Abstract
Background
Traditional approaches to pharmacovigilance center on the signal detection from spontaneous reports, e.g., the U.S. Food and Drug Administration (FDA) adverse event reporting system (FAERS). In order to enrich the scientific evidence and enhance the detection of emerging adverse drug events that can lead to unintended harmful outcomes, pharmacovigilance activities need to evolve to encompass novel complementary data streams, for example the biomedical literature available through MEDLINE.
Objectives
1) to review how the characteristics of MEDLINE indexing influence the identification of adverse drug events (ADEs); 2) to leverage this knowledge to inform the design of a system for extracting ADEs from MEDLINE indexing; and 3) to assess the specific contribution of some characteristics of MEDLINE indexing to the performance of this system.
Methods
We analyze the characteristics of MEDLINE indexing. We integrate three specific characteristics into the design of a system for extracting ADEs from MEDLINE indexing. We experimentally assess the specific contribution of these characteristics over a baseline system based on co-occurrence between drug descriptors qualified by adverse effects and disease descriptors qualified by chemically induced.
Results
Our system extracted 405,300 ADEs from 366,120 MEDLINE articles. The baseline system accounts for 297,093 ADEs (73%). 85,318 ADEs (21%) can be extracted only after integrating specific pre-coordinated MeSH descriptors and additional qualifiers. 22,889 ADEs (6%) can be extracted only after considering indirect links between the drug of interest and the descriptor that bears the ADE context.
Conclusions
In this paper, we demonstrate significant improvement over a baseline approach to identifying ADEs from MEDLINE indexing, which mitigates some of the inherent limitations of MEDLINE indexing for pharmacovigilance. ADEs extracted from MEDLINE indexing are complementary to, not a replacement for, other sources.
Keywords: MEDLINE indexing, pharmacovigilance, adverse drug events
Graphical Abstract
1 Introduction
The timely identification of adverse drug events (ADEs) during the post-approval phase is an important goal of the public health system. Undetected ADEs result in potentially preventable harm to a substantial number of patients and impose a significant burden on the healthcare system [1–4].
While the U.S. Food and Drug Administration (FDA) collects and analyzes drug safety reports through the FDA Adverse Event Reporting System (FAERS) [5], the systematic inventory and collection of ADEs in structured form remains a challenge. Moreover, given well-recognized limitations with such systems [6], pharmacovigilance activities must evolve to encompass novel complementary data streams, in order to enrich the scientific evidence and enhance the detection of emerging adverse drug events that can lead to unintended harmful outcomes (e.g., [7–9]).
Text mining techniques have been used to extract ADEs from various sources [10]. Several recent and ongoing projects have attempted to extract ADEs from the DailyMed structured product labels (e.g., [11]), unstructured clinical notes, like those in electronic health records (e.g., [12–14]), the social media (e.g., [15, 16]), from the biomedical literature, especially MEDLINE® (e.g., [17–19]), or a combination of such sources (e.g., [20]).
The biomedical literature contains valuable information about ADEs in the form of case reports, clinical studies, and observational studies. This information enables drug safety evaluators to assess potentially new ADEs, such as those identified through FAERS. In addition, the biomedical literature may contain ADEs that are not detectable through systems such as FAERS, because healthcare practitioners or researchers are more enthusiastic about publishing their ADE-related findings in scientific journals than reporting them to systems such as FAERS. Moreover, MEDLINE is one of the largest and most comprehensive biomedical literature databases with a broad diversity of human, animal, and in vitro data presented in a variety of publication types. In contrast to FAERS, MEDLINE also offers the advantage of providing scientific information that spans the entire life cycle of a drug from early pre-market drug development through useful market life. This makes the biomedical literature a very important source of information about ADEs.
Unlike systems such as FAERS, the new data sources that are now being considered for ADE detection, such as the biomedical literature, were not created specifically for ADE-related applications, and therefore necessitate different approaches to uncover ADEs.
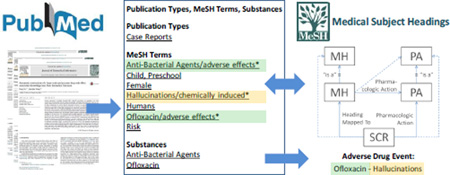
In the MEDLINE database, ADEs are not only expressed in natural language form in the title and abstract, but also in structured form through MeSH indexing. In MEDLINE, citations are indexed with Medical Subject Headings (MeSH®) descriptors (or “main headings”), e.g., for diseases or drugs, that are often enriched with qualifiers (or “subheadings”) that express the specific context in which a topic (e.g., the drug Levofloxacin) is discussed in a given citation (e.g., adverse effects). In addition to text mining techniques, researchers have also leveraged MEDLINE indexing for identifying ADEs. For example, in the context of the EU-ADR project, Avillach et al. have used co-occurring MeSH descriptors and qualifier pairs in MEDLINE citations as the basis for identifying ADEs [21]. Typically, ADEs are identified by the co-occurrence of a drug descriptor qualified by adverse effects and a disease descriptor qualified by chemically induced.
However, there are complexities and limitations to extracting ADEs from MEDLINE indexing. For example, while most ADEs are indexed through the combination of a disease descriptor and the chemically induced qualifier (e.g., Tendinopathy/chemically induced), others, because they are more frequent or important, are completely reflected by a descriptor alone (e.g., Drug-Induced Liver Injury) and would not be found by searching the corresponding combination (Liver Diseases/chemically induced). In practice, detailed knowledge about the characteristics of MEDLINE indexing is essential to complete and accurate retrieval of ADEs.
Although our approach to extracting ADEs from MEDLINE indexing is generally similar to Avillach’s, the goals of the two projects are different. Avillach et al. are primarily interested in detecting known ADEs in MEDLINE, while our goal is to systematically harvest all ADEs from MEDLINE, already known or not, in order to support signal detection in pharmacovigilance. Therefore, our strategy for identifying ADEs tends to be more aggressive and goes beyond the simple co-occurrence of descriptor-qualifier pairs pioneered by Avillach. In a pilot study designed to expand the scope of resources beyond FAERS, we assessed the feasibility of detecting drug-adverse event safety signals for fluoroquinolones through quantitative data mining of MEDLINE indexing terms [22].
The objectives of this work are 1) to review how the characteristics of MEDLINE indexing influence the identification of ADEs; 2) to leverage this knowledge to inform the design of a system for extracting ADEs from MEDLINE indexing; and 3) to assess the specific contribution of some characteristics of MEDLINE indexing to the performance of this system.
2 Background
In this section, we review some of the characteristics of MEDLINE indexing and analyze their influence on the identification of ADEs from MEDLINE indexing. In the course of our research, we have identified nine of these characteristics as potential issues, and have discussed them with MEDLINE indexing specialists at the National Library of Medicine (NLM). The list of these issues is provided in Table 1. Each issue is presented in detail later in this section. We have developed mitigation strategies for three of these issues (1–3), which we have integrated in the design of our system for identifying ADEs from MEDLINE indexing. For the other issues, we provide some recommendations in the discussion section.
Table 1.
How the characteristics of MEDLINE indexing influence the identification of ADEs
| 1 | The chemically induced qualifier is not always necessary to denote ADEs |
| 2 | The adverse effects qualifier is not the only qualifier to denote ADEs |
| 3 | The ADE context is sometimes borne by a broader term rather than the drug of interest |
| 4 | ADEs indexed with MeSH are skewed towards case reports |
| 5 | MEDLINE does not record a relation between a drug and the manifestation of an adverse event |
| 6 | MeSH descriptors sometimes conflate several drugs |
| 7 | MEDLINE indexing rules sometimes aggregate multiple drugs under a broader MeSH descriptor |
| 8 | Changes to MeSH have consequences on the retrieval of ADEs |
| 9 | MEDLINE indexing is not always immediately available at publication time |
2.1 The chemically induced qualifier is not always necessary to denote ADEs
As mentioned before, qualifiers are often used by MEDLINE indexers to describe the specific aspects of a descriptor discussed in a given article. For example, the qualifier chemically induced can be attached to terms representing biological phenomena, diseases, syndromes, congenital abnormalities, or symptoms caused by endogenous or exogenous substances. In the context of adverse drug events, any term with the chemically induced qualifier potentially represents a drug-induced manifestation. However, also applicable in this context are terms (predominantly) from the Chemically-Induced Disorders (C25 subtree) in MeSH, which represent additional disorders caused by the intentional or unintentional ingestion or exposure to chemical substances, such as pharmaceutical preparations. These descriptors are “pre-coordinated” in the sense that they implicitly denote the chemically induced qualifier (e.g., in Drug-Induced Liver Injury), and must be used in lieu of the corresponding descriptor-qualifier combination (e.g., Liver Diseases/chemically induced) used for most descriptors. It is important to note that a user searching the descriptor-qualifier combination expressed by these descriptors would not retrieve the corresponding citations. The corresponding ADEs would be missed if the search was restricted to descriptors qualified by chemically induced. Table 2 lists 19 such descriptors that can be used for indexing ADEs.
Table 2.
Pre-coordinated MeSH descriptors implicitly denoting the chemically induced qualifier
| Drug-Induced Liver Injury | Drug Eruptions | Drug-Induced Liver Injury, Chronic |
| Erythema Nodosum | Serotonin Syndrome | Hand-Foot Syndrome |
| Stevens-Johnson Syndrome | Neuroleptic Malignant Syndrome | MPTP Poisoning |
| Dyskinesia, Drug-Induced | Neurotoxicity Syndromes | Psychoses, Substance-Induced |
| Akathisia, Drug-Induced | Anticholinergic Syndrome | Acute Generalized Exanthematous Pustulosis |
| Asthma, Aspirin-Induced | Drug Hypersensitivity Syndrome | Chemotherapy-Induced Febrile Neutropenia |
| Abnormalities, Drug-Induced |
2.2 The adverse effects qualifier is not the only qualifier to denote ADEs
Sometimes multiple MeSH qualifiers can be logically grouped together. For example, the related qualifiers poisoning and toxicity are grouped under the broad qualifier adverse effects. From a searcher’s perspective this means that all the grouped qualifiers can be retrieved together when searching on the broad qualifier. However, once articles are retrieved for further processing, the exact qualifier is present in the citation. For example, a citation retrieved from a search on Levofloxacin/adverse effects may actually contain the index terms Levofloxacin/toxicity. Therefore, subsequent filtering of the ADE citations should consider not only the qualifier adverse effects, but also the qualifiers it subsumes, namely poisoning or toxicity. Along the same lines, the qualifier contraindications also often denotes ADEs and should be considered.
2.3 The ADE context is sometimes borne by a broader term rather than the drug of interest
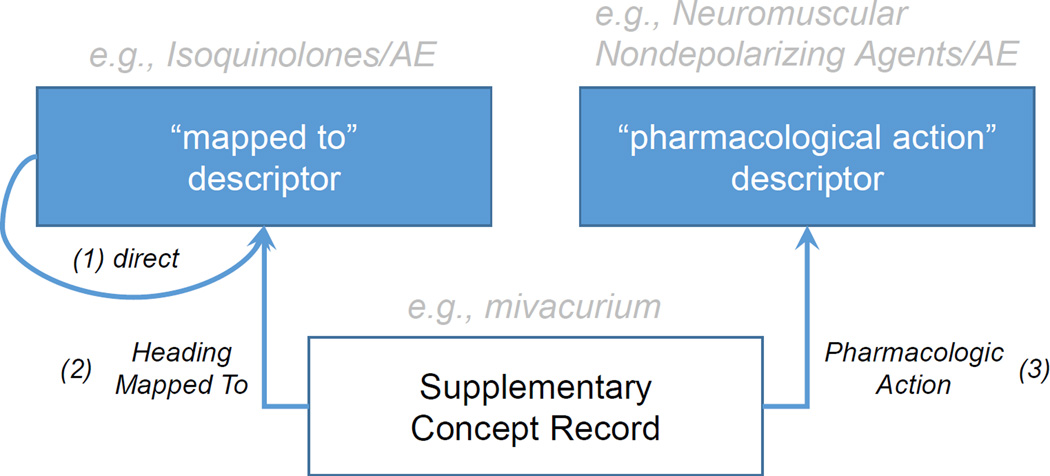
As mentioned earlier, indicative of an ADE is the presence of a drug entity with specific qualifiers (e.g. adverse effects). However, as shown in Figure 1, qualifiers can only be attached to descriptors (1), whereas many specific drugs are only represented in MeSH as supplementary concept records (SCR), to which a qualifier cannot be attached directly. SCRs corresponding to drugs can be linked to qualifiers through two different mechanisms. All SCRs have a Heading Mapped to relation to at least one descriptor in MeSH (2). For example the SCR mivacurium has a Heading Mapped to relation to the descriptor Isoquinolines. In practice, any citation index containing mivacurium will automatically be enriched with the descriptor Isoquinolines. In order to qualify the drug SCR (e.g., with adverse effects), indexers must link the qualifier to its corresponding descriptor. Therefore, qualifiers added to drug descriptors in a citation also qualify the corresponding drug SCRs in this citation. The second possible link between a drug SCR and a qualifier is through pharmacologic action relations. The pharmacological action (PA) descriptors can also be qualified, similar to the Heading Mapped to descriptors (3). For example, mivacurium might be indexed together with its PA, Neuromuscular Nondepolarizing Agents. In this case the qualifier of the PA descriptor also qualifies the corresponding drug SCR.
Figure 1.
Relations between Supplementary Concept Records, Descriptors, and Pharmacological Actions in MeSH
In some cases, qualifiers denoting ADEs can be found attached to broader descriptors, relative to the drug of interest. Leveraging these indirect associations between a drug of interest and qualifiers may be important in some cases, especially when MeSH indexing has evolved over time, resulting in the creation of more specific descriptors (see Changes to MeSH have consequences on the retrieval of ADEs).
2.4 ADEs indexed with MeSH are skewed towards case reports
Although MEDLINE contains and indexes information from various publication types, the majority of indexed ADE information originates from case reports. Case reports and clinical trials have very different foci, reflected in the indexing. While case reports usually focus on one or a few specific ADEs for a drug (which are indexed individually), clinical trial articles tend to report complete safety profiles for the drug. Moreover, efficacy, not safety, is generally the main focus of clinical trials. As a result, safety information from clinical trial articles is generally not indexed. Moreover, case reports tend to capture rather rare and unusual ADEs, in contrast to clinical trials (usually not large enough to detect rare events, since the study populations are usually sized towards efficacy). Conversely, it is unlikely that an overall safety profile for a drug can be derived from case reports alone.
2.5 MEDLINE does not record a relation between a drug and the manifestation of an adverse event
MeSH index terms for a given citation are provided as a flat list and are generally independent of one another. In fact, in the context of adverse events, it should be noted that the MEDLINE indexers have no way to annotate an ADE pair directly, i.e., to link a drug and a disease for this ADE. Instead, the index will contain a drug qualified by adverse effects and a disease term qualified by chemically induced. Co-occurrence of the two index terms in a citation is no guarantee for the existence of a direct link between them.
The implicit nature of these co-occurrence relationships is especially problematic when ADE articles are indexed with more than one drug or more than one disease term. For example, a recent study on the prevention and management of major side effects of breast cancer drugs is indexed with the adverse events cardiovascular, gastrointestinal, and skin diseases, as well as the antineoplastic agents lapatinib, bevacizumab, and trastuzumab [23]. Without further information, the simplest assumption is that each of the drugs is possibly responsible for each of the ADEs mentioned (cross-product).
2.6 MeSH descriptors sometimes conflate several drugs
MeSH descriptors, which are used for indexing documents in MEDLINE, are in fact small aggregates of concepts, grouped together as needed to support indexing and retrieval [24]. For example, the MeSH descriptor Citalopram (D015283) is used for indexing not only the drug citalopram (as expected), but also its stereoisomer, the drug Escitalopram. Although Escitalopram is arguably a separate drug (with specific brand names), it does not exist in MeSH outside the descriptor Citalopram, making it impossible to distinguish between the two drugs on the basis of the index alone.
Nowadays MeSH tends to create distinct descriptors for most major drugs. For example, a descriptor was recently developed for Levofloxacin, distinct from the descriptor Ofloxacin under which it was grouped prior to 2014. However, granularity issues still exist for some drugs, such as Abatacept / Belatacept, Dropropizine / Levodropropizine, and Sultopride / Amisulpride.
2.7 MEDLINE indexing rules sometimes aggregate multiple drugs under a broader MeSH descriptor
Although MEDLINE indexers usually select the most specific descriptors for the topics discussed in an article, according to the indexing rule “Rule of Three”, a group of three or more specific descriptors must be replaced by one more general descriptor, if these specific terms are treed under the more general one. For example, although an article about fluoroquinolone-associated myasthenia gravis exacerbation [25] mentions several individual fluoroquinolones in its abstract (levofloxacin, moxifloxacin, ciprofloxacin, ofloxacin, gatifloxacin, norfloxacin, and trovafloxacin), the article is not indexed with any of these drug terms but with the more general descriptor Fluoroquinolones instead.
One consequence of the Rule of Three is that some adverse events are captured at the class level rather than at the level of individual drugs. This problem might be less prominent in case reports, which focus often on fewer individual drugs than, for instance, research and review articles.
2.8 Changes to MeSH have consequences on the retrieval of ADEs
The MeSH terminology is not static but evolves over time to reflect changes in biomedical knowledge, as well as terminology editorial policies. As mentioned before, MeSH descriptors sometimes group several related concepts. These groupings change over time as part of the evolution of MeSH.
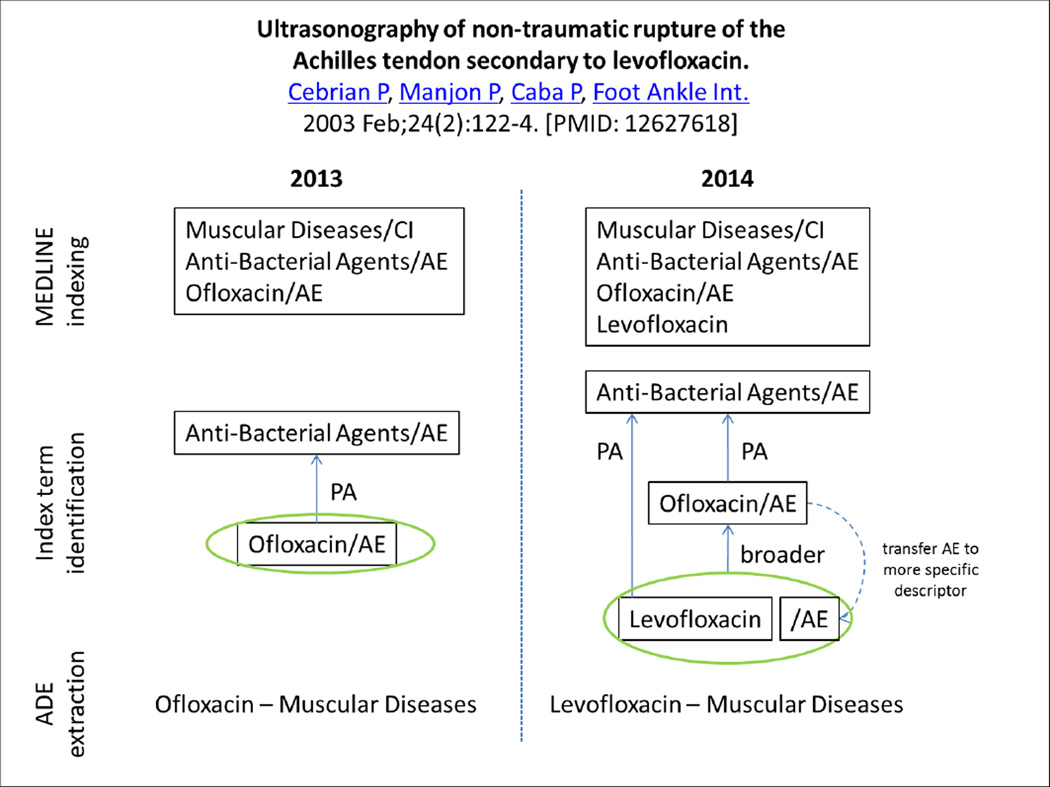
For example, MeSH treated Levofloxacin as an entry term for the descriptor Ofloxacin, until the distinct descriptor Levofloxacin was created in 2014. Articles indexed from 2014 onwards can be indexed with the descriptor Levofloxacin, but earlier articles about levofloxacin are still indexed with the descriptor Ofloxacin (see Figure 2). In the case of Levofloxacin, the new descriptor was actually added retrospectively to citations previously indexed with Ofloxacin, if the string “levofloxacin” appears in the title or abstract. In many other cases, however, the indexing of older citations is not modified to reflect descriptors recently added to MeSH.
Figure 2.
Changes to MeSH have consequences on the retrieval of ADEs
While the retrospective addition of more specific index terms is generally beneficial to retrieval, it also creates some issues. The first issue is ambiguity. In practice, in articles such as this review article from 2009 reporting on seizures associated with Levofloxacin [26], it is not possible to distinguish from the indexing between citations about levofloxacin originally indexed with Ofloxacin, and to which Levofloxacin was later added, and citations natively indexed with both Ofloxacin and Levofloxacin. Moreover, a second issue arises from the fact that the qualifiers originally attached to the broader descriptor Ofloxacin were not transferred to the more specific descriptor Levofloxacin when it was retrospectively assigned to the citation. As a consequence, in order to identify ADEs from the indexing, the adverse effects qualifier attached to the broader descriptor Ofloxacin needs to be considered when extracting the ADEs of Levofloxacin. (This issue was discussed earlier under The ADE context is sometimes borne by a broader term rather than the drug of interest).
2.9 MEDLINE indexing is not always immediately available at publication time
Indexing MEDLINE citations with appropriate MeSH terms is still a dominantly manual process conducted by the indexers and the National Library of Medicine. Although articles are usually available through PubMed at the time they are provided by the publishers (status PubMed - in process), the time for completing the indexing (status PubMed - indexed for MEDLINE) and quality control can vary from a few days or weeks for articles from journals such as the Journal of the American Medical Association, New England Journal of Medicine, Science, or Nature, to several months for articles published in other journals. As full MEDLINE indexing cannot be expected to be immediately available at publication time for all submitted citations, there may be delays in identifying ADEs from some of the most recently submitted articles.
3 Materials
3.1 MEDLINE
MEDLINE is the U.S. National Library of Medicine's (NLM) premier bibliographic database that contains currently over 23 million references to journal articles in life sciences with a concentration on biomedicine. MEDLINE contains citations from over 5,600 worldwide journals in about 40 languages. Since 2005, between 2,000 and 4,000 completed references are added each day, a total of more than 700,000 in 2013. A distinctive feature of MEDLINE is that the records are indexed with NLM Medical Subject Headings (MeSH) by human expert curators. Each year during November and December, NLM makes the transition to a new year of Medical Subject Headings (MeSH) vocabulary used to index the articles (Year-End-Processing). In this study, we access MEDLINE through PubMed and eUtilities, retrieving citation records in XML format and indexed with MeSH 2014 vocabulary (as of May 2014).
3.2 Medical Subject Headings (MeSH)
The Medical Subject Headings (MeSH) is a controlled vocabulary produced and maintained by the NLM [27]. It is used for indexing, cataloging, and searching the biomedical literature in the MEDLINE/PubMed database, and other documents. As of 2014, the MeSH thesaurus includes 27,149 descriptors organized in 16 hierarchies (e.g., Chemicals and Drugs). Additionally, MeSH provides about 210,000 supplementary concept records (SCRs), of which many represent chemicals and drugs (e.g., atorvastatin). Each SCR is linked to at least one descriptor through a Heading mapped to relation (e.g., atorvastatin is associated with Heptanoic Acids and Pyrroles). The descriptors mapped to generally denote the chemical structure of the drug. While most chemical descriptors provide a structural perspective on drugs, some descriptors play a special role as they can be used to annotate the functional characteristics of drug descriptors and SCRs through a pharmacologic action relation (e.g., atorvastatin is linked to the mechanism of action Hydroxymethylglutaryl-CoA Reductase Inhibitors and to the therapeutic use Anticholesteremic Agents). MeSH 2014 is used in this study.
3.3 RxNorm
RxNorm is a standardized nomenclature for medications produced and maintained by the U.S. National Library of Medicine (NLM) [28]. RxNorm concepts are linked by NLM to multiple drug identifiers for commercially available drug databases and standard terminologies, including MeSH and ATC. RxNorm serves as a reference terminology for drugs in the U.S. The February 2014 version of RxNorm used in this study integrates 11,788 substances, including ingredients (IN) and precise ingredients (PIN). Ingredients generally represent base forms (e.g., atorvastatin), while precise ingredients tend to represent esters and salts (e.g., atorvastatin calcium). RxNorm also represents clinical drugs, i.e., the drugs relevant to clinical medicine (e.g., atorvastatin 10 MG Oral tablet). The relations among the various drug entities are represented explicitly in RxNorm (e.g., between ingredients and clinical drugs). NLM also provides an application programming interface (API) for accessing RxNorm data programmatically [29].
4 Methods and Results
In this section, we present the system we have created for extracting ADEs from MEDLINE indexing. In addition to extracting basic co-occurrences between a drug entity, qualified by the adverse effects qualifier, and a disease descriptor, qualified by chemically induced, we introduce two types of refinement leveraging the characteristics of MEDLINE indexing discussed in the Background section. In the first refinement, we aim to extend the scope of descriptors and qualifiers by adding descriptors which, in the absence of a chemically induced qualifier, already denote adverse events (pre-coordinated MeSH descriptors) and capturing all drug entities bearing the ADE context qualified by adverse effects and by the qualifiers it subsumes, namely poisoning or toxicity, as well as contraindications. In the second major refinement, we consider indirect links between drug entities and broader drug descriptors bearing the ADE context, attempting to “transfer” the denotation of ADEs to fine-grained drug entities. These two refinements are not modular additions to the system, but rather integral to it. However, we keep provenance information about each ADE we extract, making it possible to analyze the specific contribution of each part of the system. After the system design, we present an evaluation, both quantitative and qualitative, of the specific contribution of the two refinements made to the baseline system.
4.1 System design
Our approach to extracting ADEs from MEDLINE indexing can be summarized as follows. First, we run a query against the MEDLINE database to retrieve all articles that are relevant to adverse drug events. In each article, we identify among the MeSH indexing terms those that represent the drugs and diseases involved in an ADE. Finally, we extract the drug-manifestation pairs, along with provenance and metadata information (e.g., publication type).
4.1.1 Step 1: Identifying MEDLINE citations corresponding to ADEs
We designed a broad query to be run against the MEDLINE database capturing all citations with at least one drug in the context of adverse effects (drug facet) and one chemically induced manifestation (manifestation facet). The query is shown in Figure 3.
Figure 3.
MEDLINE search query to identify citations with adverse drug events in MEDLINE
Drug facet
The first query facet uses the query term Chemicals and Drugs Category, which captures all citations indexed with any drug or chemical term. The qualifier adverse effects, which also includes the more specific qualifiers of poisoning and toxicity, restricts to citations in which the drugs or chemicals are indexed in the context of an adverse event. (Of note, PubMed automatically extends the query to the qualifiers subsumed by adverse effects.) This query facet also captures citations that discuss contraindications of drugs or chemicals. Restricting to Chemicals and Drugs Category prevents the query from capturing irrelevant articles, e.g., articles about the adverse events of medical devices.
Manifestation facet
The second facet of the search captures citations indexed with at least one manifestation caused by a drug or chemical. The qualifier chemically induced is searched unbound to any particular manifestation term. The scope of this facet is broadened by the addition of 19 hand-selected pre-coordinated MeSH descriptors that implicitly denote the chemically induced qualifier, e.g., Drug-Induced Liver Injury.
Only articles that fulfill the criteria set by both the drug and the manifestation facets of our query are captured. For example, an article indexed with both the descriptor/qualifier combinations Heparin/adverse effects and Thrombosis/chemically induced would be captured, under the assumption that it contains the ADE pair of Heparin and Thrombosis. Similarly, an article would be captured containing the descriptor/qualifier combination Acetaminophen/poisoning and the pre-coordinated MeSH descriptor Drug-Induced Liver Injury.
The query was executed using the NCBI Entrez Programming Utilities. The resulting list of PubMed IDs was used to retrieve the corresponding MEDLINE records in XML format to be further processed. The search query yielded a MEDLINE subset of 360k PMIDs updated through May, 2014.
4.1.2 Step 2: Identifying index terms for drugs and manifestations involved in ADEs
In this second step, we identify index terms for drugs and manifestations involved in ADEs among the MEDLINE citations retrieved at Step 1.
4.1.2.1 Drugs
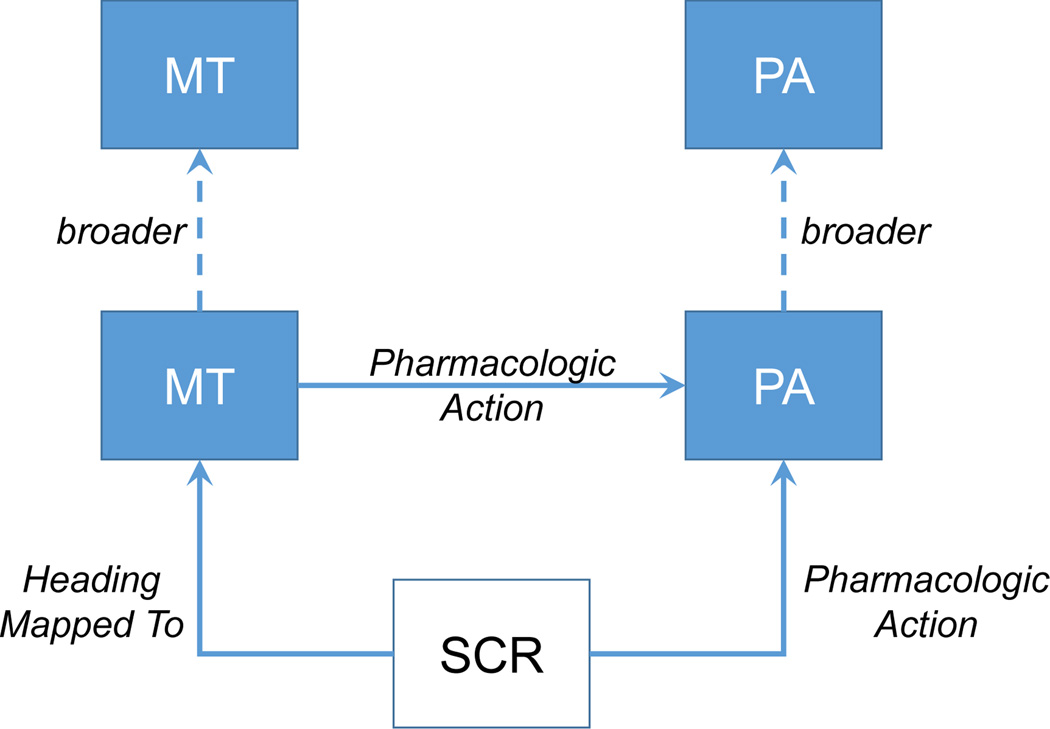
For a given citation, we consider as drug candidates all indexed descriptors that are located in the Chemicals and Drugs tree in MeSH as well as all supplementary concept records, if any, that are connected to any of these descriptors through Heading Mapped to or pharmacological action relationships. We leverage information from the MeSH terminology to reconstruct all hierarchical relationships between all drug candidates for a given citation, see Figure 4. We use the hierarchical information to report only on terms with highest specificity, i.e., the leaf nodes in the reconstructed tree, whereby information from qualifiers, i.e., the active role in adverse effects, poisoning, toxicology, or contraindications, can be passed on from higher level terms to their children. For example, if a citation contains the SCR mivacurium, the “mapped to” descriptor Isoquinolines, and the “pharmacological action” descriptor Neuromuscular Nondepolarizing Agents, with relations as depicted in Figure 1, we identify only the SCR as the ADE drug. Furthermore, this SCR “inherits” the adverse effects (AE) qualifier from the “mapped to” descriptor and the PA descriptor, respectively, and will be considered as an involved drug in this citation. Conversely, if a specific drug does not bear the ADE context (directly or indirectly), it is recorded as a concomitant drug.
Figure 4.
Relations between MESH terms representing drugs or drug classes in MEDLINE indexing (SCR: supplementary concept record; MT: “mapped to” descriptor; PA: “pharmacological action” descriptor)
4.1.2.2 AE manifestations
For a given citation, we consider as manifestations of an adverse event all descriptors from the citation’s index that are further qualified by the qualifier chemically induced, as well as any of the 19 pre-coordinated descriptors from Table 2.
4.1.3 Step 3: Extracting ADE pairs and metadata
In the third component of our system, we extract the ADE pairs based on the entities identified in Step 2, filter them for clinical relevance, and enrich the ADE pairs with metadata information.
4.1.3.1 ADE pairs
As mentioned before, ADE pairs are not explicitly given as part of the MEDLINE indexing. Instead, we reconstruct the pairs based on the drugs and event manifestations individually identified in Step 2. We derive the ADE pairs for a given citation by applying the Cartesian product between all specific drugs and all AE manifestations. The role of a given drug in an ADE pair is then classified as either involved or concomitant, based on the presence or absence of the appropriate qualifiers for this drug.
4.1.3.1.1 Filtering for clinical significance
In the context of pharmacovigilance of already approved drugs, we define as drugs of interest those clinically relevant drugs that are currently prescribable and available on the U.S. market. However, descriptors from the Chemicals and Drugs tree (D tree) and associated supplementary concept records (SCRs) can also denote chemicals (e.g., carcinogens, environmental), drugs withdrawn from the market (e.g., rimonabant), non-prescribable drugs or products (e.g., coffee or cosmetics), or drug-classes (e.g., pyridines). Mapping the MeSH drugs from ADE pairs to a drug terminology helps distinguish drugs of interest from other entities and thus improves precision of the extraction process. RxNorm is suitable for establishing such a filter, since it provides curated and regularly updated mappings to drugs in MeSH, as well as information such as prescribability and clinical relevance. Since MeSH is integrated in RxNorm, we use the RxNorm API to map the MeSH descriptor and supplementary concept record identifiers of the candidate drug entities to RxNorm concept unique identifiers (RxCUIs). Subsequently, we normalize all RxCUIs to Ingredients (IN), whenever mappings from MeSH drugs were established to precise ingredients (PIN). Finally, to assess clinical relevance of the ingredients, we require that the ingredients be associated with at least one clinical drug (SCD) in the RxNorm graph.
4.1.3.2 Metadata
Besides drugs and adverse events, we extract additional information that could be relevant in the context of signal detection for pharmacovigilance. In addition to identifying drugs as involved or concomitant, we systematically collect MeSH terms providing information about species (B01 subtree), gender (“male” or “female”), age groups (M01.060 subtree), publication types (V tree), epidemiologic methods (E05.318 subtree), and the indication of a drug (any descriptor with the qualifier drug therapy), whenever available. Additionally, we extract publication dates from various data elements in the XML file of each citation. Based on these data, we can later easily refine the analysis of our data (e.g., restricted to ADEs found in case reports), compare different cohorts within one set (male vs. female, studies in animals vs. humans), or determine when information about a specific ADE was published for the first time.
4.2 Experiments
4.2.1 Two levels of refinement over the baseline
As mentioned earlier, our approach to extracting ADEs from MEDLINE indexing integrates two levels of refinement over the baseline, namely an extension of the scope of descriptors and qualifiers for ADEs, and the possibility for a drug descriptor to “inherit” the ADE context (qualifier) placed on a broader drug descriptor. The list of features corresponding to each level of refinement is summarized in Table 3. While these refinements are already built into our system, we measure the specific contribution of each level by tracking at which level (Baseline, Extension, Inheritance) a given ADE is extracted.
Table 3.
Strategies followed at different levels
| Strategies | Feature ID | Baseline | Extension | Inheritance | |
|---|---|---|---|---|---|
| Drugs | Chem + Drug/adverse effects | 1 | + | + | + |
| Chem + Drug/poisoning | 2 | − | + | + | |
| Chem + Drug/toxicity | 3 | − | + | + | |
| Chem + Drug/contraindication | 4 | − | + | + | |
| SCR → MT descr (direct) | 5 | + | + | + | |
| SCR → PA descr (direct) | 6 | + | + | + | |
| SCR → MT descr (indirect) | 7 | − | − | + | |
| SCR → PA descr (indirect) | 8 | − | − | + | |
| MT descr | 9 | + | + | + | |
| MT descr → MT descr (direct) | 10 | − | − | + | |
| MT descr → PA descr (direct) | 11 | − | − | + | |
| MT descr → MT descr (indir.) | 12 | − | − | + | |
| MT descr → PA descr (indir.) | 13 | − | − | + | |
| Events | Descr/chemically induced | 14 | + | + | + |
| 19 pre-coordinated Descr | 15 | − | + | + | |
4.2.1.1 Baseline
This is the most restrictive of the three levels and corresponds to a naïve approach to extracting ADEs from MEDLINE indexing (basic co-occurrence of descriptor-qualifier pairs for drugs and manifestations in the context of ADEs).
The only qualifier we consider for drugs is adverse effects [Feature 1 in Table 3]. The qualifier contraindications, as well as the two qualifiers grouped under adverse effects, namely poisoning and toxicity, are not considered.
Manifestations of adverse events are only identified by descriptors qualified by the chemically induced qualifier [Feature 14 in Table 3]. The 19 pre-coordinated manifestation descriptors are not considered.
No indirect inheritance of the ADE context is allowed. In practice, drug descriptors are considered only if they are directly qualified by adverse effects [Feature 9 in Table 3]; SCRs inherit the adverse effects qualifier only from the descriptors to which they have direct Heading Mapped to or Pharmacologic Action relations [Features 5–6 in Table 3].
4.2.1.2 Extension of the scope of descriptors and qualifiers for ADEs
In addition to all the features of the baseline, we extend the scope of descriptors and qualifiers for ADEs, by allowing additional (unqualified) descriptors and additional qualifiers.
In addition to the qualifier adverse effects, we consider the qualifier contraindications, as well as the two qualifiers grouped under adverse effects, namely poisoning and toxicity [Features 2–4 in Table 3].
In addition to the descriptors qualified by the chemically induced qualifier, we consider the 19 pre-coordinated manifestation descriptors [Feature 15 in Table 3].
As for the baseline, no indirect inheritance of the ADE context is allowed.
4.2.1.3 Inheritance of the ADE context
At this level, we apply all the strategies from the Baseline and Extension levels. Additionally, inheritance of the ADE context (represented by the qualifiers adverse effects, poisoning, toxicity, and contraindications) is allowed. In practice, drug descriptors can inherit the ADE context from associated “mapped to” (MT) and “pharmacological action” (PA) descriptors [Feature 10 and 11 in Table 3]. Moreover, both drug descriptors and SCRs can inherit the ADE context from any of their direct or indirect parent descriptors [Feature 7, 8, 12 and 13 in Table 3].
For example, the descriptor Levofloxacin can inherit the ADE context from its parent descriptor Ofloxacin (Feature 10). In the index for an article about alternatives to the drug Practolol [30], the drug inherits the ADE context from the PA Adrenergic beta-Antagonists, which is among the index terms for this article, although the asserted PA in MeSH for this drug is Adrenergic beta-1 Receptor Antagonists, a child term of Adrenergic beta-Antagonists (Feature 13).
4.2.2 Specific contribution of each level
In the following we provide a quantitative evaluation of the results of our system, in terms of extracted drugs, manifestations, and ADE pairs, focusing on the specific contribution of each level. Table 4 shows the cumulative results and the relative gain for the three levels.
Table 4.
Specific contributions of each level (cumulative numbers of ADE instances and the relative gain)
| Baseline | + Extension | + Inheritance | |
|---|---|---|---|
| Number of unique Drugs or Chemicals | 9,786 | 14,268 (+46%) | 14,712 (+3%) |
| Number of unique, clinically relevant Drugs | 2,146 | 2,239 (+4%) | 2,250 (+0.5%) |
| Number of unique Manifestations | 3,007 | 3,097 (+3%) | 3,107 (+0.3%) |
| Number of unique ADE pairs | 95,911 | 113,285 (+18%) | 118,552 (+5%) |
| Number of ADE pair instances | 297,093 | 382,411 (+29%) | 405,300 (+6%) |
| Number of Citations | 152,729 | 198,676 (+30%) | 205,597 (+4%) |
4.2.2.1 Baseline
We retrieve ADEs for 9,786 unique MeSH drugs and chemicals, of which 2,146 (22%) pass our RxNorm filter for clinical relevance. The relevant drugs are paired with 3,007 unique manifestations. In total we harvest 95,911 unique ADE pairs and 297,093 ADE instances from 152,729 citations.
4.2.2.2 Extension
This refinement level shows only moderate improvement in terms of unique drugs (+93) and manifestations (+90). However, we retrieve an additional 17,374 (+18%) unique ADE pairs and 85,318 (+29%) ADE instances. More importantly, 45,978 of these ADE instances refer to drug-manifestation pairs that could not be captured at the Baseline level. We harvest information from 45,947 additional citations (+30%) that had not been considered in our baseline approach.
4.2.2.3 Inheritance
The overall contribution of this level is marginal in comparison to the first two levels. We only retrieve 15 additional drugs and 10 additional manifestations, nonetheless yielding 5,267 additional unique ADEs and 22,889 additional ADE instances. Again, 6,262 of these ADE instances refer to drug-manifestation pairs that could not be captured at the Baseline or Extension levels. We harvest information from 205,597 of the 360k citations retrieved by our broad MEDLINE query. (The remaining 160k MEDLINE citations may contain ADE pairs for drugs that are not clinically relevant, chemicals, drug combinations, or drug classes, all of which are ignored on purpose by our RxNorm filter).
4.2.3 Evaluation
By measuring the specific contribution of each level, we were able to demonstrate that the two levels of refinement over the baseline yielded significant numbers of additional ADEs, including novel ADEs that could not be captured at previous levels. While these additional levels are obviously productive, we need to evaluate if the ADEs obtained at each level are valid.
In order to evaluate the quality of the ADEs extracted at the Extension and Inheritance levels, we focus on the novel ADEs extracted at these levels, i.e., unique ADEs that could not be extracted at previous levels. (That is, we do not review additional instances of ADEs already extracted at previous levels). We also evaluate the ADEs extracted at the Baseline level. For each ADE, we need to assess whether the MEDLINE citation from which a given ADE is extracted provides evidence for (true positive) or against (false positive) the ADE. We make this determination from the information contained in the title and abstract of the MEDLINE citation and, if necessary, from the full-text article. For this reason, our evaluation was restricted to those ADEs for which the full-text article was available in PubMed Central.
In practice, we randomly selected 100 ADEs for the Baseline and Extension levels, and 50 for the less productive Inheritance level. Two of the authors (OB and AS), physicians by training, manually compared each candidate ADE against the information available in the corresponding articles. Differences in opinion were discussed and the most conservative evaluation was kept in case of disagreement.
Table 5 shows the detailed results of our evaluation. The proportion of ADEs with supporting evidence in the article is 69% for the Baseline level, 74% for the Extension level, and 48% for the Inheritance level.
Table 5.
Evaluation of the ADE pairs extracted at the three levels
| Level | # True positive (“evidence for”) |
# False positive (“evidence against”) |
Total |
|---|---|---|---|
| Baseline | 69 | 31 | 100 |
| Extension | 74 | 26 | 100 |
| Inheritance | 24 | 26 | 50 |
Together with the evaluation, we also performed a failure analysis to determine the cause of the false positive ADEs. The two most common types of error were the detection of a wrong association and the detection of an association when there was none. These two types of errors account for 73% of all false positive ADEs overall. The proportion of wrong associations is higher at the Extension level (46%), while lack of association is more frequent at the Baseline level (45%). The highest proportion of wrong associations is observed at the Inheritance level (61%).
Wrong associations tend to occur when several drugs and/or several manifestations are mentioned as index terms in a MEDLINE citation. Our strategy of computing the cross-product of the drugs and manifestations as potential ADEs may result in false positives (e.g., if a given manifestation is related to a specific drug, but not to all the drugs indexed in the article). For example, the drug Procarbazine is mentioned in the indexing of an article titled “Late cardiac toxicity of doxorubicin, epirubicin, and mitoxantrone therapy for Hodgkin's disease in adults” [31] because it is part of the MOPP chemotherapy regimen used in the control group. While our method rightly identifies a cardiac toxicity ADE for Doxorubicin, Epirubicin, and Mitoxantrone, it also wrongly identifies an ADE for Procarbazine. Lack of association primarily occurs when an article provides evidence for the absence of an ADE, because such articles are indexed similarly to those reporting evidence of an ADE. In fact, in both cases, the articles discuss (potential) adverse events of drugs, which is reflected in the indexing. For example, an article whose conclusion is “This study supports the safety of the treatment of schizophrenia with pramipexole and haloperidol as a combination therapy” [32] is appropriately indexed with Pramipexole and Dyskinesia, Drug-Induced despite the fact that the authors demonstrate the lack of worsening of extrapyramidal side effects.
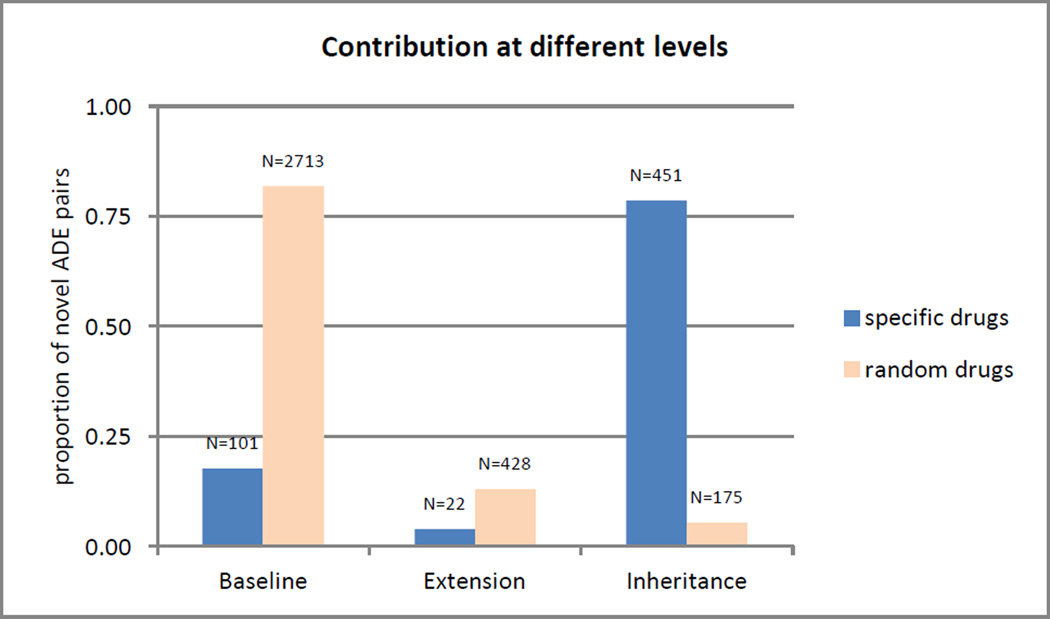
As shown in Table 5, there is a striking difference in the proportion of false positive ADEs between the Baseline and Extension levels (26–31%) on the one hand and the Inheritance level (52%) on the other. In fact, the Inheritance level was added specifically for certain types of ADEs, namely ADEs for drugs involving MeSH descriptors that had undergone recent changes (e.g., when Levofloxacin was extracted out of the descriptor Ofloxacin, with MEDLINE citations still showing the ADE context attached to the previous indexing, Ofloxacin). We decided to evaluate such ADEs specifically (15 drug descriptors with a recent mention of previous indexing notes, such as Levofloxacin) against all other ADEs captured at this level (15 randomly selected drugs), contrasting the number of novel ADEs specifically extracted at the Inheritance level in each set. As shown in Figure 5, the profiles of the two sets are completely different. For the randomly selected drugs, 82% of the ADEs are extracted at the Baseline level, whereas 79% of the ADEs are extracted at the Inheritance level for the 15 specifically selected descriptors. In other words, for drugs such as Levofloxacin, most of the ADEs would be missed if it were not for the Inheritance level. Given that the percentage of false positive ADE is higher at the Inheritance level, our recommendation is therefore to apply the Inheritance level not to all drugs, but only to those MeSH descriptors that have undergone recent changes and for which the ADE context may be borne by another descriptor as a result of these changes.
Figure 5.
Relative and absolute contribution at different levels for random and specific drugs
5 Discussion
We outline the significance of our findings, present lessons learned and recommendations, contrast text mining with data mining for ADE extraction, and discuss some limitations and future work.
5.1 Significance
In this work we investigated the properties and limitations of MEDLINE indexing for use in pharmacovigilance. We identified nine MEDLINE indexing properties that affect the manner in which ADEs can be identified and extracted. Based on these findings, we proposed a comprehensive approach for the extraction of ADEs from MEDLINE indexing.
Our evaluation reinforces the notion presented in earlier studies that the use of MEDLINE indexing is a viable approach for extracting valuable safety information from the biomedical literature. In addition, we demonstrate that our approach provides an improvement over existing methods for detecting ADEs via MEDLINE indexing. Specifically, we demonstrate that our approach is able to identify additional ADE-related citations and additional ADEs with relatively high precision that would otherwise be unidentified by existing methods. In a recent related study [33], we applied our ADE extraction approach to evaluate MEDLINE’s capability for signaling recently labeled adverse events. The findings of that study provide additional support for the value of MEDLINE indexing for pharmacovigilance. Similarly, our pilot investigation of fluoroquinolone drugs also demonstrated the feasibility of using our multi-step ADE extraction approach to generate a highly relevant, ADE-focused subset of MEDLINE data (from a variety of publication types) that could effectively support quantitative data mining for the detection of drug-adverse event safety signals [22].
More specifically, this investigation defines the role played by MEDLINE indexing in support to pharmacovigilance. On the one hand, extracting ADEs from MEDLINE indexing is relatively straightforward, fully automated and can be repeated on a regular basis to monitor the report of new ADEs in the biomedical literature. Moreover, the rich set of metadata attached to a MEDLINE citation (e.g., publication date and type and population characteristics) can easily be extracted along with the ADE and used for fine-grained analysis of the ADE dataset. On the other hand, we point out (and address) some of the inherent limitations of MEDLINE indexing for pharmacovigilance and demonstrate significant improvements over a naïve baseline approach, i.e., co-occurrence of descriptor-qualifier pairs. We also show how the characteristics of MEDLINE indexing contribute to biases in the ADE information extracted from MEDLINE indexing (e.g., underreporting of some ADE types in case reports vs. clinical trials, specific issues with certain drugs due to their representation in the MeSH vocabulary). Understanding these limitations is critical for developing mitigation strategies when possible. At a minimum, biomedical researchers need to be aware of these limitations in order to define the role ADEs from MEDLINE indexing can play in pharmacovigilance together with other sources of information (e.g., spontaneous reports and clinical trials). These biases and limitations are also the reason why an evaluation against reference lists of ADEs would not be meaningful and was not performed as part of this study.
5.2 Lessons learned and recommendations
5.2.1 From the experiment
Overall, the simple analysis of co-occurrence of descriptor-qualifier pairs (Baseline level) remains a valid strategy for extracting ADEs from MEDLINE indexing. It provides the bulk of the ADEs extracted from MEDLINE indexing, of which 69% were shown to be correct.
However, a significant number of important ADEs are denoted by pre-coordinated MeSH descriptors and could not be identified if only descriptor-qualifier pairs were used (e.g., Drug- Induced Liver Injury). We were able to retrieve 17,374 (+18%) additional unique ADE pairs. The quality of ADEs extracted at the Extension level is even slightly higher (74% correct) than for the Baseline level. These important ADEs could not have been captured at all with an approach limited to simple descriptor-qualifier pairs. A smaller number of ADEs are captured by qualifiers other than adverse effects.
For a small number of drugs, ADEs can be captured only by considering indirect links between a specific drug and the broader descriptor bearing the ADE context (Inheritance level). While this phenomenon is only marginal overall, it is very important for specific drugs (e.g., levofloxacin). However, it should not be applied across the board, as its performance is generally limited (48% correct).
5.2.2 From studying the characteristics of MEDLINE indexing
5.2.2.1 ADEs indexed with MeSH are skewed towards case reports
ADEs extracted from MEDLINE are not sufficient for signal detection for pharmacovigilance, as most of the ADEs indexed come from case reports, in which the ADEs reported are not representative of all ADEs. Therefore, the integration of multiple sources of information beyond the biomedical literature is likely to be critical in order to obtain comprehensive drug safety information. MEDLINE indexing should be considered as one source of ADE information, along with spontaneous reporting (FAERS), observational data (from electronic health records), etc.
5.2.2.2 MEDLINE does not record a relation between a drug and the manifestation of an adverse event
The overall quality (i.e., signal-to-noise ratio) of the data originating from peer-reviewed and manually indexed articles should be generally higher than, for instance, the quality of the raw data submitted to the FDA Adverse Event Reporting System (FAERS), where consumers might report entire medication lists alongside the list of symptoms they experienced. Overall, despite potential false positives, the ADEs extracted from MEDLINE indexing are expected to be more targeted than those from spontaneous reporting systems.
5.2.2.3 MeSH descriptors sometimes conflate several drugs
Although we showed this was a problem for some drugs (e.g., levofloxacin), the impact of this characteristic of MeSH indexing is limited overall. Moreover, over the past years, MeSH has created distinct descriptors for most major drugs. However, the evolution of the MeSH vocabulary for drugs that were once conflated into one descriptor should be taken into account for the analysis of specific drugs.
5.2.2.4 MEDLINE indexing rules sometimes aggregate multiple drugs under a broader MeSH descriptor
This issue is an indexing issue, independent from the organization of the MeSH vocabulary itself. Although we have not specifically measured its impact, we suspect it is limited overall. Moreover, this rule applies not to specific drugs, but to all MeSH descriptors. Therefore, it should not have introduced any bias towards specific ADEs.
5.2.2.5 Changes to MeSH have consequences on the retrieval of ADEs
This issue is a consequence of the evolution of the MeSH vocabulary, especially when drugs that were conflated under the same descriptor become distinct descriptors. As mentioned earlier, while important for specific drugs, the impact of this issue is marginal overall.
5.2.2.6 MEDLINE indexing is not always immediately available at publication time
As a consequence, ADEs extracted from MEDLINE indexing cannot be expected to be immediately available at publication time. This lag time must be taken into account when extracting ADEs (or other information) from MEDLINE indexing.
5.3 Text mining vs. data mining for ADE extraction
It is worth noting the parallel efforts for extracting ADEs from the biomedical literature, which apply natural language processing (NLP) techniques to article titles and abstracts [17–19]. Given the current state of the art, it is unclear which of the two approaches—MEDLINE indexing versus NLP—is the better approach for extracting ADEs. These two approaches are most likely complementary. Nonetheless, many of the advantages and limitations of MEDLINE indexing versus NLP are apparent. The indexing is readily available, human-curated, is based on the full text of articles (not just abstracts), and does not require the use of complex NLP techniques that are more prone to error. Conversely, applying NLP to article titles and abstracts (or full-text articles when available) is not limited by the scope and granularity of the MeSH vocabulary or by the NLM annotation rules. The use of NLP is also not limited by time delays resulting from the need for human annotation.
5.4 Limitations and future work
5.4.1 MeSH vs. other ADE vocabularies
One limitation of this study is that it does not address the comparison between ADEs extracted from MEDLINE indexing (in reference to the MeSH vocabulary) and other sources of ADEs, coded to different vocabularies. For example, the Medical Dictionary for Regulatory Activities (MedDRA) is the terminology most commonly used by regulatory authorities in the pharmaceutical industry and is endorsed for adverse event classification by the International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH) [34]. Such comparison would require a mapping between MeSH and MedDRA. While equivalences between MeSH and MedDRA are available through the Unified Medical Language System (UMLS) Metathesaurus, meaningful comparison between the MEDLINE and FAERS ADEs would require a significant curation effort in order to bridge granularity differences between the two vocabularies. This effort was beyond the scope of this investigation, but will be the object of future work. In addition, the hierarchical structure of MedDRA allows for vertical aggregation of manifestation terms up to the System Organ Class (SOC) level or horizontal aggregation through Standardised MedDRA Queries (SMQs). MeSH only supports vertical aggregation through its hierarchical structure, and is organized to support indexing and retrieval, not pharmacovigilance. Groupings of MeSH descriptors would need to be defined in order to meaningfully aggregate the ADEs extracted from MEDLINE indexing.
Of note, the indexing of drugs with MeSH does not offer the same challenges. Through the integration of MeSH drugs in RxNorm, these drugs are already aligned with other drug classification systems, such as the Anatomical Therapeutic Chemical (ATC) classification system developed by the World Health Organization (WHO). ATC supports the aggregation of drugs into classes on four levels of granularity. It is commonly used for pharmacoepidemiology and in research projects (e.g., EU-ADR). Comparing classes between MeSH and ATC is not as straightforward [35], and exploiting the ADEs extracted from MEDLINE indexing at the level of drug classes (as opposed to individual drugs) remains somewhat challenging.
5.4.2 ADE selection for signal generation
It was beyond the scope of this investigation to determine which statistical techniques would be best for analyzing the signal generated from MEDLINE indexing. Similarly, we chose to follow a search and extraction strategy that retrieves a broad set of ADEs for a diverse set of chemicals and drugs from various types of publications and studies. While Avillach filtered out some publication types as non-contributory [21], and Gurulingappa only considered case-reports [17], we chose to be inclusive of all publication types and to capture provenance information, rather than making an a priori selection. This strategy provides greater flexibility and supports refinements of the statistical analysis as needed.
6 Conclusions
To enhance the detection of emerging adverse drug events that can lead to unintended harmful outcomes, pharmacovigilance activities needs to evolve to encompass novel complementary data streams, for example the biomedical literature available through MEDLINE.
In this investigation, we focused on the extraction of ADEs from MEDLINE indexing. We confirmed that the analysis of co-occurrence of descriptor-qualifier pairs remains a valid strategy. We proposed significant improvements over a baseline approach, in order to mitigate some of the inherent limitations of MEDLINE indexing for pharmacovigilance. The system we created successfully extracted 405,300 ADE instances from 205,597 MEDLINE citations vs. 297,093 ADE instances from 198,676 citations for the baseline system. We verified that the majority of these additional ADE instances are correct.
ADEs extracted from MEDLINE indexing for pharmacovigilance purposes are complementary to, not a replacement for, other sources. ADEs could not be reliably extracted from MEDLINE indexing if MEDLINE did not provide fine-grained indexing, not only at the level of individual drugs and manifestations, but also reflecting the specific ADE context for these index terms.
Acknowledgements
This work was supported by the Intramural Research Program of the NIH, National Library of Medicine (NLM). This work also received support from the US Food and Drug Administration (FDA) through the Center for Drug Evaluation and Research (CDER) Critical Path Program [interagency agreement with NLM (XLM12011 001)] and from the Office of Translational Sciences at CDER. RW was supported by an appointment to the NLM Research Participation Program administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the National Library of Medicine. RH was employed by Stanford University while conducting this research, and acknowledges support by NIH grant U54-HG004028 for the National Center for Biomedical Ontology and by NIGMS grant GM101430- 01A1.
The authors would like to thank Lou S. Knecht, Deputy Chief, and Susan C. Schmidt, Unit Head Indexing Section, of the Bibliographic Services Division, Library Operations, at the National Library of Medicine (NLM), and the entire Indexing Section for their tremendous help with this project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclaimer
The findings and conclusions expressed in this report are those of the authors and do not necessarily represent the views of the FDA.
The authors declare they have no conflict of interest.
References
- 1.Khan LM. Comparative epidemiology of hospital-acquired adverse drug reactions in adults and children and their impact on cost and hospital stay--a systematic review. European journal of clinical pharmacology. 2013;69(12):1985–1996. doi: 10.1007/s00228-013-1563-z. [DOI] [PubMed] [Google Scholar]
- 2.Smyth RM, Gargon E, Kirkham J, Cresswell L, Golder S, Smyth R, et al. Adverse drug reactions in children--a systematic review. PloS one. 2012;7(3):e24061. doi: 10.1371/journal.pone.0024061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meier F, Maas R, Sonst A, Patapovas A, Muller F, Plank-Kiegele B, et al. Adverse drug events in patients admitted to an emergency department: an analysis of direct costs. Pharmacoepidemiology and drug safety. 2014 doi: 10.1002/pds.3663. [DOI] [PubMed] [Google Scholar]
- 4.Hug BL, Keohane C, Seger DL, Yoon C, Bates DW. The costs of adverse drug events in community hospitals. Joint Commission journal on quality and patient safety / Joint Commission Resources. 2012;38(3):120–126. doi: 10.1016/s1553-7250(12)38016-1. [DOI] [PubMed] [Google Scholar]
- 5.FDA Adverse Event Reporting System (FAERS) http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/default.htm.
- 6.Harpaz R, DuMouchel W, Shah NH, Madigan D, Ryan P, Friedman C. Novel data-mining methodologies for adverse drug event discovery and analysis. Clinical pharmacology and therapeutics. 2012;91(6):1010–1021. doi: 10.1038/clpt.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coloma PM, Schuemie MJ, Trifiro G, Gini R, Herings R, Hippisley-Cox J, et al. Combining electronic healthcare databases in Europe to allow for large-scale drug safety monitoring: the EU-ADR Project. Pharmacoepidemiology and drug safety. 2011;20(1):1–11. doi: 10.1002/pds.2053. [DOI] [PubMed] [Google Scholar]
- 8.Platt R, Wilson M, Chan KA, Benner JS, Marchibroda J, McClellan M. The new Sentinel Network--improving the evidence of medical-product safety. The New England journal of medicine. 2009;361(7):645–647. doi: 10.1056/NEJMp0905338. [DOI] [PubMed] [Google Scholar]
- 9.Stang PE, Ryan PB, Racoosin JA, Overhage JM, Hartzema AG, Reich C, et al. Advancing the science for active surveillance: rationale and design for the Observational Medical Outcomes Partnership. Annals of internal medicine. 2010;153(9):600–606. doi: 10.7326/0003-4819-153-9-201011020-00010. [DOI] [PubMed] [Google Scholar]
- 10.Harpaz R, Callahan A, Tamang S, Low Y, Odgers D, Finlayson S, et al. Text mining for adverse drug events: the promise, challenges, and state of the art. Drug safety : an international journal of medical toxicology and drug experience. 2014;37(10):777–790. doi: 10.1007/s40264-014-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Molecular systems biology. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haerian K, Varn D, Vaidya S, Ena L, Chase HS, Friedman C. Detection of pharmacovigilance-related adverse events using electronic health records and automated methods. Clinical pharmacology and therapeutics. 2012;92(2):228–234. doi: 10.1038/clpt.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LePendu P, Iyer SV, Bauer-Mehren A, Harpaz R, Mortensen JM, Podchiyska T, et al. Pharmacovigilance using clinical notes. Clinical pharmacology and therapeutics. 2013;93(6):547–555. doi: 10.1038/clpt.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warrer P, Hansen EH, Juhl-Jensen L, Aagaard L. Using text-mining techniques in electronic patient records to identify ADRs from medicine use. British journal of clinical pharmacology. 2012;73(5):674–684. doi: 10.1111/j.1365-2125.2011.04153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leaman R, Wojtulewicz L, Sullivan R, Skariah A, Yang J, Gonzalez G. Towards Internet-Age Pharmacovigilance: Extracting Adverse Drug Reactions from User Posts in Health-Related Social Networks. Proceedings of the 2010 Workshop on Biomedical Natural Language Processing. 2010:117–125. [Google Scholar]
- 16.Nikfarjam A, Sarker A, O'Connor K, Ginn R, Gonzalez G. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J Am Med Inform Assoc. 2015;22(3):671–681. doi: 10.1093/jamia/ocu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurulingappa H, Mateen-Rajput A, Toldo L. Extraction of potential adverse drug events from medical case reports. Journal of biomedical semantics. 2012;3(1):15. doi: 10.1186/2041-1480-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shetty KD, Dalal SR. Using information mining of the medical literature to improve drug safety. Journal of the American Medical Informatics Association : JAMIA. 2011;18(5):668–674. doi: 10.1136/amiajnl-2011-000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang C, Srinivasan P, Polgreen PM. Automatic adverse drug events detection using letters to the editor. AMIA … Annual Symposium proceedings / AMIA Symposium. AMIA Symposium. 2012;2012:1030–1039. [PMC free article] [PubMed] [Google Scholar]
- 20.Yeleswarapu S, Rao A, Joseph T, Saipradeep VG, Srinivasan R. A pipeline to extract drug-adverse event pairs from multiple data sources. BMC medical informatics and decision making. 2014;14:13. doi: 10.1186/1472-6947-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avillach P, Dufour JC, Diallo G, Salvo F, Joubert M, Thiessard F, et al. Design and validation of an automated method to detect known adverse drug reactions in MEDLINE: a contribution from the EU-ADR project. Journal of the American Medical Informatics Association : JAMIA. 2013;20(3):446–452. doi: 10.1136/amiajnl-2012-001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorbello A, Harpaz R, Szarfman A, Bodenreider O, Winnenburg R, Ripple A, et al. Detecting drug-adverse event safety signals through quantitative data mining of MEDLINE indexing terms: A pilot study; 54th Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC); 2014. Poster A-055. [Google Scholar]
- 23.Metzger Filho O, Saini KS, Azim HA, Jr, Awada A. Prevention and management of major side effects of targeted agents in breast cancer. Critical reviews in oncology/hematology. 2012;84(Suppl 1):e79–e85. doi: 10.1016/j.critrevonc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 24.Nelson SJ, Johnston WD, Humphreys BL. Relationships in Medical Subject Headings (MeSH) Relationships in the organization of knowledge. 2001:171–184. [Google Scholar]
- 25.Jones SC, Sorbello A, Boucher RM. Fluoroquinolone-associated myasthenia gravis exacerbation: evaluation of postmarketing reports from the US FDA adverse event reporting system and a literature review. Drug safety : an international journal of medical toxicology and drug experience. 2011;34(10):839–847. doi: 10.2165/11593110-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 26.Bellon A, Perez-Garcia G, Coverdale JH, Chacko RC. Seizures associated with levofloxacin: case presentation and literature review. European journal of clinical pharmacology. 2009;65(10):959–962. doi: 10.1007/s00228-009-0717-5. [DOI] [PubMed] [Google Scholar]
- 27.Medical Subject Headings (MeSH) http://www.nlm.nih.gov/mesh/
- 28.RxNorm. http://www.nlm.nih.gov/research/umls/rxnorm/
- 29.RxNorm API. http://rxnav.nlm.nih.gov.
- 30.Alternatives to practolol - the argument in more detail. Drug and therapeutics bulletin. 1975;13(23):92. [PubMed] [Google Scholar]
- 31.Aviles A, Arevila N, Diaz Maqueo JC, Gomez T, Garcia R, Nambo MJ. Late cardiac toxicity of doxorubicin, epirubicin, and mitoxantrone therapy for Hodgkin's disease in adults. Leuk Lymphoma. 1993;11(3–4):275–279. doi: 10.3109/10428199309087004. [DOI] [PubMed] [Google Scholar]
- 32.Kasper S, Barnas C, Heiden A, Volz HP, Laakmann G, Zeit H, et al. Pramipexole as adjunct to haloperidol in schizophrenia. Safety and efficacy. Eur Neuropsychopharmacol. 1997;7(1):65–70. doi: 10.1016/s0924-977x(96)00393-8. [DOI] [PubMed] [Google Scholar]
- 33.Harpaz R, Odgers D, Gaskin G, DuMouchel W, Winnenburg R, Bodenreider O, et al. A time-indexed reference standard of adverse drug reactions. Sci Data. 2014;1:140043. doi: 10.1038/sdata.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ICH guideline E2B (R2), Electronic transmission of individual case safety reports message specification. Final Version 2.3. 2001 Feb 1; Document Revision ed. [Google Scholar]
- 35.Winnenburg R, Bodenreider O. A framework for assessing the consistency of drug classes across sources. Journal of biomedical semantics. 2014;5:30. doi: 10.1186/2041-1480-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]