Abstract
Purpose
Specificity protein 1 (SP1) is an oncogenic transcription factor over-expressed in various human malignancies. This study sought to examine SP1 expression in malignant pleural mesotheliomas (MPM), and ascertain the potential efficacy of targeting SP1 in these neoplasms.
Experimental Design
qRT-PCR, immunoblot and immunohistochemistry techniques were used to evaluate SP1 expression in cultured MPM cells and MPM specimens and normal mesothelial cells/pleura. MTS, chemotaxis, soft agar, ß-galactosidase and Apo-BrdU techniques were used to assess proliferation, migration, clonogenicity, senescence and apoptosis in MPM cells following SP1 knockdown, p53 over-expression, or mithramycin treatment. Murine subcutaneous and intraperitoneal (IP) xenograft models were used to examine effects of mithramycin on MPM growth in vivo. Microarray, qRT-PCR, immunoblot and chromatin immunoprecipitation techniques were used to examine gene expression profiles mediated by mithramycin and combined SP1 knockdown/p53 over-expression, and correlate these changes with SP1 and p53 levels within target gene promoters.
Results
MPM cells and tumors exhibited higher SP1 mRNA and protein levels relative to control cells/tissues. SP1 knockdown significantly inhibited proliferation, migration and clonogenicity of MPM cells. Mithramycin depleted SP1 and activated p53, dramatically inhibiting proliferation and clonogenicity of MPM cells. IP mithramycin significantly inhibited growth of subcutaneous MPM xenografts, and completely eradicated mesothelioma carcinomatosis in 75% of mice. Mithramycin modulated genes mediating oncogene signaling, cell cycle regulation, senescence and apoptosis in-vitro and in-vivo. The growth inhibitory effects of mithramycin in MPM cells were recapitulated by combined SP1 knockdown/p53 overexpression.
Conclusions
These findings provide preclinical rationale for phase II evaluation of mithramycin in mesothelioma patients.
Keywords: malignant pleural mesothelioma, epigenetics, SP1, p53, senescence, apoptosis
Introduction
Malignant pleural mesotheliomas (MPM) are highly lethal neoplasms attributable primarily to occupational or environmental exposures to asbestos and other related mineral fibers such as erionite {Bononi, 2015 3215 /id;Carbone, 2011 3220 /id;International Agency for Research on Cancer 3221 /id}. Due to the long latency associated with these neoplasms, the global incidence of MPM continues to increase, despite asbestos being banned in many countries {Bononi, 2015 3215 /id;International Agency for Research on Cancer 3221 /id}. Presently, in the United States the incidence of MPM is approximately 3000 cases annually {Henley, 2013 3213 /id}. Median overall survival of MPM patients undergoing aggressive multimodality therapy ranges from 14-22 months, depending on tumor stage and histology, extent of surgical resection, and response to chemotherapy {Leuzzi, 2015 3216 /id;Opitz, 2015 3217 /id;Shersher, 2013 2615 /id}.
Recent investigative efforts have provided new insights regarding the pathogenesis of MPM {Bononi, 2015 3215 /id}. For example, rare familial MPMs and approximately 65% of sporadic MPM exhibit mutations involving BRCA1 Associated Protein-1 (BAP1), which encodes a nuclear ubiquitin hydrolase with diverse activities including DNA repair and de-ubiquitination of the repressive histone mark H2AK119Ub {Bott, 2011 2623 /id;Nasu, 2015 3134 /id;Testa JR, 2011 2598 /id;Scheuermann, 2010 2593 /id}. Additionally, MPMs exhibit recurrent cytogenetic abnormalities including allelic loss of CDKN2A and p14 ARF {Jean, 2012 2896 /id} and amplification of MYC and PVT1 oncogenes {Riquelme, 2014 2899 /id}. Epigenomic perturbations including aberrant activity of DNA methyltransferases and over-expression of Polycomb-Repressor Complex-2, silence tumor suppressor genes and noncoding RNAs {Kemp, 2011 1750 /id;Kubo, 2011 2897 /id}. To date, clinical efforts to specifically target oncogene signaling or reverse epigenomic derangements in MPM have had limited success {Christoph, 2014 2894 /id}.
Specificity Protein 1 (SP1) is a zinc-finger transcription factor that binds to GC-rich motifs {Blume, 1991 1392 /id}, and mediates diverse physiological processes such as cell cycle regulation, apoptosis and angiogenesis, which are essential for normal embryonic development and tissue differentiation {Li, 2010 2908 /id;Safe, 2014 2883 /id}. SP1 interacts with a variety of transcription factors, histone acetyltransferases and deacetylases, as well as chromatin remodeling complexes to activate or repress transcription in a context dependent manner {Davie, 2008 2909 /id;Li, 2010 2908 /id}. SP1 is over-expressed and contributes to the malignant phenotype of a variety of human cancers by up-regulating genes that enhance proliferation, invasion, and metastasis {Li, 2010 2908 /id;Safe, 2014 2883 /id}, as well as stemness and chemoresistance {Zhang, 2012 2399 /id;Saha, 2014 2942 /id;Chen, 2014 2943 /id;Yang, 2013 2901 /id}. To date the role of SP1 in MPM has not been defined. The present study was undertaken to examine SP1 expression in MPM, and ascertain the potential clinical efficacy of targeting this transcription factor for mesothelioma therapy.
Materials and Methods
Cell lines, tumor samples and in-vitro mithramycin treatments
H28 and H2452 human MPM lines were obtained from American Type Culture Collection, and maintained in RPMI plus Pen/Strep and 10% fetal bovine serum (normal media). LP3 and LP9 normal mesothelial cell lines were obtained from the Coriell Institute for Medical Research (Camden, NJ), and cultured per vendor recommendations. MES1-9 cell lines were established in the Thoracic Epigenetics Laboratory, Thoracic and GI Oncology Branch, NCI from primary or metastatic MPM, and cultured in normal media. Validation of these cell lines, which have been continuously passaged for over three years, was performed by HLA typing as well as immunohistochemical and molecular profiling relative to the primary tumors (Hong et al, manuscript in preparation). All tumor samples were obtained from resected specimens at the NCI in accordance with Institutional Review Board (IRB)-approved protocols. Normal pleura RNA was obtained from ProteoGenex, CA. Mithramycin (MM; Sigma-Aldrich) was prepared as described previously {Zhang, 2012 2399 /id}. For in-vitro experiments, cells were seeded in tissue culture plates. The next day, media was replaced with fresh NM with or without mithramycin (MM). Twenty-four hours later media was removed, cells were washed three times with HBSS, and either harvested immediately or maintained in normal media for analysis at later time points. Cell proliferation, migration and clonogenicity assays are described in Supplementary Methods.
RNA isolation, real-time quantitative reverse transcription PCR, and microarray analysis
RNA isolation, qRT-PCR using comparative Ct method {Aarskog, 2000 3133 /id} and microarray analysis are described in Supplementary Methods.
Immunoblotting
Immunoblotting was done as previously described {Rao, 2011 1680 /id} using antibodies listed in Supplementary Methods. Cells for immunoblot analysis were harvested immediately after 24 hour MM treatment in-vitro, and three days following the last IP MM injection for xenograft experiments.
Immunohistochemistry of primary tumor samples
Murine xenograft experiments
Athymic nude mice were injected in bilateral flanks with 2×106 of MES1 or MES7 cells. After 10-15 days, mice were sorted by tumor size, and randomly assigned to receive either normal saline or MM (1 or 2 mg/kg) intraperitoneally (IP) every Monday, Wednesday and Friday for three weeks. Tumor size and mouse weights were measured twice weekly, and MM doses were held for weight loss >20% of initial body weight. After completion of three weeks of treatment (day 31, 3 days after last MM injection) or when control tumors reached maximum allowable size, mice were euthanized, tumors were excised and processed for additional studies. For the IP tumor model, 3×106 MES1 cells were injected IP into athymic nude mice. Ten days later, mice were randomized to receive saline or MM (1 mg/kg) IP every MWF for three weeks. Approximately one week later, mice were euthanized and laparotomies were performed to assess tumor burden. All animal procedures were approved by the National Cancer Institute Animal Care and Use Committee, and were in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Generation of stable cells expressing shRNA constructs
MES1 and MES7 were transfected with commercially validated short hairpin RNA (shRNA) targeting SP1 or sham sequences (Sigma) according to manufacturer’s instructions (Supplementary Methods). Cell lines were selected with puromycin (Sigma), and expanded after confirmation of knockdown by qRT-PCR and immunoblot techniques.
Chromatin immunoprecipitation experiments
Quantitative chromatin immunoprecipitation (ChIP) assays were conducted as described {Rao, 2011 1680 /id} with minor modifications using either nonspecific immunoglobulin G (IgG) or ChIP-grade antibodies as well as PCR primers listed in Supplementary Methods.
Adenoviral transduction of p53 in SP1 knockdown cells
Both sham control or SP1 knockdown MES1 and MES7 were transduced with high titer Ad-GFP or Ad-GFP-p53 (Vector labs, CA) for 48 hours. Thereafter, the cells were collected for counting, RNA, and protein analysis. For in vivo studies, 2×106 cells were injected subcutaneously into the bilateral flanks of athymic nude mice and tumor volume was measured every four days until the experiment was completed.
Senescence, cell cycle and apoptosis analysis
3×104 MPM cells were seeded in each of four-chamber slides overnight. The following day, cells were treated for 24 hours with MM (0-100nM), and senescence was assessed using the SA-β-gal staining kit from Cell BioLabs. Similar techniques were used to detect senescence in sham control or SP1 knockdown MES1 and MES7 cells 48 hours following transduction with high titer Ad-GFP or Ad-GFP-p53. For cell cycle analysis, 1×106 cells were plated overnight in serum-containing media. The following day, NM was replaced with serum free RPMI for 48 hours. Thereafter cells were exposed to NM with or without MM (0-100nM) for 24 hours. Cells were harvested at indicated time points and processed for cell cycle analysis using flow cytometry techniques {Thenappan, 2011 2918 /id}. Apoptosis was assessed by flow cytometry using reagents and protocols contained in the Apo-BrdU Kit (BD Pharmingen).
Statistical analysis
SD or SEM are indicated by bars on figures and were calculated using GraphPad Prism 6.0. All experiments were conducted with at a minimum of triplicate samples, and all p values were calculated with two-tailed t tests.
Results
SP1 expression in MPM
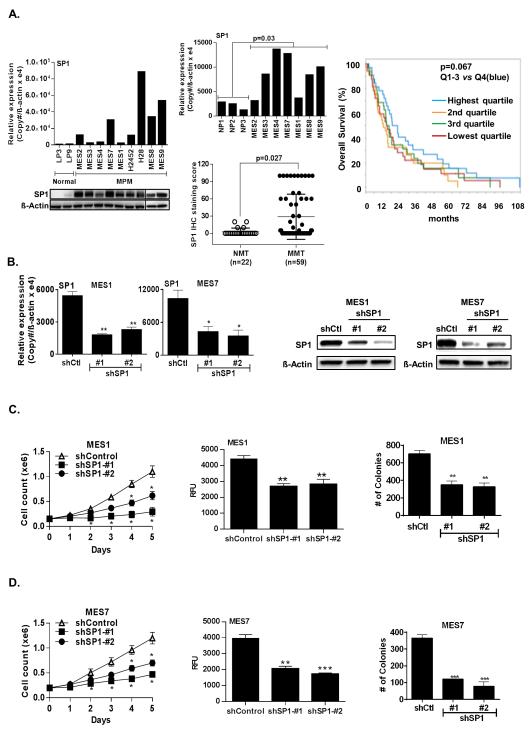
qRT-PCR and immunoblot experiments were performed to examine SP1 expression in cultured MPM cells and tumors relative to cultured normal mesothelial cells or normal pleura. Seven of nine MPM lines exhibited overexpression of SP1 mRNA relative to normal mesothelial cells (LP3 or LP9; Figure 1A; upper panel). Immunoblot experiments demonstrated markedly higher SP1 protein levels in MPM cells relative to normal mesothelial cells (Figure 1A; lower panel). Additional qRT-PCR experiments demonstrated overexpression of SP1 in 5 of 7 primary MPM specimens from which the aforementioned cell lines were derived, although the magnitude of SP1 over-expression in cell lines and primary tumors did not exactly coincide (Figure 1A; middle-upper panel). Immunohistochemistry experiments demonstrated overexpression of SP1 in 18 of 19 primary MPM specimens compared to normal pleura, including seven from which our cell lines were derived (Supplementary Figure S1A). To extend these observations, IHC techniques were used to evaluate SP1 expression in commercial tissue arrays containing 59 primary mesotheliomas and 22 normal mesothelial tissues. A spectrum of SP1 expression was detected, with significantly increased SP1 staining in MPMs compared to normal mesothelia (Figure 1A; middle-lower panel).
Figure 1.
SP1 is overexpressed in malignant pleural mesothelioma cells. (*, P < 0.05; **, P < 0.01; ***, P < 0.001.)
A). qRT-PCR (left, upper panel) and immunoblot (left, lower panel) showing that SP1 mRNA and protein expression levels are higher in cultured MPM cells relative to normal mesothelia; qRT-PCR (middle, upper panel) showing that SP1 mRNA is overexpressed in primary MPM compared to normal pleura; TMA analysis (middle, lower panel) of SP1 expression in normal mesothelial tissues (NMT, n=22) and malignant mesothelial tissues (MMT, n=59). SP1 staining was scored from 0-100 based on percentage of positive cells. The majority of the MMT had significantly increased SP1 staining compared to NMT. Correlation of increased intra-tumoral SP1 expression with overall survival in 39 patients with locally advanced MPM undergoing potentially curative resections (right panel).
B). Confirmation of stable knockdown of SP1 by qRT-PCR and immunoblot analysis in MES1 and MES7 cells (left and right panels, respectively).
C) and D). Time-dependent inhibition of cell proliferation, migration (after 72 hours) and clonogenicity (after 21 days) of SP1 knockdown in MES1 and MES7 cells, respectively. Both shRNA sequences significantly inhibit soft agar clonogenicity of MPM cells.
Additional analysis was undertaken to ascertain if intratumoral SP1 expression detected by Illumina array techniques correlated with survival in 39 patients with locally advanced MPM undergoing potentially curative resections at Brigham and Women’s Hospital. Twenty-four patients had epithelial mesotheliomas, whereas seven patients had biphasic and eight patients had sarcomatoid malignancies. Increased expression of SP1 tended to be associated with shorter survival of MPM patients, although this was not statistically significant (Figure 1A: right panel).
Effects of SP1 depletion in MPM cells
Additional experiments were performed to examine if SP1 expression modulates the malignant phenotype of pleural mesothelioma cells. Briefly, shRNA techniques were used to knockdown SP1 in cultured MPM cells. qRT-PCR and immunoblot experiments demonstrated significant decreases in SP1 expression in MES1 and MES7 cells following transfection with either of two shRNA sequences (shSP1 #1 and #2) relative to controls (Figure 1B; left and right panels). Knockdown of SP1 significantly diminished proliferation, migration, and soft agar clonogenicity of MES1 and MES7 cells (Figure 1C and 1D).
Effect of mithramycin in MPM cells
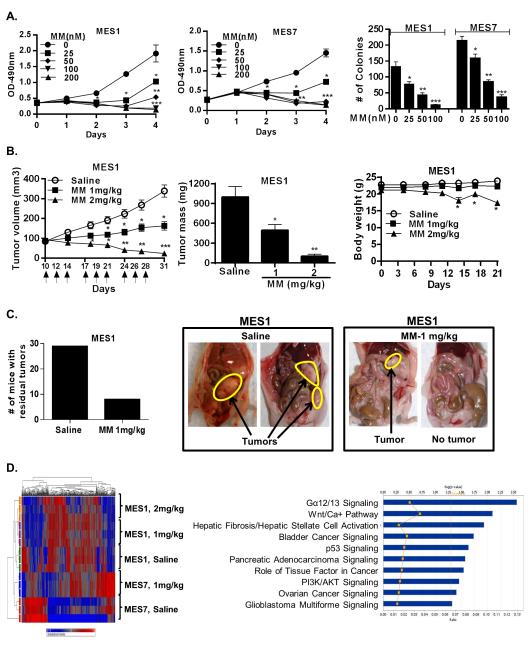
Seeking potential translation of the aforementioned findings to the clinic, additional experiments were performed to examine if mithramycin (MM), an anti-neoplastic agent that inhibits binding of SP1to DNA {Blume, 1991 1392 /id}, could similarly inhibit the malignant phenotype of pleural mesothelioma cells. Briefly, MES1 and MES7 cells were cultured in normal media with or without MM (25-100nM) for 24 hours. qRT-PCR and immunoblot experiments (Supplementary Figure S1B) demonstrated dose dependent decreases in SP1 expression in both cell lines following MM exposure. MTS assays demonstrated that 24 hr MM exposure dramatically inhibited proliferation of MES1 and MES7 cells (Figure 2A; left and middle panels). Cytotoxicity was not evident until 24 h following removal of MM, suggesting delayed effects of drug exposure. Although some recovery of cell growth was observed three to five days following MM exposures of 25nM or 50nM, treatment with doses of 100nM or more resulted in progressive decreases in cell viability over the ensuing four days. Furthermore, 24 hour exposure of MM significantly inhibited soft agar clonogenicity of MES1 and MES7 cells in a dose dependent manner (Figure 2A; right panel).
Figure 2.
Effects of MM in vitro and in vivo in MPM cells. (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
A). Effects of MM on proliferation and soft agar clonogenicity of MES1 and MES7 cells.
B). Effects of IP MM on volume and mass of established subcutaneous MES1 xenografts (left and middle panels, respectively), and body weight in tumor bearing mice (right panel).
C). Effects of IP administration of MM (1mg/kg IP) in nude mice with MES1 induced carcinomatosis. Numbers of mice with residual tumors following MM treatment vs saline injections (left panel). Representative results depicting carcinomatosis in control mice compared to no residual tumor or minimal residual tumors in mice following IP MM (right panel).
D). Heatmap (left panel) and ingenuity pathway analysis (right panel) of genes modulated in MES1 and MES7 xenografts in mice receiving MM (1mg kg).
Additional experiments were performed to examine if MM inhibited growth of MPM cells in-vivo. As shown in Figure 2B (left and middle panels), as well as Supplementary Figure S1C, MM significantly diminished growth of subcutaneous MES1 and MES7 xenografts in a dose dependent manner. The growth inhibitory effects of MM were readily apparent shortly after treatment was initiated, with statistically significant differences in xenograft volumes evident within 7 days of commencing treatment (3 IP injections). The 1 mg/kg dose was well tolerated with no significant weight loss, lethargy or decreased activity; however, the 2 mg/kg dose group experienced moderate or statistically significant weight loss and some visible toxicities (Figure 2B, right panel, and Supplementary Figure S1C). These latter findings were in contrast to our previously published experiments pertaining to IP MM treatment of lung cancer xenografts {Zhang, 2012 2399 /id}, in which 2 mg/kg MM appeared to be well tolerated, possibly because clinical grade MM was used for the lung cancer studies but not the current experiments.
In the second series of experiments, mice with intraperitoneal mesothelioma carcinomatosis were randomly allocated to receive saline or MM (1 mg/kg) IP every Monday, Wednesday and Friday for three weeks, followed by euthanasia three days later. Preliminary experiments were performed to confirm the reproducibility and growth kinetics of the carcinomatosis model, and timing of intervention. Results of three independent experiments totaling 60 mice are summarized in Figure 2C. Whereas 29 of 30 control mice (97%) had extensive carcinomatosis when euthanized, 23 of 30 MM-treated mice (77%) had no evidence of disease (p< 0.02); the remaining seven MM treated mice had minimal residual tumors.
Effects of MM on global gene expression in MPM cells
Affymetrix microarray experiments were performed to examine global gene expression profiles in cultured MES1 and MES7 cells immediately after 24 hour exposure to normal media with or without MM (25 or 100nM), and their corresponding xenografts harvested three days after the last IP saline or MM injection. MM mediated dramatic dose-dependent alterations in gene expression in cultured MPM cells. Using criteria of fold change greater than 2 for drug treatment versus control, 986 genes were commonly modulated in MES1 and MES7 cells following 25 and 100nM MM exposures, respectively (Supplementary Figure S2A); significant overlap was observed at both drug concentrations. Approximately 65% of genes modulated by MM in cultured MPM cells at either drug concentration were repressed by MM. Top canonical pathways affected by MM exposure in-vitro are depicted in Supplementary Figure S2B. Within each cell line, more overlap was observed with cells treated with 100nM rather than 25nM in-vitro and tumor xenografts (Supplementary Figure S2C). Using criteria of fold change equal to or greater than 2 relative to appropriate controls, 27 genes were differentially regulated in cultured MES1 and MES7 cells following exposure to 100nM MM as well as xenografts from mice treated with 1mg/kg MM (Supplementary Table S1). Top canonical pathways (depicted in Supplementary Figure S2D) included p53 signaling, G-alpha 12/13 and Rho signaling, as well as pathways associated with immunologic destruction of cancer cells.
More than 1200 genes and nearly 640 genes were significantly modulated by MM treatment in subcutaneous MES1 and MES7 xenografts, respectively. Over 700 genes were commonly regulated in MES1 cells exposed to MM (100nM) in vitro and MES1 xenografts from 1mg/kg and 2mg/kg treated mice (Supplementary Figure S2C). Similar analysis could not be performed for MES7 due to the limited quantity and poor quality of the RNA from xenografts from 2mg/kg treated mice. Fifty-six genes were commonly regulated in MES1 and MES7 xenografts from mice treated with 1mg/kg MM IP (heat map is depicted in Figure 2D, left panel); 55% of these genes were down-regulated (Table 1). Notably, KIAA1199, which recently has been implicated in modulating metabolism, EGFR and Wnt signaling in cancer cells {Jami, 2014 2930 /id;Evensen, 2013 2929 /id} was markedly repressed in MES1 as well as MES7 xenografts from MM treated mice. Additionally, the oncofetal receptor tyrosine kinase ROR1, which is over-expressed in a variety of human malignancies but to date has not been studied in pleural mesotheliomas, was significantly down-regulated in MPM xenografts from MM treated mice. Top canonical pathways included G-alpha 12/13 signaling, which mediates cytoplasmic beta catenin levels {Meigs, 2001 2944 /id}, Wnt/Ca++ signaling, axonal guidance signaling, which has recently been implicated recently in pluripotency and metastatic potential of cancer cells {Tang, 2014 2875 /id}, p53 signaling, as well as pathways associated with bladder, pancreas, ovarian, and brain cancer (Figure 2D; right panel).
Table 1.
Genes Induced or Repressed by Mithramycin in MES1 as well as MES7 Xenografts.
| Down-regulated | Up-regulated | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Probeset ID | Gene Symbol |
In vivo 1mg/kg |
Probeset ID | Gene Symbol |
In vivo 1mg/kg |
||||
| MES1 | MES7 | MES1 | MES7 | ||||||
| 1 | 212942_s_at | KIAA1199 | −12.6 | −10.5 | 1 | 232165_at | EPPK1 | 9.1 | 5.2 |
| 2 | 209183_s_at | C10orf10 | −5.2 | −6 | 2 | 236646_at | C12orf59 | 2.4 | 3.3 |
| 3 | 202688_at | TNFSF10 | −3.8 | −4.5 | 3 | 1560683_at | BCL8 | 6.6 | 3.3 |
| 4 | 213931_at | ID2 /// ID2B | −6.8 | −3.7 | 4 | 221577_x_at | GDF15 | 5.3 | 3.1 |
| 5 | 227955_s_at | EFNA5 | −2.6 | −3.5 | 5 | 207761_s_at | METTL7A | 4.9 | 3 |
| 6 | 201438_at | COL6A3 | −3.2 | −3.5 | 6 | 228293_at | DEPDC7 | 2.5 | 2.9 |
| 7 | 205199_at | CA9 | −4 | −3.2 | 7 | 235230_at | PLCXD2 | 3.6 | 2.9 |
| 8 | 224771_at | NAV1 | −2.2 | −3.1 | 8 | 237737_at | LOC100289026 | 8.5 | 2.8 |
| 9 | 210601_at | CDH6 | −2.8 | −2.9 | 9 | 224566_at | NEAT1 | 2.2 | 2.7 |
| 10 | 205681_at | BCL2A1 | −3 | −2.8 | 10 | 1556194_a_at | LOC100507455 | 2.3 | 2.7 |
| 11 | 46142_at | LMF1 | −2.2 | −2.7 | 11 | 205543_at | HSPA4L | 2.4 | 2.6 |
| 12 | 232060_at | ROR1 | −3.4 | −2.7 | 12 | 204286_s_at | PMAIP1 | 2 | 2.5 |
| 13 | 216222_s_at | MYO10 | −3.6 | −2.6 | 13 | 202672_s_at | ATF3 | 2 | 2.4 |
| 14 | 1561615_s_at | SLC8A1 | −2.8 | −2.5 | 14 | 234303_s_at | GPR85 | 2.9 | 2.2 |
| 15 | 221029_s_at | WNT5B | −2 | −2.5 | 15 | 215440_s_at | BEX4 | 2.2 | 2.2 |
| 16 | 1554452_a_at | C7orf68 | −3.8 | −2.5 | 16 | 226103_at | NEXN | 2.7 | 2.2 |
| 17 | 200632_s_at | NDRG1 | −8.3 | −2.4 | 17 | 1554020_at | BICD1 | 2.7 | 2.2 |
| 18 | 234994_at | TMEM200A | −3 | −2.3 | 18 | 202704_at | TOB1 | 2 | 2.1 |
| 19 | 203397_s_at | GALNT3 | −4.4 | −2.2 | 19 | 230748_at | SLC16A6 | 2.3 | 2.1 |
| 20 | 202821_s_at | LPP | −2.1 | −2.2 | 20 | 206066_s_at | RAD51C | 2.2 | 2 |
| 21 | 204298_s_at | LOX | −4.8 | −2.1 | 21 | 205386_s_at | MDM2 | 2.9 | 2 |
| 22 | 228570_at | BTBD11 | −2.1 | −2.1 | 22 | 228855_at | NUDT7 | 2.5 | 2 |
| 23 | 213506_at | F2RL1 | −2.1 | −2.1 | 23 | 219179_at | DACT1 | 2 | 2 |
| 24 | 220227_at | CDH4 | −4.6 | −2 | 24 | 209481_at | SNRK | 2 | 2 |
| 25 | 210512_s_at | VEGFA | −2.9 | −2 | 25 | 1557765_at | LOC643401 | 5.3 | 2 |
| 26 | 43544_at | MED16 | −2.1 | −2 | |||||
| 27 | 224646_x_at | H19 | −3.8 | −2 | |||||
| 28 | 242301_at | CBLN2 | −2.1 | −2 | |||||
| 29 | 204348_s_at | AK4 | −5.6 | −2 | |||||
| 30 | 33778_at | TBC1D22A | −8.2 | −2 | |||||
| 31 | 226899_at | UNC5B | −2.8 | −2 | |||||
Because p53 signaling was observed to be a top pathway affected by MM in-vitro and in-vivo, genes from Table 1 and Supplementary Table S1 were screened for SP1and p53 binding sites within their respective promoters using software guided analysis (DECODE: DECipherment Of DNA Elements or UCSC Genome Browser). Approximately 50% of these genes had recognition elements for either SP1, p53, or both within their promoters.
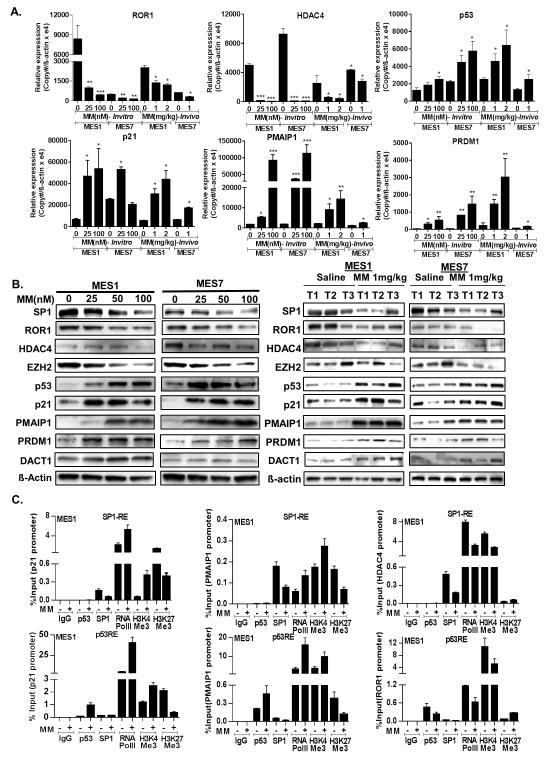
qRT-PCR and immunoblot techniques were used to validate Affymetrix array results using selected genes modulated by MM in-vitro and/or in-vivo. qRT-PCR analysis (Figure 3A) confirmed that MM decreased ROR1 expression in-vitro and in-vivo. Furthermore, although not identified as a commonly regulated gene by microarray analysis, HDAC4- a well-established SP1 target {Liu, 2006 1710 /id}, was repressed by MM in-vitro and in-vivo in MES1 and MES7 cells. MM also decreased expression of SP1 as well as EZH2, which previously has been shown to be an epigenetic driver of malignancy in pleural mesotheliomas {Kemp, 2011 1750 /id} (Supplementary Figure S3A); this phenomenon was not seen in corresponding xenografts possibly due to the timing and intermittent nature of the IP drug treatments and interval from last drug treatment to euthanasia. In addition, MM mediated dose-dependent increases in p53, p21, PMAIP1, and PRDM1 in-vitro and in-vivo (Figure 3A). Subsequent immunoblot experiments (Figure 3B) confirmed results of qRT-PCR experiments.
Figure 3.
Validation of microarray data. (*, P < 0.05; **, P < 0.01; ***, P < 0.001.)
A). qRT-PCR showing the effects of MM on ROR1, HDAC4, p53, p21, PMAIP1 and PRDM1 in MES1 and MES7 cells harvested immediately after 24 hour exposure, as well as corresponding xenografts harvested 3 days following the last IP saline or MM injection.
B) Immunoblot analysis (left panel) showing the dose-dependent effects of MM for 24 hours on SP1, ROR1, HDAC4, EZH2, p53, p21, PMAIP1, PRDM1 protein levels in cultured MPM cells; representative immunoblot analysis (right panel) demonstrating the effects of MM on SP1, ROR1, HDAC4, EZH2, p53, p21, PMAIP1, PRDM1 protein levels in subcutaneous MPM xenografts from MM-treated and control mice. Three tumors (T1, T2 and T3) were selected from saline control vs MM 1mg/kg. Tumors were harvested 3 days following the last IP saline or MM injection.
C). Quantitative ChIP analysis of various gene promoters in MES1 cells cultured in normal media with or without MM (100nM, 24 hours).
MM activates p53 in MPM cells
Following stress or DNA damage, p53 undergoes various post translational modifications such as acetylation or phosphorylation, mechanisms which are known to activate and stabilize this transcription factor {Marouco, 2013 2919 /id}. To examine this issue, MES1 and MES7 cells, which express wt p53 were treated with MM at various doses for 24 hr. Immunoblot experiments demonstrated dose dependent increases in ɤH2AX in MES1 and MES7 cells indicative of DNA damage, which coincided with increases in total p53 as well as acetylated p53-K320, K373, K382 and phosphorylated p53-S15 levels in MES7 cells, and to a lesser extent, MES1 cells following MM exposure (Supplementary Figures S3B, S3C and S3D). Collectively, these data suggest that DNA damage contributes to MM-mediated activation of p53 in MPM cells.
Effects of MM on target gene promoters
Chromatin immunoprecipitation (ChIP) experiments were conducted to further investigate the mechanisms by which MM regulates oncogene and tumor suppressor gene expression in MPM cells. ChIP was performed using primers flanking p53 or SP1 response elements to quantitate occupancy of p53, SP1 or RNA Pol II, as well as levels of H3K4Me3 and H3K27Me3 (euchromatin and heterochromatin histone marks, respectively) within the promoter regions of p21, PMAIP1, HDAC4, ROR1, DACT1, and PRDM1. As shown in Figure 3C and Supplementary Figures S4A and S4B, activation of p53 or repression of SP1 by MM increased p53 and decreased SP1 occupancy, respectively within the p21, PMAIP1 and PRDM1 promoters, which coincided with increased occupancy of RNA Pol II, increased H3K4Me3, and decreased levels of H3K27Me3 in the respective promoters in MES1 and MES7 cells. MM appeared to decrease occupancy of p53 as well as SP1 and RNA Pol II and diminish H3K4Me3 levels, while increasing H3K27Me3 levels within the ROR1 promoter. Furthermore, MM decreased occupancy of SP1 and RNA PolII, coinciding with decreased H3K4Me3, and increased H3K27Me3 within the HDAC4 promoter. Lastly, MM increased occupancy of RNAPolII, and increased H3K4Me3 levels while decreasing H3K27Me3 within the DACT1 promoter region, which contains no prototypic SP1 or p53 recognition elements (Supplementary Figures S4A and S4B), suggesting an indirect mechanism of activation of this tumor suppressor gene by MM.
Effects of SP1 knockdown and p53 over-expression in MPM cells
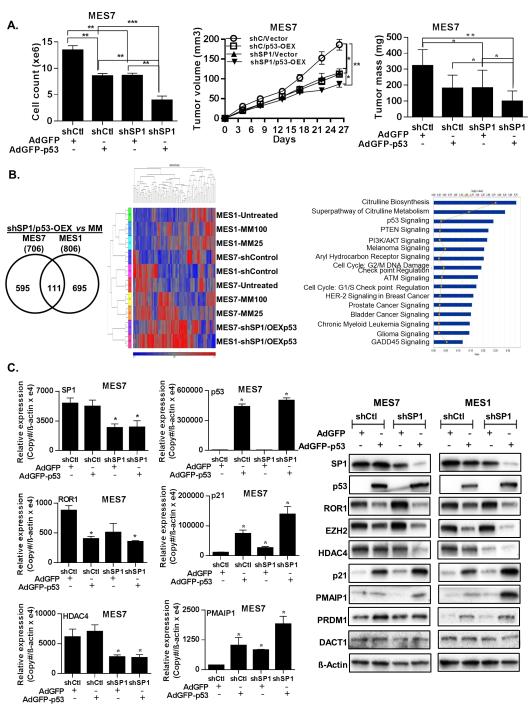
Additional experiments were undertaken to ascertain if SP1 knockdown (shSP1) and p53 over-expression (p53-OEX) could mimic the effects of MM in MPM cells. MES1 and MES7 cells exhibiting either knockdown of SP1, or respective vector controls, were transduced with adenovirus GFP-p53-WT or control adenovirus. Cell count assays for 48 hr post-adenoviral transduction demonstrated a significant additive effect of shSP1 and p53-OEX in MPM cells (representative results pertaining to MES7 cells are depicted in Figure 4A; left panel). Further experiments demonstrated that shSP1 and p53-OEX significantly diminished growth of subcutaneous MPM xenografts (Figure 4A; middle and right panels); the in-vivo growth inhibitory effects of combined shSP1/p53-OEX were more pronounced than either shSP1 or p53-OEX alone in MES7 cells. Similar results were observed in MES1 cells (Supplementary Figure S5A).
Figure 4.
Effects of SP1 knockdown with or without overexpression of p53 in MPM cells. (*, P < 0.05; **, P < 0.01; ***, P < 0.001.)
A). Left panel: SP1 depletion and p53 over-expression significantly inhibits in-vitro proliferation (left panel), and decreases volume and mass of MES7 xenografts (middle and right panels, respectively).
B). Venn diagram (left panel), and corresponding heatmap (middle panel) corresponding to genes regulated by MM and overexpression of p53 in SP1-depleted MES1 and MES7 cells. Ingenuity pathway analysis of genes commonly regulated by in MES1 and/or MES7 cells by MM and SP1KD/p53OEX (right panel).
C). qRT-PCR and immunoblot analysis (left and right panels, respectively) demonstrating the effects of SP1 knockdown or overexpression of p53 on genes targeted by MM in cultured MES1 and MES7 cells.
Gene expression profiles were obtained from additional microarray experiments in MPM cells following combined shRNA mediated knockdown of SP1 and adenoviral mediated overexpression of p53 (shSP1/p53-OEX). As shown in Figure 4B (left panel), 706 and 806 genes were commonly regulated by combined shSP1/p53-OEX in MES1 and MES7 cells, respectively. Additional analysis was performed to compare gene expression profiles in MM-treated and shSP1/p53-OEX MPM cells. The heatmap for this analysis is shown in Figure 4B, middle panel. Fifty-three genes were differentially regulated by MM and shSP1/p53-OEX in either MES1 or MES7 (Supplementary Table S2). Subsequent ingenuity pathway analysis demonstrated that these commonly regulated genes mediated p53 signaling, G1/S, G2/M and DNA checkpoint regulation, ATM and GADD45 signaling, as well as citrulline, glutamine and arginine metabolism (Figure 4B; right panel). Results of this analysis as well as other aforementioned experiments were further validated by qRT-PCR and immunoblot experiments (Figure 4C and Supplementary Figures S5B and S5C). Knock-down of SP1 had no effect on endogenous levels of p53 in MES1 or MES7 cells (Supplementary Figure S5D), but modestly induced expression of p21 (Figure 4C). Over-expression of p53 markedly increased p21 levels in these cells. These findings suggest that up-regulation of p21 in MPM cells by MM occurs via p53 dependent as well as independent mechanisms. Modulation of other targets such as ROR1 and EZH2 appeared to be determined by SP1 depletion as well as activation of p53.
Effects of MM on senescence and apoptosis in MPM cells
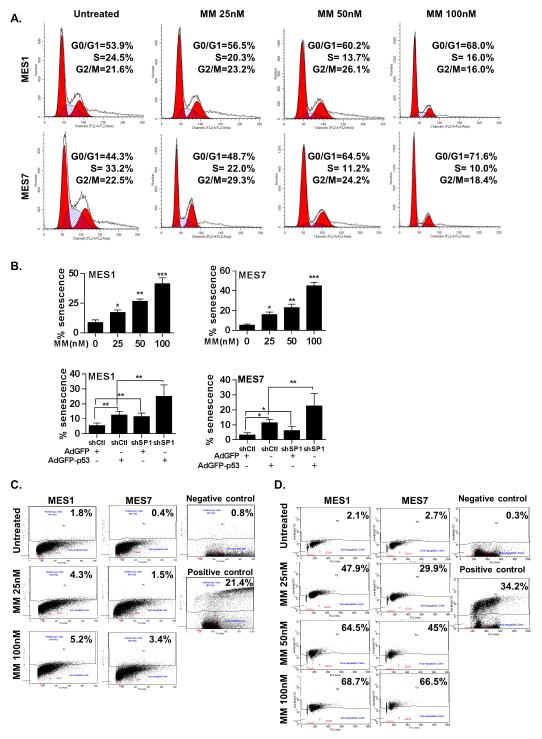
Because MM modulated a variety of genes including p53, p21, PMAIP1, PRDM1 SP1, HDAC4 and EZH2 that either promote or inhibit cell cycle arrest, senescence and apoptosis in cancer cells, additional experiments were performed to further characterize the mechanisms by which MM mediated cytotoxicity in MPM cells. Flow cytometry experiments (Figure 5A) demonstrated dose dependent accumulation of MPM cells in G0/G1 without a sub-G0 fraction consistent with a G0/G1 arrest immediately following 24 hour MM exposure. Histochemistry experiments performed at this time point demonstrated that MM mediated dose-dependent increases in ß-galactosidase expression indicative of senescence in MPM cells; similarly, shSP1 and p53-OEX had additive effects on senescent phenotype in MPM cells (Figure 5B; additional data available upon request). Apo-BrdU experiments demonstrated no significant increase in apoptosis in MPM cells immediately following 24 hour MM treatment (Figure 5C). However, significant dose dependent apoptosis was observed 48 hours following completion of MM treatment in MES1 as well as MES7 cells (Figure 5D). These findings were consistent with results of MTT assays depicted in Figure 2A.
Figure 5.
Effects of MM on cell cycle progression, senescence and apoptosis in MPM cells. (*, P < 0.05; **, P < 0.01; ***, P < 0.001.)
A). Propidium iodide staining demonstrating that 24 hour MM treatment induces dose dependent G0/G1 arrest in MPM cells.
B). ß-galactosidase staining assays demonstrating that 24 hour MM treatment induces dose dependent senescence in MES1 and MES7 cells.
C) and D). Apo-BrdU analysis demonstrating minimal apoptosis in MPM cells immediately following 24 hour MM exposure (C), but significant dose-dependent apoptosis 48 hours following drug treatment (D).
Discussion
Despite being relatively rare, pleural mesotheliomas continue to challenge clinicians due to relentless growth and resistance to conventional treatment modalities as well as novel therapeutics {Bononi, 2015 3215 /id}. As such, there is an urgent need for innovative treatment regimens targeting specific genetic/epigenetic drivers in pleural mesotheliomas, and a more thorough appreciation of drug delivery to these neoplasms in clinical settings.
In the present study, we sought to examine the potential efficacy of targeting SP1 expression in MPM. We observed over-expression of SP1 in the majority of cultured MPM lines and primary pleural mesotheliomas relative to cultured normal mesothelial cells or normal pleura. Knockdown of SP1 inhibited growth, migration and tumorigenicity of MPM cells, strongly suggesting that SP1 functions as an oncogene in MPM. Whereas SP1 over-expression has been associated with decreased survival of patients with lung and esophageal cancers {Sun, 2014 2914 /id;Cao, 2014 2915 /id}, our micro-array experiments did not reveal an association between SP1 mRNA expression and survival in MPM patients, possibly due to the limited number of samples analyzed, as well as potential discrepancies between mRNA and protein levels detected in MPM specimens using various techniques. Additional experiments demonstrated that MM markedly diminished growth of MPM cells in-vitro and in-vivo via induction of DNA damage, cell cycle arrest and senescence with subsequent apoptosis. To the best of our knowledge, these experiments are the first to demonstrate over-expression of SP1 in MPM, and the potential efficacy of MM for mesothelioma therapy.
MM is a naturally occurring polyauroleic acid isolated from Streptomyces, which was originally evaluated as a chemotherapeutic agent in patients with a variety of malignancies during the 1960s and 70s; although complete responses were observed in approximately 10-15% of patients with sarcomas and germ cell tumors, the drug was discontinued because of excessive systemic toxicities that were poorly characterized {CURRERI, 1960 1583 /id;Sewell, 1966 1586 /id}. Recently there has been renewed interest in clinical development of MM and its analogues because of their ability to specifically inhibit binding of SP1 to GC-rich DNA resulting in repression of numerous genes mediating proliferation, invasion, and metastasis of cancer cells {Ohgami, 2010 2917 /id;Zhang, 2012 2399 /id;Yang, 2013 2901 /id;Zhao, 2013 2921 /id;Choi, 2013 2928 /id}. In an ongoing phase II trial at the NCI using drug of higher purity than previously available, MM has been surprisingly well tolerated in cancer patients when administered at the previously recommended dose and schedule (25-30 mcg/kg IV over six hours × 7 days q 4 weeks). Specifically, no nausea, vomiting, bleeding or myelosuppression has been observed in twelve adult patients with various malignancies; however, nine of these individuals developed dose-limiting transaminitis, which resolved spontaneously following cessation of drug. Affymetrix Drug Metabolizing Elimination and Transport (DMET) microarray experiments demonstrated that MM-induced hepatotoxicity correlated with single nucleotide polymorphisms (SNP) in several genes encoding transporter proteins regulating bile flow (Schrump et al, manuscript in preparation). Based on these findings as well as review of PK data from this trial, the protocol has been amended to enroll only those patients with favorable genotypes while intensifying the treatment regimen to recapitulate drug exposure conditions achieved in our preclinical studies.
Although we initially used MM to target SP1 expression, our micro-array and gene manipulation experiments revealed that activation of p53 is a major mechanism by which MM inhibits growth of MPM cells. These findings are consistent with recent studies demonstrating that the anti-angiogenic effects of MM in myeloma cells in-vivo are mediated not by inhibition of SP1 signaling, but rather by activation of p53 {Otjacques, 2013 2916 /id}, and that knockdown of p53 significantly attenuates MM-mediated cytotoxicity in endometrial carcinoma cells {Ohgami, 2010 2917 /id}.
The majority of pleural mesotheliomas retain wt p53 expression and exhibit functional disruption of p53 activity via allelic loss of p14 ARF {Bononi, 2015 3215 /id}; as such, our experiments did not specifically delineate the effects of MM in MPM cells bearing p53 mutations. However, our recent studies have demonstrated dramatic cytotoxic effects of MM irrespective of p53 mutation status in lung and esophageal cancer cells {Zhang, 2012 2399 /id}; these latter observations suggest that p53 activation may not be essential for growth arrest induced by MM in MPM cells. Nevertheless, p53 status may affect extent of cell cycle arrest and propensity for apoptosis in cancer cells following SP1 depletion by mithramycin {Enge, 2009 2923 /id}. Recent elegant studies have demonstrated that highly complex interactions between p53, SP1 and mdm2 regulate cell cycle arrest and apoptosis in cancer cells following exposure to chemotherapeutic agents which activate p53 {Li, 2014 2936 /id}. SP1 is a critical determinant of apoptosis but not cell cycle arrest mediated by p53. Furthermore, although dispensable for induction of pro-apoptotic genes, SP1 is required for pro-apoptotic transcriptional repression by p53 {Li, 2014 2936 /id}. Mdm2, which is up-regulated by p53, facilitates proteosomal degradation of SP1, and negatively regulates p53 {Wade, 2013 2938 /id;Lin, 2010 2939 /id}. Conceivably, early cell cycle arrest/senescence and subsequent apoptosis of MPM cells following MM exposure is attributable, at least in part, to time and dose dependent fluctuations of SP1, p53 and mdm2. The fact that combined SP1 knockdown/p53 over-expression did not fully recapitulate growth inhibition and gene expression profiles observed in MM-treated MPM cells suggests that additional as yet uncharacterized mechanisms contribute to MM-mediated cell cycle arrest, senescence and apoptosis in MPM cells. For example, recently described interactions of MM with core histones could account, in part, for repression of genes lacking prototypic SP1 or p53 recognition elements within their respective promoters {Banerjee, 2014 3218 /id}. Experiments are underway to examine these issues, and to ascertain if small molecule mdm2-p53 inhibitors {Zhang, 2015 3222 /id} or agents which inhibit p21 expression {Nguyen, 2004 1774 /id} can augment MM-mediated apoptosis in MPM cells.
Our studies demonstrated that 24 hour MM treatment was sufficient to mediate progressive decreases in MPM cell viability in-vitro; furthermore, repeated IP administration of MM significantly inhibited growth of subcutaneous MPM xenografts and eradicated established intraperitoneal mesothelioma xenografts in nearly 75% of mice. These findings strongly suggest that the anti-proliferative effects of MM are not primarily due to acute cytotoxicity, but are instead related to transcriptional reprogramming, which persists long after MM exposure. Our micro-array experiments demonstrated considerable overlap between in-vitro and in-vivo MM exposures, which have been helpful regarding delineation of the mechanisms of MM-mediated cytotoxicity in MPM cells. Some of the genes modulated by MM (particularly KIAA1199, ROR1, HDAC4, p21, PMAIP1, and PRDM1) may be relevant pharmacodynamic endpoints for clinical trials evaluating MM in mesothelioma patients.
Although MM was administered to approximately 1500 cancer patients during the 1960’s and early 1970’s, no mesothelioma patients were included in these trials. Furthermore, no pharmacokinetic data were available from any of the initial trials, since methods for analyzing MM levels have only recently been developed {Roth, 2014 3142 /id}. Data presented in this manuscript, as well as our previously published studies {Zhang, 2012 2399 /id} indicate that MM-mediated cytotoxicity occurs in-vitro following 24 hour MM exposure at concentrations > 50nM; MM tissue concentrations in mice with regressing tumor xenografts range from 50-100nM for >24 hr following IP MM injections {Kennedy, 1967 1708 /id}. These exposure conditions are potentially achievable in clinical settings by systemic infusions or regional perfusion techniques. Collectively these observations support evaluation of MM for mesothelioma therapy using precision medicine techniques to identify patients for treatment, and pharmacokinetic modeling to optimize dose and scheduling to maximize clinical efficacy.
Supplementary Material
Clinical Relevance.
Pleural mesotheliomas are highly lethal neoplasms for which there are no effective treatments. Experiments described in this manuscript demonstrate that under exposure conditions potentially achievable in clinical settings, mithramycin depletes SP1 and activates p53 signaling to mediate profound growth arrest associated with senescence and subsequent apoptosis of pleural mesothelioma cells in-vitro and in-vivo. These finding provide the preclinical rationale for evaluation of mithramycin administered systemically or by regional perfusion techniques in mesothelioma patients.
Acknowledgments
The authors express their gratitude to Jan Pappas for assistance regarding manuscript preparation.
Funding Information
This work was supported by NCI Intramural grants ZIA BC 011122 (to D.S. Schrump) and ZIA BC 011418 (to D.S. Schrump).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: M. Rao, S. Atay, V. Shukla, D.S. Schrump.
Development of methodology: M. Rao, S. Atay, V. Shukla, R.T. Ripley, Y. Hong, T. Upham, P. Fetsch, M. Miettenen C.J. Peer, T Sissung, W.D. Figg, D.S. Schrump.
Acquisition of data (provided animals acquired and managed patients, provided facilities, etc.): M. Rao, S. Atay, V. Shukla, Y. Hong, R.T. Ripley, P. Fetsch, M. Miettenen, X. Li, C.J. Peer, T Sissung, W.D. Figg, D.S. Schrump.
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): M. Rao, S. Atay, V. Shukla, Y. Hong, R.T. Ripley, P. Fetsch, M. Miettenen, X. Li, C.J. Peer, T Sissung, W.D. Figg, D.S. Schrump.
Writing, review, and/or revision of the manuscript: M. Rao, S. Atay, V. Shukla, Y. Hong, R.T. Ripley, J.A. Hong, M. Zhang, E. Reardon, P. Fetsch, M. Miettenen, X. Li, D.S. Schrump
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M. Rao, C.J. Peer, T Sissung, W.D. Figg, D.S. Schrump.
Study supervision: M. Rao, D.S. Schrump
References
- (1).Bononi A, Napolitano A, Pass HI, Yang H, Carbone M. Latest developments in our understanding of the pathogenesis of mesothelioma and the design of targeted therapies. Expert Rev Respir Med. 2015;9:633–54. doi: 10.1586/17476348.2015.1081066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Carbone M, Baris YI, Bertino P, Brass B, Comertpay S, Dogan AU, et al. Erionite exposure in North Dakota and Turkish villages with mesothelioma. Proc Natl Acad Sci USA. 2011;108:13618–23. doi: 10.1073/pnas.1105887108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).International Agency for Research on Cancer . IARC monographs on the evaluation of carcinogenic risks of humans: Volume 100C: a review of human carcinogens: arsenic, metals, fibres, and dust. International Agency for Research on Cancer; Lyon: 2011. [Google Scholar]
- (4).Henley SJ, Larson TC, Wu M, Antao VC, Lewis M, Pinheiro GA, et al. Mesothelioma incidence in 50 states and the District of Columbia, United States, 2003-2008. Int J Occup Environ Health. 2013;19:1–10. doi: 10.1179/2049396712Y.0000000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Leuzzi G, Rea F, Spaggiari L, Marulli G, Sperduti I, Alessandrini G, et al. Prognostic score of long-term survival after surgery for malignant pleural mesothelioma: a multicenter analysis. Ann Thorac Surg. 2015;100:890–7. doi: 10.1016/j.athoracsur.2015.04.087. [DOI] [PubMed] [Google Scholar]
- (6).Opitz I, Friess M, Kestenholz P, Schneiter D, Frauenfelder T, Nguyen-Kim DL, et al. A new prognostic score supporting treatment allocation for multimodality therapy for malignant pleural mesothelioma- A review of 12 years' experience. J Thorac Oncol. 2015 Aug 27; doi: 10.1097/JTO.0000000000000661. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- (7).Shersher DD, Liptay MJ. Multimodality treatment of pleural mesothelioma. Surg Oncol Clin N Am. 2013;22:345–55. doi: 10.1016/j.soc.2012.12.004. [DOI] [PubMed] [Google Scholar]
- (8).Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–72. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Nasu M, Emi M, Pastorino S, Tanji M, Powers A, Luk H, et al. High incidence of somatic BAP1 alterations in sporadic malignant mesothelioma. J Thorac Oncol. 2015;10:565–76. doi: 10.1097/JTO.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Testa JR, Cheung M, Pei J, Below JE, Tan Y, Sementino E, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genetics. 2011;43:1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–7. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Jean D, Daubriac J, Le Pimpec-Barthes F, Galateau-Salle F, Jaurand MC. Molecular changes in mesothelioma with an impact on prognosis and treatment. Arch Pathol Lab Med. 2012;136:277–93. doi: 10.5858/arpa.2011-0215-RA. [DOI] [PubMed] [Google Scholar]
- (13).Riquelme E, Suraokar MB, Rodriguez J, Mino B, Lin HY, Rice DC, et al. Frequent coamplification and cooperation between C-MYC and PVT1 oncogenes promote malignant pleural mesothelioma. J Thorac Oncol. 2014;9:998–1007. doi: 10.1097/JTO.0000000000000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kemp CD, Rao M, Xi S, Inchauste S, Mani H, Fetsch P, et al. Polycomb Repressor Complex-2 is a Novel Target for Mesothelioma Therapy. Clin Cancer Res. 2012;18:77–90. doi: 10.1158/1078-0432.CCR-11-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kubo T, Toyooka S, Tsukuda K, Sakaguchi M, Fukazawa T, Soh J, et al. Epigenetic silencing of microRNA-34b/c plays an important role in the pathogenesis of malignant pleural mesothelioma. Clin Cancer Res. 2011;17:4965–74. doi: 10.1158/1078-0432.CCR-10-3040. [DOI] [PubMed] [Google Scholar]
- (16).Christoph DC, Eberhardt WE. Systemic treatment of malignant pleural mesothelioma: new agents in clinical trials raise hope of relevant improvements. Curr Opin Oncol. 2014;26:171–81. doi: 10.1097/CCO.0000000000000053. [DOI] [PubMed] [Google Scholar]
- (17).Blume SW, Snyder RC, Ray R, Thomas S, Koller CA, Miller DM. Mithramycin inhibits SP1 binding and selectively inhibits transcriptional activity of the dihydrofolate reductase gene in vitro and in vivo. J Clin Invest. 1991;88:1613–21. doi: 10.1172/JCI115474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Li L, Davie JR. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann Anat. 2010;192:275–83. doi: 10.1016/j.aanat.2010.07.010. [DOI] [PubMed] [Google Scholar]
- (19).Safe S, Imanirad P, Sreevalsan S, Nair V, Jutooru I. Transcription factor Sp1, also known as specificity protein 1 as a therapeutic target. Expert Opin Ther Targets. 2014;18:759–69. doi: 10.1517/14728222.2014.914173. [DOI] [PubMed] [Google Scholar]
- (20).Davie JR, He S, Li L, Sekhavat A, Espino P, Drobic B, et al. Nuclear organization and chromatin dynamics--Sp1, Sp3 and histone deacetylases. Adv Enzyme Regul. 2008;48:189–208. doi: 10.1016/j.advenzreg.2007.11.016. [DOI] [PubMed] [Google Scholar]
- (21).Zhang M, Mathur A, Zhang Y, Xi S, Atay S, Hong JA, et al. Mithramycin represses basal and cigarette smoke-induced expression of ABCG2 and inhibits stem cell signaling in lung and esophageal cancer cells. Cancer Res. 2012;72:4178–92. doi: 10.1158/0008-5472.CAN-11-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Saha S, Mukherjee S, Mazumdar M, Manna A, Khan P, Adhikary A, et al. Mithramycin A sensitizes therapy-resistant breast cancer stem cells toward genotoxic drug doxorubicin. Transl Res. 2015;165:558–77. doi: 10.1016/j.trsl.2014.10.011. [DOI] [PubMed] [Google Scholar]
- (23).Chen DQ, Huang JY, Feng B, Pan BZ, De W, Wang R, et al. Histone deacetylase 1/Sp1/microRNA-200b signaling accounts for maintenance of cancer stem-like cells in human lung adenocarcinoma. PLoS One. 2014;9:e109578. doi: 10.1371/journal.pone.0109578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yang WJ, Song MJ, Park EY, Lee JJ, Park JH, Park K, et al. Transcription factors Sp1 and Sp3 regulate expression of human ABCG2 gene and chemoresistance phenotype. Mol Cells. 2013;36:368–75. doi: 10.1007/s10059-013-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Aarskog NK, Vedeler CA. Real-time quantitative polymerase chain reaction. A new method that detects both the peripheral myelin protein 22 duplication in Charcot-Marie-Tooth type 1A disease and the peripheral myelin protein 22 deletion in hereditary neuropathy with liability to pressure palsies. Hum Genet. 2000;107:494–8. doi: 10.1007/s004390000399. [DOI] [PubMed] [Google Scholar]
- (26).Rao M, Chinnasamy N, Hong JA, Zhang Y, Zhang M, Xi S, et al. Inhibition of histone lysine methylation enhances cancer-testis antigen expression in lung cancer cells: implications for adoptive immunotherapy of cancer. Cancer Res. 2011;71:4192–204. doi: 10.1158/0008-5472.CAN-10-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Thenappan A, Shukla V, Abdul Khalek FJ, Li Y, Shetty K, Liu P, et al. Loss of transforming growth factor beta adaptor protein beta-2 spectrin leads to delayed liver regeneration in mice. Hepatology. 2011;53:1641–50. doi: 10.1002/hep.24111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Jami MS, Hou J, Liu M, Varney ML, Hassan H, Dong J, et al. Functional proteomic analysis reveals the involvement of KIAA1199 in breast cancer growth, motility and invasiveness. BMC Cancer. 2014;14:194. doi: 10.1186/1471-2407-14-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Evensen NA, Kuscu C, Nguyen HL, Zarrabi K, Dufour A, Kadam P, et al. Unraveling the role of KIAA1199, a novel endoplasmic reticulum protein, in cancer cell migration. J Natl Cancer Inst. 2013;105:1402–16. doi: 10.1093/jnci/djt224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Meigs TE, Fields TA, McKee DD, Casey PJ. Interaction of Galpha 12 and Galpha 13 with the cytoplasmic domain of cadherin provides a mechanism for beta -catenin release. Proc Natl Acad Sci U S A. 2001;98:519–24. doi: 10.1073/pnas.021350998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Tang H, Wei P, Duell EJ, Risch HA, Olson SH, Bueno-de-Mesquita HB, et al. Axonal guidance signaling pathway interacting with smoking in modifying the risk of pancreatic cancer: a gene- and pathway-based interaction analysis of GWAS data. Carcinogenesis. 2014;35:1039–45. doi: 10.1093/carcin/bgu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Liu F, Pore N, Kim M, Voong KR, Dowling M, Maity A, et al. Regulation of histone deacetylase 4 expression by the SP family of transcription factors. Mol Biol Cell. 2006;17:585–97. doi: 10.1091/mbc.E05-08-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Marouco D, Garabadgiu AV, Melino G, Barlev NA. Lysine-specific modifications of p53: a matter of life and death? Oncotarget. 2013;4:1556–71. doi: 10.18632/oncotarget.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Sun Z, Wang L, Eckloff BW, Deng B, Wang Y, Wampfler JA, et al. Conserved recurrent gene mutations correlate with pathway deregulation and clinical outcomes of lung adenocarcinoma in never-smokers. BMC Med Genomics. 2014;7:32. doi: 10.1186/1755-8794-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Cao HH, Zheng CP, Wang SH, Wu JY, Shen JH, Xu XE, et al. A molecular prognostic model predicts esophageal squamous cell carcinoma prognosis. PLoS One. 2014;9:e106007. doi: 10.1371/journal.pone.0106007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Curreri AR, Ansfield FJ. Mithramycin-human toxicology and preliminary therapeutic investigation. Cancer Chemother Rep. 1960;8:18–22. [PubMed] [Google Scholar]
- (37).Sewell IA, Ellis H. A trial of mithramycin in the treatment of advanced malignant disease. Br J Cancer. 1966;20:256–63. doi: 10.1038/bjc.1966.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Ohgami T, Kato K, Kobayashi H, Sonoda K, Inoue T, Yamaguchi S, et al. Low-dose mithramycin exerts its anticancer effect via the p53 signaling pathway and synergizes with nutlin-3 in gynecologic cancers. Cancer Sci. 2010;101:1387–95. doi: 10.1111/j.1349-7006.2010.01543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Zhao Y, Zhang W, Guo Z, Ma F, Wu Y, Bai Y, et al. Inhibition of the transcription factor Sp1 suppresses colon cancer stem cell growth and induces apoptosis in vitro and in nude mouse xenografts. Oncol Rep. 2013;30:1782–92. doi: 10.3892/or.2013.2627. [DOI] [PubMed] [Google Scholar]
- (40).Choi ES, Chung T, Kim JS, Lee H, Kwon KH, Cho NP, et al. Mithramycin A induces apoptosis by regulating the mTOR/Mcl-1/tBid pathway in androgen-independent prostate cancer cells. J Clin Biochem Nutr. 2013;53:89–93. doi: 10.3164/jcbn.13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Otjacques E, Binsfeld M, Rocks N, Blacher S, Vanderkerken K, Noel A, et al. Mithramycin exerts an anti-myeloma effect and displays anti-angiogenic effects through up-regulation of anti-angiogenic factors. PLoS One. 2013;8:e62818. doi: 10.1371/journal.pone.0062818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Enge M, Bao W, Hedstrom E, Jackson SP, Moumen A, Selivanova G. MDM2-dependent downregulation of p21 and hnRNP K provides a switch between apoptosis and growth arrest induced by pharmacologically activated p53. Cancer Cell. 2009;15:171–83. doi: 10.1016/j.ccr.2009.01.019. [DOI] [PubMed] [Google Scholar]
- (43).Li H, Zhang Y, Strose A, Tedesco D, Gurova K, Selivanova G. Integrated high-throughput analysis identifies Sp1 as a crucial determinant of p53-mediated apoptosis. Cell Death Differ. 2014;21:1493–502. doi: 10.1038/cdd.2014.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Lin RK, Wu CY, Chang JW, Juan LJ, Hsu HS, Chen CY, et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 2010;70:5807–17. doi: 10.1158/0008-5472.CAN-09-4161. [DOI] [PubMed] [Google Scholar]
- (46).Banerjee A, Sanyal S, Kulkarni KK, Jana K, Roy S, Das C, et al. Anticancer drug mithramycin interacts with core histones: An additional mode of action of the DNA groove binder. FEBS Open Bio. 2014;4:987–95. doi: 10.1016/j.fob.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Zhang B, Golding BT, Hardcastle IR. Small-molecule MDM2-p53 inhibitors: recent advances. Future Med Chem. 2015;7:631–45. doi: 10.4155/fmc.15.13. [DOI] [PubMed] [Google Scholar]
- (48).Nguyen DM, Schrump WD, Chen GA, Tsai W, Nguyen P, Trepel JB, et al. Abrogation of p21 expression by flavopiridol enhances depsipeptide-mediated apoptosis in malignant pleural mesothelioma cells. Clin Cancer Res. 2004;10:1813–25. doi: 10.1158/1078-0432.ccr-0901-3. [DOI] [PubMed] [Google Scholar]
- (49).Roth J, Peer CJ, Widemann B, Cole DE, Ershler R, Helman L, et al. Quantitative determination of mithramycin in human plasma by a novel, sensitive ultra-HPLC-MS/MS method for clinical pharmacokinetic application. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;970:95–101. doi: 10.1016/j.jchromb.2014.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Kennedy BJ, Sandberg-Wollheim M, Loken M, Yarbro JW. Studies with tritiated mithramycin in C3H mice. Cancer Res. 1967;27:1534–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.