and encouraged us to proceed with isolating the HLA-A*11:01–restricted TCRs responsible for this reactivity.
Figure 2.
Murine T cells reactive to KRAS G12V or KRAS G12D generated from HLA-A*11:01 transgenic mice by in vivo peptide immunization. (A) IFNγ production of murine T cells from splenocytes or draining lymph node lymphocytes (LN) from peptide-immunized HLA-A*11:01 transgenic mice. Spleen and LN from immunized mice were harvested, and stimulated with different concentrations (1, 0.1 or 0.01μM) of KRAS G12V7-16 peptide once in vitro. 7 days after in vitro stimulation, T cells were co-cultured with COS7 stably transduced with HLA-A*11:01 (COS7/A11) and KRAS minigenes encoding the 23 N-terminal amino acids of wild type KRAS (WT), mutation variants KRAS G12D and KRAS G12V, and 3 HLA-A*11:01–transduced pancreatic tumor lines carrying KRAS G12V mutations. After overnight incubation, supernatants were harvested and IFNγ production was measured. (B) IFNγ production of murine T cells from splenocytes or LN from HLA-A*11:01 transgenic mice immunized three times with KRAS G12D7-16 peptide. Spleen and LN from immunized mice were harvested, and stimulated with different concentrations (1, 0.1 or 0.01μM) of KRAS G12D7-16 peptide once in vitro. Seven days after in vitro stimulation, T cells were cocultured with COS7/A11 transduced with KRAS minigenes, and four HLA-A*11:01–positive pancreatic tumor lines carrying KRAS G12D mutations. After overnight incubation, the supernatant was harvested and IFNγ production was measured.
Identification of HLA-A11*01–restricted KRAS G12V-reactive TCRs
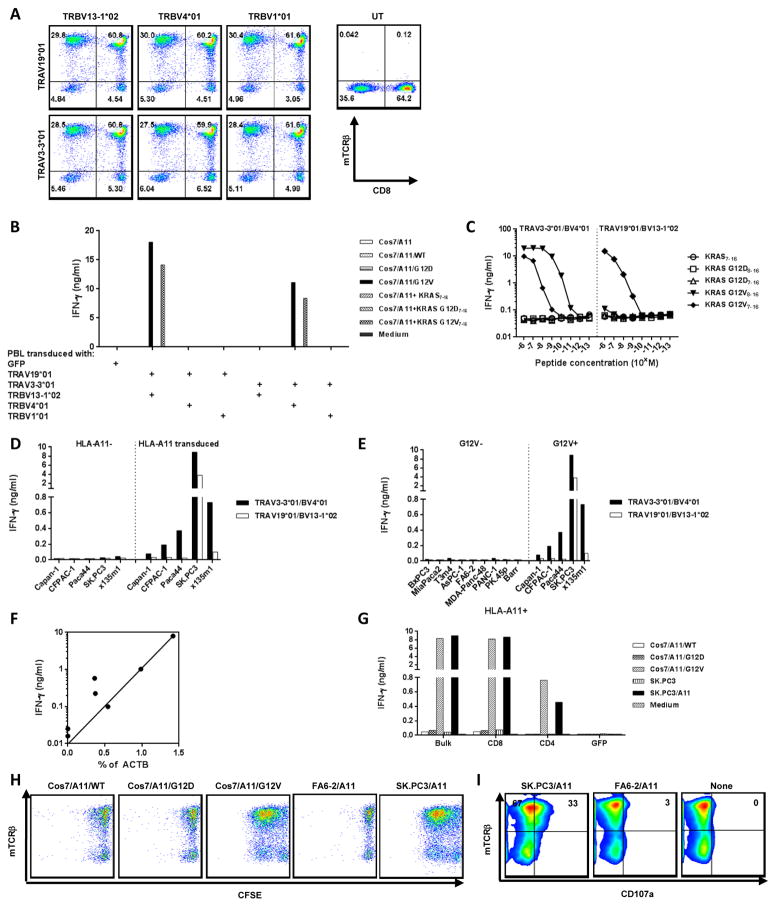
Two dominant TCR α chains and three dominant β chains were identified by 5′RACE from KRAS G12V-reactive splenocytes (Table 1). Retroviral vectors were constructed for each individual chain and screened for correct pairing by co-transducing alpha and beta chains into anti-CD3 stimulated HLA-A*11:01 positive PBL (Fig. 3A). Similar expression of murine TCRs (by mouse anti-TCRβ staining) was obtained with all six candidate combinations. Two of these candidate TCR combinations, TRAV19*01/AJ53 paired with TRBV13-1*02/BD2*01/BJ2-1*01 (TRAV19*01/BV13-1*02), and TRAV3-3*01/AJ17*01 paired with TRBV4*01/BD2*01/BJ2-1*01 (TAV3-3*01/BV4*01), had specific reactivity to the KRAS G12V7-16 peptide (pulsed onto COS7/A11), and COS7/A11/G12V (Fig. 3B).
Table 1.
Oligoclonal TCRs identified from murine KRAS G12V-specific splenocytes (1μM) by 5′RACE
| V Region | D/J Region | CDR3 | Frequency | |
|---|---|---|---|---|
| Alpha chains | TRAV19*01 | 53*01 | CAAGDSGGSNYKLTF | 31% |
| TRAV3-3*01 | 17*01 | CAVSGGTNSAGNKLTF | 14% | |
|
| ||||
| Beta chains | TRBV13-1*02 | 2*01/2-1*01 | CASASWGGYAEQFF | 23% |
| TRBV4*01 | 2*01/2-1*01 | CASSRDWGPAEQFF | 15% | |
| TRBV1*01 | 1*01/2-3*01 | CTCSADRGAETLYF | 12% | |
Figure 3.
Characteristics of HLA-A*11:01–restricted KRAS G12V-specific murine TCRs. (A) Expression of human PBL cotransduced with candidate TCR α and β chains. Two oligoclonal α chains and three oligoclonal β chains were identified from murine KRAS G12V-reactive splenocytes (1 μM) by 5′RACE (Table 1). All of them were constructed to retroviral vector, pMSGV1, separately. Allogeneic PBLs were stimulated with anti-CD3 (50 ng/ml) for 2 days and cotransduced twice with retroviruses encoding oligoclonal TCR α and β chains at 0.5 × 106 cells per well in a 24-well plate. Three days after transduction, T cells transduced with all six possible TCR pairs were labeled with antibodies to CD3, CD8, and mouse TCRβ, and analyzed on a FACS Canto II. Data was gated on the live CD3+ population. (B) Reactivity of PBL cotransduced with oligoclonal TCR α and β chains. Anti-CD3 stimulated human PBL cotransduced with six pairs of α and β chains were cocultured with COS7/A11 transduced with WT, G12D, or G12V minigenes, or pulsed with KRAS wildtype7-16 (WT7-16), KRAS G12D7-16, and KRAS G12V7-16 10-mer peptides. (C) Affinity comparison of two KRAS G12V-reactive TCRs. Anti-CD3 stimulated human PBL were transduced with retroviruses encoding either TRAV3-3*01/BV4*01 or TRAV19*01/BV13-1*02 TCR as described above. Three days after transduction, TCR-transduced cells were cocultured with COS7/A11 pulsed with 1:10 serial diluted peptides starting from 10−6 M. (D) Both KRAS G12V-reactive TCRs were HLA-A*11:01–restricted. T cells transduced with either TRAV3-3*01/BV4*01 or TRAV19*01/BV13-1*02 were cocultured with KRAS G12V-positive pancreatic tumor lines transduced with HLA-A*11:01 and their parental HLA-A*11:01–negative tumor lines. (E) Both TCRs were KRAS G12V specific. T cells transduced with either TCR were cocultured with a panel of HLA-A*11:01–positive pancreatic tumor lines with or without the KRAS G12V mutation. (F) Correlation between mutated KRAS expression and IFNγ production by T cells transduced with TRAV3-3*01/BV4*01 and tested against a panel of pancreatic tumor lines with or without G12V mutation (R2 = 0.68, P = 0.02). (G) TRAV3-3*01/BV4*01 had CD8 coreceptor–independent reactivity. CD8 or CD4 enrichment was performed on T cells transduced with retrovirus encoding TRAV3-3*01/BV4*01, and then cocultured with COS7/A11 KRAS transfectants and pancreatic tumor lines. From (B) to (G), all functional analysis was done by assessing IFNγ production from the coculture supernatant after overnight incubation. (H) TRAV3-3*01/BV4*01 proliferated upon antigen-specific stimulation. T cells transduced with TRAV3-3*01/BV4*01 were labeled with CFSE, cocultured with various targets for 3 days, and further labeled with antibodies to human CD3 and murine TCRβ, and then analyzed on a FACS Canto II. Data was gated on the live CD3+ population. (I) Antigen-specific degranulation of TRAV3-3*01/BV4*01. T cells transduced with TRAV3-3*01/BV4*01 were cocultured with various targets in the presence of anti-CD107a-FITC for 2 hr, labeled with antibodies to human CD3 and to murine TCRβ, and then analyzed on FACS Canto II. Data was gated on live CD3+CD8+ populations.
A biscistronic retroviral vector was then made for each reactive TCR and their anti-KRAS G12V reactivity was evaluated in anti-CD3–stimulated T cells. These TCR-transduced PBLs specifically recognized COS7/A11 pulsed with G12V7-16 peptide at a concentration of10−9 M, but not control peptides including a wild-type 10-mer (Fig. 3C). The TCR combination TRAV3-3*01/BV4*01 could also confer reactivity to the 9-mer KRAS G12V8-16 peptide pulsed on COS7/A11 at a concentration of 10−11 M, suggesting KRAS G12V8-16 was the minimal determinant for this TCR. Further characterization of these two TCRs confirmed HLA-A11*01 restriction, and KRAS G12V-specificity (Fig. 3D and E). T cells transduced with either TCR only recognized KRAS G12V-positive pancreatic tumors that stably expressed HLA-A*11:01, with no IFNγ secretion on coculture with HLA-A11–negative or KRAS G12V-negative tumor lines. When comparing these two TCRs, TRAV3-3*01/BV4*01 could confer reactivity against multiple HLA-A11–positive and KRAS G12V-expressing tumor lines, whereas only one of five tumor lines (SK.PC3/A11) was recognized by T cells retrovirally-transduced with TRAV19*01/BV13-1*02. Overall, these results showed that TRAV3-3*01/BV4*01 was a higher avidity TCR than TRAV19*01/BV13-1*02, and IFNγ release was significantly correlated with the quantitative KRAS G12V expression of the target cell lines (R2=0.68, P=0.02) (Fig. 3F).
Both CD8- and CD4-enriched T cells expressing TRAV3-3*01/BV4*01 recognized COS7/A11/G12V, and SK.PC3/A11, although the recognition of CD4-enriched T cells was much lower than CD8-enriched T cells (Fig. 3G). T cells transduced with TRAV3-3*01/BV4*01 also had KRAS G12V-specific proliferation and degranulation. TRAV3-3*01/BV4*01–transduced T cells stimulated with target cells that harbor KRAS G12V (COS7/A11/G12V and SK.PC3/A11), but not irrelevant target cells, resulted in dilution of CFSE (Fig. 3H). CD107a degranulation, a surrogate marker for cytolytic activity of T cells, was evaluated in TCR-transduced T cells cocultured with various targets (Fig. 3I). Coculture with SK.PC3/A11 led to 33% of TCR-transduced T cells (identified by mTCRβ staining) becoming CD107a positive. The mTCRβ− cells did not show any CD107a upregulation when cocultured with SK.PC3/A11, implying that degranulation was mediated by this TCR only. TCR-transduced T cells alone, or cocultured with irrelevant target cells (FA6-2/A11), did not show upregulation of CD107a. These data show TRAV3-3*01/BV4*01 also mediates antigen-specific proliferation and lytic activities.
Identification of HLA-A11*01–restricted KRAS G12D-reactive TCR
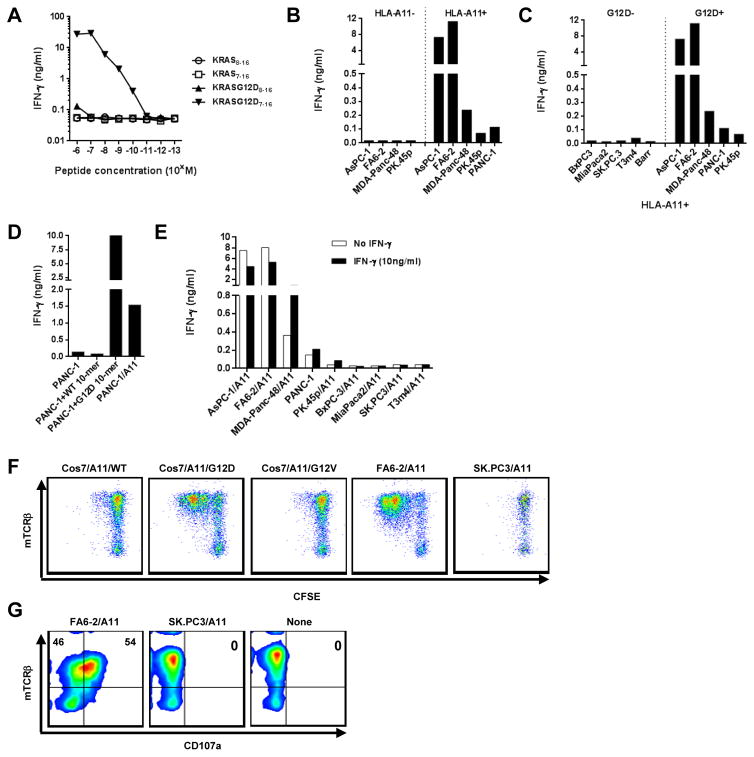
In parallel experiments, two dominant TCR α chains and one β chain were identified by 5′RACE from the splenocytes of mice vaccinated with the KRAS G12D7-16 peptide (Table 2). The two dominant TCR α chains identified share the same V/J regions, but differed slightly in CDR3 regions [henceforth designated as TRAV4-4*01/AJ49*01 (1) and TRAV4-4*01/AJ49*01 (2)]. Using similar approaches to screen for KRAS G12D-reactivity, the pairing of TRAV4-4/AJ49*01 (1) with TRBV12-2*01/BD1*01/BJ1-2*01 (TRAV4-4*01/BV12-2*01) conferred specific reactivity against COS7/A11/G12D (Supplementary Fig. S1A and B). We further identified the 10-mer KRAS7-16 peptide as the minimal determinant, recognized by TCR transduced T cells at a concentration of 10−10 M when pulsed onto COS7/A11 (with no recognition of the 9-mer KRAS G12D8-16 peptide nor 9-mer or 10-mer wild-type peptide; Fig. 4A). Similar to the KRAS G12V-specific TCRs, the recognition by this TCR was of a naturally processed and presented epitope, was HLA-A*11:01–restricted and was KRAS G12D-specific, as shown by its reactivity to a panel of pancreatic tumor lines (Fig. 4B and C). Tumor lines that were HLA-A11 negative, or did not harbor KRAS G12D mutations were not recognized by TRAV4-4*01/BV12-2*01–transduced T cells. Some lines that were HLA-A*11:01 and KRAS G12D were not well recognized. To investigate this, we overexpressed HLA-A*11:01 by transfecting the poorly-recognized HLA-A*11:01 positive line, PANC-1, or simply pulsing that line with exogenous KRAS G12D7-16 peptide, and either intervention alone resulted in brisk recognition, indicating that a combination of low HLA plus low antigen expression was limiting immune recognition of this tumor (Fig. 4D). In addition, when pancreatic tumor lines were pretreated with IFNγ to augment antigen processing and MHC class I expression, modest enhancement of recognition was also observed in the less-recognized tumor lines (Fig. 4E). Furthermore, as with the previous anti-KRAS G12V TCR, recognition of pancreatic tumor lines was highly correlated with KRAS G12D expression by the target tumor lines (comparing IFNγ production versus copy number of KRAS G12D, R = 0.98, P < 0.0001; Supplementary Fig. S1C). These data imply that amounts of either MHC class I or mutated KRAS could be limiting for these TCRs. Similar to anti-KRAS G12V TCR, TRAV4-4*01/BV12-2*01–transduced T cells proliferated only when T cells were stimulated with target cells carrying KRAS G12D mutations (COS7/A11/G12D and FA6-2/A11), but not control targets cells (Fig. 4F). Also, 54% of TRAV4-4*01/BV12-2*01–transduced T cells were CD107a-positive when T cells were co-cultured with FA6-2/A11, compared to 0% when co-cultured with SK.PC3/A11, or T cells alone (Fig. 4G). Taken together, proliferation and CD107a degranulation of anti-KRAS G12D TCR–transduced T cells were highly antigen-specific.
Table 2.
Oligoclonal TCRs identified from murine KRAS G12D-specific splenocytes (1μM) by 5′RACE:
| V Region | D/J Region | CDR3 | Frequency | |
|---|---|---|---|---|
| Alpha chains | TRAV4-4/DV10*01(1) | 49*01 | CAADSSNTGYQNFYF | 30% |
| TRAV4-4/DV10*01 (2) | 49*01 | CAALNTGYQNFYF | 10% | |
|
| ||||
| Beta chains | TRBV12-2*01 | 1*01/1-2*01 | CASSLTDPLDSDYTF | 18% |
Figure 4.
Characteristics of HLA-A*11:01–restricted KRAS G12D-reactive murine TCR, TRAV4-4*01/BV12-2*01. (A) Affinity of the KRAS G12D-reactive TCR. Anti-CD3 stimulated human allogeneic PBL were transduced with retrovirus encoding TRAV4-4*01/BV12-2*01. Three days after transduction, TCR-transduced cells were co-cultured with COS7/A11 pulsed with 1:10 serial diluted peptides. (B) TRAV4-4*01/BV12-2*01 was HLA-A*11:01–restricted. TCR-transduced T cells were cocultured with KRAS G12D-positive pancreatic tumor lines with or without HLA-A*11:01 expression. (C) TRAV4-4*01/BV12-2*01 was KRAS G12D specific. TCR-transduced T cells were cocultured with a panel of HLA-A*11:01 expressing pancreatic tumor lines with or without KRAS G12D mutation. (D) Reactivity of KRAS G12D-specific TCR against PANC-1. TCR-transduced T cells were cocultured with PANC-1, PANC-1 pulsed with 10-mer peptides, or PANC-1 transduced to overexpress HLA-A*11:01. (E) Reactivity of KRAS G12D-specific TCR against IFNγ treated pancreatic tumor lines. Pancreatic tumor lines were pre-treated with IFNγ (10ng/ml) for 48hrs, and then cocultured with TCR-transduced T cells. From (A) to (E), supernatant of cocultures were harvested and IFNγ production was assessed. (F) T cells transduced with TRAV4-4*01/BV12-2*01 proliferated upon antigen-specific stimulation. T cells transduced with TRAV4-4*01/BV12-2*01 were labeled with CFSE, cocultured with various targets. Three days after coculture, T cells were labeled with antibodies to human CD3 and to murine TCRβ, and then analyzed on a FACS Canto II. Data was gated on the live CD3+ population. (G) Antigen-specific degranulation of TRAV4-4*01/BV12-2*01–transduced T cells. T cells transduced with TRAV4-4*01/BV12-2*01 were cocultured with various targets in the presence of anti-CD107a-FITC for 4 hr, labeled with antibodies to human CD3 and to murine TCRβ, and then analyzed on FACS Canto II. Data was gated on live CD3+CD8+ populations.
Treatment efficacy of KRAS G12D-reactive TCR in a xenograft model
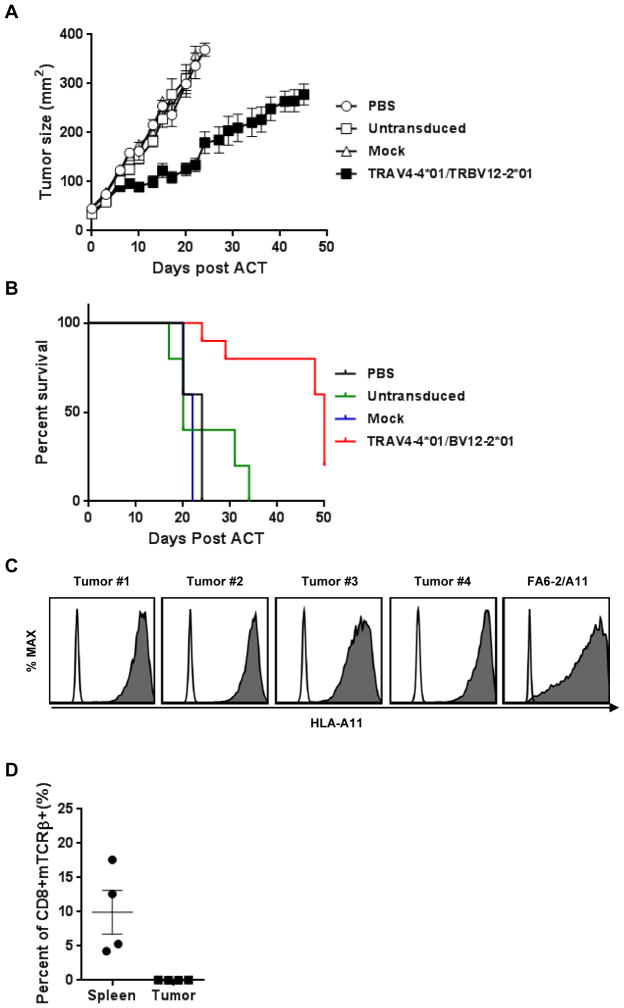
To test the treatment efficacy of a mutated KRAS–reactive TCR, we implanted the HLA-A*11:01–positive, KRAS G12D-positive human pancreatic tumor line, FA6-2/A11, into immunodeficient NSG mice and treated them with human T cells transduced with TRAV4-4*01/BV12-2*01 when tumors became palpable. The growth of tumors in mice treated with TRAV4-4*01/BV12-2*01–transduced T cells was significantly delayed compared to all control groups (compared to treatment with mock-transduced T cells (P = 0.002; Fig. 5A). Although regression of these large established tumors was not achieved, these mice survived significantly longer than control groups (P = 0.001; Fig. 5B). We were unable to identify a tumor line naturally expressing the G12V mutation that would propagate in NSG mice. To study the mechanisms of tumor escape, MHC class I expression of tumors and persistence of T cells in spleens and tumors from treated mice were analyzed. All relapsing tumors had similar expression of HLA-A11 compared to the parental tumor, FA6-2/A11 (Fig. 5C) and were recognized by mutated KRAS-directed TCRs in vitro. Although an average of 10% of transferred T cells, shown as CD8+, murine TCRβ+, could be detected in the spleen, no transferred tumor-reactive T cells could be detected in tumor infiltrating lymphocytes 50 days after cell transfer (Fig. 5D). Our results suggest that one transfer of T-cells engineered with high avidity mutated KRAS-reactive TCRs can significantly affect the growth of tumors carrying the appropriate mutation and prolong survival.
Figure 5.
Adoptive cell transfer of TRAV4-4*01/BV12-2*01-transduced cells to NSG mice. (A) Treatment efficacy of TRAV4-4*01/BV12-2*01. Pancreatic tumor line, FA6-2/A11, was injected into NSG mice subcutaneously, and 10 days after inoculation, 1 × 107 T cells transduced with TRAV4-4*01/BV12-2*01 were injected intravenously, following by daily intraperitoneal IL2 injection for 3 days. Mice given no treatment, untransduced T cells, or mock-transduced T cells, served as controls. Serial tumor measurements were obtained, and tumor area calculated. Control groups had 5 mice and the treatment group10 mice. Center Bar = Mean; Error Bars = SEM. (B) Kaplan-Meier analysis of survival in tumor-bearing mice receiving adoptive transferred T cells transduced with TRAV4-4*01/BV12-2*01 versus controls; (TCR-tranduced T cells versus mock tranduced T cells; P <0.0001). “ACT” represents “adoptive cell transfer”. (C) HLA-A11 expression of tumors from treated mice. Tumors from mice which were treated with TRAV4-4*01/BV12-2*01–transduced cells were labeled with antibody to HLA-A11 and analyzed on FACS CantoII; Open: isotype control, Shaded: HLA-A11. FA6-2/A11 was used as the positive control. (D) Presence of transferred cells in treated mice. Spleens and tumors from 4 mice treated as described above were labeled with antibodies to human CD3, human CD8 and to mouse TCR β and analyzed on FACS CantoII. Data was gated on live CD3+ T cells.
Discussion
In this study, we were able to generate HLA-A*11:01–restricted mouse T cells that recognize naturally processed and presented epitopes in G12D- and G12V-mutated KRAS and clone the TCRs responsible for this reactivity. Our results indicate that either 9-mer or 10-mer peptides could be the minimal determinant of mutated KRAS in the context of HLA-A*11:01. Approximately 25% of all human cancers harbor mutations in KRAS, and for GI cancers, the G12D and G12V mutations are present in the majority of cases. While the frequency of HLA-A*11:01 is approximately 14% in U.S. Caucasians and 23% in Asian-Americans (28), it has been reported to be the most frequent Class I HLA allele in southern Chinese, with frequencies up to 40% (28, 29). For pancreatic adenocarcinoma with a 70% frequency of one of these two KRAS mutations, even a 14% incidence of HLA-A*11:01 means one in ten patients could be eligible for protocols utilizing these two receptors. RAS family proteins share complete amino acid homology at their N-termini, making these anti-KRAS G12V and G12D TCRs potential treatments for other cancers that harbor these RAS mutation variants.
Enormous effort and expense have been expended trying to develop therapies targeting mutated KRAS, with no real success. Naturally occurring T-cell responses to mutated KRAS have been reported for 15 years (21,22). In addition, multiple groups have tested vaccines targeting the most common mutated KRAS variants (30–33). None of the vaccination studies have produced evidence of efficacy and the finding of T-cell responses to mutated KRAS has not provided ways to translate these findings into effective immunotherapies. Our study, however, does demonstrate an efficacious treatment with T cells that recognize mutated KRAS. When retrovirally engineered into donor PBLs, the TCRs we identified can recognize their cognate peptides at concentrations of 10−10 to 10−11 M, recognize antigen and HLA appropriate tumors, and show no recognition of wild-type KRAS epitopes. However, several tumors were not recognized, or only weakly recognized, by these TCRs. Our data suggest that the amount of mutated KRAS was critical for tumor recognition, although in some cases MHC class I expression was also limiting. In contrast, we did not encounter pancreatic tumors with defective antigen processing. It may prove necessary to evaluate MHC and mutated KRAS expression to optimally select patients for T-cell therapy trials.
Treatment with TCR-transduced T cells of a human pancreatic tumor expressing the appropriate HLA allele and the KRAS G12D mutation in an immunodeficient mouse model significantly inhibited tumor growth, but not regression. Species incompatibilities in cytokine, homing, and chemotactic receptors, and in the kinetics of tumor growth, all could affect the ability to directly translate treatment results in this model to patients. The inability to identify persisting T cells in tumor at 50 days may illuminate one of the areas requiring additional investigation. Approaches to enhancing T-cell penetration into solid tumors may be of benefit (34). In patients, the adoptive transfer of native tumor-reactive T cells can cause durable, complete regressions of metastatic melanoma in up to 20% of patients. Results with receptor-engineered T cells have also shown high response rates with durable complete responses when targeting the NY-ESO-1 and the CD19 antigens (15, 16). Coupled with new data supporting the value of tumor-specific neoantigens in responses to a variety of immunotherapies, the opportunity to target mutated KRAS with adoptive T-cell therapy is of great interest and these two receptors alone would pertain to over ten thousand patients dying of cancer every year. These TCRs represent a direct method of generating unlimited T cells against a key driver mutation, and they will be a test of this approach to treating a variety of advanced cancers.
Supplementary Material
Acknowledgments
Financial support: This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The authors thank Drs. Steven A Rosenberg and Paul Robbins for thoughtful discussions. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Conflict of interest: There is no conflict of interest to disclose.
References
- 1.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS medicine. 2005;2(1):e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo T, Masson K, Jaffe JD, Silkworth W, Ross NT, Scherer CA, et al. STK33 kinase inhibitor BRD-8899 has no effect on KRAS-dependent cancer cell viability. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(8):2860–5. doi: 10.1073/pnas.1120589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultz NA, Roslind A, Christensen IJ, Horn T, Hogdall E, Pedersen LN, et al. Frequencies and prognostic role of KRAS and BRAF mutations in patients with localized pancreatic and ampullary adenocarcinomas. Pancreas. 2012;41(5):759–66. doi: 10.1097/MPA.0b013e31823cd9df. [DOI] [PubMed] [Google Scholar]
- 5.Hruban RH, van Mansfeld AD, Offerhaus GJ, van Weering DH, Allison DC, Goodman SN, et al. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. The American journal of pathology. 1993;143(2):545–54. [PMC free article] [PubMed] [Google Scholar]
- 6.Laghi L, Orbetegli O, Bianchi P, Zerbi A, Di Carlo V, Boland CR, et al. Common occurrence of multiple K-RAS mutations in pancreatic cancers with associated precursor lesions and in biliary cancers. Oncogene. 2002;21(27):4301–6. doi: 10.1038/sj.onc.1205533. [DOI] [PubMed] [Google Scholar]
- 7.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. The New England journal of medicine. 2008;359(17):1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 8.Russo AL, Borger DR, Szymonifka J, Ryan DP, Wo JY, Blaszkowsky LS, et al. Mutational analysis and clinical correlation of metastatic colorectal cancer. Cancer. 2014;120(10):1482–90. doi: 10.1002/cncr.28599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coulie PG, Lehmann F, Lethe B, Herman J, Lurquin C, Andrawiss M, et al. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(17):7976–80. doi: 10.1073/pnas.92.17.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. The New England journal of medicine. 2014;371(23):2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(13):4550–7. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344(6184):641–5. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114(3):535–46. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kochenderfer JN, Dudley ME, Feldman SA, Wilson WH, Spaner DE, Maric I, et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood. 2012;119(12):2709–20. doi: 10.1182/blood-2011-10-384388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robbins PF, Kassim SH, Tran TL, Crystal JS, Morgan RA, Feldman SA, et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(5):1019–27. doi: 10.1158/1078-0432.CCR-14-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen CJ, Zheng Z, Bray R, Zhao Y, Sherman LA, Rosenberg SA, et al. Recognition of fresh human tumor by human peripheral blood lymphocytes transduced with a bicistronic retroviral vector encoding a murine anti-p53 TCR. Journal of immunology. 2005;175(9):5799–808. doi: 10.4049/jimmunol.175.9.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkhurst MR, Yang JC, Langan RC, Dudley ME, Nathan DA, Feldman SA, et al. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19(3):620–6. doi: 10.1038/mt.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen CJ, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer research. 2006;66(17):8878–86. doi: 10.1158/0008-5472.CAN-06-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linard B, Bezieau S, Benlalam H, Labarriere N, Guilloux Y, Diez E, et al. A ras-mutated peptide targeted by CTL infiltrating a human melanoma lesion. Journal of immunology. 2002;168(9):4802–8. doi: 10.4049/jimmunol.168.9.4802. [DOI] [PubMed] [Google Scholar]
- 21.Gjertsen MK, Saeterdal I, Saeboe-Larssen S, Gaudernack G. HLA-A3 restricted mutant ras specific cytotoxic T-lymphocytes induced by vaccination with T-helper epitopes. Journal of molecular medicine. 2003;81(1):43–50. doi: 10.1007/s00109-002-0390-y. [DOI] [PubMed] [Google Scholar]
- 22.Kubuschok B, Neumann F, Breit R, Sester M, Schormann C, Wagner C, et al. Naturally occurring T-cell response against mutated p21 ras oncoprotein in pancreatic cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12(4):1365–72. doi: 10.1158/1078-0432.CCR-05-1672. [DOI] [PubMed] [Google Scholar]
- 23.Lang AH, Drexel H, Geller-Rhomberg S, Stark N, Winder T, Geiger K, et al. Optimized allele-specific real-time PCR assays for the detection of common mutations in KRAS and BRAF. The Journal of molecular diagnostics : JMD. 2011;13(1):23–8. doi: 10.1016/j.jmoldx.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang QJ, Hanada K, Robbins PF, Li YF, Yang JC. Distinctive features of the differentiated phenotype and infiltration of tumor-reactive lymphocytes in clear cell renal cell carcinoma. Cancer research. 2012;72(23):6119–29. doi: 10.1158/0008-5472.CAN-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-3. 0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic acids research. 2008;36(Web Server issue):W509–12. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkhurst MR, Joo J, Riley JP, Yu Z, Li Y, Robbins PF, et al. Characterization of genetically modified T-cell receptors that recognize the CEA:691–699 peptide in the context of HLA-A2. 1 on human colorectal cancer cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15(1):169–80. doi: 10.1158/1078-0432.CCR-08-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chinnasamy N, Wargo JA, Yu Z, Rao M, Frankel TL, Riley JP, et al. A TCR targeting the HLA-A*0201-restricted epitope of MAGE-A3 recognizes multiple epitopes of the MAGE-A antigen superfamily in several types of cancer. Journal of immunology. 2011;186(2):685–96. doi: 10.4049/jimmunol.1001775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Middleton D, Menchaca L, Rood H, Komerofsky R. New allele frequency database: http://www.allelefrequencies.net. Tissue antigens. 2003;61(5):403–7. doi: 10.1034/j.1399-0039.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- 29.Shen Y, Cao D, Li Y, Kulski JK, Shi L, Jiang H, et al. Distribution of HLA-A, -B, and -C alleles and HLA/KIR combinations in Han population in China. Journal of immunology research. 2014;2014:565296. doi: 10.1155/2014/565296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toubaji A, Achtar M, Provenzano M, Herrin VE, Behrens R, Hamilton M, et al. Pilot study of mutant ras peptide-based vaccine as an adjuvant treatment in pancreatic and colorectal cancers. Cancer immunology, immunotherapy : CII. 2008;57(9):1413–20. doi: 10.1007/s00262-008-0477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weden S, Klemp M, Gladhaug IP, Moller M, Eriksen JA, Gaudernack G, et al. Long-term follow-up of patients with resected pancreatic cancer following vaccination against mutant K-ras. International journal of cancer Journal international du cancer. 2011;128(5):1120–8. doi: 10.1002/ijc.25449. [DOI] [PubMed] [Google Scholar]
- 32.Chaft JE, Litvak A, Arcila ME, Patel P, D’Angelo SP, Krug LM, et al. Phase II study of the GI-4000 KRAS vaccine after curative therapy in patients with stage I-III lung adenocarcinoma harboring a KRAS G12C, G12D, or G12V mutation. Clinical lung cancer. 2014;15(6):405–10. doi: 10.1016/j.cllc.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Rahma OE, Hamilton JM, Wojtowicz M, Dakheel O, Bernstein S, Liewehr DJ, et al. The immunological and clinical effects of mutated ras peptide vaccine in combination with IL-2, GM-CSF, or both in patients with solid tumors. Journal of translational medicine. 2014;12:55. doi: 10.1186/1479-5876-12-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caruana I, Savoldo B, Hoyos V, Weber G, Liu H, Kim ES, et al. Heparanase promotes tumor infiltration and antitumor activity of CAR-redirected T lymphocytes. Nature medicine. 2015;21(5):524–9. doi: 10.1038/nm.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.