Abstract
The mammalian inner ear consists of the cochlea and the vestibular labyrinth (utricle, saccule, and semicircular canals), which participate in both hearing and balance. Proper development and life-long function of these structures involves a highly complex coordinated system of spatial and temporal gene expression. The characterization of the inner ear transcriptome is likely important for the functional study of auditory and vestibular components, yet, primarily due to tissue unavailability, detailed expression catalogues of the human inner ear remain largely incomplete.
We report here, for the first time, comprehensive transcriptome characterization of the adult human cochlea, ampulla, saccule and utricle of the vestibule obtained from patients without hearing abnormalities. Using RNA-Seq, we measured the expression of >50,000 predicted genes corresponding to approximately 200,000 transcripts, in the adult inner ear and compared it to 32 other human tissues. First, we identified genes preferentially expressed in the inner ear, and unique either to the vestibule or cochlea. Next, we examined expression levels of specific groups of potentially interesting RNAs, such as genes implicated in hearing loss, long non-coding RNAs, pseudogenes and transcripts subject to nonsense mediated decay (NMD). We uncover the spatial specificity of expression of these RNAs in the hearing/balance system, and reveal evidence of tissue specific NMD. Lastly, we investigated the non-syndromic deafness loci to which no gene has been mapped, and narrow the list of potential candidates for each locus.
These data represent the first high-resolution transcriptome catalogue of the adult human inner ear. A comprehensive identification of coding and non-coding RNAs in the inner ear will enable pathways of auditory and vestibular function to be further defined in the study of hearing and balance. Expression data are freely accessible at https://www.tgen.org/home/research/research-divisions/neurogenomics/supplementary-data/inner-ear-transcriptome.aspx
Keywords: Hearing, Balance, cochlea, vestibule, RNA-Seq, transcriptome
1. Introduction
The inner ear consists of auditory and vestibular components that involve a highly complex coordinated system of spatial and temporal gene expression to control the connectivity of neurons and circuits. The vestibular components are further divided into the utricle, saccule and three semicircular ducts each with a terminal enlargement called an osseous ampulla. The large number of genetic mutations and loci that have been implicated in hearing and balance disorders further illustrate structural and functional complexity of this organ. Yet, much of what is known about the genetics of hearing in humans comes from the study of Mendelian disorders. More than 100 loci were identified in familial studies of nonsyndromic hearing loss, with ~30% of the studies failing to identify the specific gene or mutation implicated (http://hereditaryhearingloss.org/). The genetic basis for the more common and complex forms of hearing and balance disorders, such as presbycusis and disequilibrium and falls in the elderly, has been established (Fransen et al., 2014) but remains poorly understood.
Over the past decade, microarray technology has proven to be a useful tool for performing high throughput gene expression analyses of the inner ear. While initially used primarily to characterize the level of expression of mRNAs in the wild type tissue, it was later used to identify complex gene networks, decipher regulatory pathways and characterize response to various stimuli. Numerous studies have profiled the inner ear gene expression using microarray technology, focusing on model organisms such as mouse and zebrafish, both in normal and pathological conditions (Hertzano and Elkon, 2012; Scheffer et al., 2015). In contrast to the wealth of data available for model organisms, very little is known about gene expression in the human inner ear, mainly due to difficulties in obtaining the relevant tissues. In fact the only available genome wide profiling of the transcriptome in human inner ear was published in 1999 and 2002 by the laboratory of Dr. Cynthia Morton, using expressed sequence tags in human fetal cochlear tissue (Resendes et al., 2002; Skvorak et al., 1999). We believe that knowledge of the elementary expression patterns of the hearing/balance system in the functioning human ear will be a valuable tool for gaining systems-level understanding of basic biological processes during normal development, or development of pathological conditions and will allow significant progress in identification of new genes functionally important for human hearing.
Recent advances in whole genome expression analyses, namely RNA expression profiles produced by next-generation sequencing technology (RNA-Seq) allow comprehensive interrogation of transcribed sequences within cells or tissues, using very small amounts of total RNA. Advantages of this approach include an improved sensitivity to detect transcripts with low copy number, a significantly broader dynamic range resulting in enhanced sensitivity for detection of differential expression, detection of non-coding RNA's and isoform specific expression and identification of novel transcripts (Hertzano and Elkon, 2012; Scheffer et al., 2015). RNA expression profiles of tissues that are relevant to a particular trait are recognized as an important aspect of systematic genetic analysis of these traits. In addition, transcriptional data may be used in genetic mapping of functional genetic variants where large intervals that include multiple genes are identified, and often are difficult to narrow down to a specific gene of interest. Expression profiles of the specialized tissues were shown to be instrumental in narrowing the researchers' focus on genes that are functionally relevant to the trait (Vahedi et al., 2010). Last, regulatory non-coding RNAs display tissue-specific expression profiles and contribute to tissue patterning and to the control of different cellular programs, such as cell proliferation, differentiation, migration, or apoptosis. They have recently been implicated in the development of sensory organs and disorders (Conte et al., 2013). A recent paper focused on next generation sequencing of miRNAs in the mouse inner ear (Rudnicki et al., 2014), but a transcriptional inventory of long non-coding RNAs such as long intervening non-coding (lincRNAs) of the inner ear still remains incomplete. Thus, the lack of a well-characterized and in-depth analysis of the transcriptome of the human inner ear remains an impediment to progress in the field of hearing loss.
In this manuscript we describe, for the first time, RNA-Sequencing of the cochlea, ampulla, saccule and utricle of the adult human inner ear derived from fresh surgically resected tissue from four adult donor patients with normal hearing. We compare it to the transcriptome of 32 other human tissues, and focus on classes of RNAs potentially relevant to human hearing, such as specific mRNA isoforms and long non-coding RNAs. These data provide a rich, freely-accessible resource for understanding the inner ear and may prove useful as a resource for prioritization of candidate genes for the study of hearing loss.
2. Material and Methods
2.1 Sample collection
Specimens were collected by trans-labyrinthine and trans-cochlear approaches to tumors of the skull base of three donor patients, where hearing preservation was not an option. The six specimens are as following: Individual 1: Ampulla, Utricle and Cochlea_3; Individual 2: Saccule and Cochlea_1; Individual 3: Cochlea_2. The trans-labyrinthine approach provides access to the structures of the vestibular labyrinth including the ampulla of the semicircular canals and the utricle and saccule. Extension of the inferior dissection of the internal auditory canal provides direct access to the cochlear lumen and easy removal of the cochlear duct. Tissues were removed under microscopic vision and immediately placed in Trizol (TRIzol (Life Technologies, Carlsbad, CA) and RNA was extracted following the manufacturer's instruction. Samples were collected from three patients between the ages of 45 and 60 years old after obtaining informed consent. The experimental protocol and informed consent were approved by the institutional review board of St. Vincent's Medical Center, Los Angeles CA.
2.2 RNA-preparation and RNA-Seq
10 – 50ng input of total RNA extracted from inner ear tissues was used for Ovation Total RNA (NuGEN, San Carlos, CA), followed by sample preparation (NEBNext, New England biolabs, Ipswish, MA). For the 32 other human tissues, total RNA was purchased from Asterand (Detroit, MI). RNA-Seq libraries were prepared for sequencing using Illumina's TruSeq mRNA Sample Preparation Kit (v2) using 0.1-1ug total RNA input, following the manufacturer's protocol (Illumina Inc, San Diego, USA). All libraries were unstranded and rRNA depletion was not done. Sequencing was done by 104bp paired-end sequencing on a HiSeq 2000 instrument (Illumina Inc, San Diego, USA), or 50bp paired-end sequencing for the non-inner ear tissues. Inner ear samples were not barcoded and each ran on a single lane (18pM). Other tissue samples were indexed and 4 samples were pooled on a flow cell, 20-22pM. PhiX was spiked in on each lane (at the same pM).
2.3 RNA-Seq analysis and reference data
Analysis was performed in parallel by members of two groups (I.S. and Y.H.). Quality control (QC) of the unaligned data were done using FastQC (Andrews et al.), and based on mean quality by cycle and the base frequency report (to detect skewing), data were trimmed.
Inner ear samples were aligned with RNAstar v2.3.0.1 after trimming to 75bp based on QC (Dobin et al., 2013). A STAR reference was built using human genome annotation from ENCODE (Bernstein et al., 2012) with –sjdbGTFfile option, and setting – sjdbOverhang to 74bp (as recommended for 75bp reads). STAR alignment was performed setting the maximum number of mismatches to 5, with --outSAMstrandField intronMotif and --outFilterIntronMotifs RemoveNoncanonicalUnannotated options, as recommended for subsequent analysis by Cufflinks (Pollier et al., 2013). Next, cufflinks was used to assemble the aligned reads into transcripts and to estimate their abundances. Fragments per kilobase of exon per million reads mapped (FPKM) values were calculated as described (Pollier et al., 2013). In a rough scale, 1 FPKM corresponds to weak expression, 10 FPKM to moderate expression and 100 FPKM to high expression (Flegel et al., 2013). Custom scripts in R or ggplot2 were used to visualize output.
To calculate differential expression within inner ear samples and between inner ear samples and other tissues, data was analyzed using the bcbio-nextgen toolkit (v0.8.8) (Guimera, 2012). Data were trimmed to 80 and 50 bp respectively based on QC before entering the bcbio pipeline. In short, reads were trimmed further with cutadapt (Martin, 2011), aligned to the reference genome GRCh37 using RNAstar 2.3.0.1 (Dobin et al., 2013), and gene and exon read counts were generated using HTSeq (Anders et al., 2014). Normalization, differential expression analysis and correction for multiple testing were done with DESeq2 (Love et al., 2014) and PoissonSeq (Li et al., 2012) for gene expression and DEXSeq for differential exon usage (Anders et al., 2012).
Human inner ear tissues (6×) were compared to other human tissues (32×), to detect genes preferentially expressed in the inner ear. Only poly-adenylated transcripts (mRNA) were included because the sample preparation of the 32 human non inner ear tissues included a poly-A selection. Histone-coding genes were removed as well as a subset of poly(A)- histone mRNAs exist (Yang et al., 2011). NormalizationFactors (NF) were calculated instead of SizeFactors to allow gene-specific normalization and to account for differences in library preparation (Love et al., 2014). Next, inner ear cochlea (3×) was compared against vestibular tissues (3×). rRNA, tRNA contaminants were removed and short non-coding RNAs were removed as well since they were not captured well by the sample preparation method. Normalized counts were plotted as regularized logarithm (rLog) (Love et al., 2014). Custom scripts in R or ggplot2 were used to visualize output.
A further QC of the data was done using Cummerbund (Trapnell et al., 2012). Original ENCODE annotation was used to identify biological gene type (Bernstein et al., 2012).
Last, a comparison of count expression data of the human cochlea (N= 3) to microarray data (SurePrint G3 Mouse Exon Microarrays (Agilent Technologies)) from cochlear RNA extracted from 6-week old C57BL/6 mice was done (N=12) (Yoshimura et al 2014), by a spearman correlation of normalized count values of the RNA-seq data to the average expression level of the 12 cochlear samples of as analyzed by Yoshimura et al, 2014.
Nonsyndromic deafness associated loci and genes were downloaded from http://hereditaryhearingloss.org. For both loci with an unknown causative gene and genes implicated in nonsyndromic deafness, genomic coordinates for GRCh37 (hg19) were extracted from OMIM.
3. Results
Recent high throughput RNA-Seq studies identified tens of thousands of expressed regions in various human tissues and cell types. We used the combined annotation of >50,000 predicted protein coding and non-coding genes (~ 200,000 transcripts) available from these studies to investigate gene expression in distinct anatomical parts of the human inner ear. Specifically we characterized gene expression in three cochlea samples, one saccule, one utricle, and one ampulla derived from fresh surgically resected tissues. We found that the dynamic range of gene expression in the different parts of the ear ranges nine orders of magnitude, which is comparable to other human tissues profiled with RNA-Seq. The expression of gene biotype per sample is summarized in Figure S1 in Additional file 1.
3.1 Genes and transcript types expressed in the inner ear
3.1.1 Differential expression between the vestibule and the cochlea
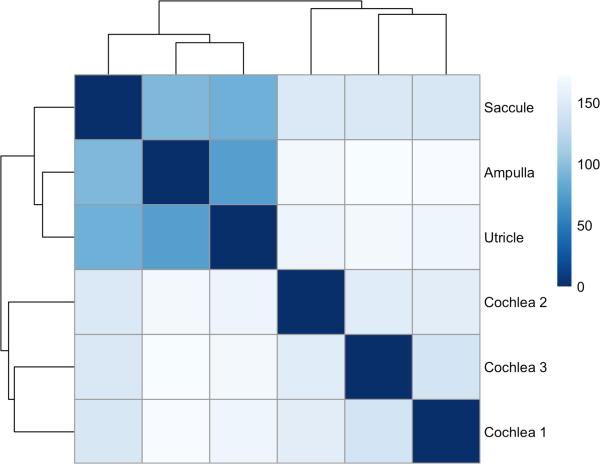
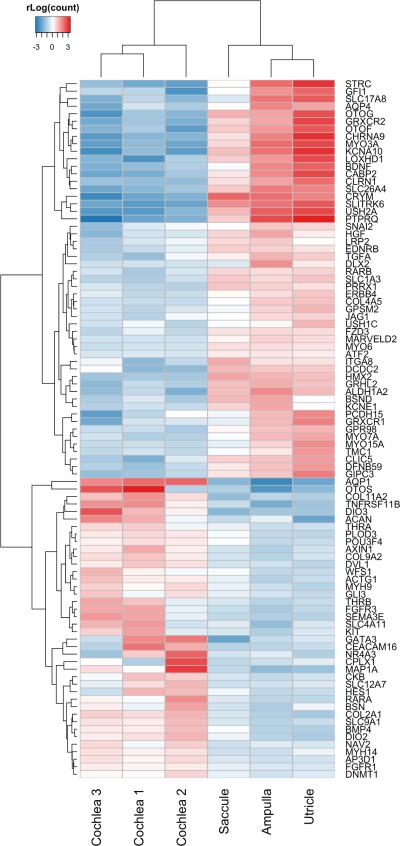
Spatial expression patterns in human tissues represent the diversity of cell types in tissue compartments and are important in revealing pathways directly related to the function of this sub-tissue in human sensory function. To explore spatial expression differences between the vestibule and cochlea, we compared expression levels of genes in the cochlea to those in the vestibule (consisting of ampulla, utricle and saccule samples), and found a relatively high correlation between these tissues (Figure S2 in Additional file 1). However, exon expression is less clearly correlated between both tissues (Figure S3 in Additional file 1), especially for less abundantly expressed exons. This is might be due to sampling noise, which has a stronger impact on low expression levels. We also show that the vestibule is transcriptionally distinct from the cochlea, and that the utricle and saccule are more similar to each other than the ampulla (Figure 1 and Figure S4 in Additional file 1). Last, we investigated differentially expressed genes between the cochlea and the vestibule. We found that >3500 genes are differentially expressed between both compartments (padj < 0.05 (corrected for multiple testing); Table S8 in Additional file 4). 93 of these are known hearing/balance genes (Figure 2) (Brownstein et al., 2011).
Figure 1.
Heatmap showing the Euclidean distances between the 6 human tissues studied by RNA-Seq including 3 cochlear and 3 vestibular samples (ampulla, saccule and utricle). Cochlear and vestibular tissue transcriptional environment can be clearly distinguished from each other. Regularized log transformation of counts generated in HTSeq were used to create this plot.
Figure 2.
Differentially expressed known hearing/balance disorder genes between the cochlea and vestibular system of the inner ear (padj < 0.05). Ward's minimum variance method was used for hierarchical clustering. This plot shows Regularized log transformation of counts generated in HTSeq.
3.1.2 Long non-coding RNAs in the inner ear
Long non-coding RNAs play an important role in regulating multiple aspects of gene transcription, and often show a spatial and temporal expression pattern. We identified several long non-coding RNAs in the inner ear. Most abundant are lincRNAs. Over 7000 lincRNAs were investigated (Table S1 in Additional file 2) and 253 are differentially expressed in the inner ear (Table S8 in Additional file 4).
3.1.3 Pseudogenes, nonsense mediated decay and putative gene expression
Several recent studies suggested genome wide expression of pseudogenes, with potentially functional consequences (Kalyana-Sundaram et al., 2012). Pseudogene transcripts can provide a novel tier of gene regulation through generation of endogenous siRNAs or miRNA-binding sites (Kalyana-Sundaram et al., 2012). We looked at expression of 12,231 pseudogenes annotated in the human genome by GENCODE V19, of which 156 were expressed in at least 1 sample, and 153 were expressed in all inner ear samples. Average expression of pseudogenes was comparable to other biotypes (Figure S1 in additional file 1). In terms of mapping pseudogenes had an average coverage >40,000 reads per pseudogene (in all inner ear samples combined), with >60% of the reads mapped uniquely to the pseudogene (Figure S5 in Additional file 1, Table S2 in Additional file 2). Of these, 50% (77) pseudogenes showed >10 fold difference between average expression in cochlea to average expression in the other inner ear samples, suggesting potential specificity and regulation of their expression (Figure S6 in Additional file 1).
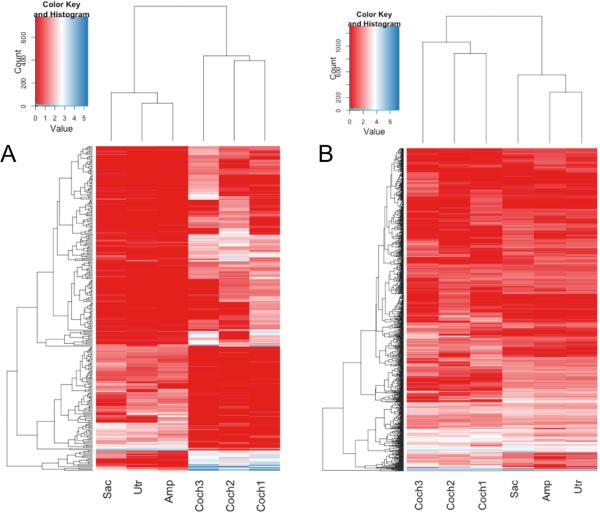
Another potential interesting group of RNA's are transcripts that are predicted to go through nonsense mediated decay (NMD), but escape this fate for some reason. Out of 12,940 transcripts predicted to go through NMD by GENCODE V19 annotation, 2,089 are expressed (FPKM>1) in at least one of the inner ear tissues, and 360 are expressed in all six samples. Interestingly, 285 NMD transcripts are >10 fold over or under expressed in cochlea, relative to the other inner ear tissues, implying potential tissue specificity of either NMD escape mechanisms or transcript expression (Figure 3a). However another 1,133 non NMD transcripts of the same genes that do not show any cochlear specificity (Figure 3b), further supporting potential tissue specificity of NMD degradation rather than expression regulation.
Figure 3.
Nonsense mediated decay may be regulated in a tissue specific manner. A. Transcripts annotated as “nonsense mediated decay” that show >10 fold difference in expression between cochlea and other samples. B. Heat map of 1,133 transcripts that correspond to the same genes as transcripts in A, but are not annotated as “nonsense mediated decay”. All annotations are based on GENCODE V19 annotation of the human genome. Count and Value on the key scale reflect the count of features fitting that value (bright blue line in the key), and the color code of the expression value used in the heatmap, respectively. Plot is based on FPKM values.
3.1.4 Human and mouse cochlea
To correlate the expression of genes in the human and mouse cochlea, we compared expression of genes in the human cochlea (N=3) analyzed in our study to microarray data (SurePrint G3 Mouse Exon Microarrays (Agilent Technologies)) from cochlear RNA extracted from 6-week old C57BL/6 mice (N=12) (Yoshimura et al., 2014) (Figure S7 in Additional file 1). When comparing all genes investigated in both studies we calculated a Spearman's correlation of 0.52. Genes that correlate less well between mouse and human cochlea (defined as the top 5% of differentially expressed genes) include genes important in signal transduction (29%), metabolism (28%) and the immune system (19%), and might represent more fundamental differences between human and mouse physiology rather than specific differences in hearing mechanisms.
3.2 mRNA Transcripts preferentially expressed in the inner ear
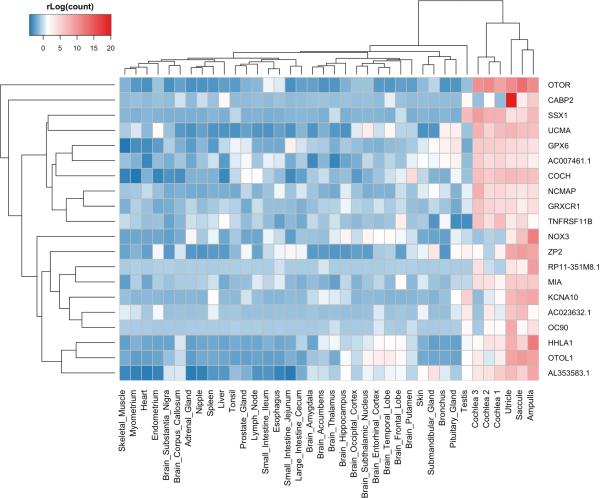
To investigate transcripts that are preferentially expressed in the inner ear, we compared inner ear expression patterns to the expression profile of 32 other human tissues (Figure 4). When correcting for multiple testing, we identified 420 protein coding genes that reach genome wide significance with both DESeq2 and PoissonSeq analysis that are expressed at a higher level in the inner ear compared to other tissues (Figure 4, Table S2 in Additional file 4). 34 (7.98%) of these genes are known hearing/balance disorder genes (Figure S8 in Additional file 1). This is a significant enrichment (8.1%) compared the ratio of deafness genes in all genes investigated (1.5%; p < 1 × 10−4).
Figure 4.
Genome wide significant protein coding genes expressed significantly higher in the inner ear compared to 32 other human tissues (padj < 0.05). The 20 genes with the highest difference in expression level between the inner ear and other tissues (DESeq2), and with a mean normalized count of at least 5 are shown here. Ward's minimum variance method was used for hierarchical clustering. This plot shows Regularized log transformation of counts generated in HTSeq.
Differential exon usage between inner ear and other tissues was computed as well to identify differential isoform expression between tissues (Table S11 in Additional file 1). Figure S9 in additional file 1 shows the differential exon usage of the MITF gene in the cochlea, vestibule and other human tissues included in this study.
3.3. Inner ear expression of genes implicated in hearing loss
The database of hereditary hearing loss (http://hereditaryhearingloss.org/) lists 146 autosomal non-syndromic loci associated with hearing loss that were identified in familial linkage studies. However, some of these loci overlap, do not have exact genomic location, were merged with other loci, or identify the same gene as another locus. After removing redundancies, we examined 100 distinct loci (Additional file 3). For 70 loci, the causative gene had been identified; while for 30 other loci it remains unknown. First, we focused specifically on expression of the hearing loss related genes in our data, and show that 57 (>80%) are expressed in at least one of the examined inner ear tissues (Additional file 3 and Figure S10 in Additional file 1).
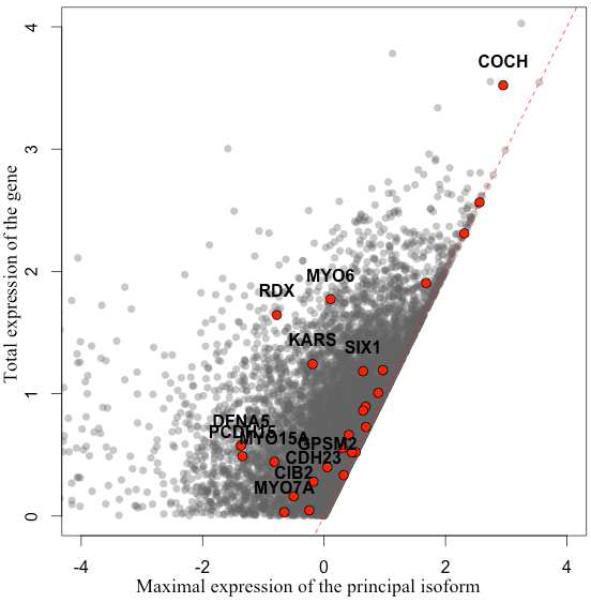
An additional advantage of RNA-Seq data is that one can quantify expression of different isoforms of the same gene. Based on GENCODE V19 annotation, at least 20,878 human genes have more than one annotated transcript, and ~5,000 have more than 10. However, in most biological conditions, only one or few isoforms typically account for the majority of expression of the gene, and these transcripts are annotated as “principal” in the same annotation source. Nevertheless, transcript prevalence can be tissue specific, therefore the principal transcript in other tissues will not necessarily represent the principal transcript in the inner ear. Of 57 hearing loss related genes, 38 have multiple annotated transcripts, with at least one of them designated as principal, and at least one transcript expressed in our sample (FPKM>1). In total we examined 67 principal transcripts and only 22 of them are indeed the most highly expressed isoform of the relevant genes in the inner ear. Moreover, for 13 out of 38 genes, the principal transcript expression accounted for less than half of the total expression of the gene (Figure 5).
Figure 5.
Expression of hearing loss related genes in the inner ear. Expression of principal versus total gene expression, for hearing loss related genes (red) and all expressed genes (grey). Hearing loss genes, for which the principle isoform accounts for less than 50% of the total expression are named. Plot is based on FPKM values.
Twenty-nine of the autosomal loci implicated in hereditary hearing loss do not have an identified causative mutation or specific gene. Notably, these are large genomic intervals that range from 2 to 35Mb in size, encompassing up to hundreds of genes. The size and number of genes in a locus significantly hamper progress in identification of the relevant variants. Therefore, in hope of narrowing the list of potential candidates, we examined the expression pattern in inner ear samples for these 29 loci, identifying which of the annotated genes are expressed (Table 1). While we were not able to identify single candidates for any of the loci, we could on average reduce the list of potential candidates by 75%. For example, locus DFNA43 harbors 74 known genes, but only 11 are expressed in any of our samples (Figure S11 in Additional file 1). However, this filtering is based on our expression cutoff (FPKM>1) and the data we present are only valid in a very limited number of samples, differing by many other biological parameters - such as age, or environment. Therefore this expression data can only be considered in conjunction with other evidence, such as genetic mapping, functional studies and model sytems.
Table 1.
Non-syndromic hearing loss loci (dominant DFNA, recessive DFNB and X-linked DFNX) for which no causative gene has been identified. We present here the number of genes expressed in the inner ear in each locus (i.e FPKM>1 in at least 1 sample).
| Locus Name | Chr | Locus start | Locus End | Locus size (Mb) | N annotated genes in locus | N expressed genes (FPKM >1) |
|---|---|---|---|---|---|---|
| DFNB96 | 1 | 5400000 | 20400000 | 15.0 | 386 | 119 |
| DFNA7 | 1 | 142600000 | 165500000 | 22.9 | 852 | 282 |
| DFNB45 | 1 | 236600000 | 249250621 | 12.7 | 232 | 45 |
| DFNB47 | 2 | 7100000 | 16700000 | 9.6 | 138 | 35 |
| DFNA43 | 2 | 75000000 | 83300000 | 8.3 | 74 | 11 |
| DFNA16 | 2 | 148700000 | 169700000 | 21.0 | 200 | 64 |
| DFNA18 | 3 | 129200000 | 138700000 | 9.5 | 150 | 49 |
| DFNA27 | 4 | 52700000 | 66600000 | 13.9 | 155 | 38 |
| DFNA52 | 4 | 123800000 | 139500000 | 15.7 | 127 | 23 |
| DFNA24 | 4 | 183200000 | 191154296 | 8.0 | 153 | 37 |
| DFNA21 | 6 | 11600000 | 25200000 | 13.6 | 162 | 53 |
| DFNB38 | 6 | 161000000 | 171115067 | 10.1 | 128 | 25 |
| DFNB44 | 7 | 37200000 | 72200000 | 35.0 | 571 | 108 |
| DFNB14 | 7 | 107400000 | 127100000 | 19.7 | 189 | 49 |
| DFNB13 | 7 | 138200000 | 159138663 | 20.9 | 442 | 122 |
| DFNB71 | 8 | 12700000 | 23300000 | 10.6 | 159 | 55 |
| DFNA47 | 9 | 14200000 | 33200000 | 19.0 | 210 | 52 |
| DFNA59 | 11 | 26100000 | 63400000 | 37.3 | 783 | 193 |
| DFNB51 | 11 | 31000000 | 43500000 | 12.5 | 136 | 44 |
| DFNB20 | 11 | 130800000 | 135006516 | 4.2 | 47 | 12 |
| DFNB62 | 12 | 10100000 | 27800000 | 17.7 | 281 | 97 |
| DFNA53 | 14 | 19100000 | 33300000 | 14.2 | 519 | 122 |
| DFNB85 | 17 | 10700000 | 31800000 | 21.1 | 639 | 174 |
| DFNB46 | 18 | 0 | 7100000 | 7.1 | 132 | 33 |
| DFNB68 | 19 | 6900000 | 13900000 | 7.0 | 349 | 126 |
| DFNB65 | 20 | 49800000 | 63025520 | 13.2 | 249 | 61 |
| DFNB40 | 22 | 17900000 | 29600000 | 11.7 | 490 | 100 |
| DFNX3 | X | 29300000 | 61500000 | 32.2 | 23 | 5 |
| DFNX5 | X | 108700000 | 147100000 | 38.4 | 537 | 95 |
4. Discussion
The aim of this study was to build a comprehensive catalogue of the adult human inner ear transcriptome, distributed across 4 individual regions, including the cochlea and 3 different parts of the vestibule: the saccule, utricle and ampulla. Each region in the inner ear has a unique and distinguishable transcriptional microenvironment (Figure 1; Figure S4 in Additional file 1). We found that gene expression between the cochlea and vestibule correlates relatively well, though exon usage is less correlated. This might be due to sampling error affecting less abundant expressed genes, or this might also suggest that isoform specific expression is important in defining specific regulatory pathways important in the shaping and functioning of the hearing or balance system.
To identify genes important in these unique local environments, we looked at differentially expressed genes in the different compartments of the inner ear, and found >3500 genes differentially expressed between the cochlea and vestibule. 83 of these are known hearing/balance disorder genes (Figure 2). For example, MYO6, USH1C and CRYM genes are expressed at a higher level in the vestibule. MYO6 and USH1C are shown to be associated with vestibular defects in human and/or mice in addition to deafness (Avraham et al., 1995; Verpy et al., 2000) and CRYM was previously shown to be highly expressed in the vestibule (Abe et al., 2003), confirming these results. In addition, OTOG is expressed at a higher level in vestibular tissue compared to cochlea, replicating previous expression studies in the mouse (El-Amraoui et al., 2001). OTOG mice present deafness and severe imbalance (Simmler et al., 2000). In contrast, AQP1, COL11A2, OTOS, and many others are expressed much higher in the cochlea than in the vestibule (Figure 2). In addition, we identified several genes and exons unique to the cochlea and to vestibular tissue (Additional file 4). These and all other differentially expressed genes in the inner ear shape the unique transcriptional sub-environments in the inner ear.
Subsequently, we also performed RNA-Seq on 32 other human tissues to identify transcripts preferentially expressed in the inner ear (Additional file 4). Figure 4 shows the 20 genes with the highest difference in expression level between the inner ear and other tissues. This includes known human deafness genes COCH, GRXCR1 and CABP2 (Brownstein et al., 2011; Schrauwen et al., 2012), in addition to genes involved in hearing/vestibular dysfunction in mice: OC90, NOX3, KCNA10 and TNFRSF11B (Lee et al., 2013; Paffenholz et al., 2004; Zehnder et al., 2006; Zhao et al., 2008). NOX3 has recently also been identified as a risk factor for susceptibility to noise-induced hearing loss (Lavinsky et al., 2015). The gene that shows the highest differential expression between in the inner ear and other tissues, OTOR, has been previously described as preferentially expressed in the inner ear (Robertson et al., 2000), thus confirming these results.
Specific isoforms associated with preferential inner ear expression were also identified (Table S11 in Additional file 4). For example, MITF shows differential exon usage between different tissues and within the inner ear (Figure S9 in additional file 1). Mutations in this gene cause microphthalmia and early onset hearing loss (Shibahara et al., 2001). MITF has many known isoforms, and indeed, some MITF isoforms are widely expressed in many cell types, and others show a very restricted expression pattern (Shibahara et al., 2001), suggesting that the defective function of specific isoforms leads to the tissue restricted disease phenotype seen with mutations.
In addition to protein coding genes, genomes produce many thousands of regulatory non-coding RNAs (ncRNAs) (Conte et al., 2013). These include housekeeping ncRNAs (tRNA, rRNA, snoRNA and snRNA) and regulatory RNAs, the latter including lincRNAs, miRNA, piRNAs, natural antisense transcripts (NATs) and transcribed ultraconserved regions (T-UCRs). Several of these ncRNAs are involved in the development of many different human disorders (Esteller, 2011), including hearing loss. Especially miRNAs and long non-coding RNA have received increasing attention because of their implication in the function of chromatin-modifying complexes and the regulation of transcriptional and post-transcriptional events (Conte et al., 2013). In the vertebrate inner ear, miRNAs have been studied most extensively. miRNAs are essential for controlling development and survival of hair cells (Ushakov et al., 2013), and recently, a detailed transcriptional series of miRNA's in the inner ear has been published (Rudnicki et al., 2014). However, not much is known about long non-coding RNAs of the inner ear, and neither has a detailed expression catalogue been made available. Two studies have reported the importance of lincRNAs in the inner ear so far (Manji et al., 2006; Roberts et al., 2012). Maternally expressed gene 3 (Meg3/Gtl2), an imprinted noncoding RNA, is highly expressed in the cochlea, brain, and eye (Manji et al., 2006), and was suggested to be important in inner ear pattern specification and differentiation of cells during otocyst development, as well as in the maintenance of a number of terminally differentiated cochlear cell types (Manji et al., 2006). We confirmed a high expression of MEG3 in all parts of the human inner ear, with the highest expression present in the cochlea (Table S1 in additional file 2).
Additional groups of transcripts that recently came to light are transcripts that are predicted to go through nonsense mediated decay (NMD) and transcribed pseudogenes. NMD is a cellular mechanism that targets and degrades potentially protein coding transcripts with premature stop codons, which sometimes arise due to genetic variation. A number of such transcripts are annotated in the human genome, and studies in mouse and human suggested that while many of these variants are detected at very low levels, presumably due to degradation, others escape this fate through unknown mechanisms (Hasin-Brumshtein et al., 2014; Lappalainen et al., 2013). We have identified several NMD transcripts that escape this fate in specific parts of the inner ear (Figure 5), and show evidence supporting potential tissue specificity of NMD degradation rather than expression regulation. However, it is important to keep in mind that these transcripts are only predicted to be targeted by NMD, by sequence based criteria. Without experimental confirmation, it is unclear whether these transcripts are indeed a true target of the degradation process. Several recent studies also reported genome wide transcription of pseudogenes, and their potential functionality (Kalyana-Sundaram et al., 2012). We report a subset of pseudogenes that are also transcribed in the human inner ear (Figure S6 in Additional file 1), in a tissue specific manner, however the actual implication of these remains unclear.
While the genetic code of most cells in any given person is, with few exceptions, identical, the tightly regulated transcription process gives rise to a wide spectrum of functional cell types and tissues. Multiple examples of tissue specific transcription and regulation are well known, and believed to play a role in many complex phenotypes as well as in Mendelian conditions. Therefore, we carefully examined the expression of genes implicated in hearing loss by genetic mapping, and show that for many of these the major isoform in the context of human hearing accounts for less than 50% of the gene expression in the inner ear. For example the primary isoform of the cochlin (COCH) gene accounts only for ~26% of its total expression (Figure 5; Table S4 in Additional file 2). Thus, our findings underline the importance of examining gene expression in the relevant tissue and provide useful guidance for further molecular and functional studies of these genes. We further investigated hearing loss loci, for which no causative gene had been identified, and were able to show that only ~25% of the genes were expressed in our samples, suggesting use of these as candidates in further investigation of these loci.
Finally, the recent advent of RNA-Seq technology allowed generation of an unprecedented amount of transcriptome data, and improvement of computational tools for genome annotation. On the other side, this wealth of data precludes careful manual curation of genomic features. Indeed out of 57,820 genes in the human annotation 9,848 (17%) are automatically annotated loci, lacking experimental evidence. We detected expression of only 244 of these loci in the inner ear samples, suggesting that experimental verification might be a crucial step in assessment of computational tools.
One of the main limitations of our study is the very small number of available samples. Expression in human tissues is affected by many environmental as well as genetic factors, and we suggest keeping this limitation in mind when drawing conclusions from these data, especially concerning lack of expression of certain genes. Since human ear tissue is extremely difficult to obtain, we were not able to augment our analysis with additional samples.
5. Conclusion
Our data provides for the first time a unique glimpse of the transcriptional landscape of the adult human inner ear. Our results offer a useful reference for neurobiologists studying any candidate gene that might be important in our hearing/balance system. We highlight specific genes that are preferentially expressed in the inner ear, and show spatial expression patterns of coding and non-coding genes in the inner ear. The data set(s) supporting the results of this article are freely available at https://www.tgen.org/home/research/research-divisions/neurogenomics/supplementary-data/inner-ear-transcriptome.aspx.
Supplementary Material
Study highlights.
- We performed RNA-seq of the human cochlea, ampulla, saccule and utricle
- We have built a comprehensive database of transcripts of the adult human inner ear
- We describe a first detailed inventory of long non-coding RNA's in the inner ear
- Our data supports potential tissue specificity of NMD in the inner ear
- The expression of genes in deafness loci without gene identification is described
Acknowledgements
This research was supported by funding from R01 grant DC010215 and the Belgian Science Policy Office Interuniversity Attraction Poles (BELSPO-IAP) program through the project IAP P7/43-BeMGI. IS is a postdoctoral fellow of the FWO Vlaanderen. This manuscript is also dedicated to the memory of our dear friend and colleague Jason Corneveaux. His contributions to our laboratory were innumerable and significant and he will be sorely missed.
List of Abbreviations
- NMD
nonsense mediated decay
- lincRNAs
long intervening non-coding RNAs
- FPKM
Fragments per kilobase of exon per million reads mapped
- NATs
Natural antisense transcripts
- ncRNAs
non-coding RNAs
- T-UCRs
transcribed ultraconserved regions
- AD
Autosomal dominant
- AR
Autosomal recessive
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The author(s) declare that they have no competing interests.
References
- Abe S, Katagiri T, Saito-Hisaminato A, Usami S, Inoue Y, Tsunoda T, Nakamura Y. Identification of CRYM as a candidate responsible for nonsyndromic deafness, through cDNA microarray analysis of human cochlear and vestibular tissues. Am J Hum Genet. 2003;72:73–82. doi: 10.1086/345398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq A Python framework to work with high-throughput sequencing data, bioRxiv. Cold Spring Harbor Labs Journals. 2014 doi: 10.1093/bioinformatics/btu638. doi:10.1101/002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–17. doi: 10.1101/gr.133744.111. doi:10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC a quality control tool for high throughput sequence data. 2010 http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Avraham KB, Hasson T, Steel KP, Kingsley DM, Russell LB, Mooseker MS, Copeland NG, Jenkins NA. The mouse Snell's waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat Genet. 1995;11:369–75. doi: 10.1038/ng1295-369. doi:10.1038/ng1295-369. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. doi:10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein Z, Friedman LM, Shahin H, Oron-Karni V, Kol N, Rayyan A, Parzefall T, Lev D, Shalev S, Frydman M, Davidov B, Shohat M, Rahile M, Lieberman S, Levy-Lahad E, Lee MK, Shomron N, King M-C, Walsh T, Kanaan M, Avraham KB. Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in middle eastern families. Genome Biol. 2011;12:R89. doi: 10.1186/gb-2011-12-9-r89. doi:10.1186/gb-2011-12-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte I, Banfi S, Bovolenta P. Non-coding RNAs in the development of sensory organs and related diseases. Cell Mol Life Sci. 2013;70:4141–55. doi: 10.1007/s00018-013-1335-z. doi:10.1007/s00018-013-1335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. doi:10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Amraoui A, Cohen-Salmon M, Petit C, Simmler MC. Spatiotemporal expression of otogelin in the developing and adult mouse inner ear. Hear Res. 2001;158:151–9. doi: 10.1016/s0378-5955(01)00312-4. [DOI] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. doi:10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Flegel C, Manteniotis S, Osthold S, Hatt H, Gisselmann G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS One. 2013;8:e55368. doi: 10.1371/journal.pone.0055368. doi:10.1371/journal.pone.0055368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen E, Bonneux S, Corneveaux JJ, Schrauwen I, Di Berardino F, White CH, Ohmen JD, Van de Heyning P, Ambrosetti U, Huentelman MJ, Van Camp G, Friedman RA. Genome-wide association analysis demonstrates the highly polygenic character of age-related hearing impairment. Eur J Hum Genet. 2014 doi: 10.1038/ejhg.2014.56. doi:10.1038/ejhg.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimera RV. bcbio-nextgen: Automated, distributed next-gen sequencing pipeline. EMBnet.journal. 2012;17:30. doi:10.14806/ej.17.B.286. [Google Scholar]
- Hasin-Brumshtein Y, Hormozdiari F, Martin L, van Nas A, Eskin E, Lusis AJ, Drake TA. Allele-specific expression and eQTL analysis in mouse adipose tissue. BMC Genomics. 2014;15:471. doi: 10.1186/1471-2164-15-471. doi:10.1186/1471-2164-15-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzano R, Elkon R. High throughput gene expression analysis of the inner ear. Hear Res. 2012;288:77–88. doi: 10.1016/j.heares.2012.01.002. doi:10.1016/j.heares.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu Y-M, Cao X, Asangani IA, Kothari V, Prensner JR, Lonigro RJ, Iyer MK, Barrette T, Shanmugam A, Dhanasekaran SM, Palanisamy N, Chinnaiyan AM. Expressed pseudogenes in the transcriptional landscape of human cancers. Cell. 2012;149:1622–34. doi: 10.1016/j.cell.2012.04.041. doi:10.1016/j.cell.2012.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedländer MR, 't Hoen PAC, Monlong J, Rivas MA, Gonzàlez-Porta M, Kurbatova N, Griebel T, Ferreira PG, Barann M, Wieland T, Greger L, van Iterson M, Almlöf J, Ribeca P, Pulyakhina I, Esser D, Giger T, Tikhonov A, Sultan M, Bertier G, MacArthur DG, Lek M, Lizano E, Buermans HPJ, Padioleau I, Schwarzmayr T, Karlberg O, Ongen H, Kilpinen H, Beltran S, Gut M, Kahlem K, Amstislavskiy V, Stegle O, Pirinen M, Montgomery SB, Donnelly P, McCarthy MI, Flicek P, Strom TM, Lehrach H, Schreiber S, Sudbrak R, Carracedo A, Antonarakis SE, Häsler R, Syvänen A-C, van Ommen G-J, Brazma A, Meitinger T, Rosenstiel P, Guigó R, Gut IG, Estivill X, Dermitzakis ET. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–11. doi: 10.1038/nature12531. doi:10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavinsky J, Crow AL, Pan C, Wang J, Aaron KA, Ho MK, Li Q, Salehide P, Myint A, Monges-Hernadez M, Eskin E, Allayee H, Lusis AJ, Friedman RA. Genome-wide association study identifies nox3 as a critical gene for susceptibility to noise-induced hearing loss. PLoS Genet. 2015;11:e1005094. doi: 10.1371/journal.pgen.1005094. doi:10.1371/journal.pgen.1005094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SI, Conrad T, Jones SM, Lagziel A, Starost MF, Belyantseva IA, Friedman TB, Morell RJ. A null mutation of mouse Kcna10 causes significant vestibular and mild hearing dysfunction. Hear Res. 2013;300:1–9. doi: 10.1016/j.heares.2013.02.009. doi:10.1016/j.heares.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Witten DM, Johnstone IM, Tibshirani R. Normalization, testing, and false discovery rate estimation for RNA-sequencing data. Biostatistics. 2012;13:523–38. doi: 10.1093/biostatistics/kxr031. doi:10.1093/biostatistics/kxr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2, bioRxiv. Cold Spring Harbor Labs Journals. 2014 doi: 10.1186/s13059-014-0550-8. doi:10.1101/002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manji SSM, Sørensen BS, Klockars T, Lam T, Hutchison W, Dahl H-HM. Molecular characterization and expression of maternally expressed gene 3 (Meg3/Gtl2) RNA in the mouse inner ear. J Neurosci Res. 2006;83:181–90. doi: 10.1002/jnr.20721. doi:10.1002/jnr.20721. [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10. doi:10.14806/ej.17.1.200. [Google Scholar]
- Paffenholz R, Bergstrom RA, Pasutto F, Wabnitz P, Munroe RJ, Jagla W, Heinzmann U, Marquardt A, Bareiss A, Laufs J, Russ A, Stumm G, Schimenti JC, Bergstrom DE. Vestibular defects in head-tilt mice result from mutations in Nox3, encoding an NADPH oxidase. Genes Dev. 2004;18:486–91. doi: 10.1101/gad.1172504. doi:10.1101/gad.1172504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollier J, Rombauts S, Goossens A. Analysis of RNA-Seq Data with TopHat and Cufflinks for Genome-Wide Expression Analysis of Jasmonate-Treated Plants and Plant Cultures. Methods Mol Biol. 2013;1011:305–15. doi: 10.1007/978-1-62703-414-2_24. doi:10.1007/978-1-62703-414-2_24. [DOI] [PubMed] [Google Scholar]
- Resendes BL, Robertson NG, Szustakowski JD, Resendes RJ, Weng Z, Morton CC. Gene discovery in the auditory system: characterization of additional cochlear-expressed sequences. J Assoc Res Otolaryngol. 2002;3:45–53. doi: 10.1007/s101620020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KA, Abraira VE, Tucker AF, Goodrich LV, Andrews NC. Mutation of Rubie, a novel long non-coding RNA located upstream of Bmp4, causes vestibular malformation in mice. PLoS One. 2012;7:e29495. doi: 10.1371/journal.pone.0029495. doi:10.1371/journal.pone.0029495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson NG, Heller S, Lin JS, Resendes BL, Weremowicz S, Denis CS, Bell AM, Hudspeth AJ, Morton CC. A novel conserved cochlear gene, OTOR: identification, expression analysis, and chromosomal mapping. Genomics. 2000;66:242–8. doi: 10.1006/geno.2000.6224. doi:10.1006/geno.2000.6224. [DOI] [PubMed] [Google Scholar]
- Rudnicki A, Isakov O, Ushakov K, Shivatzki S, Weiss I, Friedman LM, Shomron N, Avraham KB. Next-generation sequencing of small RNAs from inner ear sensory epithelium identifies microRNAs and defines regulatory pathways. BMC Genomics. 2014;15:484. doi: 10.1186/1471-2164-15-484. doi:10.1186/1471-2164-15-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer DI, Shen J, Corey DP, Chen Z-Y. Gene Expression by Mouse Inner Ear Hair Cells during Development. J Neurosci. 2015;35:6366–80. doi: 10.1523/JNEUROSCI.5126-14.2015. doi:10.1523/JNEUROSCI.5126-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen I, Helfmann S, Inagaki A, Predoehl F, Tabatabaiefar MA, Picher MM, Sommen M, Seco CZ, Oostrik J, Kremer H, Dheedene A, Claes C, Fransen E, Chaleshtori MH, Coucke P, Lee A, Moser T, Van Camp G. A mutation in CABP2, expressed in cochlear hair cells, causes autosomal-recessive hearing impairment. Am J Hum Genet. 2012;91:636–45. doi: 10.1016/j.ajhg.2012.08.018. doi:10.1016/j.ajhg.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara S, Takeda K, Yasumoto K, Udono T, Watanabe K, Saito H, Takahashi K. Microphthalmia-associated transcription factor (MITF): multiplicity in structure, function, and regulation. J Investig Dermatol Symp Proc. 2001;6:99–104. doi: 10.1046/j.0022-202x.2001.00010.x. doi:10.1046/j.0022-202x.2001.00010.x. [DOI] [PubMed] [Google Scholar]
- Simmler MC, Zwaenepoel I, Verpy E, Guillaud L, Elbaz C, Petit C, Panthier JJ. Twister mutant mice are defective for otogelin, a component specific to inner ear acellular membranes. Mamm Genome. 2000;11:960–6. doi: 10.1007/s003350010197. [DOI] [PubMed] [Google Scholar]
- Skvorak AB, Weng Z, Yee AJ, Robertson NG, Morton CC. Human cochlear expressed sequence tags provide insight into cochlear gene expression and identify candidate genes for deafness. Hum Mol Genet. 1999;8:439–52. doi: 10.1093/hmg/8.3.439. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. doi:10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushakov K, Rudnicki A, Avraham KB. MicroRNAs in sensorineural diseases of the ear. Front Mol Neurosci. 2013;6:52. doi: 10.3389/fnmol.2013.00052. doi:10.3389/fnmol.2013.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi S, Rajabian M, Misaghian A, Grbec D, Simon HH, Alavian KN. Parkinson's disease candidate gene prioritization based on expression profile of midbrain dopaminergic neurons. J Biomed Sci. 2010;17:66. doi: 10.1186/1423-0127-17-66. doi:10.1186/1423-0127-17-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpy E, Leibovici M, Zwaenepoel I, Liu XZ, Gal A, Salem N, Mansour A, Blanchard S, Kobayashi I, Keats BJ, Slim R, Petit C. A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet. 2000;26:51–5. doi: 10.1038/79171. doi:10.1038/79171. [DOI] [PubMed] [Google Scholar]
- Yang L, Duff MO, Graveley BR, Carmichael GG, Chen L-L. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. doi:10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura H, Takumi Y, Nishio S, Suzuki N, Iwasa Y, Usami S. Deafness gene expression patterns in the mouse cochlea found by microarray analysis. PLoS One. 2014;9:e92547. doi: 10.1371/journal.pone.0092547. doi:10.1371/journal.pone.0092547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehnder AF, Kristiansen AG, Adams JC, Kujawa SG, Merchant SN, McKenna MJ. Osteoprotegrin knockout mice demonstrate abnormal remodeling of the otic capsule and progressive hearing loss. Laryngoscope. 2006;116:201–6. doi: 10.1097/01.mlg.0000191466.09210.9a. doi:10.1097/01.mlg.0000191466.09210.9a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Jones SM, Yamoah EN, Lundberg YW. Otoconin-90 deletion leads to imbalance but normal hearing: a comparison with other otoconia mutants. Neuroscience. 2008;153:289–99. doi: 10.1016/j.neuroscience.2008.01.055. doi:10.1016/j.neuroscience.2008.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.