Abstract
Advances in optical manipulation and observation of neural activity have set the stage for widespread implementation of closed-loop and activity-guided optical control of neural circuit dynamics. Closing the loop optogenetically (i.e., basing optogenetic stimulation on simultaneously observed dynamics in a principled way) is a powerful strategy for causal investigation of neural circuitry. In particular, observing and feeding back the effects of circuit interventions on physiologically relevant timescales is valuable for directly testing whether inferred models of dynamics, connectivity, and causation are accurate in vivo. Here we highlight technical and theoretical foundations as well as recent advances and opportunities in this area, and we review in detail the known caveats and limitations of optogenetic experimentation in the context of addressing these challenges with closed-loop optogenetic control in behaving animals.
Introduction
Closed-loop control theory refers to a large body of work in the engineering literature concerned with using an error signal— that is, the difference between measured output and a desired target trajectory—to guide changes in control inputs to a system. Automobile cruise control is a familiar example: the car measures its speed and uses error-sensing negative feedback to accelerate or decelerate in order to maintain a target output speed. Inherent to such closed-loop control is the availability of a target (goal speed), some system control inputs (acceleration/deceleration), and measured system outputs (measured speed). Applied to neuroscience, closed-loop control could guide perturbations of neural systems (neurons and circuits) to achieve sophisticated, real-time control over neural dynamics and animal behavior and would generate, refine and confirm circuit-based models of the underlying system in the process.
Optogenetics, a methodology that allows millisecond-scale optical control of neural activity in defined cell types during animal behavior (in some cases at single-cell resolution in living mammals; Prakash et al., 2012; Rickgauer et al., 2014; Packer et al., 2015), is well-suited for closed-loop control in biological systems (for a history and overview of optogenetics, see Fenno et al., 2011). We therefore define “closed-loop optogenetics” as closed-loop control theory applied to optogenetic stimulation, inhibition, and modulation. In closed-loop optogenetics, the control input is a structured, time-varying light stimulus that is automatically modulated based on the difference between desired and measured outputs, which may include behavioral, electrophysiological, or optical readouts of activity generated by the biological system. In neural systems, closed-loop optogenetics could allow important basic-science investigations of adaptation, plasticity, and neural state changes, as well as online tuning of optogenetic inputs in vivo (to achieve specific output parameters), and online system-identification of neural circuits (that is, choosing optogenetic stimuli online to “reverse engineer” neural circuits by causally establishing functional circuit architecture), among other possibilities. Moreover, clinical closed-loop control of optical stimulation will likely be necessary to build and maintain models used to inform next-generation retinal, cochlear, motor, and neuropsychiatric prosthetics that adaptively interface with neural circuitry subject to changing internal and environmental dynamics. Indeed, preclinical evidence for closed-loop optogenetics in seizure detection and prevention has already been demonstrated (Paz et al., 2013; Krook-Magnuson et al., 2014, 2015).
To fully leverage the potential of closed-loop optogenetics, optical interventions must continue to grow more similar in scale and complexity to natural circuit activity dynamics, a need that has already spurred significant optogenetic technology development. Nevertheless, few studies have achieved closed-loop optogenetic control to date. Indeed, most published optogenetic experiments in behaving animals can be categorized instead as open loop (Figure 1A), even if still activity-guided or activity-informed in various offline ways. In the latter class of experiments, information from the literature or from direct neural recording is used to guide selection of light stimulus parameters (for example, pulse frequency), but without directly observing and feeding back the neural effects of the optogenetic stimulation online. For example, stimulation delivered as phasic bursts of light was chosen prior to experimentation to evoke high levels of dopamine release from ventral tegmental area (VTA) neurons (Gunaydin et al., 2014; Tsai et al., 2009) with behavioral impact compared to pulse number-matched low frequency (tonic) stimulation (Tsai et al., 2009), all directly informed by previously published recordings of dopamine neuron activity (Schultz, 2007) and quantitative mapping of dopamine release to carefully titrated pulse frequency (Adamantidis et al., 2011). This activity-informed, open-loop stimulation pattern was used to stimulate behaving animals, and the resulting changes in behavior quantified without recourse to feedback (Figure 1B). With some exceptions, this open-loop but activity-guided control is currently the standard in the optogenetics literature.
Figure 1. Open-Loop and Closed-Loop Optogenetic Control.
(A) Open-loop control (as a box diagram; Åström and Murray, 2010): control inputs to the system are chosen beforehand to reach a target system response, and are not affected by the measured output.
(B) A recent example of activity-guided open-loop optogenetics; control inputs (light pulses chosen to evoke short, separated phasic bursts of dopamine neuron activity in the ventral tegmental area) were chosen beforehand based on previous electrophysiological and optogenetic literature (left). Behavioral outputs were measured once and quantified (right). Adapted with permission from Gunaydin et al. (2014).
(C) Closed-loop control box diagram: control inputs are chosen online to minimize the difference between the measured system output and the target outcome this difference is (the “error”).
(D) An example of closed-loop optogenetics. Control inputs to inhibitory, fast-spiking (FS), parvalbumin-positive interneurons were generated online conditional on measured pyramidal (PY) neuron spike times. The figure shows PY cell responses to nonrhythmic synthetic EPSCs (center trace) with and without this closed-loop optical feedback inhibition. Closed-loop feedback was critical to inducing gamma oscillations based on PY firing. Adapted with permission from Sohal et al. (2009).
In contrast, closed-loop optogenetics uses simultaneous readout of neural activity or behavior to make real-time decisions about how and when to stimulate optogenetically, using measurements to guide stimulation in a closed feedback loop (Figure 1C). For example, in the initial closed-loop optogenetics experiment, Sohal et al. (2009) used dynamic clamp (a closed-loop control method based on electrophysiological methods; Sharp et al., 1993; Prinz et al., 2004) to drive inhibitory parvalbumin-positive interneurons with optogenetic stimulation triggered by observed pyramidal neuron spikes, thereby implementing circuit-level feedback inhibition (Figure 1D). Closed-loop optogenetic technology was crucial in demonstrating that the natural firing patterns of pyramidal cells could directly drive an increase in gamma-frequency power (Sohal et al., 2009). Although open-loop stimulation at gamma frequencies could also evoke increases in gamma in measured local field potentials (Cardin et al., 2009), closing the loop with real-time feedback to trigger circuit inhibition conditional on native activity established a plausible circuit-level mechanism for gamma oscillations mediated by interactions between fast-spiking inhibitory parvalbumin neurons and pyramidal cells. Further, closed-loop stimulation showed a causal effect of gamma on the efficient flow of information through the circuit, whereas randomly removing an equivalent number of spikes (i.e., implementing inhibition without real-time feedback) had no such effect (Sohal et al., 2009).
Subsequently, Paz et al. (2013) closed the loop to target thalamocortical neurons in injured epileptic cortex of awake rats, successfully interrupting seizures defined by EEG and behavior using real-time, closed-loop, optical inhibition. Using online detection of seizures near the time of onset to conditionally hyperpolarize targeted neurons using the optogenetic inhibitor eNpHR3.0 (Gradinaru et al., 2010), this study provided initial evidence that thalamocortical neuronal activity is necessary for poststroke epilepsy and suggested a therapeutic direction for otherwise untreatable epilepsies (Paz et al., 2013); conditional stimulation based on real-time readout of neural activity was necessary for effective timing of optogenetic intervention. Krook-Magnuson et al. (2014) further demonstrated that closed-loop excitation or inhibition of parvalbumin-expressing neurons in the cerebellum resulted in a decrease in temporal lobe seizure duration, and that closed-loop optogenetic hyperpolarization of granule cells in dentate gyrus efficiently terminated spontaneous temporal lobe seizures while activation of the same cells significantly worsened spontaneous seizures (Krook-Magnuson et al., 2015).
Similar closed-loop manipulations have recently been used to better understand the causal role of theta oscillations in information encoding and retrieval (Siegle and Wilson, 2014) and of genetically targeted cell types in high-frequency ripple oscillations in the hippocampus (Stark et al., 2014). Siegle and Wilson used millisecond-timescale, closed-loop control of inhibitory neurons in mouse dorsal hippocampus to gate hippocampal outputs at specific phases of the hippocampal theta cycle during a spatial navigation task. Closing the loop in vivo was necessary to provide adequate temporal precision and accuracy within the theta cycle to target optogenetic manipulations to particular phases of naturally generated theta rhythms on a trial-by-trial basis (Siegle and Wilson, 2014). This causally demonstrated that the falling or rising phases of theta had different effects depending on behavioral context: hippocampal CA1 inhibition at the peak of theta improved navigational accuracy when external cues were available, while hippocampal inhibition at the trough improved accuracy when behavioral guidance was based on internal signals alone (Siegle and Wilson, 2014). Stark et al. used high-density electrical recordings and multisite optogenetic stimulation, leveraging the speed of closed-loop methods to define causal roles for targeted pyramidal and interneuron types in maintaining and pacing sharp-wave ripple events. Pyramidal cell activity was reported to be necessary for sustaining ripple events while parvalbumin-positive interneurons were found to pace but not to cause ensemble spiking; closed-loop optogenetic stimulation based on online detection of sharp-wave ripples was needed to determine cell-type roles during these brief, dynamic events (Stark et al., 2014).
In the above examples of closed-loop optogenetics, stimulation or inhibition was achieved using real-time hardware systems to process electrophysiological data online and then conditionally modulate the light source following specific on-off control rules. Carefully measured behavioral, rather than electrophysiological, variables may be used in a similar fashion. O’Connor et al. (2013) elegantly demonstrated such an approach by targeting optogenetic stimulation to single barrels of somatosensory cortex and using real-time measurements to optogenetically mimic touch-evoked activity in layer 4 neurons during whisking. In this case, closed-loop photostimulation was sufficient to evoke behavior consistent with illusory perception of an object if stimulation occurred during a bout of whisking. Yoking precisely timed and calibrated optogenetic stimulation to milli-second-timescale whisker position allowed the authors to determine that instantaneous whisker position was not required for object localization (O’Connor et al., 2013).
All of the investigations mentioned thus far effectively utilized on-off control, that is, turning on or off a control input conditional on some event occurring. However, there exists a much broader class of closed-loop control strategies that have not been, but could be, built into optogenetic experiments (see examples in Table 1). These have been developed and applied in a vast engineering literature with examples stretching back to the 19th century (James Clerk Maxwell’s “On Governors,” 1868; reprinted in Maxwell, 2003), and some have recently been theoretically extended to the control of individual neurons (Schiff, 2012; Dasanayake and Li, 2011; Ahmadian et al., 2011; Danzl et al., 2009) and neural populations (Ching and Ritt, 2013; Liu et al., 2010; Schiff and Sauer, 2008). These strategies, coupled with the emerging technologies reviewed below, could have a profound influence on the conduct of optical physiology, allowing realtime adaptation to animal state, enforcement of physiological constraints on evoked patterns, calibrated control with cellular resolution, and a variety of important experimental controls that were previously inaccessible.
Table 1.
Control Table
| Control Type | Description | Common Example |
Pros | Cons | Neuroscience Applications |
Key References |
|---|---|---|---|---|---|---|
| On-off | When a certain condition (e.g., temperature set point), is reached, turns control input on or off |
Bimetallic domestic thermostat |
Very simple, easy to implement |
Nonadaptive, overly simplistic for many applications |
Conditional inhibition for seizure prevention (Paz et al., 2013); Motion- dependent optical stimulation (O’Connor et al., 2013) |
Sohal et al., 2009; Paz et al., 2013; O’Connor et al., 2013; Krook-Magnuson et al., 2014; Siegle and Wilson, 2014; Stark et al., 2014; Krook-Magnuson et al., 2015 |
| Proportional | Adjusts control input in direct proportion to current error; classical, model- free approach used in simple SISO systems |
Fly-ball governor, toilet bowl float proportioning valve |
Simple, fast, relatively easy to implement |
Unstable at rapid response times |
Very fast, but unlikely to be as useful as PI/PID control except under extreme time or computational constraints |
Maxwell, 2003; Åström and Murray, 2010 |
| Proportional integral (PI) and proportional integral derivative (PID) control |
Adjusts control input in direct proportion to current error, as well as to the error’s time integral and derivative; classical, model- free approach used in most SISO systems |
Automobile cruise control |
Simple, scalable, optimal for first- order (PI) and second-order (PID) linear processes without time delays; widely applied in real- world applications |
Does not account for time delays, switching dynamics, or time varying parameters |
Potential application: fast, real-time all- optical control for SISO and SIMO systems (Figures 2A–2F and 4C–4F) |
Åström and Hagglund, 2006; Åström and Murray, 2010 |
| Model predictive control (MPC) |
Uses a model of the system being controlled to accounts for time delays by predicting future states; modern and model-based |
Most industrial process control; autonomous vehicles |
Can be multivariable, robust, and nonlinear; accounts for time lags in the control process |
Requires a model of the system obtained by system identification |
Potential application: neural microcircuit control (Figure 5D) |
Rawlings, 2000; Maciejowski, 2002; Qin and Badgwell, 2003; Bertsekas, 2005a; Wirsching et al., 2007; Wang and Boyd, 2011b |
| Switching dynamical system (hybrid system) |
MPC that switches between different control models based on changes in measured dynamics |
Provably safe flight-mode switching algorithms for autopilot in commercial aircraft |
Models large changes in system dynamics that are hard to capture in one model |
Requires estimation of multiple models and change point detection |
Potential application: large changes in brain dynamics like sharp wave ripples versus theta in hippocampus, sleep/wake, etc. |
Branicky, 1998; Egerstedt, et al., 2003 |
| Robust control | MPC that allows control without knowing the distribution of error and is insensitive to modest parameter changes |
Chemical process control |
Allows control of worst case deviations from the target trajectory; does not require noise assumptions |
Can be more computationally expensive and tedious to implement; can degrade performance to increase robustness |
Potential application: accounting for large, intermittent disturbances |
Dullerud and Paganini, 2005 |
| Adaptive control | Common robust control type that adjusts model parameters as the system changes over time |
Airplane roll dynamics |
“Self tuning”; multivariable; inherently nonlinear |
Best for smoothly or slowly changing parameters, not rapid state changes |
Potential application: online adaptation to habituation or plasticity effects |
Bertsekas, 2005a; Ogunfunmi, 2007; Åström and Wittenmark, 2013 |
| Stochastic model predictive control |
MPC that models unobservable disturbances in the state evolution of the system |
Modern building climate control (based on occupancy, weather, and changing electricity costs) |
Less pessimistic than robust control; can allow for modest disturbances while maintaining performance |
Hard to solve in practice; approximate solutions are often necessary |
Potential application: accounting for modest intermittent disturbances |
Mesbah and Streif, 2014; Paulson et al., 2014 |
| Optimal control | General approach to solving control problems using optimization theory |
Aircraft performance optimization; time optimal satellite launching |
General framework for solving control problems using constrained optimization |
Can be computationally intensive |
Potential application: single neuron or small subset of neuron control with safety and/or physiological constraints |
Bertsekas, 2005a; Ogata, 2010 |
| Suboptimal control |
Stops short of the ideal optimal solution or makes approximations in order to speed up computations |
Autonomous helicopter flight; large, distributed systems control |
Deals with “curse of dimensionality,” time constraints, and imperfect state information |
Solution is suboptimal; performance guarantees are limited |
Potential application: neural microcircuit control (Figure 5D) for large numbers of neurons with constraints |
Kosut, 1970; Bertsekas, 2005b; Zeilinger et al., 2011; Wang and Boyd, 2011a |
It is perhaps surprising that while closed-loop optogenetic control has been possible for several years using either electrical recording or behavior to modulate optogenetic stimulation, to the best of our knowledge only a few papers have utilized feedback control in this way (e.g., Sohal et al., 2009; Paz et al., 2013; O’Connor et al., 2013; Krook-Magnuson et al., 2014, 2015; Siegle and Wilson, 2014; Stark et al., 2014). This is unlikely to be due to the technical and experimental challenges involved in undertaking such investigations, since neurobiologists are accustomed to the design and implementation of experiments characterized by computational and technical complexity. There may be, however, a cultural gap between biologists and engineers regarding available tools, techniques, and motivation for closed-loop optical control and related technologies in systems engineering. Here we seek to address the latter challenge by helping to unite the relevant literatures on optical actuators, optical sensors, electrophysiology, genetic and optical targeting strategies, and the engineering literature on system identification and control, all from the perspective of closed-loop optogenetics. Throughout, we seek to frame biological applications in the language of systems and control theory, as already used effectively in engineering for understanding and controlling complex dynamical systems. Considered along the way are the multiple technical limitations and potential confounds of optogenetic experimentation; we have previously described these caveats and challenges in detail along with relevant experimental design guidelines (e.g., Gradinaru et al., 2007; Yizhar et al., 2011a; Mattis et al., 2012; Ferenczi and Deisseroth, 2012; Deisseroth, 2014), but activity-guided and closed-loop methods now substantially augment these approaches for careful and rigorous conduct of optogenetics.
Electrical/Optical Devices Enabling Closed-Loop Control in Rodents and Primates
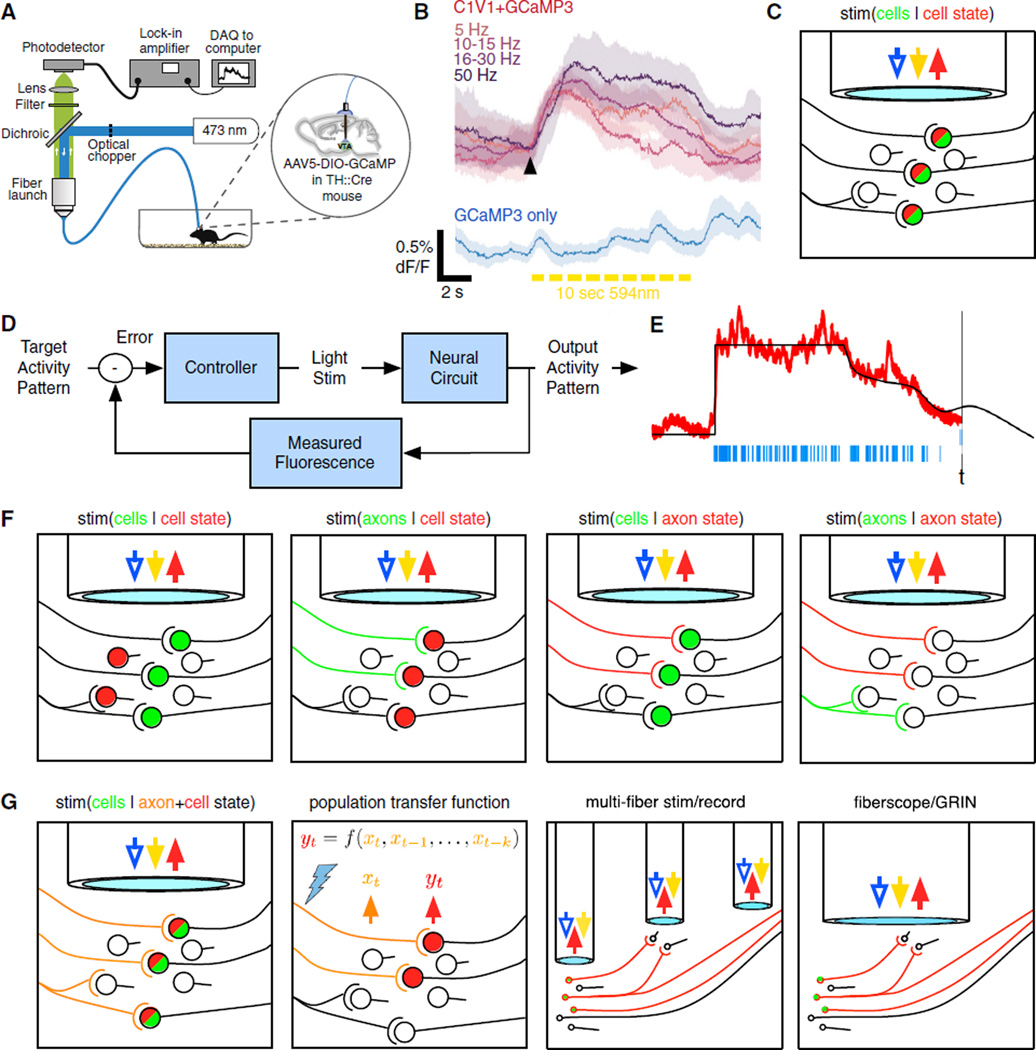
In the papers described above, digitized electrophysiological measurements provided a readily utilizable, submillisecond output source for closed-loop optogenetic control because real-time systems already exist for electrophysiological applications (Paz et al., 2013; Prinz et al., 2004). For in vitro electrophysiology, local stimulation with a guided light source (e.g., Tye et al., 2011) or integration of optical fibers into patch pipettes (Katz et al., 2013) can allow for relatively precise targeting of light as the modulated feedback signal, and various structured light approaches have already been applied for optogenetic manipulations in slice and culture (discussed in detail in a later section). For in vivo applications, the optrode (Gradinaru et al., 2007) is the simplest and most widely used device for integrated electrical recording and optical feedback and has seen several design improvements including a coaxial, tapered design (Zhang et al., 2009), a glass-coating optrode application for deep structures in primates, and an integrated µLED optrode designed with closed-loop optogenetic applications in mind (Cao et al., 2013). In systems engineering, these devices would be classified as single-input single-output (SISO) systems (Levine, 1999; Åström and Murray, 2010), allowing a single electrical measurement of the system (output) and a single optical control input to use in controlling the system (Figures 2A–2C).
Figure 2. Combination Electrical-Optical Devices for Closed-Loop Optogenetic Control.
(A–C) Single-input, single-output (SISO) systems: optrode (Gradinaru et al., 2007), optopatcher (Katz et al., 2013), and integrated µLED optrode for chronic implantation (Cao et al., 2013).
(D–F) Single-input, multiple-output (SIMO) systems: optetrode (in cross-section, fiber optic is blue, electrodes gold; Anikeeva et al., 2012), 16-site neural probe with a single optical waveguide (Cho et al., 2010), Utah array modified to include a single tapered optrode (Zhang et al., 2009).
(G–J) Multiple-input, multiple output (MIMO) systems: multishank silicon probes with integrated diodes (Stark et al., 2012), multipoint emitting tapered optical fibers combined with a silicon probe (Pisanello et al., 2014), optical fiber bundle with multiple electrodes (in cross-section, fiber bundles are blue, electrodes gold; Hayashi et al., 2012), glass optrode array (Abaya et al., 2012b). Gray box (implant innovations).
(K) Highly flexible biomimetic all-polymer fiber probes appropriate for the spinal cord and peripheral nervous system (inset shows example cross-section; Lu et al., 2014).
(L) Ultrathin, mechanically compliant, deep-brain-compatible electrodes with multiple embedded miniaturized µILEDs (Kim et al., 2013b).
(M) Fiber probe that allows for simultaneous optical stimulation, neural recording and drug delivery in behaving mice (“Ch,” the coaxial drug delivery channel; Canales et al., 2015).
(N) Wireless powering of intracranial (Kim et al., 2013b) or external skull-mounted (Wentz et al., 2011) devices has been described, though requiring a bulky head-mountable power receiver (reviewed in Warden et al., 2014); however, wireless devices for optogenetic stimulation that are fully implantable within the organism have now been designed as illustrated (Yeh et al., 2014; Yeh et al., 2014, Society for Neuroscience abstract).
A variety of strategies have been employed to increase the number of available electrical measurements while maintaining a single optical input, including optetrodes (Anikeeva et al., 2012), 16-site neural probes with a single waveguide (Cho et al., 2010), commercially available 16-recording-site, single-fiber probes (Kravitz et al., 2010), and Utah arrays modified to include a tapered optrode (Zhang et al., 2009), yielding various single-input, multiple output (SIMO) systems (Figures 2D–2F). To stimulate and record from multiple sites, multiple-input multiple output (MIMO) systems for electrical readout and optogenetic control now include multishank silicon probes with integrated light guides or diodes (Stark et al., 2012; Royer et al., 2010), Utah-slant optrode arrays (Abaya et al., 2012a), glass optrode arrays (Abaya et al., 2012b), optical fiber bundles bundled with multiple electrodes (Hayashi et al., 2012), and multipoint emitting tapered optical fibers combined with silicon probes (Pisanello et al., 2014; Figures 2G–2J). Feasibility for spike-detecting, closed-loop SIMO control has recently been demonstrated (Nguyen et al., 2014) using template matching to do online spike detection on 32-channel tetrode recordings (system outputs) and using detected spikes to control optogenetic stimulation through a single fiber optic (system input) at ~8 ms closed-loop latency in awake rats.
Categorization of systems into SISO, SIMO, MISO, and MIMO systems (Figure 2) is useful for deciding which control strategies should and can be employed. For example, with just one input and one output, SISO systems do not require consideration of correlations between inputs and outputs and allow parameters to be fit very rapidly (Åström and Hägglund, 2006; Åström and Murray, 2010). In contrast, MIMO systems generally model the effect of each input on each output, resulting in potentially increased flexibility and accuracy of control at the cost of greater computational complexity (naively viewed, this complexity is combinatorial in the number of possible relationships between inputs and outputs; but see the section on closed-loop control of microcircuits for other approaches). For the purposes of analysis, it may be useful to reduce more complicated systems to the simpler cases, for example, treating a SIMO system as multiple SISO components, or a MIMO system as multiple MISO systems. For example, the distances between individual optrodes on a Utah optrode array may allow treating each optrode as a separate SISO system, allowing much faster online control. The theory for SISO systems is by far the most developed (Åström and Hägglund, 2006). However, if evoked correlations between shanks are important, a more complicated MIMO model will be necessary, requiring more modern multivariate control strategies (Bertsekas, 2005a; Ogata, 2010). Finally, we note that although MISO systems are not currently represented in terms of optical control inputs and electrical outputs of extant devices, they remain important; for example, controlling a single behavioral output with multiple optogenetic control inputs would be a highly interesting MISO system.
Beyond the number of optical inputs and electrical outputs, several advances in optoelectrical devices for closed-loop optogenetics are relevant for determining the best device for a given application (Figures 2K–2N). First, the rigidity of implanted elements—including electrodes, fiberoptics, and other waveguides commonly employed for combined electrical recording and optogenetic stimulation—can damage tissue and may be too inflexible to use in small, more mobile structures such as the spinal cord or peripheral nerves (Lu et al., 2014; Llewellyn et al., 2010; Pashaie et al., 2014). To address these limitations, both highly flexible, biomimetic, all-polymer fiber probes appropriate for the spinal cord and peripheral nervous system (Lu et al., 2014; Towne et al., 2013) as well as ultrathin, mechanically compliant, deep-brain-compatible electrodes with multiple embedded miniaturized µILEDs one-thousandth the size of conventional LEDs (Kim et al., 2013b) have been developed to facilitate simultaneous optical stimulation and electrical recording during behavior (Figures 2K and 2L). Related flexible polymer technologies have enabled the development of fiber probes that allow for simultaneous optical stimulation, neural recording, and drug delivery in behaving mice (Figure 2M; Canales et al., 2015) as well as largely transparent, flexible electrocorticography (ECoG) grids that conform to the folds of the brain and are compatible with wide-field or structured optogenetic stimulation (Richner et al., 2014; Minev et al., 2015).
Recent developments in remote wireless powering of devices (Figure 2N) have resulted in receivers the size of peppercorns (Yeh et al., 2014), raising the tantalizing possibility of miniature, biocompatible, self-contained implants where power receiver, recording transmitter, miniature LEDs and electrodes could all be subcutaneously implanted (Yeh et al., 2014, Society for Neuroscience abstract). This would enable closed-loop optogenetics in behaving animals unhindered by large headmounted electronics, or even by lightweight flexible connectors (although systems neurobiology in rodents and monkeys has been successfully built upon such flexible connectors, with complex behaviors carried out by animals linked to readout systems by long, lightweight wires or even thinner fiberoptics, spanning the full range of well-validated motor, cognitive, social, neuropsychiatric, and other behavioral domains (reviewed by Moser et al., 2015; Deisseroth, 2014; Shenoy and Carmena, 2014; Wilson and McNaughton, 1994). Indeed, despite the abundance of possibilities, adoption of multichannel, conformal, and wireless devices has so far been slow, perhaps since the fiberoptic neural interface (Adamantidis et al., 2007) has been widely adaptable in biological discovery and also enables the two crucial capabilities of deep brain projection targeting (Deisseroth, 2014), and readout of neural activity in cells and projections during free behavior (Gunaydin et al., 2014).
Implantable devices like miniaturized LEDs not only cannot alone provide such readout capability, but can also emit substantial heat, the effects of which must be carefully measured and/or controlled for in vivo (Yizhar et al., 2011a; Li et al., 2013b; Yeh et al., 2014, Society for Neuroscience abstract). LED-based devices can be designed with more inputs and outputs, but any associated increase in size and complexity may lead to more damage to tissue when implanted (a caveat not unique to electro-optical devices). Difficulty in the fabrication of more complicated devices may also hinder adoption of the technology without productive industrial partnerships, which in turn can be slow to develop for the research community (although device designs are typically made broadly available by the originating labs). And if these devices are placed not inside but outside the brain with no fiberoptic interface (as in the initial noninvasive optogenetic control of motor output through the intact adult mouse skull; Gradinaru et al., 2007), the resulting surface interfaces (though functional) can provide neither of the two key functions of versatile projection targeting nor deep brain activity readout.
Although they are often integrated with optical control hardware and allow exquisite temporal precision, electrical recording methods exhibit well-documented limitations relative to optical approaches for readout of neural circuits. Electrical readout of activity cannot readily be genetically specified, only active cells can be observed, electrode arrangement may severely limit sampling of neural activity (especially spatially), and it is difficult or impossible in most cases to relate recorded cells to detailed anatomy or molecular phenotype. Although new all-optical approaches (discussed next) are beginning to address these gaps, electrical methods still have some strong advantages including the speed of electrical recordings, the ground-truth status of the electrically measured spike readout as fundamental to neuronal communication, the availability of commercially available real-time systems for spike waveform identification and analysis, and the current utility of electrical devices in the clinical setting.
All-Optical Closed-Loop Optogenetics: General Principles and Constraints
Optical technologies provide unique capability for precisely targeting neurons specified by type and wiring for both measurement and perturbation. Already, optical measurements of neural activity with single-cell resolution in dense populations have extended our understanding of neural activity beyond the sparse and activity-biased measurements achievable with electrode-based approaches. For example, imaging of neural activity in vivo has revealed that while activity can be sparse at any one instant (that is, with activity restricted to a small fraction of the entire neural population), correlated activation occurs in small subsets of neurons—often termed ensembles—that are scattered throughout the brain volume. Optical recording studies have revealed ensemble-like activity in sensory-evoked responses (e.g., similarly tuned neurons; Ko et al., 2011), motor-related activity (Komiyama et al., 2010), spatial navigation (Dombeck et al., 2010), and even spontaneous activity (Ko et al., 2011). Moreover, correlations within ensembles appear to increase in magnitude in relevant neurons during learning (Komiyama et al., 2010). It has also been revealed that brain wiring is not random (in the Erdös-Rényi sense), but demonstrates rules of specificity even at the microcircuit level that can map onto ensemble identity (Ko et al., 2011). Activity patterns in these ensembles have the potential for distinct influences on downstream targets (for example, higher-order cortical areas receive specific subsets of information from lower areas; Glickfeld et al., 2013), likely sampling from specific ensembles of neurons (Sato and Svoboda, 2010). Further, projection-targeting optogenetic experiments have shown that different efferent pathways from the same anatomical structure can have very different behavioral outcomes, since they arise from distinct populations that are anatomically intermixed at the cellular level (e.g., Kim et al., 2013a). But separating these populations experimentally is not always easy because distinct neural ensembles, especially of excitatory neurons, often belong to a similar genetic class and are thus difficult or impossible to target separately without including additional strategies based on function or wiring. Given that these ensembles are dynamically active in time and change with animal state, online targeting based on rapid observation and analysis of functional patterns and behavior will be required to accurately play back observed patterns of endogenous activity. This level of control will be essential for testing the causal role of specific activity patterns in generating subsequent activity patterns and behavior.
The importance of optogenetically targeting neurons based on functional ensemble identity has been recently demonstrated by using activity-dependent labeling of neurons with an inducible system based on activity-regulated c-fos promoter elements (Liu et al., 2012). With this approach it was possible to use fear conditioning in a specific context to selectively drive expression of channelrhodopsin in neurons of the dentate gyrus that were strongly active during a pharmacologically induced time window (~1 day), and to then reactivate the fear response in a different, habituated context using only optogenetic stimulation of the opsin expressing neurons in dentate gyrus (Liu et al., 2012; Figure 3F). This finding (along with a number of elegant controls in the paper) highlights the importance of targeting a specific ensemble of neurons active during behavior. While relatively little is known about the cellular identity of these activity-defined ensembles, it is possible that additional information about cell type, including molecular detail (Micheva and Smith, 2007) and wiring information (see circuit-targeting strategies reviewed below and Bock et al., 2011) could be further obtained (for example, leveraging recent advances in multiround molecular and anatomical analysis in large, intact cleared brains; Chung et al., 2013; Tomer et al., 2014). While of great value, these activity-dependent opsin expression approaches have limited temporal resolution compared with the relevant neural activity timescales. This temporal resolution also extends the time over which additional neurons less related to the specific behavior under study could be labeled by this approach.
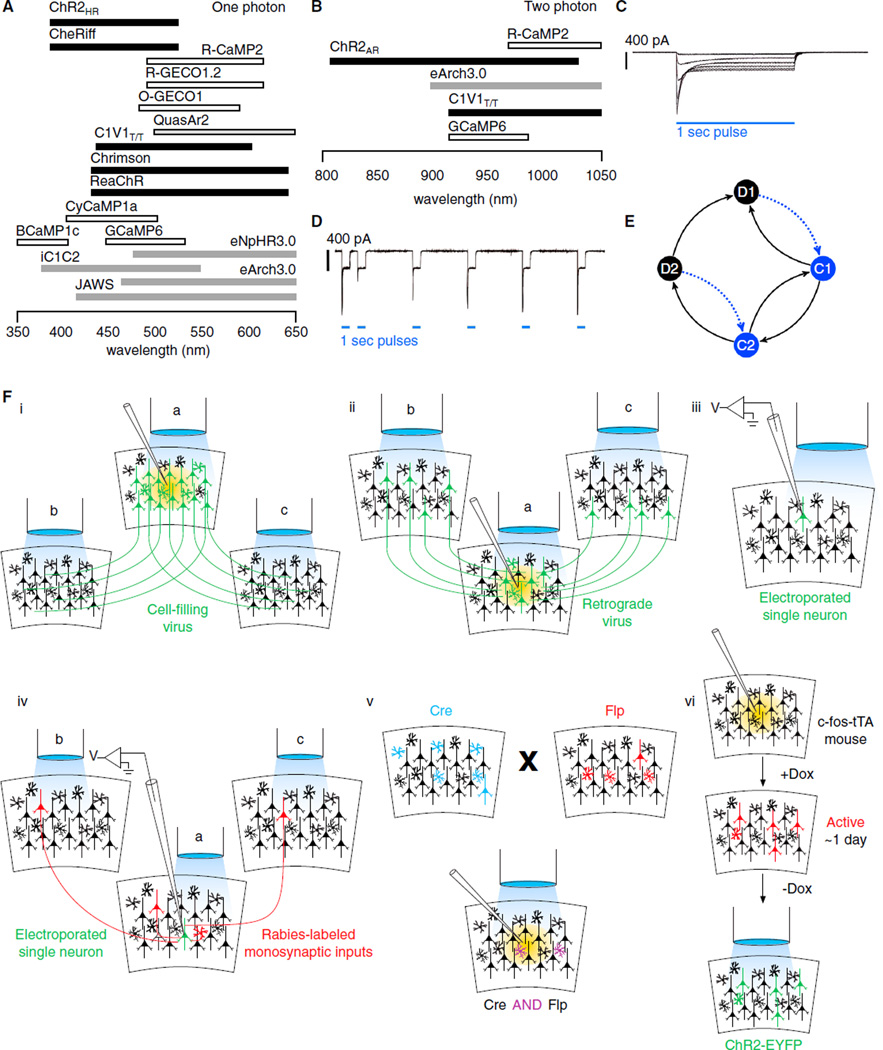
Figure 3. Consideration of Sensor, Actuator, and Targeting Parameters for Closed-Loop Optogenetics.
(A) Bars showing action spectral ranges of potentially compatible opsins and sensors (thresholded at 20% of peak; note that this is just a cross-section of representative proteins—there are many more in the literature) for one-photon closed-loop optogenetic control. Ranges for each protein are shown as horizontal bars, with color indicating excitatory actuators (black), inhibitory actuators (gray), calcium sensors (white) or voltage sensors (gray with black border; adapted from Mattis et al., 2012; Hochbaum et al., 2014; Inoue et al., 2015; Wu et al., 2013; Yizhar et al., 2011b; Klapoetke et al., 2014; Lin et al., 2013; Akerboom et al., 2012, 2013; Chen et al., 2013c; Berndt et al., 2014; Gradinaru et al., 2010; Chuong et al., 2014). Note that this is high-level comparison and that it is critical to examine nonnormalized spectra and other published details for each protein (for a developing resource in this area, see http://actionspectra.org).
(B) Bars like those in (A), but showing thresholded action spectra ranges for proteins compatible with two-photon closed-loop optogenetic control (adapted from Inoue et al., 2015; Prakash et al., 2012; Chen et al., 2013c; Akerboom et al., 2012).
(C and D) (C) Peak-to-steady-state ratio of a typical channelrhodopsin changes with stimulation light intensity (D) and with history of stimulation over time (J. Mattis and K.D., unpublished data); scale bars, 400 pA; light pulses, 1 s.
(E) These nonstationary effects can be efficiently modeled using simple linear dynamical systems (Nikolic et al., 2009; Nagel et al., 2003; Hegemann et al., 2005), e.g., describing the photocycle dynamics in terms of two conducting states that conduct at different rates (C1 and C2) and two dark states that are differentially excited to the high (C1) or low (C2) conducting states when photoexcited (D1 and D2, respectively; see equations 1a-1d of Hegemann et al., 2005). Here arrows denote transitions states governed by rate constants, and blued dotted lines show excitation from dark to conducting states by photostimulation.
(F) Targeting strategies for mammalian gene expression in specific neurons, cell types, and circuits. (i) Gene expression can be restricted to a particular brain area by localized viral injection; the virus is engineered to introduce a specific gene(s) of interest (e.g., an opsin or GECI), and may be biased to specific cell types by combinations of promoter used and viral tropism (Nathanson et al., 2009). Optogenetic light stimulation at the site of the injection (fiber a) will potentially perturb multiple outputs from the source brain area. Stimulating opsins located in projections (optical fibers b and c) from the source area can distinctly perturb specific pathways from the source brain area. (ii) Retrograde tracing viruses can infect local axon terminals and label inputs to a given brain area. Optogenetic light stimulation can be restricted to the site of injection (optical fiber a), or specific inputs (optical fibers b and c). (iii) Gene expression specificity can be restricted even to a single neuron in vivo by single-cell transfection, e.g., by single-cell electroporation (Kitamura et al., 2008). (iv) Transsynaptic tracing rabies virus can restrict expression to monosynaptic inputs to a specific population of neurons (Wickersham et al., 2007b), cell type labeled with Cre (and targeted by expression from a Cre-dependent helper virus; Wall et al., 2010), or to a single neuron by single cell transfection methods (e.g., electroporation, Marshel et al., 2010; whole-cell configuration, Rancz et al., 2011). Light-restriction strategies are similar to as in (ii) in terms of addressing specific inputs (optical fibers b and c), with the additional advantage that the transfected neuron (or Cre-defined population) at the site of the optical fiber a can be targeted for specific gene expression, separate from and in addition to the gene(s) expressed by the rabies virus. (v) A multitude of transgenic mouse lines exist for targeting specific cell types with recombinases to restrict gene expression in recombinase dependent fashion (for example, see Taniguchi et al., 2011). Intersectional strategies can increase specificity through “Boolean logic” operations of combinations of Cre and/or Flp recombinase expression (including AND, NOT, OR, NAND, NOR, XNOR, and XOR) (Fenno et al., 2014). (vi) Targeting gene expression to neurons activated in a specific time window (hours to ~1 day) is possible by taking advantage of immediate early gene expression and inducible genetic targeting systems (Liu etal., 2012). Importantly, for all of these examples, closed-loop control strategies can be implemented using all-optical methods (e.g., imaging and stimulating through the same fiber), as further elaborated in Figure 4. See Huang (2014); Luo et al. (2008), and Packer et al. (2013) for recent reviews including additional gene-targeting strategies.
Despite these and other recent advances in targeting gene expression to specific cell types based on genetics, wiring, and functional properties, matching controlled real-time dynamics to native dynamical patterns will require new complementary closed-loop approaches to ultimately understand the causal underpinnings of neural computation and behavior. Recent advances in optical methods for both observation and control, combined with precise genetic targeting, now offer a promising set of approaches for reaching these goals. While all-optical, closed-loop control (using both optical sensors of neural activity and optogenetic tools as actuators of circuit activity feedback control) has yet to be demonstrated, all of the technologies necessary are now developed and available. Further, all-optical open-loop control at cellular resolution has already been enabled using approaches defined in vivo (Prakash et al., 2012; Rickgauer et al., 2014; Szabo et al., 2014; Packer et al., 2015) as well as in vitro (Nikolenko et al., 2007; Rickgauer and Tank, 2009; Dal Maschio et al., 2010; Prakash et al., 2012; Packer et al., 2012). In this section we review three key technologies critical for achieving all-optical, closed-loop optogenetic control: the compatible optogenetic actuators and optical sensors, the tools for targeting both actuators and sensors to genetically or anatomically defined cells, and the mathematical and computational tools necessary for closed-loop control of neurons and neural circuits.
Optogenetic Actuation and Optical Sensing of Neural Activity
Here we focus on the specific optogenetic tools that are compatible with available optical sensors of neural activity, and identify parameters of these tools that will be important in the context of control. By compatible, we mean with limited spectral overlap between the illumination wavelengths necessary for imaging the activity sensor and for actuating the optogenetic tool—a critical (and historically limiting) issue for all-optical optogenetic approaches. Although optogenetic constructs and reporters of neural activity can be used together (Zhang et al., 2007; Airan et al., 2007), because most opsins have broad excitation spectra all-optical applications will require either limits on the light intensity that can be used for imaging illumination or other means to minimize cross-stimulation (e.g., limiting light duration or using patterned illumination to avoid certain cells or excitable regions). Figures 3A and 3B show a selection of opsins and sensors with windows in their spectral overlap that could allow for combination in closed-loop optogenetic control. Although we show simple bars generated by thresholding spectra in order to visually accommodate a number of sensors and actuators, we note that it is important to consider the full, non-normalized action spectra when planning a particular experiment (see http://actionspectra.org for an interactive resource for comparing sensor and actuator action spectra).
In the one-photon case, new genetically encodable Ca2+ indicators (GECIs) that are red-shifted (Inoue et al., 2015; Akerboom et al., 2013; Wu et al., 2013, 2014; Hochbaum et al., 2014; Zhao et al., 2011) are compatible with blue light-activated control tools such as the Chlamydomonas channelrhodopsin ChR2 and its variants (Akerboom et al., 2013; Inoue et al., 2015). Conversely, the initial demonstration in neurons of red light excitation (C1V1T/T; Yizhar et al., 2011b) and inhibition (eNpHR3.0; Gradinaru et al., 2010) with microbial opsins raised the prospect of combination with blue light-activated GECIs (Figure 3A,B). Such red-shifted actuation or readout will allow deeper onephoton circuit interrogation due to reduced scattering at the longer illumination wavelength. However, for deeper imaging and greater spatial restriction of photostimulation to the single-cell level, two-photon laser scanning microscopy (TPLSM; Denk et al., 1990) for activity imaging and optogenetic control (Prakash et al., 2012; Rickgauer and Tank, 2009) will likely be necessary. Two-photon methods have been shown to be effective in combination with GCaMP variants for short wavelength-driven, two-photon imaging and C1V1 opsin variants for long wavelength-driven control (Rickgauer et al., 2014; Packer et al., 2015).
Although rapidly developing with many advances still to be made, genetically encoded voltage indicators (GEVIs; Siegel and Isacoff, 1997; Ataka and Pieribone, 2002; Sakai et al., 2001) could allow for similar targeting strategies to be applied to optical membrane voltage readout with higher temporal precision (for reviews see Knöpfel et al., 2006; Peterka et al., 2011; Mutoh et al., 2011; Perron et al., 2009a); note also that many promising non-genetically targeted strategies also exist (Peterka et al., 2011). Again, minimizing direct stimulation of coexpressed optogenetic tools by imaging light will be critical to determining which opsin-GEVI combinations are viable, making low imaging illumination intensities and minimally overlapping action spectra critical. Much progress in higher signal-to-noise GEVIs has been made in recent years, including for fluorescent protein-based (Akemann et al., 2012; Jin et al., 2012; St-Pierre et al., 2014; Gong et al., 2014), and microbial-opsin-based (Kralj et al., 2012; Flytzanis et al., 2014) reporters, although the high light intensities required to image at fast frame rates with currently available probes still present a challenge for all-optical applications. GEVIs with blue range (Lundby et al., 2008; Perron et al., 2009a) and red range (Perron et al., 2009b) action spectra have been developed, and opsin-GEVI pairs have been shown to work together in cultured cells in vitro (Hochbaum et al., 2014); continuing progress in this area may also eventually allow alloptical voltage readout and photostimulation in vivo.
It is also important to note that the choice of actuator will constrain the range of firing rates that can be evoked (and the latency and jitter), defining the range of patterns that can be reliably controlled (Mattis et al., 2012; Gunaydin et al., 2010). From the systems engineering perspective, how well-actuated the system is can have a strong impact on whether the system is controllable—meaning that the chosen system can be driven from any starting condition to any desired final state in a finite amount of time. This controllability concept along with observability (see below) together play a central role in the design of control systems in state space (Åström and Murray, 2010; Kalman, 1960). In neuroscience applications of optogenetics, controllability relates to whether patterns of optogenetic stimulation can evoke desired target patterns of neural activity or behavior (e.g., emulating endogenously observed patterns). As a simple example, consider a single fast-spiking, opsin-expressing neuronto be the system under study. If certain patterns of activity (e.g., 40 Hz activity) cannot be reliably evoked using a particular opsin because of slow opsin photocycle dynamics, then the system is not controllable for that application. More generally, systems themselves may or may not be controllable depending on actuator properties, neural state space dynamics (Paninski et al., 2010; Kemere et al., 2008) and the arrangement of actuators and sensors (Summers and Lygeros, 2014; Ching and Ritt, 2013). Indeed, while parameters of optogenetic tools chosen can strongly influence whether the system will be controllable, at the circuit level interactions between cells and projections will result in more complicated dynamics than in the single neuron case. For example, it is possible to evoke gamma oscillations at the circuit level using a relatively slow opsin variant ChR2(H134R) (Sohal et al., 2009; Cardin et al., 2009), although the same construct cannot always reliably drive individual pyramidal cells at such frequencies (Gunaydin et al., 2010). It is notable that the classical controllability definition may require some relaxation for some neural systems, in particular for underactuated cases (like single optical fiber preparations; Ching and Ritt, 2013). We return to these issues below in the section on closed-loop control of circuit dynamics.
Figures 3C and 3D demonstrate another well-known property of optogenetic control tools important for effective closed-loop control: steady-state to peak current ratios change both with light intensity (top panel) and over time based on stimulation history (bottom panel; reviewed in Mattis et al., 2012). This effect can be limited by using any of the several opsins with steady-state/peak ratios approaching 1 (reviewed in Mattis et al., 2012). If this is not feasible, for example due to opsin/sensor compatibility concerns, nonstationary effects of opsin photocycle dynamics can be modeled by using observations to fit parameters in three- or four-state linear models that closely approximate opsin photocycle dynamics and would be easily run in real time (Figure 3E; Nikolic et al., 2009; Hegemann et al., 2005) such that they could be used to vary light intensity online to result in stationary control combined with other methods (e.g., Ahmadian et al., 2011).
The case of hyperpolarizing optogenetic tools is interesting from the closed-loop control perspective. First, an engineered hyperpolarizing ion pump (Mattis et al., 2012) has been shown to be effective for single-cell resolution two-photon inhibition in scattering mammalian brain tissue (Prakash et al., 2012), but light-driven chloride and proton pumps from archaeal halobacteria conduct only a single ion per photon, making responses (though quite linear with light intensity) significantly more inefficient than those of channel opsins. Recently developed chloride-selective hyperpolarizing channels (Berndt et al., 2014; Wietek et al., 2014) so far have only been employed in the onephoton regime; these are more efficient because they directly conduct many ions per photon, but this fact also complicates dynamics since conductance direction (as with native inhibition mechanisms) is conditional on membrane potential and ion gradients. Closing the loop in this case could allow more guided modulation of light-activated chloride conductance based on neural activity level, as well as allowing adaptive modulation of light to achieve complex waveforms. It is notable that placing inhibition in a closed-loop control framework would allow specifying target levels of inhibition that may decrease activity to a desired nonzero set point rather than aiming to simply silence neural firing, allowing a more nuanced approach to optogenetic inhibition.
In choosing optical sensors for closed-loop optogenetics, there are several important parameters to consider (beyond limiting crosstalk with complementary optogenetic control tools); these considerations share some similarity to those of concern in imaging-only applications (Peterka et al., 2011; Wilms and Häusser, 2014) but with new significance in the control framework. In particular, temporal resolution (e.g., on/off kinetics) and signal-to-noise over a sufficient dynamic range are important for observability. As an example, if we are interested in evoking or suppressing individual action potentials in a particular temporal pattern, but our sensor is not sensitive enough to effectively report single action potentials, then we will not know if our optogenetic manipulation has succeeded and will not be able to use feedback to reliably control the system to this level of precision. If, however, our goal is to evoke bursting of a particular magnitude or to limit firing rates below a certain observable level then the same indicator may be sufficient. In the case of GECIs, various kinds of saturation and buffering can have similar effects, thereby decreasing spike resolution at high firing rates (Vogelstein et al., 2009). Theoretical analyses, such as those for limits of detection of spikes with calcium and voltage sensors (Wilt et al., 2013; Sjulson and Miesenböck, 2007), can be used to set reasonable bounds on expected detection given collection and sensor statistics.
In mammalian tissue, scattering sets limits on both controllability (Yizhar et al., 2011a) and observability by attenuating signal and stimulation light in a depth- and tissue-dependent fashion (Svoboda et al., 1997). Although forward models of attenuation in principle can be used to alter illumination intensity in an open-loop, depth-dependent manner (Vellekoop et al., 2008) and potentially at real-time rates (Conkey et al., 2012), closed-loop modulation using direct feedback from observed activity will be more robust to model errors and more corrective of model mis-specification in terms of measured neural activity. Since both observability and controllability will degrade with tissue depth, tissue scattering should be explicitly taken into account and calibrated for in open- and closed-loop feedback models whenever possible. Further, protein expression levels from cell to cell can be variable, requiring cell-wise calibration of light stimulus intensity to evoked activity—a step more effectively accomplished using rapid optical feedback. Finally, in vivo applications can experience motion-related fluorescence changes when there has been no actual change in neural activity. Genetically encoded ratiometric sensors (Thestrup et al., 2014) or online optical correction (Chen et al., 2013b) and motion modeling may help to avoid artifactual signals from corrupting feedback control inputs. In the case of actuation, particularly at the single-cell level as described below, motion can also lead to mistargeting of light patterns away from desired neurons without closed-loop adjustment of light patterns based on detected motion.
Circuit and Cell-Type Targeting Strategies
Beyond optical actuator and sensor parameters, continued refinement in targeting expression of these proteins to prespecified populations of neurons using genetic tools will be critical for making stimulation and feedback possible in defined cell types and circuits. Several approaches are available to target specific subsets of neurons based on cell type or wiring (Figure 3F; for review, see Huang, 2014; Luo et al., 2008; Packer et al., 2013). For example, transgenic mouse and rat lines expressing recombinases in subsets of neurons may be combined with recombinase-dependent gene delivery systems such as viral approaches or crosses with other mouse lines (e.g., Witten et al., 2011; Taniguchi et al., 2011) to restrict expression of control and readout proteins to cells of interest. The combined use of multiple recombinases (e.g., Cre and Flp) has recently enabled multiple-feature “Boolean logic” neuron targeting based on multiple genetic or topological parameters (Fenno et al., 2014), greatly improving potential targeting specificity. An expanding number of line crosses are possible for recombinase-dependent expression of both activity indicators and optogenetic actuators, allowing specific cell types to be reliably targeted for all-optical interrogation.
Cell types have also been preferentially targeted based on specific developmental stage (e.g., birth date) as in the case of in utero electroporation to target specific layers in cortex (Saito and Nakatsuji, 2001; Gradinaru et al., 2007; Petreanu et al., 2007) and by promoter-based strategies and viral tropism (Adamantidis et al., 2007; Nathanson et al., 2009). Applying these approaches has been integral to observations that genetically defined cell types have distinct influences on circuit processing, as in the case of interneuron subtypes (Isaacson and Scanziani, 2011; Luo et al., 2008). As discussed above, activity-dependent gene expression systems have enabled labeling and interrogation of activity-defined ensembles of neurons in vivo (Liu et al., 2012; Guenthner et al., 2013). Finally, targeting based on wiring is possible with circuit-tracing viruses (reviewed in Luo et al., 2008). For example, rabies virus infects neurons trans-synaptically in the retrograde direction and has been modified to carry genes for fluorescently labeling neurons and expressing activity sensors and optogenetic actuators (Wickersham et al., 2007a; Osakada et al., 2011). Furthermore, rabies has been modified to spread only to monosynaptic inputs to a given brain region defined by injection site (Wickersham et al., 2007a), to genetically targeted cells (Wickersham et al., 2007b) such as a given cell type defined by recombinase-dependent infection (Wall et al., 2010), or even to a single neuron defined by in vivo single-cell electroporation (Marshel et al., 2010) or patching (Rancz et al., 2011). All of these methods on their own are powerful for targeting interrogation-tool function (e.g., in the case of single cell electroporation: Pala and Petersen, 2015; Judkewitz et al., 2009; Kitamura et al., 2008), alongside other targeting methods involving anterograde projection targeting with lentivirus or adeno-associated virus (AAV, discussed below) and trans-synaptic targeting with HSV (Lo and Anderson, 2011) or wheat germ agglutinin (WGA) in AAV (Gradinaru et al., 2010; Braz et al., 2002; Xu and Südhof, 2013; Gunaydin et al., 2014).
Important considerations in selection of targeting strategy are the gene expression timescale needed and the strength of promoter to be used because—as with any transgene–genetically encoded fluorescent proteins, optogenetic actuators, and optical sensors all carry the risk of possible toxic effects of high or long-term protein expression. In the case of opsins, this risk is now routinely addressed with use of appropriate promoters and viruses suitable for expression timing and strength (reviewed in Yizhar et al., 2011a), protein modifications including routine addition of short molecular motifs borrowed from mammalian membrane proteins that allow these evolutionarily distant proteins to be efficiently and safely trafficked within the cell over the experimental timescales required (Gradinaru et al., 2008, 2010), and proper experimental design controls including incorporation of light on/off controls and behavioral and physiological comparisons with control-transduced (non-opsin expressing) animals at baseline (Yizhar et al., 2011a). Such controls are now standard practice for any such experimental intervention in neuroscience and are extended to the fluorescent activity reporters and structural markers as well; even with native proteins, overexpression over time causes toxicity. Integration of activity sensing with control also facilitates monitoring and testing for typical activity patterns at baseline and in response to perturbation in the presence or absence of different expressed markers.
Control Theory and System Identification for Neurons
Once appropriate optogenetic actuators and sensors have been targeted to cells of interest, online algorithms are needed for designing stimulation with light conditional on observed neural activity or behavior (Figure 1C). Optical sensors measure activity, and this information is used by the controller to estimate the current state of the neural system. This neural state estimate is then used as input to algorithms that compute the necessary control action (e.g., light input) to achieve a target activity level or pattern. Finally, this control action is carried out and the reaction of the system is again recorded by the sensors, closing the loop.
To provide some general background on the types of control algorithms, Table 1 reviews existing approaches in closed-loop control theory. Note these broad categories of closed-loop control are not exclusive and can be combined, nor are these all the categories that could be highlighted (the systems engineering and control literature is quite substantial). However, a few major distinctions are worth keeping in mind when considering methods for feedback control. In general, such methods can be categorized according to linearity (linear versus nonlinear), time representation (continuous versus discrete), and domain representation (frequency versus time) (Aström and Murray, 2010).
Continuous frequency-domain approaches generally tend to take a more classical view and have a powerful and deeply developed theory for SISO systems going back more than two centuries (Åström and Hägglund, 2006). Time-domain, or “state-space” methods (Kalman, 1960), which have already seen wide application for modeling dynamic systems in neuroscience (Paninski et al., 2010; Shenoy and Carmena, 2014), are a cornerstone of modern control theory and are well-suited to MIMO systems. We anticipate classical methods like the fast proportional-integral-derivative (PID) control used in automobile cruise control (Table 1) will be most useful in single fiber/electrode SISO or SIMO applications, while state-space approaches will typically be more appropriate for MIMO experiments involving arrays, fiber bundles, or imaging with structured illumination—consistent with applications in existing control literature (Table 1).
Effective integration of closed-loop control theory with neuroscience will be highly interdisciplinary, even beyond the advanced optics and physiology involved, ideally extending to the involvement of computational and anatomical expertise. First, because neurons and neural circuits are complicated, nonlinear, nonstationary systems composed of heterogeneous cell types that change dynamically on millisecond timescales, and because safety and physiological constraints are important, tools from modern and nonlinear control theory that are robust, adaptive, and allow formulation as an optimization problem with constraints will be most appropriate in all but the simplest or most time-limited cases (Ogata, 2010; Bertsekas, 2005a; Kuo, 1982). Further, because closed-loop depends on realtime computation to keep up with rapid ongoing dynamics, there is always a computational budget that places limitations on model complexity. In many modern applications such constraints may lead to only partially solving optimization problems at each time step, resulting in “suboptimal” control (Bertsekas, 2005a) which nonetheless can be very effective in real-world applications where time budgets are limited (Boyd et al., 2014; Wang and Boyd, 2010, 2011b; Bertsekas, 2005a; Wirsching et al., 2007). On the anatomical side, genetic-targeting strategies and post hoc molecular phenotyping (with, for example, coregistration to high-resolution anatomical data; Tomer et al., 2014) will be necessary for identifying cell-type roles in dynamics, as well as aid in understanding projection patterns alongside circuit tracing technologies (which will bring its own computational challenges).
Recent work on closed-loop control for more effective and safer electrical microstimulation for electrical deep-brain stimulation (EDBS) in Parkinson’s disease has developed continuous-time nonlinear control tools for both SISO (Danzl et al., 2009) and MIMO (Liu et al., 2010) electrical recording and stimulation devices. Although validated only via simulation, several important points arise in this work. Danzl et al. (2009) demonstrated in simulation that synchronized activity can be actively disrupted using minimal intensity inputs chosen online using constrained nonlinear control (solving a constrained optimization problem) to use minimal electrical inputs in a SISO system. Liu, Oweiss, and Khalil simulated closed-loop control in an all-electrical MIMO system for EDBS and raised key points directly relevant to closed-loop optogenetics for MIMO systems, perhaps most importantly showing that a properly designed MIMO feedback controller can control a subset of simulated neurons to follow a prescribed spatiotemporal firing pattern despite the presence of unobserved disturbances (an inevitability in most neural systems of interest, as most of the brain will remain unobserved. Furthermore, this paper showed that a simplified linear-nonlinear model can be quite effective in controlling firing rates, despite strong simplifying assumptions (this is important for systems where speed dictates hard computational constraints). In addition to the practical goal of safer, more effective deep-brain stimulation, the resulting spatiotemporal patterns identified could themselves be of intrinsic value in providing new insights into how neural circuits process information.
Additional theoretical work involves optimal control theory to design control inputs that evoke desired spike patterns with minimum-power stimuli in single neurons (Dasanayake and Li, 2011) and ensembles of neurons (Ahmadian et al., 2011) using electrical current injection. Robust computational models using similar methods have been developed for optimal control of simple models of spiking neural networks (Li et al., 2013a) and more abstractly, individually controlling coupled oscillators using multilinear feedback (Kano and Kinoshita, 2010). Given that converging evidence suggests that abnormalities in synchronized oscillatory activity of neurons may have a role in the pathophysiology of some psychiatric disease (Uhlhaas and Singer, 2006) and considering their established role in epilepsy, it seems fruitful to continue considering oscillations themselves as a direct target of closed-loop optogenetic control (Sohal et al., 2009; Witt et al., 2013) alongside control of spiking neurons.
In a seminal paper, Ahmadian et al. (2011) presented a fast (convex), discrete-time approach finding the best time-dependent modulation of electrical or optogenetic inputs to cause a neuron to follow a target spike pattern as closely as possible (subject to hardware limitations and physiologically inspired safety measures). Importantly, the method was validated in vitro (using electrical stimulation), demonstrating optimal control with biological constraints directly applied to spike control in single neurons. In simulation, this paper also showed extensions to multicell stimulation including modeling of crosstalk. Although this treatment of optogenetic stimulation did not include the particular dynamics of photoexcitable channels and pumps nor model the effects of optical recording, it could be readily extended to include photocycle dynamics (as pointed out by the authors) and combined with existing methods for spike estimation from optical physiology data.
So far, all discussion here of closed-loop control has implicitly assumed existence of a model relating optical inputs to effects on the neural system that the controller uses to choose these optogenetic inputs (this model is called the “input transfer function” for classical systems or the “input equation” for state-space models; Åström and Murray, 2010). However, in all but the simplest cases we usually start an experiment with inexact knowledge of how light inputs will perturb the system. Previous work has addressed this by mapping stimulus parameters, varying light intensity and/or frequency to gain insight into the relationship between optogenetic stimulation and behavior (e.g., Adamantidis et al., 2007; Tsai et al., 2009; Cardin et al., 2009) and thoroughly characterizing the response of individual cells expressing optogenetic constructs to light impulses in vitro (Mattis et al., 2012). In systems engineering, estimating this relationship between inputs and outputs of a system is known as system identification (Ljung, 1998, 2010; Zadeh, 1956). System identification is a critical step for any control application and from the perspective of “reverse engineering” the brain, a major end in itself for understanding neural circuits. Indeed from this reverse-engineering perspective control is in some sense a means of validating the quality of system identification (which is generally a model of how the system functions). Of course, in some applications like brain machine interfaces and prosthetics, closed-loop control performance may be more important than whether the identified model best approximates the true neural system.
In the SISO case in particular, it is possible to effectively achieve “system identification” without directly modeling the system. For example, in a PID controller, closed-loop control can be obtained by iteratively fitting three model parameters with no explicit model of the system being controlled (Åström and Hägglund, 2006). This is called “black box” modeling (to differentiate from “white box” modeling in which one is given a full and accurate physical model of the system) and is used quite widely (Åström and Hägglund, 2006; Ljung, 1998, 2010). An example of black box modeling mentioned above would be fitting the relationship between light inputs for optogenetic control and simultaneous fiber photometry measurements (see below). In this case, limited system knowledge and imperfect control (e.g., optogenetic underactuation; Ching and Ritt, 2013) will likely lead to a “model-free” approach like PID control. More common in MIMO systems where we have some limited physical information are “gray box” models, for which we can build a parametric model based on our imperfect physical knowledge, and fit the parameters of the model using observed input-output data (Ljung, 1998, 2010). In general, it is important to consider system identification both as a means to understanding the system (“reverse engineering”) and as a precursor to controlling the system, whether or not the control application is for basic science purposes like causal model validation, or for practical purposes like neural prosthetics.
For those interested in learning more about system identification and control, many excellent references are available on systems engineering and control theory that are relevant to neural control with optogenetics, including a survey of neural control engineering (Schiff, 2012), a recent introduction to systems and control theory for biologists (Control Theory for Bioengineers, H.M. Sauro, 2015, Ambrosius Publishing ISBN-13: 978–0982477380; available online), general texts on feedback control (Åström and Murray, 2010), detailed engineering texts on modern control theory (Franklin et al., 2015; Kuo, 1982; Ogata, 2010) and nonlinear control theory (Vidyasagar, 2002; Khalil, 2002), and an overview (Ljung, 2010) and textbook (Ljung, 1998) on system identification. A brief nonmathematical introduction to control theory can be found in Mitra and Bokil (Mitra and Bokil, 2007, chapter 3). For those with more engineering background the two-volume treatment of optimal control and dynamic programming by Bertsakas (Bertsekas, 2005a) is both accessible and comprehensive.
Observing and Controlling Population and Projection Dynamics in Behaving Animals
Specification of defined neural pathways for optogenetic perturbation has been achieved in a number of ways (reviewed in Deisseroth, 2014; Packer et al., 2013; Zalocusky and Deisseroth, 2013). One approach (called projection targeting) relies on optogenetic actuator expression in an upstream neuronal population defined by focal virus injection; a subset of these neurons (defined by having efferent connections to a spatially-separated downstream brain area) is then selected by restricting light delivery to excite or inhibit the axons of this neuronal subpopulation in the target brain region (or, more generally, in a location that distinguishes the pathway of interest) in vivo during behavior (Gradinaru et al., 2009; Tye et al., 2011; Stuber et al., 2011). This approach depends on functional expression of optogenetic actuator in the axons of targeted neurons (Gradinaru et al., 2007; Petreanu et al., 2007), which may require longer expression times or axon-targeting expression strategies to achieve adequate expression levels (Mattis et al., 2012; Gradinaru et al., 2010). Other approaches use various forms of retrograde tracing to target actuator expression in neurons that project to the target brain area (e.g., Gradinaru et al., 2010; Gunaydin et al., 2014), which can be useful for comparing multiple projections to a specific target (see also Britt et al., 2012) and can help avoid concerns about stimulating only fibers of passage in a target brain region (from the same source but to different, unintended targets). Analysis and understanding of projection anatomy are also key to best design and interpretation of projection-targeting experiments.
For in vivo experiments, the fact that during behavior microbial opsin-expressing projections can be either excited (Gradinaru et al., 2009; Tye et al., 2011; Stuber et al., 2011) or inhibited (Tye et al., 2011; Stuber et al., 2011) is useful for establishing necessity and sufficiency of anatomically defined cells in driving specific behaviors. It is also important to note that in many cases, excitation of opsin-containing axons can lead to antidromic (reverse-propagating) action potentials potentially reaching the cell body and/or other axon collaterals of an excited neuron (Deisseroth, 2014). In general this is a desired effect, for recruiting a cell type in its entirety defined by connectivity; the wiring-defined cell type is more likely to be a functional unit in nervous system processing than excitation of a specific subbranch of an axon, which will not typically happen in isolation.
However, it may also be desired in certain cases to isolate the influence of a specific collateral projection in a specific brain region, for example to gain knowledge of finer-scale organization of neural pathways and in certain clinically oriented applications (Deisseroth, 2014; Li et al., 2012; Gradinaru et al., 2009). Here, it is helpful that optogenetic inhibition of the axon or its branch will remain local to the site of light delivery, and can be used to provide that level of specificity where desired. In the context of excitation, control experiments with local pharmacological blockade (e.g., Schneider et al., 2014; Znamenskiy and Zador, 2013) or direct modulation of other known pathways can help determine if those projections influence the same or distinct output responses (Kim et al., 2013a), as needed. All of these methods are now widely used in optogenetics when axon collaterals, rather than projection-defined cell types, are the circuit element of interest.
By utilizing closed-loop optogenetics additional approaches become possible, since stimulation light intensity, duration, and frequency could be tuned to the level sufficient to attain the desired excitatory output pattern while also minimizing side effects. For example, simultaneously observing activity in the upstream brain area containing cell bodies of the projection or in other recipient areas during optogenetic stimulation of the projection could confirm whether antidromic or collateral stimulation occurs (e.g., by imaging or electrically recording action-potential-generated activity in the cell bodies, in collateral branches, or in other recipient areas). If such activity is observed, the same measurements could be used to further determine whether specific light delivery parameters as needed promote or reduce the effect. In a similar vein, light delivery patterns could be calibrated online to minimize overall light delivery needed to achieve a desired activity pattern in the target brain region, for example, as measured by fiber photometry (Gunaydin et al., 2014) or when a specific behavioral outcome is achieved.
More generally, without real-time observation of activity it is not clear for most interventions (including electrical and optogenetic stimulation) whether the intervention provides stronger or weaker, or more or less synchronous, activity in the target population than naturally occurs. However, natural activity patterns recorded using an optical fiber could be used in a closed-loop optogenetic framework in order to evoke target activity levels similar to those already observed in the same population of cells, keeping the evoked activity within physiological ranges and potentially allowing replay of naturally occurring patterns. Such fiber-based, all-optical approaches would enable the all-optical closed-loop experiments described here (among other opportunities), but would require new methodological developments to be realizable.
To reach this goal, methods for population recording from a genetically specified cell population and genetically and topologically defined projections during behavior have recently been demonstrated (Gunaydin et al., 2014), dovetailing naturally with standard optogenetic approaches for open-loop control (Figure 4A). Figure 4B illustrates the initial simultaneous fiber photometry and optogenetic stimulation of a genetically targeted neural population (in this case, tyrosine hydroxylase-expressing VTA neurons) in a freely behaving animal; the red-excited opsin is the C1V1 variant C1V1T/T (Yizhar et al., 2011b) alongside the blue-excited indicator GCaMP3, targeted as described earlier (Gunaydin et al., 2014) but here using a titer-matched mixture of two recombinase-dependent (DIO) viruses (I. Kauvar, L.G., K. Zalocusky, and K.D, unpublished data). Although the low illumination intensities enabled by fiber photometry allow this opsin-sensor combination, recently developed red sensor variants may be even more suitable when combined with blue-light-sensitive opsins (Inoue et al., 2015; Akerboom et al., 2013; Wu et al., 2013, 2014; Hochbaum et al., 2014; Zhao et al., 2011), as these will more effectively limit opsin-sensor spectral overlap and allow a large range of optogenetic actuators to be used. Still, care must be taken to avoid (or model) artifacts during optogenetic stimulation since fluorescence from these sensors can fluctuate in blue light and may resemble neural activity (Akerboom et al., 2013; Wu et al., 2013). Nevertheless, in a manner crucial for closed-loop and activity-guided optogenetics, all-optical minimally invasive single-fiber recording and optogenetic stimulation in a genetically targeted deep brain cell population during behavior is now possible (Figure 4B).
Figure 4. Opportunities for Fiber-Based Closed-Loop Optogenetics.
(A) Single fiber observation and control device. (Left) Fiber photometry setup showing light path for fluorescence excitation and emission through a single 400 micron fiber optic implanted in the ventral tegmental area (VTA). (Right) Recombinase-dependent viral targeting of GCaMP5 to VTA dopamine neurons. Adapted with permission from Gunaydin et al. (2014).
(B) All-optical interrogation of frequency-dependent effects using simultaneous fiber photometry and optogenetic stimulation of targeted neurons in dopaminergic neurons in the VTA; traces recorded using fiber photometry from a freely behaving animal expressing C1V1T/T and GCaMP3 (red-toned traces; n = 3 animals) or GCaMP3 only (blue trace; n = 1) targeted as previously described (Gunaydin et al., 2014); photostimulation of C1V1 with 594 nm laser light at 7.9 mW/mm2 (measured at tip of fiber) was administered at 5, 10, 12.5, 16, 25, or 50 Hz at 50% duty cycle (so total light power over the 10 s of stimulation was the same on each trial), and VTA dopaminergic activity was recorded simultaneously by illuminating GCaMP3 with 473 nm light at 0.2 mW/mm2 (leveraging the sensitivity of fiber photometry to keep illumination intensity low to minimize cross talk); dF/F traces grouped by 5, 10–15, 16–30, and 50 Hz (3 animals with 15, 14, 29, and 14 trials per group, respectively) are shown with bootstrapped 68% confidence intervals for the mean, baseline-corrected to coincide 250 ms prior to photostimulation onset (black arrow) for clearer comparison of poststimulation effects (stimulation effects add to a naturally time-varying baseline); recording GCaMP3 expression alone with the same stimulation and recording protocol is shown in blue (1 animal, 46 stimulation trials); yellow bar indicates period of photostimulation (I. Kauvar, L.G., and K.D., unpublished data).
(C) The results shown in (B) open up the possibility of stimulating cells conditional on their simultaneously measured activity. Here and below, open blue arrow indicates excitation light, closed yellow arrow indicates imaging excitation light, and closed red arrow indicates imaging emission light. A single transduced cell body population is represented with red and green circles; stimulation may be made conditional upon parameters of observed activity state in this population below the fiber.
(D) Block diagram of closed-loop optogenetic stimulation through a single fiber; the target activity pattern is compared to current fluorescence measurements of activity, and used to choose light inputs in real time to bring the observed activity more in line with the target activity.
(E) Illustration of feedback control to achieve a theoretically desired activity waveform (black), using activity information (red) fed back from neural circuitry as modulated light pulse rate changes (blue dashes); the light pulses would be chosen online based on current observed activity; for example, at time t a pulse has been algorithmically chosen online to try and increase activity to return decreasing activity to the (arbitrary) target trajectory.
(F) Using genetic and projection-specific targeting strategies: conditional optogenetic control of one cell type based on the activity of another cell type (left), conditional control of targeted axonal projections conditional on local cell state (left center), conditional control of local cells based on axon activity (right center), and even conditional control of one axon projection based on the activity of another projection (right), are now possible.
(G) (Left) Careful use of more than one sufficiently spectrally separated GECI (e.g., those in Wu et al., 2013) or GEVI could allow separately targeted pre- and postsynaptic fiber photometry with closed-loop optogenetic stimulation. (Left center) By allowing selective stimulation of genetically and topologically targeted projections while reading out activity in stimulated and postsynaptic neurons, such a configuration would enable system identification of population synaptic effects yielding transfer functions between projections and their postsynaptic targets. More elaborate configurations might involve (right center) multiple-fiber recordings from targeted cells and projections in different brain regions, or (right) fiberscope or GRIN lenses for image-forming applications.
For genetically-targeted, closed-loop optogenetic control in vivo, the most straightforward example would be with this configuration, in which optical stimulation could be constrained by optical recording through a single fiber in the same targeted cell population: an all-optical SISO system (Figure 4C). The simplest closed-loop control in this case would resemble the cruise control example: given a set point or target trajectory for the observed activity (based on observed endogenous activity, for example), the controller tunes stimulation parameters to evoke the desired magnitude and time course of activity using online recording as feedback for how well error is minimized between a target level or time-varying trajectory and the observed activity (proportional-integral-derivative, or PID, control; Åström and Hägglund, 2006; Figures 4D and 4E; Table 1). Conditional inhibition of a genetically targeted subpopulation using the simultaneously recorded activity of that subpopulation is another straightforward example. Because fiber photometry readout is a univariate, time-varying scalar, submillisecond processing with a real-time operating system (Sohal et al., 2009; O’Connor et al., 2009; Paz et al., 2013; Krook-Magnuson et al., 2014; Siegle and Wilson, 2014; Stark et al., 2014; Laxpati et al., 2014; Krook-Magnuson et al., 2015) could be used to optogenetically clamp activity in the cells below a target level with minimal intensity inputs (Ahmadian et al., 2011). Because a constant online stream of optical information about how targeted cells are responding to the photoinhibition is available in a single channel, illumination could, in theory, be adjusted in real-time to be no more intense than needed, to adaptively increase to avoid escape from the optical clamp, to silence potential rebound activity if desired, and to reveal which time-varying pattern of inhibition was most effective at achieving these goals (itself useful informative about circuit dynamics). As mentioned above, the best control strategies for this kind of application are likely to be the fast classical proportional-integral (PI) or PID control approaches (Åström and Häg-glund, 2006) based on “black box” models of the system (that is, statistically derived models that assume little about the biophysics of the system; Ljung, 1998).
It is of course important to consider this use of optogenetics in the context of prior methods for stimulation or inhibition (lesions, pharmacology, and electrical stimulation). Proper conduct of optogenetics has long capitalized on its relative speed, reversibility, and flexibility; for example, the extent of optogenetic modulation can be smoothly varied in parametric fashion even in the same animals (from below detection limit to near seizure-promotion level) by varying intensity and/or frequency of the laser light delivered by fiberoptic or objective (e.g., Adamantidis et al., 2007). To track the effects of such mapping, it is often best practice in optogenetic circuit analysis to conduct real-time recording from circuitry (whether optically or electrically), capitalizing on the opportunity that was not present with electrical stimulation in terms of recording simultaneity and cell type targeting, nor for lesions and pharmacology in terms of temporal precision. From this perspective, closed-loop and activity-based optogenetics is moving to fully utilize natural advantages of optogenetics in terms of speed and simultaneity of observation and control. The ability to observe and evoke activity in the same genetically and topologically targeted population in a behaving animal is a new opportunity, which can be achieved even with the same versatile fiberoptic device for both quantitative photometry and control during behavior (Gunaydin et al., 2014; Figure 4B). Although it is possible to separately image native dynamics and then try to evoke a similar response in open loop fashion by designing light stimuli before the experiment, such an approach is highly sensitive to model misspecification, calibration, and state changes in the system (habituation, plasticity, motor state, etc.), and without simultaneous measurement it cannot be confirmed that the response was accurately evoked. Closed-loop feedback control now allows real-time adjustment of input parameters to keep the observed output as close as possible to a target level or time-varying trajectory (Figure 4E).
A key limitation of fiber photometry is its design for recording from populations of neurons and their processes rather than single cells, resulting in target-element averaged responses. However, leveraging the targeting strategies discussed above greatly improves the effective resolution of the method; the ability to record optically from both genetically specified cell bodies as well as topographically or genetically defined projections, and to rapidly use the resulting signal to conditionally modify optogenetic stimulation online, now enables interesting experiment types when coupled with emerging intersectional targeting (Fenno et al., 2014) and projection-based strategies. Conditional optogenetic control of one cell type based on the activity of another cell type, conditional control of targeted axonal projections to a region conditional on local cell state, conditional control of local cells based on axon recordings, and even conditional control of one axon projection based on the activity of another, all could now be done through the single fiber implant already used in standard optogenetics experiments (Figure 4F). For example, by using more than one spectrally separated GECI (such as a combination of orange and red or far-red indicators) with sufficient care it would be possible to separately record from targeted pre- and postsynaptic circuit elements (and thereby infer population-defined and averaged synaptic weights during behavior) while including closed-loop optogenetic stimulation of either the post- (Figure 4G, left) or presynaptic (Figure 4G, left middle) population.
Similar targeting strategies could be applied with multiple implants allowing readout and control at several potentially connected locations, using modified fibers to spatially modulate optogenetic stimulation while optically recording with fiber photometry, or with image-forming devices such as fiber bundles (Szabo et al., 2014) or implantable GRIN optics (Ghosh et al., 2011; Flusberg et al., 2005) that can allow near-cellular resolution imaging with optogenetic stimulation (Figure 4G; right middle and right panels). Indeed, in areas that accommodate larger implants comparable in size to those used for hippocampal imaging in vivo (Ziv et al., 2013; Barretto and Schnitzer, 2012; Dombeck et al., 2010; ~2–3 mm implant outer diameter), fiberscope imaging using a fiber bundle and implanted GRIN lens has recently been used for simultaneous imaging and photostimulation in freely moving animals (Szabo et al., 2014). In particular, Szabo et al. (2014) pioneered the use of a fiberscope combined with a phase spatial light modulator (SLM) for structured stimulation in the illumination arm and a digital micromirror device (DMD) in the imaging arm to demonstrate simultaneous structured optogenetic stimulation and near-cell-resolution imaging in behaving animals (Szabo et al., 2014). We discuss this innovation further in the section on observing and controlling circuit dynamics below.
Closed-Loop Optical Control: Implementation at the Cellular and Microcircuit Level
As described above, in vivo neural activity involves spatiotemporal patterns of cells and ensembles, with active cells interspersed among inactive cells in scattered locations throughout the brain volume. Information sent from one area to another may vary over time in ensemble-specific fashion, and activity will likely be integrated and further refined by specific wiring linking different ensembles in each area, with important rules of connectivity at the neuronal level. A straightforward example for the relevance of this point of view is sensory processing, where the existence of “parallel pathways” has been well-studied for decades (e.g., Nassi and Callaway, 2009 for a review of parallel pathways in the primate visual system). This organization is apparent from the periphery (e.g., functionally specific retinal ganglion cells) up through high-order cortical areas, where complex receptive fields and multimodal response patterns are found and thought to derive in part from combinations of specific feedforward input. In addition, these responses are further influenced by top-down and modulatory inputs as well as local circuit dynamics. Evidence suggests that computations can occur at the level of population dynamics (Mante et al., 2013; Shenoy et al., 2013), but causal understanding of how these dynamics emerge and how computations are achieved at the circuit level is largely lacking. To enable testing of causal relationships in neural circuits at this level of detail, observational and perturbational approaches should be designed to access together correspondingly fine levels of spatial and temporal resolution. Specifically for the single-cell resolution subtype of closed-loop optogenetics neural ensembles would need to be selected and perturbed in real time based on observation of behaviorally relevant activity patterns at single-cell resolution across multiple neurons in vivo.
Recent years have witnessed the development and application of a number of promising tools for reaching cellular or near-cellular resolution—both for optogenetic stimulation and for optical imaging of neural activity. When combined, these tools, along with imaging modalities like two-photon laser scanning microscopy (TPLSM; Denk et al., 1990), enable activity-dependent optogenetic control at cellular or near-cellular resolution. This in turn provides the possibility of functional measurement and conditional perturbation of cortical or subcortical microcircuits to better understand dynamics and to relate these dynamics to sensation, behavior, and internal states.
A natural categorization of these tools is once again in terms of illumination type—specifically one- versus two-photon—with direct consequences for the choice of opsin, sensor, and illumination strategy. One-photon fluorescence imaging modalities used for activity observation can be subdivided into wide-field (Ziv et al., 2013; Grienberger and Konnerth, 2012), fiber-bundle (Szabo et al., 2014; Hayashi et al., 2012), light sheet (Bouchard et al., 2015; Ahrens et al., 2013), and light field (Grosenick et al., 2009; Broxton et al., 2013; Prevedel et al., 2014; Cohen et al., 2014) approaches. One-photon structured illumination methods usable for multicell perturbational approaches include laser scanning (Wilson et al., 2012), micro-LED array (Grossman et al., 2010), digital micromirror device (DMD; Dhawale et al., 2010), light field (Levoy et al., 2009; Figure 5A), and holographic (Lutz et al., 2008; Szabo et al., 2014) illumination. In the case of two-photon imaging approaches used for activity observation, TPLSM (Denk et al., 1994), two-photon extended depth of field (EDoF; Quirin et al., 2013), and two-photon 3D random access scanning (Fernández-Alfonso et al., 2014; Cotton et al., 2013; Katona et al., 2012; Grewe et al., 2010; Duemani Reddy et al., 2008; Otsu et al., 2008; Iyer et al., 2006) all provide two and three-dimensional approaches to functional imaging. Two-photon-based methods for optical perturbation have used either laser scanning (Prakash et al., 2012; Rickgauer and Tank, 2009), temporal focusing (Rickgauer et al., 2014; Andrasfalvy et al., 2010), digital holography (Packer et al., 2012, 2015; Nikolenko et al., 2007), or a combination of digital holography and temporal focusing (Oron et al., 2012; Papagiakoumou, 2013; Bègue et al., 2013) for patterned optogenetic stimulation.
Figure 5. Multiple-Cell Control for Population-Level Closed-Loop Optogenetics.
(A and B) Promising one- and two-photon methods for simultaneous structured volumetric photostimulation. (A) Light-field illumination (Levoy et al., 2009; L.G., M. Broxton, and K.D., unpublished data; see the light field illumination section of http://clarityresourcecenter.org/functional3D.html for movies and references) allows millisecond formation of structured one-photon volumes for optical stimulation. Shown are two views of a volume rendering of a light field imaging reconstruction (Broxton et al., 2013; reconstructed with 3 3 3 3 5 micron voxels) of a volume containing multiple localized peaks generated in a fluorescent volume (a hydrogel densely populated with submicron beads using light field illumination; imaging parameters: 403 0.8; 203 0.5 NA objective; 125 micron pitch, 100% fill-factor rectangular microlens arrays; 1:0.7 demagnifying telecentric relay; DMD was a TI DLP9500, and illumination path used a matched tube lens and the same microlens array parameters; setup modeled on Levoy et al., 2009).
(B) Structured two-photon excitation with a phase SLM has been shown to stimulate multiple cells simultaneously at different planes in a volume with cellular resolution and has been validated in vitro with paired patch electrophysiology (reproduced with permission from Packer et al., 2012).
(C) System identification in a neural microcircuit: patterned stimulation is chosen online in iterative fashion, guided by optical observation of neural activity and online estimation of microcircuit connectivity.
(D) Model-predictive control in a neural microcircuit. A target pattern of cellular activity is compared to a predicted outcome of past optogenetic stimulation and the difference (error) is used to choose the next photostimulation pattern subject to constraints on the photostimulation parameters in order to minimize a user-specified cost function (e.g., squared error). The model estimated in (C) is used to make predictions about the time evolution of the system, which is subject to unobserved disturbances and measurement noise.
Given all of these options for imaging and stimulation, in addition to the potential for combinations of one-photon imaging and two-photon photostimulation, there is a dizzying array of design choices to be made in constructing a system for all-optical closed-loop cellular or near-cellular optogenetic control. We therefore focus here on fundamental trade-offs and synergies for closed-loop optogenetics in behaving animals. Driving many of these considerations are the expected trade-offs among field of view, spatial resolution, and temporal resolution. To make gains in one of these areas, it is generally necessary to sacrifice in another. Ultimately, these trade-offs are set by the information capacity of the optical system (Cox and Sheppard, 1986), where degrees of freedom are traded between space and time given a fixed optical bandwidth. Although recent developments in compressed sensing and use of prior information about sample structure are increasing the efficiency with which this information can be used (Pnevmatikakis and Paninski, 2013; Studer et al., 2012; Fitzgerald et al., 2012), fundamental spatiotemporal limitations and trade-offs remain.
In terms of in vivo imaging with single-cell resolution, two-photon laser scanning methods allow precise localization in three dimensions and optical sectioning up to the diffraction limit, but at the cost of scanning in time. Light sheet methods for functional imaging (Ahrens et al., 2013; Keller and Ahrens, 2015) improve imaging speed by scanning a sheet of light over time, and are capable of achieving high spatial resolution (Tomer et al., 2014). However, this approach requires scanning a sheet of light orthogonal to the objective, complicating optical access and largely limiting in vivo applications to relatively transparent samples imaged at a few Hz. Recently, promising methods for imaging and creating a light sheet through the same objective were introduced for mammalian imaging (Bouchard et al., 2015), although at reduced optical quality relative to standard light sheet and with depth-limitations in scattering tissue, where light sheet quality degrades with depth. Planar sampling with a light sheet provides significant speed over point-scanning modalities, but still divides the frame rate of the camera by the number of planes imaged. Extended depth of field two-photon methods give a single two-dimensional projection through a scattering volume, gaining access to more neurons over the volume for simultaneous imaging at the cost of having ambiguous axial information—although structural images can help to disambiguate axial sources (Quirin et al., 2013). One-photon functional light field imaging allows fully volumetric imaging at camera frame rates and integrates information at the sensor throughout each camera frame, giving high speeds and SNR (Levoy et al., 2006; Grosenick et al., 2009; Broxton et al., 2013; Prevedel et al., 2014; Cohen et al., 2014). However, light field methods trade spatial resolution for improved depth and temporal resolution (although in our experience, single-neuron resolution over large volumes in scattering mammalian tissue may still be obtained).
Similar trade-offs exist for photoactivation with optogenetics. Scanned diffraction-limited two-photon spots can be used to reach single neuron resolution even in vivo (Prakash et al., 2012), but do so sequentially, one cell at a time. Using 3 mm galvanometer mirrors and optimized spiral scan parameters for C1V1 activation, on-cell scan time to induce maximal photocurrent (and a single spike) takes ~2 ms, and switching between neuron locations in a 400 χ 400 micron field of view takes less than 200 µs between the most distant neuron pairs (significantly less for closer pairs), leading to an approximate addressable set of 50 neurons at 10 Hz (J.H.M. and K.D., unpublished data). The rate of sequentially stimulating groups of neurons could potentially be improved by using AODs to switch between neuron locations in tens of microseconds. However, on-cell scan time involving optimized scan velocity and scan line density to efficiently drive opsin-mediated conductances across the cell body is likely to remain the rate limiting step for sequential scanning approaches, and to achieve stimulation of larger numbers of neurons at higher rates will most likely require methods for simultaneous stimulation of multiple neurons. For example, phase spatial light modulations (SLMs) focusing multiple diffraction limited two-photon spots in three dimensions simultaneously can scan those spots over cell bodies of at least ten neurons to stimulate at the same time with single cell resolution in vitro (Packer et al., 2012; Figure 5B) and in vivo (Packer et al., 2015), as discussed further below.
As a potential fast alternative to scanning, temporal focusing (TF; Oron et al., 2012) allows the axial beam profile to be controlled independently of its lateral distribution. For two-photon optogenetic stimulation this obviates the need for scanning to stimulate a single cell and may result in faster membrane depolarization by opening a larger numbers of conductances per pulse than a diffraction-limited spot, as has been demonstrated in vitro (Andrasfalvy et al., 2010). Further, TF may be more robust to scattering than focused spots (Bègue et al., 2013; Papagiakoumou 2013; Dana and Shoham, 2011) and has been shown to work for cellular or near-cellular resolution optogenetic stimulation of single neurons within densely labeled populations in vivo (Rickgauer et al., 2014). TF in combination with digital holography has been demonstrated for multispot optogenetic stimulation in mammalian tissue in vitro (Bègue et al., 2013; Papagiakoumou, 2013; Oron et al., 2012), and a recent design in principle allows volumetric scanning of TF for sequential optogenetic stimulation (Mayblum et al., 2015). However, it is still an open question how well combined TF and digital holography approaches could scale for stimulation of larger numbers of neurons.
Scaling up multineuron stimulation approaches while maintaining precise single-cell resolution is a challenge of growing interest. Simultaneously scanned, diffraction-limited spots generated with a phase SLM have been shown to excite at least ten cells simultaneously in vivo with single-spike precision (confirmed with simultaneous optical readout, and spikes were detected with electrophysiology from one neuron when up to 20 cells were stimulated simultaneously; Packer et al., 2015). These approaches should therefore scale to at least tens of neurons, for example with improved SLMs and lasers. However, in scaling to many neurons obstacles arise in terms of achieving and maintaining rigorous multiple single-cell resolution, and avoiding tissue heating or overcoming damage effects may be needed. Observed increases in nontargeted local circuit activity may result from synaptic activation of connected neurons, direct cross-stimulation of immediately neighboring nontargeted neurons, or a combination of the two (Packer et al., 2015). It is important to note that the spatial resolution will depend crucially on whether the scanned stimulation pattern overlaps any neighboring neurons laterally, a parameter that can readily be adjusted to be more conservative (J.H.M. and K.D., unpublished data; A. Packer and M. Hausser, personal communication). In general, parameter tuning and improved optics could be used to generate more spatially restricted spots and trajectories, although more laterally limited scan patterns could result in reduced stimulation efficacy, and physical device characteristics of currently available SLMs force a tradeoff between targetable field of view and resolution (which may be adjusted depending on the experiment). Of course, it is possible that more efficient stimulation scan patterns and improvements in target cell light-sensitivity could also help scale up these technologies. More neurons could be targeted using higher power as well as more efficient (higher peak power) lasers, ultimately constrained in principle by tissue heating and nonlinear damage effects (although such limits thus far have not been reached in published work of this type). More studies and new technologies are needed to quantify and limit these effects, and to compare spot-scanning SLM and combined TF/SLM approaches. Furthermore, closed-loop approaches can be used to help determine effective stimulation patterns to achieve the desired goal (e.g., desired population dyanamics trajectory, or behavior) taking into account the controllability of the system and physiological contraints as previously described.
Patterned one-photon, two-dimensional illumination with digital micromirror devices (DMDs; Dhawale et al., 2010) and one-photon, three-dimensional illumination with light field illumination (Levoy et al., 2009; Figure 5A) are unlikely to yield true single-neuron stimulation resolution as scattered and out-of-focus light could both effectively drive opsins in neurons and dendrites adjacent to the target cell. However, these approaches would allow simultaneous patterned illumination of many neurons switchable at tens of kilohertz using current DMD technology. Such one-photon methods can also drive low-jitter spikes with considerably less power than two-photon approaches over wide fields of view. Further, these one-photon approaches are known to be compatible with most microbial opsins, including those appropriate for driving fast spiking (Gunaydin et al., 2010) and inhibitory channel opsins (Berndt et al., 2014; Wietek et al., 2014) and therefore may promise superior temporal controllability. It also remains to be seen which system identification and closed-loop control applications require absolute single-cell resolution from the photostimulation side and which do not. The answer will likely be circuit and question dependent, with one-photon methods allowing rapid differential photostimulation or photoinactivation of many neurons while two-photon methods stimulate fewer cells (based on higher power requirements per cell) at slower speeds for more refined microcircuit mapping when cross-stimulation cannot be modeled or is otherwise unacceptable to the experimental design. Finally, one-photon methods are currently much more easily powered and miniaturized (Ziv et al., 2013; Ghosh et al., 2011; Kim et al., 2013b; Yeh et al., 2014), and are therefore better candidates for commercial and prosthetic applications. As always, questions of the spatial and temporal resolution needed for desired levels of control will have practical consequences, here for selecting optimal closed-loop optical approaches.
Fast neuronal activity imaging with light field microscopy (Grosenick et al., 2009; Broxton et al., 2013; Prevedel et al., 2014; Cohen et al., 2014), and fast delivery of control light with light field illumination (Levoy et al., 2009; Figure 5A), currently promise the greatest speeds for simultaneous imaging and photostimulation of many neurons over large volumes, but in both cases spatial resolution must be traded to gain camera-limited or DMD-limited rates of imaging and stimulation. Further, both are one-photon techniques, limiting their performance in turbid mammalian tissue. Although light field imaging can provide single-neuron resolution over large volumes, thanks to advanced deconvolution methods (Cohen et al., 2014; Broxton et al., 2013), light field illumination is purely a physical process and cannot be improved using statistical deconvolution. It thus provides a rapid (up to tens of kHz) method for creating dynamic volumes of light, with the critical caveat that the spatial restriction of the light will always be worse than that of two-photon photostimulation methods. As the speed of phase SLMs improves, phase modulating devices may also be used for fast one-photon optogenetic stimulation building on existing work using one-photon digital holography for photostimulation (Lutz et al., 2008; Szabo et al., 2014).
System Identification for Neural Microcircuits
In the case of systems identification for spiking neurons observed with single-cell resolution (that is, a MIMO application), we can utilize existing biophysical knowledge to construct parameterized gray box models where cells are treated as separate variables of the system interacting through estimated synaptic connections (Dahlhaus et al., 1997; Brillinger et al., 1997). The end goal in this case would be to find differential equations describing the evolution of interactions between neurons in terms of their connections and estimated synaptic weights, conditional on animal internal state, sensory inputs, and motor behavior.
Of course, before cell-resolution time series can be modeled, cells must be localized in optical physiology data and clean time series extracted for each cell. For image data, a variety of automated methods for motion correction (Greenberg and Kerr, 2009; Dombeck et al., 2010) and cell region-of-interest (ROI) detection have been developed. The latter include template matching approaches (Ahrens et al., 2013; MacLean et al., 2005; Cossart et al., 2003); local correlation heuristics (Smith and Häusser, 2010); independent-components analysis applied to volume data (ICA) (Grosenick et al., 2009); spatiotemporal ICA applied to image data (Mukamel et al., 2009); and sparse, structured matrix factorization techniques applied to image data (Andilla and Hamprecht, 2013, 2014; Maruyama et al., 2014; Pachitariu et al., 2013). Although effective in specific applications, these methods are still evolving, in part through community-driven competitions like the Neurofinder challenge (http://codeneuro.org/neurofinder/).
Once cell ROIs have been found and validated, the individual traces often require additional processing. In the state-space formulation for neural data (Paninski et al., 2010), actual spiking activity of the neurons is hidden through a measurement process that involves both measurement noise and other measurement limitations, such as convolution of the spikes with slower calcium dynamics in the case of calcium imaging data. In this situation, if one is explicitly interested in the spiking data, it is necessary to infer the times or probabilities of individual action potentials using deconvolution (Andilla and Hamprecht, 2014; Vogelstein et al., 2009, 2010; Yaksi and Friedrich, 2006), template matching (Lütcke et al., 2013; On˜ ativia et al., 2013; Grewe et al., 2010; Greenberg et al., 2008; Kerr et al., 2007), or supervised learning algorithms trained on labeled data (Theis et al., 2014; Sasaki et al., 2008).
Several studies have shown that at high frame rates (>30 Hz) network information can be estimated from such inferred population spike dynamics (Fletcher and Rangan, 2014; Lütcke et al., 2013; Stetter et al., 2013; Mishchenko et al., 2011; Vogelstein et al., 2010). Indeed, simulations by Lutke et al. suggest that connectivity between neurons can be partially inferred from limited observational calcium imaging data, limited sampling from the population, and in the context of fluctuating, unobserved, common input (Lütcke et al., 2013). However, these important theoretical studies were focused primarily on the recovery of estimated synaptic connectivity offline under somewhat idealized conditions and did not optimize for speed, or test how well these estimates performed in recapitulating circuit dynamics—critical considerations for model validation and control. Finally, dynamical models that can estimate nonlinear relationships between cells online (Luo et al., 1996); can give good system estimates in the presence of highly correlated and nonlinear noise; and are capable of modeling potential neural dynamical processes including chaos, bifurcations, and subharmonics are available (Isermann and Münchhof, 2011; Billings 2013; Fan and Yao, 2008). These should be used alongside standard GLM models (Kass et al., 2005, 2014). Improved models of neuronal variability that account for changing animal state should also be integrated with these and other existing models of dynamic variability (Lai and Xing, 2010; Goris et al., 2014).
Finally, information about the structure of microcircuit dynamics can be used to inform and improve system identification. For example, models of imaging data that include “low-rank” models of dynamics (Soudry et al., 2013; Buesing et al., 2014; Pfau et al., 2013) are desirable for their consistency with current models of low-rank state evolution and common-input properties in observed brain dynamics (Shenoy et al., 2013; Mante et al., 2013; Kaufman et al., 2014; Churchland et al., 2012; Harvey et al., 2012; Yu et al., 2006; Sahani, 1999). Once estimated, these models offer computational advantages in the form of a smaller state space to consider, along with better stability and generalization to new data (Katayama, 2006). Similarly, sparsity priors or constraints on models are consistent with existing data on synaptic connectivity in microcircuits and have been shown to improve and accelerate model estimation procedures for microcircuit imaging data (Fletcher and Rangan, 2014; Shababo et al., 2013; Pfau et al., 2013; Mishchenko et al., 2011; Grosenick et al., 2009). These can be further combined with low-rank models (Pfau et al., 2013) and models able to include prior information about connectivity and structured spatial correlations (Watanabe et al., 2014; Grosenick et al., 2013; Allen et al., 2013).
Online Experiment Design for Neural Microcircuits
Given availability of the inputs described above, the next question is which inputs should be chosen to yield good system identification results. This can be categorized as a problem of experiment design—an area of inquiry dating back to the early days of statistics (Fisher, 1925). Basic sequential experimental designs are already in use in neuroscience; for example, in vitro studies of mammalian microcircuits have used imaging in hippocampal brain slices to screen for rare, highly connected “hub” neurons that appear to be important for engaging the larger network in oscillations (Bonifazi et al., 2009). In this study, analysis of functional calcium imaging data during the experiment (which was limited in time by the longevity of the sliced tissue) allowed the identification of small subsets of neurons that had strong directional correlations with many other neurons in the population. These correlational data were then used to target single candidate “hub” neurons in the same slices for stimulation by patch-clamp electrophysiology to test the role of these neurons in driving network synchronization. Importantly, this sequential experiment design used a first stage of online correlational modeling to enable further rounds of more refined experimentation on a time budget, and thereby was able to establish synaptic connectivity details in what would otherwise have been very unlikely neurons to patch at random.
In the field of online experiment design, this type of staged approach is taken to a mathematical and algorithmic level able to estimate the best stimulation inputs to make at each stage, based on models fit to the observations and stimulation made at previous stages (Figure 5C). Such algorithms have been developed for neurophysiology experiments (Lewi et al., 2009; Paninski, 2005), and most recently for mapping neural microcircuits with optical stimulation (Shababo et al., 2013). The latter work considers the problem of finding the minimal set of optical inputs to identify synaptic strengths by stimulating a set of presynaptic neurons while recording electrophysiologically from a postsynaptic neuron. As in Bonifazi et al. (2009), the authors were motivated by the need to efficiently find connections during a time-sensitive experiment. They therefore developed an online method for finding a sequence of such inputs that (on simulated data) significantly improved performance over random inputs and over previous work using random inputs with compressed sensing (Hu et al., 2009). In addition to the improvements over random inputs, Shababo et al. (2013) also presented general methods for modeling variability in stimulation efficacy and a clear overall motivation for the online identification problem. However, extension of this model to the optogenetic case—by incorporating more realistic physiological details (e.g., from Ahmadian et al., 2011), as well as adjusting to the realities of optical observation of data and changes in animal state while maintaining real-time performance, remains for future work.
As highlighted by Shababo et al. (2013), algorithm speed is critical for online neurostimulation applications due to potentially rapid changes in preparation state and health. This is particularly true for less-stable in vitro applications. In contrast, chronic in vivo imaging allows for the possibility of imaging the same circuit over multiple days with cellular or near-cellular resolution in behaving animals (Ziv et al., 2013). Minimizing imaging light intensity to avoid photodamage to cells under study could enable multiple rounds of system identification and control over days, potentially tiled over multiple fields of view or simultaneously from different regions (Lecoq et al., 2014). This outcome would greatly expand the number of computational approaches that could be tried, as more computationally intensive steps would be left for offline analysis between experimental sessions and used as starting points for online procedures during experiments. In anticipation of multiple field-of-view imaging data, computational work has already begun to explore methods for combining multiple fields of view to estimate low-rank dynamics shared across fields that would be characteristic of, for example, common inputs (Soudry et al., 2013; Turaga et al., 2013).
Still, online adjustments will clearly be necessary to account for variations in the preparation, errors in coregistration to data from previous experiments, and changing state of the animal. As a result, microcircuit-appropriate methods for online system identification, streaming clustering and factor analysis (for online updating low-rank estimates embedded in dynamical systems like those in Buesing et al., 2014; Pfau et al., 2013), and change-point detection algorithms to identify rapid shifts in animal brain state (for example, those seen during sharp wave ripples as compared to during theta oscillations in hippocampus (Buzsaki, 2006) will all be useful. Various relevant streaming clustering and factor analytic approaches have already been developed (Akhtar et al., 2012; Mairal et al., 2010; O’Callaghan et al., 2002). Change-point identification methods have also been developed for spiking neural data (Pillow et al., 2011), and could help identify large state changes in system dynamics in such a way that several state-appropriate models could be fit and switched among. There is a strong precedent in the control and time series literatures for such procedures when switching dynamics are present (Fan and Yao, 2008; Liu and Gong, 2014; Zappavigna et al., 2010; Egerstedt et al., 2003; Xu and Antsaklis, 2002) as they allow for the fitting of several (often simpler; for example, locally linear) models appropriate under different conditions. System identification for such switching models can be accomplished by explicit identification of different regimes (e.g., running versus sitting quietly for rodent hippocampal recordings) or in a data-driven fashion using “sum-of-norms” methods (Ohlsson and Ljung, 2013; Ohlsson et al., 2010).
Implementing Closed-Loop Control for Neural Microcircuits
Once tentatively satisfied with a model describing a microcircuit’s dynamics as well as a model relating optogenetic inputs to neural activity (e.g., evoked spikes or firing rates), the closed-loop-capable neuroscientist will be ready to use optimal or suboptimal control theory (Bertsekas, 2005a) to choose appropriate inputs to try achieving a target pattern of activity in the microcircuit online (Figure 5D). This process will be similar to the goals set forth and applied in Ahmadian et al. (2011), but now applied to the more difficult task of optogenetically stimulating multiple neurons and predicting and manipulating interactions online (instead of controlling a single neuron). Still, on the level of tens of cells this control problem is not clearly more difficult than some solved in a variety of engineering applications. Therefore, given the remarkable technological advances in patterned illumination, optical sensing, and optogenetic control reviewed above, and given the many labs now working toward this overarching goal, we expect achieving all-optical microcircuit feedback control to be only a matter of time.
We therefore briefly review the requisite toolkit from the control literature that might be most useful and promising for the case of all-optical, multineuron feedback control in actual experiments, assuming existence of time series data coming from identified cell locations (as discussed above). Latencies introduced by data acquisition (e.g., scanning-image reconstruction time, frame-grabber latency, operating system limitations), online image processing (e.g., online motion correction, cell ROI application), the optical sensors themselves (e.g., rise time), computational time to choose inputs (e.g., solving a constrained optimization problem), and delays in hardware actuation (e.g., loading images to spatial light modulators, galvanometer or AOD travel and settling time) will likely together sum to milliseconds of delay between detecting an event and delivering a targeted light stimulus (Laxpati et al., 2014).
Such delays, and the need to test the quality of the microcircuit model in situ, suggest that fast model predictive control (MPC) with constraints (Wang and Boyd, 2010; Bemporad, 2006; Qin and Badgwell, 2003; Maciejowski, 2002; Camacho and Bordons, 2004) may be important to achieve control at the speeds allowed by optogenetic tools while utilizing the model of connectivity found during system identification and including constraints on input power and evoked responses. MPC (and stochastic MPC) uses estimated system dynamics to forecast where the system will be in several time steps and then chooses control inputs that will bring that forecast more in line with the target trajectory over that time horizon (Figure 5D; Table 1; Rawlings, 2000; Cheng et al., 2014). Various fast strategies have been developed to solve such problems including precomputed lookup tables (Rauov et al., 2009; Maurovic et al., 2011), suboptimal control strategies (Wang and Boyd, 2010; Bertsekas, 2005a), and fast explicit solutions that may be carried out at each step (Wang and Boyd, 2011b). New computational architectures like Apache Spark and Spark Streaming (as applied in Freeman et al.,2014) as well as GPU and FPGA computing could aid in accelerating on-and offline computation for optogenetic MPC.
It is well-documented that the overall dynamics of the microcircuit may change, potentially dramatically, with changing motor activity, sensory inputs, and internal state of the animal (see examples below). Robust and adaptive control (Bertsekas, 2005a; Ogunfunmi, 2007; Dullerud and Paganini, 2005) and switching systems (Egerstedt et al., 2003) can be combined with the methods above to account for such changes (Table 1). Adaptive control tends to work on slower timescales whereas switching models fit different models of dynamics for different regimes and switch among them rapidly when appropriate (Ogunfunmi, 2007). Switching models are particularly promising for state change adaptation under strict time constraints as the separate models can be fit offline but still allow online adjustment to changing conditions. In situations with strong random disturbances driving the system, stochastic versions of the above approaches will be needed (Cannon et al., 2010; Couchman et al., 2006; Table 1). Given that most current methods for cell localization, spike inference, and system identification operate in less than real time, we fully expect hybrid offline/online, open/closed-loop strategies will be the norm in multicell applications for some time. A clear example of such an offline/online, activity-guided approach is the recent all-optical work by Rickgauer et al. (2014), wherein offline processing of calcium imaging data was used to identify place cell fields in a mouse navigating a virtual linear track. Subsequently, this model of place fields could be used to optogenetically impose place-cell-like activity online as mice traversed the appropriate segments of a virtual environment. In this way, computationally slow but necessary preprocessing and fitting could be performed offline, yielding a model useful for online conditional stimulation of neural activity.
Finally, as the number of neurons recorded grows, models of larger systems will require decomposition into smaller subsystems or simpler low-rank models if the models are to be run in real time. Recent progress in dynamic low-rank models for microcircuits recorded with optical sensors of neural activity (Buesing et al., 2014; Pfau et al., 2013), as well as recent developments in sparse modeling and screening rules for decomposition of graphical models into subsystems (Witten et al., 2011; Voorman et al., 2013; Mazumder and Hastie, 2012) are promising avenues for developing lower dimensional or local submodels that will run in parallel at millisecond timescales.
Closed-Loop and Activity-Guided Optogenetics across Brain Scales
Here, we have focused on closed-loop and activity-guided optogenetics at the circuit level involving connections between and within specific brain areas. Brain dynamical processes span a remarkable range of spatiotemporal scales, from millisecond dynamics in dendrites and spines to slow, many-seconds-time-scale synchronization and desynchronization across brain regions. Neuroimaging modalities have rapidly developed to encompass this wide range, from fast local calcium imaging of dendritic spines to slow and global methods such as functional magnetic resonance imaging (fMRI). By applying closed-loop optogenetic strategies across scales, dynamics spanning the subcellular (Tischer and Weiner, 2014), the microcircuit, the interregional, and the whole-brain level could be similarly investigated across scales. At the finest scale, fast light-targeting methods have been developed in dendritic imaging and glutamate uncaging experiments (Branco et al., 2010; Lutz et al., 2008; Nikolenko et al., 2008; Svoboda et al., 1997; Yuste, 2010; Denk et al., 1996; reviewed in Grienberger et al., 2015). At the same time, mathematical and computational machinery necessary for system identification and control on dendrites using observation with optical voltage and calcium sensors has been developed (Pakman et al., 2014; Pnevmatikakis et al., 2012; Huggins and Paninski, 2012; Paninski, 2010), rendering optogenetic system identification and control of dendritic trees a promising area for future investigations. Recently, holographic light shaping and SLM point-scanning optogenetic manipulations have been explored using tools defined at the subcellular and cellular scale (Prakash et al., 2012; Packer et al., 2012; Vaziri and Emiliani, 2012; Anselmi et al., 2011; Yang et al., 2011; Papagiakoumou et al., 2008).
At the mesoscopic scale, optical investigation across brain regions has been achieved through large cranial window, multisite, and clear-skull preparations (Lecoq et al., 2014; Andermann et al., 2011; Marshel et al., 2011; Guo et al., 2014). Bridging scales to the whole-brain level in mammals, fiber-based optical activity recording (Schulz et al., 2012), and optogenetic stimulation (Lee et al., 2010) have been combined with fMRI (optogenetic fMRI, or ofMRI) to simultaneously control local cell population activity and record whole-brain dynamics (Lee et al., 2010). Combining all-optical closed-loop optogenetics through one or multiple fibers with fMRI would put real-time online readout and control of targeted mammalian cell populations in a whole-brain context. Further, recent development of real-time ofMRI using graphics processing units (GPUs) to reconstruct, motion correct, and analyze fMRI volume data in under 15 ms (Fang and Lee, 2013) puts fMRI processing on a timescale amenable to closed-loop optogenetic manipulations conditional on whole-brain state or interregional dynamics.
Outlook
Brain activity is an emergent phenomenon depending on many subcategories of activity including previous history, input signals, ongoing internal dynamics, and neuromodulatory or brain-state regulation mechanisms. A comprehensive understanding of neural function would require that all of these factors be considered and, ideally, measured and precisely perturbed. One of the hallmarks of optogenetics compared to more traditional methods of neural perturbation (e.g., electrical stimulation, pharmacology, and lesion) is the opportunity for increased temporal and cellular specificity in this effort, by controlling these different classes of activity during behavior in the various responsible neurons and projections, via genetic targeting schemes, localized viral injections, circuit labeling techniques, and precise light guidance. As a result, traditional (open-loop) optogenetics, building upon a long history of electrophysiological, pharmacological, imaging, and lesion studies, has already identified activity patterns in numerous neural cell populations and pathways important for specific behavioral outcomes (reviewed in Deisseroth, 2014). Yet even with this enormous progress, there is no question that our understanding is still lacking in critical dimensions ranging from information representation and population coding to detailed wiring implementations of information transmission in the circuit elements that are likely to underlie natural behavior in all its complexity. As technologies continue to mature for closed-loop optogenetics, new opportunities will emerge to causally reveal and confirm increasingly detailed neural circuit mechanisms underlying behavior.
As just one example, the advent of in vivo all-optical approaches at the single-cell level has the potential to illuminate the importance of ensemble wiring and activity on behavioral outcomes (Rickgauer et al., 2014; Packer et al., 2015). These approaches are currently capable of optogenetic stimulation with ~10–20 ms onset latencies and imaging rates of 15–30 Hz when relying on two-photon C1V1 stimulation (Prakash et al., 2012) and GCaMP6 imaging (Chen et al., 2013c). Used in combination (Rickgauer et al., 2014; Packer et al., 2015), it is possible to all-optically stimulate and record at least ten neurons simultaneously every 10–20 ms (Packer et al., 2015) using the scanning, multifocus-phase SLM approach described above. However, even for just ten cells, the number of possible connectivity patterns is combinatorial (35,184,372,088,832 different possible undirected graphs could describe the basic binary connectivity relations for ten cells, ignoring projection direction and synaptic strength/type). To make matters worse, connectivity is difficult to predict a priori, and the behavior of the network is bound to change conditional on previous activity, inputs, internal dynamics, and neuromodulation. In order to robustly identify likely connections between neurons within the timescale of an experiment and without damaging or altogether changing microcircuit behavior, application of online system identification methods that adaptively and minimally stimulate in closed-loop to reduce model uncertainty will almost certainly be necessary. Of course, multiple, animal-state-dependent estimates may be needed and the quality of the estimated relationships should be tested by demonstrating that naturally observed activity patterns can be evoked in the cells under appropriate conditions.
Imagine, for example, that our ten optically controlled and observed cells are pyramidal neurons in the dorsal hippocampus of an awake, head-fixed mouse (as in Rickgauer et al., 2014). We know that pyramidal cell spiking activity should advance to earlier phases of the theta cycle as the mouse passes through the cell’s place field (O’Keefe and Recce, 1993), that changes in GABAergic and cholinergic tone can alter the amplitude and frequency of the theta oscillation (Lee et al., 1994; Yoder and Pang, 2005), that the same cell might be involved in rapid replay events during sharp-wave ripples when the mouse is not running (Skaggs and McNaughton, 1996), that these cells are constantly updating their tuning conditional on the animal’s experiences (Muller and Kubie, 1987; Bostock et al., 1991; Markus et al., 1995), and that causal interventions have different effects depending both on the specific phase of endogenous oscillations and on available sensory information (Siegle and Wilson, 2014). Thus the responsiveness of the ten cells to the same pattern of optogenetic stimulation is likely to vary significantly with contextual changes, locomotion, theta cycle phase, neuromodulation, and other variables. So, just as commercial airliners adjust the responses of their control surfaces online to account for rapidly changing wind and weather conditions, the correct pattern of optogenetic stimulation in a behaving animal should be chosen online to accurately evoke a particular pattern of behavior while taking into account changes in observed animal behavioral and brain state. Closed-loop control is almost universally the engineering approach taken to solve such dynamic and conditional control problems.
In most cases, the integrated technologies described above in their current forms cannot reliably achieve the higher temporal precision (~1–4 ms) that has been observed in certain brain circuits (Wehr and Zador, 2003; Pouille and Scanziani, 2001) and that could be essential for coding and—from an experimental standpoint—for optimal integration and controllability of optogenetically driven activity into ongoing circuit dynamics. However, future integration of faster and more sensitive activity sensors and actuators, more efficient stimulation patterns, higher-speed sequencing between stimulated neurons, and improved illumination sources all promise to improve the speed and scale of single-cell, all-optical approaches. Further, it is important to note that manipulation of local populations with single-cell resolution at the scale made possible by recent published studies is already within the range of the number of neurons that have driven behavioral outcomes in mammals (~1–50 neurons) in pioneering studies of the value of individual or small sets of neurons on learning and behavior (Kwan and Dan, 2012; Clancy et al., 2014; Vallbo et al., 1984; Papadopoulou et al., 2011; Li et al., 2009; Huber et al., 2008; Houweling and Brecht, 2008; Doron et al., 2014; Brecht et al., 2004; Bonifazi et al., 2009).
Even prior to single-cell resolution control of large numbers of cells, a current capability of closed-loop optogenetics is population-scale emulation of naturalistic activity patterns while accounting for the fact that neural system properties are highly nonlinear and nonstationary. Neural activity operates in distinct regimes within a specific, broad dynamic range (Pouille et al., 2009), for example, maintaining a critical balance of excitation and inhibition (Shu et al., 2003; Haider and McCormick, 2009; Okun and Lampl, 2008; Wehr and Zador, 2003; Xue et al., 2014; Yizhar et al., 2011b; Isaacson and Scanziani, 2011). The processes that regulate dynamic range and the balance of excitation and inhibition involve carefully orchestrated circuit mechanisms, including feedforward and feedback inhibitory circuits (Isaacson and Scanziani, 2011). These are particularly relevant when considering how natural or experimental perturbations are integrated into the network. Local feed forward inhibitory circuits within region A that are normally activated by afferent excitatory inputs from region B, presumably in part to balance excitation with inhibition, will not be as precisely activated by direct experimental stimulation (whether electrical or optical) of neuronal cell bodies in region A (highlighting the unique opportunity provided by projection-targeted optogenetics which can directly control the inputs from region B to region A). In general, however, much more investigation is required to understand how precise afferent activity is integrated into local circuits. Without ongoing measurements of circuit activity, it is difficult or impossible to know how native feedback excitatory and inhibitory circuit mechanisms are engaged, and with what circuit-level outcome. All-optical, closed-loop optogenetic experiments have the potential to reveal and tune these neural dynamics.
Beyond the fact that more specific and potent behavioral effects can be seen with optogenetic projection targeting compared with focal regional stimulation (e.g., Tye et al., 2011; Warden et al., 2012; Britt et al., 2012; Znamenskiy and Zador, 2013; reviewed in Deisseroth, 2014), it is also the case that different afferent projections of the same general class (e.g., excitatory/glutamatergic) to the same target region can have very different behavioral effects from each other or from direct modulation (such as excitation) of the target region (e.g., Warden et al., 2012; Britt et al., 2012). Of course, if the different afferent excitatory projections to the target region connected with exactly the same synapse types upon the same local ensembles and were recruited with identical dynamics and strength, presumably the same behavioral effects would result; but the fact is that different projections do have different strengths, dynamics, and local wiring essential to their function, and optogenetic projection targeting provides a handle on this diversity (Britt et al., 2012). For example, increasingly elegant closed-loop and activity-guided experiments will allow dynamical patterns of projection activation to be matched to natural patterns with increasing precision, with accessible parameters now including activity level (Figure 4B), timing with regard to environmental or brain events (Sohal et al., 2009; Paz et al., 2013; O’Connor et al., 2013; Krook-Magnuson et al., 2014; Siegle and Wilson, 2014; Stark et al., 2014; Krook-Magnuson et al., 2015), and even addition or deletion of individual members of the neuronal ensemble (Prakash et al., 2012; Rickgauer et al., 2014; Packer et al., 2015). These more natural and nuanced experiments will be important for addressing potential caveats of experimental design and intervention, and for achieving and testing predictive models of neural activity patterns on specific downstream and behavioral outputs.
Closed-loop optogenetics has the potential also to help understand how circuits change with stimulation and learning; and to recruit in vivo plasticity mechanisms for desired effects. Several studies have recently used projection-recruitment methods to show causal significance for plasticity in defined projections in mammalian behavior (Nabavi et al., 2014; Creed et al., 2015). This capability also represents a unique advantage of optogenetic projection targeting because electrodes cannot specifically recruit a single projection defined by origin and target, which has real experimental consequences. For example, it is has been reported that DBS-like electrical stimulation of the nucleus accumbens does not fully recruit native plasticity at the normally plastic cortico-accumbens glutamatergic synapses due to spurious coactivation of local dopaminergic terminals by the electrode (Creed et al., 2015). To achieve greater plasticity, it has been reported that the local dopamine receptors should be blocked pharmacologically at the same time—an experimental complication that is not needed with the more precise optogenetic projection-targeting driven plasticity (Creed et al., 2015). At the single-neuron, microcircuit, or projection level, activity-dependent plasticity mechanisms, such as those involving spike timing-dependent plasticity (STDP) or other LTP/D (long-term potentiation/depression) effects, could be guided by closed-loop methods to test the causal relevance of circuit element-specific activity-dependent plasticity mechanisms in development, learning, memory, and computation. For example, online monitoring of correlation coefficients between repetitively optogenetically stimulated neurons that are also imaged over time could be used in closed-loop fashion to set constraints on promoting or minimizing detected plasticity in the population, and population synaptic weights could be tracked and tuned in real time using fiber photometry (Figures 4A and 4G). Closed-loop control has already been used in vitro to investigate plasticity at the microcircuit level, to examine the effects of reducing AMPAergic glutamate receptor transmission on plasticity independent of spiking levels in the local circuit (by optogenetically keeping population spiking stable during AMPA blockade; Fong et al., 2015).
More generally, online analysis and closed-loop methods may help in determining photostimulation protocols that are efficacious with the least amount of intervention, addressing a concern common to all approaches (including electrophysiological and optical) for perturbing brain function—that of eliciting undesired effects, such as heating, membrane damage, and cell-health changes. In optogenetics, investigators have long had the unique opportunity to include (where indicated) the powerful control conditions of “light but no opsin” and “opsin but no light,” accounting for concerns common to many kinds of intervention but uniquely addressable in this way in optogenetics—for example, tissue heating (Yizhar et al., 2011a) and cell health related to viral transduction or long-term overexpression of transgenes (Gradinaru et al., 2008, 2010). But more subtly, closed-loop methods allow tracking, detection, and minimization/elimination (if needed) of specific cellular activity changes in response to the intervention, which may include effects of postintervention rebound, ion redistribution and biochemical adaptation (Gradinaru et al., 2007; Ferenczi and Deisseroth, 2012), and other short or long-term plasticity associated with altered activity (which are seen in response to even naturally shifting activity levels). It is helpful to consider these factors when designing and interpreting optogenetic (and other) experiments, and closed-loop methods provide new ways for achieving high specificity of neural circuit dissection in terms of both observation and perturbation.
While electrical stimulation will tend to drive local cells, afferent axons, and fibers of passage with synchrony that is difficult to control or measure, and is unlikely to reflect native patterns, optogenetic methods can bring this aspect of experimental stimulation (depending on the need) under more flexible control. The absence of photosensitivity in diverse off-target afferent fibers when typical AAV-based projection targeting is carried out already addresses a major synchrony confound of electrical stimulation (Creed et al., 2015). Though natural population firing aligned with brain rhythm phase (and other population-wide synchronous events) are common features of native neural activity, cell bodies within a region are not usually driven with tight synchrony by typical moderate optogenetic somatic or projection stimulation due to variability of cell history, photon flux, opsin expression, and synaptic properties—and therefore of spike latency (e.g., Lee et al., 2010; Anikeeva et al., 2012; Ching and Ritt, 2013). However, synchrony of experimental drive can easily be increased where desired with high-intensity and briefer light or diminished with reduced light intensity (Cardin et al., 2009), or entirely eliminated in favor of subthreshold biasing of excitability of the target population (which then fire asynchronously as driven by native timing). The latter effect is readily achieved by providing steady low-intensity light, or by using step function bistable opsins (Yizhar et al., 2011b; Berndt et al., 2009), which are now in wide use across many laboratories (e.g., Tanaka et al., 2012; Bepari et al., 2012; Haikala et al., 2013; Carter et al., 2012; Schultheis et al., 2011). Beyond synchrony, in general the level of induced population activity can be easily parametrically mapped with high resolution, from undetectably low to seizure levels, simply by varying light intensity and/or pulse frequency while mapping behavior or physiology effects in the same animal (as was carried out even in very early optogenetics studies; Adamantidis et al., 2007). It would not (a priori) be clear if an experimental manipulation were providing more or less synchrony, or greater or lower levels of activity, than native dynamics which span a broad range of activity and synchrony levels. However, in a unique advantage of optogenetics, monitoring targeted neuron activity online and in closed-loop fashion facilitates understanding the amount of activity, and level of synchrony, produced by stimulation, thereby enabling the experimenter to promote or diminish synchronous patterns or activity level depending on the experimental goal.
Having a precise understanding of how individual cells, circuits and cell types influence local and brain-wide dynamics and plasticity in real time is both central to understanding diseases of brain circuitry and to developing smarter interventions to improve or repair their function. Some of the first successful closed-loop optogenetic experiments have been applied in clinically motivated experiments to quickly thwart epileptiform activity using online analysis and optogenetic intervention (Paz et al., 2013; Krook-Magnuson et al., 2014, 2015). Another important application of closed-loop optogenetics will likely be in the development and application of neuroprosthetics, to potentially improve brain-machine interfaces and close the loop with sensory feedback for neuroprosthetic devices (Shenoy and Carmena, 2014; O’Connor et al., 2013). Closed-loop optogenetics may help improve circuit targeting for prosthetic devices to help manage paralysis, given already intriguing progress based on closed-loop electrical epidural stimulation to recover walking behavior in paralyzed rats (Wenger et al., 2014). Moreover, closed-loop optogenetics with single-cell resolution may be able to track and modulate learning and behavior even by targeting small numbers of neurons. Along these lines, a recent brain-machine interface study relying on two-photon calcium imaging in mice of dense populations of layer 2/3 motor and somatosensory cortical neurons showed rapid learning by the mice to volitionally control the firing rate of specific, small subsets of neurons (1–11 total neurons) selected by the experimenter a priori, in order to satisfy the task goal of varying the frequency of an auditory stimulus and receive a reward (Clancy et al., 2014).
Finally, though speculative, closed-loop optogenetics could also play an essential role in determining precise circuit perturbations that lead to effective treatments of psychiatric illness, with side effects decreased by including appropriate feedback to keep them minimized in the model. Insights from optogenetics have already been put forth in the form of strategies to design novel nonoptogenetic interventions (Chen et al., 2013a; Creed et al., 2015; Gradinaru et al., 2009). As is the case for a growing number of illnesses, once relevant areas, cell types, pathways, and basic activity patterns have been identified, functional circuit models can be further refined and better tested with closed-loop approaches. Together, the closed-loop and activity-guided approaches outlined here may help realize a promise of understanding and applying systems neuroscience to improve our understanding of both normal brain function and neuropsychiatric disease symptoms by distilling circuit activity patterns down to the most elemental but effective features and thereby improving understanding of how any causal intervention (including pharmacological, optogenetic, electrical or otherwise) operates on the brain.
Acknowledgments
L.G., J.H.M., and K.D. wrote the paper. We thank Darcy Peterka, Forrest Collman, Benjamin Grewe, Polina Anikeeva, Adam Packer, Michael Hausser, Eftychios Pnevmatikakis, Yiyang Gong, Lief Fenno, Joshua Vogelstein, Michael Broxton, Samuel Yang, Isaac Kauvar, Tom Davidson, and Liam Paninski for comments on the manuscript and the members of the Deisseroth lab for helpful discussions. J.H.M. is a Simons Foundation Fellow of the Life Sciences Foundation; K.D. is supported by the DARPA Neuro-FAST program; NIMH; NIDA; NSF; the Simons Foundation; the Gatsby Foundation; the Wiegers Family Fund; and the Grosfeld, Snyder, Woo, and Albert Yu and Mary Bechman Foundations.
REFERENCES
- Abaya TVF, Diwekar M, Blair S, Tathireddy P, Rieth L, Clark GA, Solzbacher F. Characterization of a 3D optrode array for infrared neural stimulation. Biomed. Opt. Express. 2012a;3:2200–2219. doi: 10.1364/BOE.3.002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abaya TVF, Blair S, Tathireddy P, Rieth L, Solzbacher F. A 3D glass optrode array for optical neural stimulation. Biomed. Opt. Express. 2012b;3:3087–3104. doi: 10.1364/BOE.3.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamantidis AR, Tsai H-C, Boutrel B, Zhang F, Stuber GD, Budygin EA, Touriño C, Bonci A, Deisseroth K, de Lecea L. Optogenetic interrogation of dopaminergic modulation of the multiple phases of reward-seeking behavior. J. Neurosci. 2011;31:10829–10835. doi: 10.1523/JNEUROSCI.2246-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian Y, Packer AM, Yuste R, Paninski L. Designing optimal stimuli to control neuronal spike timing. J. Neurophysiol. 2011;106:1038–1053. doi: 10.1152/jn.00427.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods. 2013;10:413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- Airan RD, Hu ES, Vijaykumar R, Roy M, Meltzer LA, Deisseroth K. Integration of light-controlled neuronal firing and fast circuit imaging. Curr. Opin. Neurobiol. 2007;17:587–592. doi: 10.1016/j.conb.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akemann W, Mutoh H, Perron A, Park YK, Iwamoto Y, Knöpfel T. Imaging neural circuit dynamics with a voltage-sensitive fluorescent protein. J. Neurophysiol. 2012;108:2323–2337. doi: 10.1152/jn.00452.2012. [DOI] [PubMed] [Google Scholar]
- Akerboom J, Chen T-W, Wardill TJ, Tian L, Marvin JS, Mutlu S, Calderón NC, Esposti F, Borghuis BG, Sun XR, et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerboom J, Carreras Calderón N, Tian L, Wabnig S, Prigge M, Tolö J, Gordus A, Orger MB, Severi KE, Macklin JJ, et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front. Mol. Neurosci. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar MT, Jung T-P, Makeig S, Cauwenberghs G. Recursive independent component analysis for online blind source separation. 2012 IEEE International Symposium on Circuits and Systems (ISCAS) 2012:2813–2816. [Google Scholar]
- Allen GI, Grosenick L, Taylor J. A Generalized Least-Square Matrix Decomposition. J. Am. Stat. Assoc. 2013;109:145–159. [Google Scholar]
- Andermann ML, Kerlin AM, Roumis DK, Glickfeld LL, Reid RC. Functional specialization of mouse higher visual cortical areas. Neuron. 2011;72:1025–1039. doi: 10.1016/j.neuron.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andilla FD, Hamprecht FA. Learning multi-level sparse representations. In: Burges CJC, Bottou L, Welling M, Ghahramani Z, Weinberger KQ, editors. Advances in Neural Information Processing Systems 26. Red Hook, NY: Curran Associates, Inc; 2013. pp. 818–826. [Google Scholar]
- Andilla FD, Hamprecht FA. Sparse space-time deconvolution for calcium image analysis. In: Ghahramani Z, Welling M, Cortes C, Lawrence ND, Weinberger KQ, editors. Advances in Neural Information Processing Systems 27. Red Hook, NY: Curran Associates, Inc.; 2014. pp. 64–72. [Google Scholar]
- Andrasfalvy BK, Zemelman BV, Tang J, Vaziri A. Two-photon single-cell optogenetic control of neuronal activity by sculpted light. Proc. Natl. Acad. Sci. USA. 2010;107:11981–11986. doi: 10.1073/pnas.1006620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anikeeva P, Andalman AS, Witten I, Warden M, Goshen I, Grosenick L, Gunaydin LA, Frank LM, Deisseroth K. Optetrode: a multichannel readout for optogenetic control in freely moving mice. Nat. Neurosci. 2012;15:163–170. doi: 10.1038/nn.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselmi F, Ventalon C, Bègue A, Ogden D, Emiliani V. Three-dimensional imaging and photostimulation by remote-focusing and holographic light patterning. Proc. Natl. Acad. Sci. USA. 2011;108:19504–19509. doi: 10.1073/pnas.1109111108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åström KJ, Hägglund T. Advanced PID Control. Research Triangle Park, NC: ISA—The Instrumentation, Systems, and Automation Society; 2006. [Google Scholar]
- Åström KJ, Murray RM. Feedback Systems: An Introduction for Scientists and Engineers. Princeton: Princeton University Press; 2010. [Google Scholar]
- Åström KJ, Wittenmark B. Adaptive Control. Second. Courier Corporation; 2013. [Google Scholar]
- Ataka K, Pieribone VA. A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys. J. 2002;82:509–516. doi: 10.1016/S0006-3495(02)75415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto RPJ, and Schnitzer MJ. In vivo microendoscopy of the hippocampus. Cold Spring Harb. Protoc. 2012;2012:1092–1099. doi: 10.1101/pdb.prot071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bègue A, Papagiakoumou E, Leshem B, Conti R, Enke L, Oron D, Emiliani V. Two-photon excitation in scattering media by spatiotemporally shaped beams and their application in optogenetic stimulation. Biomed. Opt. Express. 2013;4:2869–2879. doi: 10.1364/BOE.4.002869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemporad A. Model Predictive Control Design: New Trends and Tools. 2006 45th IEEE Conference on Decision and Control; 2006. pp. 6678–6683. [Google Scholar]
- Bepari AK, Sano H, Tamamaki N, Nambu A, Tanaka KF, Takebayashi H. Identification of optogenetically activated striatal medium spiny neurons by Npas4 expression. PLoS ONE. 2012;7:e52783. doi: 10.1371/journal.pone.0052783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat. Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- Berndt A, Lee SY, Ramakrishnan C, Deisseroth K. Structure-guided transformation of channelrhodopsin into a light-activated chloride channel. Science. 2014;344:420–424. doi: 10.1126/science.1252367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsekas DP. Dynamic Programming and Optimal Control. Nashua, NH: Athena Scientific; 2005a. [Google Scholar]
- Bertsekas DP. Dynamic Programming and Suboptimal Control: A Survey from ADP to MPC*. Eur. J. Control. 2005b;11:310–334. [Google Scholar]
- Billings SA. Nonlinear System Identification: NARMAX Methods in the Time, Frequency, and Spatio-Temporal Domains. New York: John Wiley & Sons; 2013. [Google Scholar]
- Bock DD, Lee W-CA, Kerlin AM, Andermann ML, Hood G, Wetzel AW, Yurgenson S, Soucy ER, Kim HS, Reid RC. Network anatomy and in vivo physiology of visual cortical neurons. Nature. 2011;471:177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifazi P, Goldin M, Picardo MA, Jorquera I, Cattani A, Bianconi G, Represa A, Ben-Ari Y, Cossart R. GABAergic hub neurons orchestrate synchrony in developing hippocampal networks. Science. 2009;326:1419–1424. doi: 10.1126/science.1175509. [DOI] [PubMed] [Google Scholar]
- Bostock E, Muller RU, Kubie JL. Experience-dependent modifications of hippocampal place cell firing. Hippocampus. 1991;1:193–205. doi: 10.1002/hipo.450010207. [DOI] [PubMed] [Google Scholar]
- Bouchard MB, Voleti V, Mendes CS, Lacefield C, Grueber WB, Mann RS, Bruno RM, Hillman EMC. Swept confocally-aligned planar excitation (SCAPE) microscopy for high speed volumetric imaging of behaving organisms. Nat. Photonics. 2015;9:113–119. doi: 10.1038/nphoton.2014.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd S, Mueller MT, O’Donoghue B, Wang Y. Performance bounds and suboptimal policies for multi-period investment. Found Trends Optim. 2014;1:1–72. [Google Scholar]
- Branco T, Clark BA, Häusser M. Dendritic discrimination of temporal input sequences in cortical neurons. Science. 2010;329:1671–1675. doi: 10.1126/science.1189664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branicky MS. Multiple Lyapunov functions and other analysis tools for switched and hybrid systems. IEEE Trans. Autom. Control. 1998;43:475–482. [Google Scholar]
- Braz JM, Rico B, Basbaum AI. Transneuronal tracing of diverse CNS circuits by Cre-mediated induction of wheat germ agglutinin in transgenic mice. Proc. Natl. Acad. Sci. USA. 2002;99:15148–15153. doi: 10.1073/pnas.222546999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht M, Schneider M, Sakmann B, Margrie TW. Whisker movements evoked by stimulation of single pyramidal cells in rat motor cortex. Nature. 2004;427:704–710. doi: 10.1038/nature02266. [DOI] [PubMed] [Google Scholar]
- Brillinger DR, Villa AEP, Villa REP. Assessing connections in networks of biological neurons. In: Tukey W, Brillinger D, Fernholtz L, Morgenthaler S, editors. The Practice of Data Analysis: Essays in Honor of John. Princeton: Princeton University Press; 1997. pp. 77–91. [Google Scholar]
- Britt JP, Benaliouad F, McDevitt RA, Stuber GD, Wise RA, Bonci A. Synaptic and behavioral profile of multiple glutamatergic inputs to the nucleus accumbens. Neuron. 2012;76:790–803. doi: 10.1016/j.neuron.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxton M, Grosenick L, Yang S, Cohen N, Andalman A, Deisseroth K, Levoy M. Wave optics theory and 3-D deconvolution for the light field microscope. Opt. Express. 2013;21:25418–25439. doi: 10.1364/OE.21.025418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buesing L, Machado TA, Cunningham JP, Paninski L. Clustered factor analysis of multineuronal spike data. In: Ghahramani Z, Welling M, Cortes C, Lawrence ND, Weinberger KQ, editors. Advances in Neural Information Processing Systems 27. Red Hook, NY: Curran Associates, Inc; 2014. pp. 3500–3508. [Google Scholar]
- Buzsaki G. Rhythms of the Brain. New York: Oxford University Press; 2006. [Google Scholar]
- Camacho EF, Bordons C. Model Predictive Control. London: Springer London; 2004. [Google Scholar]
- Canales A, Jia X, Froriep UP, Koppes RA, Tringides CM, Selvidge J, Lu C, Hou C, Wei L, Fink Y, Anikeeva P. Multifunctional fibers for simultaneous optical, electrical and chemical interrogation of neural circuits in vivo. Nat. Biotechnol. 2015;33:277–284. doi: 10.1038/nbt.3093. [DOI] [PubMed] [Google Scholar]
- Cannon M, Kouvaritakis B, Rakovic SV, Cheng Q. Stochastic tubes in model predictive control with probabilistic constraints. IEEE Trans. Automatic Control. 2010;56:194–200. [Google Scholar]
- Cao H, Gu L, Mohanty SK, Chiao J-C. An integrated µLED optrode for optogenetic stimulation and electrical recording. IEEE Trans. Biomed. Eng. 2013;60:225–229. doi: 10.1109/TBME.2012.2217395. [DOI] [PubMed] [Google Scholar]
- Cardin JA, Carle´n M, Meletis K, Knoblich U, Zhang F, Deisseroth K, Tsai L-H, Moore CI. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–667. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter ME, Brill J, Bonnavion P, Huguenard JR, Huerta R, de Lecea L. Mechanism for Hypocretin-mediated sleep-to-wake transitions. Proc. Natl. Acad. Sci. USA. 2012;109:E2635–E2644. doi: 10.1073/pnas.1202526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AC, Oathes DJ, Chang C, Bradley T, Zhou ZW, Williams LM, Glover GH, Deisseroth K, Etkin A. Causal interactions between fronto-parietal central executive and default-mode networks in humans. Proc. Natl. Acad. Sci. USA. 2013a;110:19944–19949. doi: 10.1073/pnas.1311772110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Pfäffli OA, Voigt FF, Margolis DJ, Helmchen F. Online correction of licking-induced brain motion during two-photon imaging with a tunable lens. J. Physiol. 2013b;591:4689–4698. doi: 10.1113/jphysiol.2013.259804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013c;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Cannon M, Kouvaritakis B, Evans M. Stochastic MPC for systems with both multiplicative and additive disturbances. In: Boje E, Xiaohua X, editors. Proceedings of the 19th IFAC World Congress; 2014. pp. 2291–2296. [Google Scholar]
- Ching S, Ritt JT. Control strategies for underactuated neural ensembles driven by optogenetic stimulation. Front. Neural Circuits. 2013;7:54. doi: 10.3389/fncir.2013.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I-J, Won Baac H, Yoon E. A 16-site neural probe integrated with a waveguide for optical stimulation. 2010 IEEE 23rd International Conference on Micro Electro Mechanical Systems (MEMS); 2010. pp. 995–998. [Google Scholar]
- Chung K, Wallace J, Kim S-Y, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong AS, Miri ML, Busskamp V, Matthews GAC, Acker LC, Sørensen AT, Young A, Klapoetke NC, Henninger MA, Kodandaramaiah SB, et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat. Neurosci. 2014;17:1123–1129. doi: 10.1038/nn.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchland MM, Cunningham JP, Kaufman MT, Foster JD, Nuyujukian P, Ryu SI, Shenoy KV. Neural population dynamics during reaching. Nature. 2012;487:51–56. doi: 10.1038/nature11129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy KB, Koralek AC, Costa RM, Feldman DE, Carmena JM. Volitional modulation of optically recorded calcium signals during neuroprosthetic learning. Nat. Neurosci. 2014;17:807–809. doi: 10.1038/nn.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen N, Yang S, Andalman A, Broxton M, Grosenick L, Deisseroth K, Horowitz M, Levoy M. Enhancing the performance of the light field microscope using wavefront coding. Opt. Express. 2014;22:24817–24839. doi: 10.1364/OE.22.024817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conkey DB, Brown AN, Caravaca-Aguirre AM, Piestun R. Genetic algorithm optimization for focusing through turbid media in noisy environments. Opt. Express. 2012;20:4840–4849. doi: 10.1364/OE.20.004840. [DOI] [PubMed] [Google Scholar]
- Cossart R, Aronov D, Yuste R. Attractor dynamics of network UP states in the neocortex. Nature. 2003;423:283–288. doi: 10.1038/nature01614. [DOI] [PubMed] [Google Scholar]
- Cotton RJ, Froudarakis E, Storer P, Saggau P, Tolias AS. Three-dimensional mapping of microcircuit correlation structure. Front. Neural Circuits. 2013:7. doi: 10.3389/fncir.2013.00151. http://dx.doi.org/10.3389/fncir.2013.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couchman PD, Cannon M, Kouvaritakis B. Stochastic MPC with inequality stability constraints. Automatica. 2006;42:2169–2174. [Google Scholar]
- Cox IJ, Sheppard CJR. Information capacity and resolution in an optical system. J. Opt. Soc. Am. A. 1986;3:1152–1158. [Google Scholar]
- Creed M, Pascoli VJ, Lüscher C. Addiction therapy. Refining deep brain stimulation to emulate optogenetic treatment of synaptic pathology. Science. 2015;347:659–664. doi: 10.1126/science.1260776. [DOI] [PubMed] [Google Scholar]
- Dahlhaus R, Eichler M, Sandkühler J. Identification of synaptic connections in neural ensembles by graphical models. J. Neurosci. Methods. 1997;77:93–107. doi: 10.1016/s0165-0270(97)00100-3. [DOI] [PubMed] [Google Scholar]
- Dal Maschio M, Difato F, Beltramo R, Blau A, Benfenati F, Fellin T. Simultaneous two-photon imaging and photo-stimulation with structured light illumination. Opt. Express. 2010;18:18720–18731. doi: 10.1364/OE.18.018720. [DOI] [PubMed] [Google Scholar]
- Dana H, Shoham S. Numerical evaluation of temporal focusing characteristics in transparent and scattering media. Opt. Express. 2011;19:4937–4948. doi: 10.1364/OE.19.004937. [DOI] [PubMed] [Google Scholar]
- Danzl P, Hespanha J, Moehlis J. Event-based minimum-time control of oscillatory neuron models: phase randomization, maximal spike rate increase, and desynchronization. Biol. Cybern. 2009;101:387–399. doi: 10.1007/s00422-009-0344-3. [DOI] [PubMed] [Google Scholar]
- Dasanayake I, Li J-S. Optimal design of minimum-power stimuli for phase models of neuron oscillators. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2011;83:061916. doi: 10.1103/PhysRevE.83.061916. [DOI] [PubMed] [Google Scholar]
- Deisseroth K. Circuit dynamics of adaptive and maladaptive behaviour. Nature. 2014;505:309–317. doi: 10.1038/nature12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Denk W, Delaney KR, Gelperin A, Kleinfeld D, Strowbridge BW, Tank DW, Yuste R. Anatomical and functional imaging of neurons using 2-photon laser scanning microscopy. J. Neurosci. Methods. 1994;54:151–162. doi: 10.1016/0165-0270(94)90189-9. [DOI] [PubMed] [Google Scholar]
- Denk W, Yuste R, Svoboda K, Tank DW. Imaging calcium dynamics in dendritic spines. Curr. Opin. Neurobiol. 1996;6:372–378. doi: 10.1016/s0959-4388(96)80122-x. [DOI] [PubMed] [Google Scholar]
- Dhawale AK, Hagiwara A, Bhalla US, Murthy VN, Albeanu DF. Non-redundant odor coding by sister mitral cells revealed by light addressable glomeruli in the mouse. Nat. Neurosci. 2010;13:1404–1412. doi: 10.1038/nn.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombeck DA, Harvey CD, Tian L, Looger LL, Tank DW. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat. Neurosci. 2010;13:1433–1440. doi: 10.1038/nn.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron G, von Heimendahl M, Schlattmann P, Houweling AR, Brecht M. Spiking irregularity and frequency modulate the behavioral report of single-neuron stimulation. Neuron. 2014;81:653–663. doi: 10.1016/j.neuron.2013.11.032. [DOI] [PubMed] [Google Scholar]
- Duemani Reddy G, Kelleher K, Fink R, Saggau P. Three-dimensional random access multiphoton microscopy for functional imaging of neuronal activity. Nat. Neurosci. 2008;11:713–720. doi: 10.1038/nn.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dullerud GE, Paganini F. A Course in Robust Control Theory: A Convex Approach. New York: Springer; 2005. [Google Scholar]
- Egerstedt M, Wardi Y, Delmotte F. Optimal control of switching times in switched dynamical systems. 42nd IEEE Conference on Decision and Control Proceedings; 2003. pp. 2138–2143. [Google Scholar]
- Fan J, Yao Q. Nonlinear Time Series: Nonparametric and Parametric Methods. New York: Springer-Verlag; 2008. [Google Scholar]
- Fang Z, Lee JH. High-throughput optogenetic functional magnetic resonance imaging with parallel computations. J. Neurosci. Methods. 2013;218:184–195. doi: 10.1016/j.jneumeth.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu. Rev. Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat. Methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenczi E, Deisseroth K. When the electricity (and the lights) go out: transient changes in excitability. Nat. Neurosci. 2012;15:1058–1060. doi: 10.1038/nn.3172. [DOI] [PubMed] [Google Scholar]
- Fernandez-Alfonso T, Nadella KMNS, Iacaruso MF, Pichler B, Ros H, Kirkby PA, Silver RA. Monitoring synaptic and neuronal activity in 3D with synthetic and genetic indicators using a compact acousto-optic lens two-photon microscope. J. Neurosci. Methods. 2014;222:69–81. doi: 10.1016/j.jneumeth.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. Statistical Methods For Research Workers. New York: Oliver and Boyd; 1925. [Google Scholar]
- Fitzgerald JE, Lu J, Schnitzer MJ. Estimation theoretic measure of resolution for stochastic localization microscopy. Phys. Rev. Lett. 2012;109:048102. doi: 10.1103/PhysRevLett.109.048102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher AK, Rangan S. Scalable inference for neuronal connectivity from calcium imaging. In: Ghahramani Z, Welling M, Cortes C, Lawrence ND, Weinberger KQ, editors. Advances in Neural Information Processing Systems 27. Red Hook, NY: Curran Associates, Inc; 2014. pp. 2843–2851. [Google Scholar]
- Flusberg BA, Cocker ED, Piyawattanametha W, Jung JC, Cheung ELM, Schnitzer MJ. Fiber-optic fluorescence imaging. Nat. Methods. 2005;2:941–950. doi: 10.1038/nmeth820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flytzanis N, Bedbrook C, Gradinaru V. Neuronal activity sensing and modulation with Archers. SPIE Newsroom. 2014 http://dx.doi.org/10.1117/2.1201411.005708. [Google Scholar]
- Fong MF, Newman JP, Potter SM, Wenner P. Upward synaptic scaling is dependent on neurotransmission rather than spiking. Nat. Commun. 2015;6:6339. doi: 10.1038/ncomms7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin GF, Powell JD, Emami-Naeini A. Feedback control of dynamic systems. New York: Prentice Hall; 2015. [Google Scholar]
- Freeman J, Vladimirov N, Kawashima T, Mu Y, Sofroniew NJ, Bennett DV, Rosen J, Yang C-T, Looger LL, Ahrens MB. Mapping brain activity at scale with cluster computing. Nat. Methods. 2014;11:941–950. doi: 10.1038/nmeth.3041. [DOI] [PubMed] [Google Scholar]
- Ghosh KK, Burns LD, Cocker ED, Nimmerjahn A, Ziv Y, Gamal AE, Schnitzer MJ. Miniaturized integration of a fluorescence microscope. Nat. Methods. 2011;8:871–878. doi: 10.1038/nmeth.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickfeld LL, Andermann ML, Bonin V, Reid RC. Corticocortical projections in mouse visual cortex are functionally target specific. Nat. Neurosci. 2013;16:219–226. doi: 10.1038/nn.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Wagner MJ, Zhong Li J, Schnitzer MJ. Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors. Nat. Commun. 2014:5. doi: 10.1038/ncomms4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris RLT, Movshon JA, Simoncelli EP. Partitioning neuronal variability. Nat. Neurosci. 2014;17:858–865. doi: 10.1038/nn.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Thompson KR, Zhang F, Mogri M, Kay K, Schneider MB, Deisseroth K. Targeting and readout strategies for fast optical neural control in vitro and in vivo. J. Neurosci. 2007;27:14231–14238. doi: 10.1523/JNEUROSCI.3578-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36:129–139. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K. Optical deconstruction of parkinsonian neural circuitry. Science. 2009;324:354–359. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, Diester I, Goshen I, Thompson KR, Deisseroth K. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DS, Kerr JND. Automated correction of fast motion artifacts for two-photon imaging of awake animals. J. Neurosci. Methods. 2009;176:1–15. doi: 10.1016/j.jneumeth.2008.08.020. [DOI] [PubMed] [Google Scholar]
- Greenberg DS, Houweling AR, Kerr JND. Population imaging of ongoing neuronal activity in the visual cortex of awake rats. Nat. Neurosci. 2008;11:749–751. doi: 10.1038/nn.2140. [DOI] [PubMed] [Google Scholar]
- Grewe BF, Langer D, Kasper H, Kampa BM, Helmchen F. High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nat. Methods. 2010;7:399–405. doi: 10.1038/nmeth.1453. [DOI] [PubMed] [Google Scholar]
- Grienberger C, Konnerth A. Imaging calcium in neurons. Neuron. 2012;73:862–885. doi: 10.1016/j.neuron.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Grienberger C, Chen X, Konnerth A. Dendritic function in vivo. Trends Neurosci. 2015;38:45–54. doi: 10.1016/j.tins.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Grosenick L, Anderson T, Smith SJ. Elastic source selection for in vivo imaging of neuronal ensembles. IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2009. 2009:1263–1266. [Google Scholar]
- Grosenick L, Klingenberg B, Katovich K, Knutson B, Taylor JE. Interpretable whole-brain prediction analysis with GraphNet. Neuroimage. 2013;72:304–321. doi: 10.1016/j.neuroimage.2012.12.062. [DOI] [PubMed] [Google Scholar]
- Grossman N, Poher V, Grubb MS, Kennedy GT, Nikolic K, McGovern B, Berlinguer Palmini R, Gong Z, Drakakis EM, Neil MAA, et al. Multi-site optical excitation using ChR2 and micro-LED array. J. Neural Eng. 2010;7:16004. doi: 10.1088/1741-2560/7/1/016004. [DOI] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron. 2013;78:773–784. doi: 10.1016/j.neuron.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Yizhar O, Berndt A, Sohal VS, Deisseroth K, Hegemann P. Ultrafast optogenetic control. Nat. Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, et al. Natural neural projection dynamics underlying social behavior. Cell. 2014;157:1535–1551. doi: 10.1016/j.cell.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ZV, Li N, Huber D, Ophir E, Gutnisky D, Ting JT, Feng G, Svoboda K. Flow of cortical activity underlying a tactile decision in mice. Neuron. 2014;81:179–194. doi: 10.1016/j.neuron.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider B, McCormick DA. Rapid neocortical dynamics: cellular and network mechanisms. Neuron. 2009;62:171–189. doi: 10.1016/j.neuron.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haikala V, Joesch M, Borst A, Mauss AS. Optogenetic control of fly optomotor responses. J. Neurosci. 2013;33:13927–13934. doi: 10.1523/JNEUROSCI.0340-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey CD, Coen P, Tank DW. Choice-specific sequences in parietal cortex during a virtual-navigation decision task. Nature. 2012;484:62–68. doi: 10.1038/nature10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Tagawa Y, Yawata S, Nakanishi S, Funabiki K. Spatio-temporal control of neural activity in vivo using fluorescence microendoscopy. Eur. J. Neurosci. 2012;36:2722–2732. doi: 10.1111/j.1460-9568.2012.08191.x. [DOI] [PubMed] [Google Scholar]
- Hegemann P, Ehlenbeck S, Gradmann D. Multiple photocycles of channelrhodopsin. Biophys. J. 2005;89:3911–3918. doi: 10.1529/biophysj.105.069716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochbaum DR, Zhao Y, Farhi SL, Klapoetke N, Werley CA, Kapoor V, Zou P, Kralj JM, Maclaurin D, Smedemark-Margulies N, et al. Alloptical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods. 2014;11:825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houweling AR, Brecht M. Behavioural report of single neuron stimulation in somatosensory cortex. Nature. 2008;451:65–68. doi: 10.1038/nature06447. [DOI] [PubMed] [Google Scholar]
- Hu T, Leonardo A, Chklovskii DB. Reconstruction of Sparse Circuits Using Multi-neuronal Excitation (RESCUME) In: Bengio Y, Schuurmans D, Lafferty JD, Williams CKI, Culotta A, editors. Advances in Neural Information Processing Systems 22. Red Hook, NY: Curran Associates, Inc; 2009. pp. 790–798. [Google Scholar]
- Huang ZJ. Toward a genetic dissection of cortical circuits in the mouse. Neuron. 2014;83:1284–1302. doi: 10.1016/j.neuron.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber D, Petreanu L, Ghitani N, Ranade S, Hromádka T, Mainen Z, Svoboda K. Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice. Nature. 2008;451:61–64. doi: 10.1038/nature06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins JH, Paninski L. Optimal experimental design for sampling voltage on dendritic trees in the low-SNR regime. J. Comput. Neurosci. 2012;32:347–366. doi: 10.1007/s10827-011-0357-5. [DOI] [PubMed] [Google Scholar]
- Inoue M, Takeuchi A, Horigane S, Ohkura M, Gengyo-Ando K, Fujii H, Kamijo S, Takemoto-Kimura S, Kano M, Nakai J, et al. Rational design of a high-affinity, fast, red calcium indicator R-CaMP2. Nat. Methods. 2015;12:64–70. doi: 10.1038/nmeth.3185. [DOI] [PubMed] [Google Scholar]
- Isaacson JS, Scanziani M. How inhibition shapes cortical activity. Neuron. 2011;72:231–243. doi: 10.1016/j.neuron.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isermann R, Mönchhof M. Identification of Dynamic Systems: An Introduction with Applications. Berlin: Springer-Verlag; 2011. [Google Scholar]
- Iyer V, Hoogland TM, Saggau P. Fast functional imaging of single neurons using random-access multiphoton (RAMP) microscopy. J. Neurophysiol. 2006;95:535–545. doi: 10.1152/jn.00865.2005. [DOI] [PubMed] [Google Scholar]
- Jin L, Han Z, Platisa J, Wooltorton JRA, Cohen LB, Pieribone VA. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. 2012;75:779–785. doi: 10.1016/j.neuron.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judkewitz B, Rizzi M, Kitamura K, Häusser M. Targeted single-cell electroporation of mammalian neurons in vivo. Nat. Protoc. 2009;4:862–869. doi: 10.1038/nprot.2009.56. [DOI] [PubMed] [Google Scholar]
- Kalman RE. A New Approach to Linear Filtering and Prediction Problems. J. Fluids Eng. 1960;82:35–45. [Google Scholar]
- Kano T, Kinoshita S. Control of individual phase relationship between coupled oscillators using multilinear feedback. Phys. Rev. E Stat. Nonlin. Soft Matter Physiol. 2010;81:026206. doi: 10.1103/PhysRevE.81.026206. [DOI] [PubMed] [Google Scholar]
- Kass RE, Ventura V, Brown EN. Statistical issues in the analysis of neuronal data. J. Neurophysiol. 2005;94:8–25. doi: 10.1152/jn.00648.2004. [DOI] [PubMed] [Google Scholar]
- Kass RE, Eden UT, Brown EN. Analysis of Neural Data. New York: Springer; 2014. [Google Scholar]
- Katayama T. Subspace Methods for System Identification. London: Springer-Verlag; 2006. [Google Scholar]
- Katona G, Szalay G, Maák P, Kasza´s A, Veress M, Hillier D, Chiovini B, Vizi ES, Roska B, Rózsa B. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat. Methods. 2012;9:201–208. doi: 10.1038/nmeth.1851. [DOI] [PubMed] [Google Scholar]
- Katz Y, Yizhar O, Staiger J, Lampl I. Optopatcher—an electrode holder for simultaneous intracellular patch-clamp recording and optical manipulation. J. Neurosci. Methods. 2013;214:113–117. doi: 10.1016/j.jneumeth.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Kaufman MT, Churchland MM, Ryu SI, Shenoy KV. Cortical activity in the null space: permitting preparation without movement. Nat. Neurosci. 2014;17:440–448. doi: 10.1038/nn.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller PJ, Ahrens MB. Visualizing whole-brain activity and development at the single-cell level using light-sheet microscopy. Neuron. 2015;85:462–483. doi: 10.1016/j.neuron.2014.12.039. [DOI] [PubMed] [Google Scholar]
- Kemere C, Santhanam G, Yu BM, Afshar A, Ryu SI, Meng TH, Shenoy KV. Detecting neural-state transitions using hidden Markov models for motor cortical prostheses. J. Neurophysiol. 2008;100:2441–2452. doi: 10.1152/jn.00924.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JND, de Kock CPJ, Greenberg DS, Bruno RM, Sakmann B, Helmchen F. Spatial organization of neuronal population responses in layer 2/3 of rat barrel cortex. J. Neurosci. 2007;27:13316–13328. doi: 10.1523/JNEUROSCI.2210-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil HK. Nonlinear Systems. New York: Prentice Hall; 2002. [Google Scholar]
- Kim S-Y, Adhikari A, Lee SY, Marshel JH, Kim CK, Mallory CS, Lo M, Pak S, Mattis J, Lim BK, et al. Diverging neural pathways assemble a behavioural state from separable features in anxiety. Nature. 2013a;496:219–223. doi: 10.1038/nature12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TI, McCall JG, Jung YH, Huang X, Siuda ER, Li Y, Song J, Song YM, Pao HA, Kim R-H, et al. Injectable, cellular-scale optoelectronics with applications for wireless optogenetics. Science. 2013b;340:211–216. doi: 10.1126/science.1232437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Judkewitz B, Kano M, Denk W, Häusser M. Targeted patch-clamp recordings and single-cell electroporation of unlabeled neurons in vivo. Nat. Methods. 2008;5:61–67. doi: 10.1038/nmeth1150. [DOI] [PubMed] [Google Scholar]
- Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, et al. Independent optical excitation of distinct neural populations. Nat. Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knöpfel T, Díez-García J, Akemann W. Optical probing of neuronal circuit dynamics: genetically encoded versus classical fluorescent sensors. Trends Neurosci. 2006;29:160–166. doi: 10.1016/j.tins.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Ko H, Hofer SB, Pichler B, Buchanan KA, Sjöström PJ, Mrsic-Flogel TD. Functional specificity of local synaptic connections in neocortical networks. Nature. 2011;473:87–91. doi: 10.1038/nature09880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama T, Sato TR, O’Connor DH, Zhang Y-X, Huber D, Hooks BM, Gabitto M, Svoboda K. Learning-related fine-scale specificity imaged in motor cortex circuits of behaving mice. Nature. 2010;464:1182–1186. doi: 10.1038/nature08897. [DOI] [PubMed] [Google Scholar]
- Kosut R. Suboptimal control of linear time-invariant systems subject to control structure constraints. IEEE Trans. Autom. Control. 1970;15:557–563. [Google Scholar]
- Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat. Methods. 2012;9:90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravitz AV, Freeze BS, Parker PRL, Kay K, Thwin MT, Deisseroth K, Kreitzer AC. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622–626. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krook-Magnuson E, Szabo GG, Armstrong C, Oijala M, Soltesz I. Cerebellar directed optogenetic intervention inhibits spontaneous hippocampal seizures in a mouse model of temporal lobe epilepsy. Eneuro. 2014;1 doi: 10.1523/ENEURO.0005-14.2014. ENEURO.0005-0014.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krook-Magnuson E, Armstrong C, Bui A, Lew S, Oijala M, Soltesz I. vivo evaluation of the dentate gate theory in epilepsy. J. Physiol. 2015 doi: 10.1113/JP270056. http://dx.doi.org/10.1113/JP270056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo BC. Automatic Control Systems. New York: Prentice Hall; 1982. [Google Scholar]
- Kwan AC, Dan Y. Dissection of cortical microcircuits by single-neuron stimulation in vivo. Curr. Biol. 2012;22:1459–1467. doi: 10.1016/j.cub.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai TL, Xing H. Statistical Models and Methods for Financial Markets. New York: Springer New York; 2010. [Google Scholar]
- Laxpati NG, Mahmoudi B, Gutekunst C-A, Newman JP, Zeller-Town-son R, Gross RE. Real-time in vivo optogenetic neuromodulation and multielectrode electrophysiologic recording with NeuroRighter. Front. Neuroeng. 2014;7:40. doi: 10.3389/fneng.2014.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoq J, Savall J, Vučinić D, Grewe BF, Kim H, Li JZ, Kitch LJ, Schnitzer MJ. Visualizing mammalian brain area interactions by dual-axis two-photon calcium imaging. Nat. Neurosci. 2014;17:1825–1829. doi: 10.1038/nn.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Chrobak JJ, Sik A, Wiley RG, Buzsáki G. Hippocampal theta activity following selective lesion of the septal cholinergic system. NeuroScience. 1994;62:1033–1047. doi: 10.1016/0306-4522(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Lee JH, Durand R, Gradinaru V, Zhang F, Goshen I, Kim D-S, Fenno LE, Ramakrishnan C, Deisseroth K. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature. 2010;465:788–792. doi: 10.1038/nature09108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine WS. Control System Fundamentals. Boca Raton, FL: CRC Press; 1999. [Google Scholar]
- Levoy M, Ng R, Adams A, Footer M, Horowitz M. ACM SIGGRAPH 2006 Papers. New York: ACM; 2006. Light field microscopy; pp. 924–934. [Google Scholar]
- Levoy M, Zhang Z, McDowall I. Recording and controlling the 4D light field in a microscope using microlens arrays. J. Microsc. 2009;235:144–162. doi: 10.1111/j.1365-2818.2009.03195.x. [DOI] [PubMed] [Google Scholar]
- Lewi J, Butera R, Paninski L. Sequential optimal design of neurophysiology experiments. Neural Comput. 2009;21:619–687. doi: 10.1162/neco.2008.08-07-594. [DOI] [PubMed] [Google Scholar]
- Li CY, Poo MM, and Dan Y. Burst spiking of a single cortical neuron modifies global brain state. Science. 2009;324:643–646. doi: 10.1126/science.1169957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ke Y, Chan DCW, Qian Z-M, Yung KKL, Ko H, Arbuthnott GW, Yung W-H. Therapeutic deep brain stimulation in Parkinsonian rats directly influences motor cortex. Neuron. 2012;76:1030–1041. doi: 10.1016/j.neuron.2012.09.032. [DOI] [PubMed] [Google Scholar]
- Li L, Park IM, Brockmeier A, Chen B, Seth S, Francis JT, Sanchez JC, Príncipe JC. Adaptive inverse control of neural spatiotemporal spike patterns with a reproducing kernel Hilbert space (RKHS) framework. IEEE Trans. Neural Syst. Rehabil. Eng. 2013a;21:532–543. doi: 10.1109/TNSRE.2012.2200300. [DOI] [PubMed] [Google Scholar]
- Li Y, Shi X, Song J, Lu C, Kim T, McCall JG, Bruchas MR, Rogers JA, Huang Y. Thermal analysis of injectable, cellular-scale optoelectronics with pulsed power. Proc. R. Soc. Lond. Math. Phys. Eng. Sci. 2013b;469:20130142. doi: 10.1098/rspa.2013.0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat. Neurosci. 2013;16:1499–1508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Gong Z. Optimal Control of Switched Systems Arising in Fermentation Processes. Beijing: Tsinghua Press; Berlin: Springer Berlin Heidelberg; 2014. [Google Scholar]
- Liu J, Oweiss KG, Khalil HK. Feedback control of the spatiotemporal firing patterns of neural microcircuits. 2010 49th IEEE Conference on Decision and Control (CDC); 2010. pp. 4679–4684. [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, Tonegawa S. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung L. System Identification: Theory for the User. New York: Pearson Education; 1998. [Google Scholar]
- Ljung L. Perspectives on system identification. Annu. Rev. Contr. 2010;34:1–12. [Google Scholar]
- Llewellyn ME, Thompson KR, Deisseroth K, Delp SL. Orderly recruitment of motor units under optical control in vivo. Nat. Med. 2010;16:1161–1165. doi: 10.1038/nm.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L, Anderson DJ. A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron. 2011;72:938–950. doi: 10.1016/j.neuron.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Froriep UP, Koppes RA, Canales A, Caggiano V, Selvidge J, Bizzi E, Anikeeva P. Polymer fiber probes enable optical control of spinal cord and muscle function in vivo. Adv. Funct. Mater. 2014;24:6594–6600. [Google Scholar]
- Lundby A, Mutoh H, Dimitrov D, Akemann W, Knöpfel T. Engineering of a genetically encodable fluorescent voltage sensor exploiting fast Ci-VSP voltage-sensing movements. PLoS ONE. 2008;3:e2514. doi: 10.1371/journal.pone.0002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Billings SA, Tsang KM. On-line structure detection and parameter estimation with exponential windowing for nonlinear systems. Eur. J. Control. 1996;2:291–304. [Google Scholar]
- Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütcke H, Gerhard F, Zenke F, Gerstner W, Helmchen F. Inference of neuronal network spike dynamics and topology from calcium imaging data. Front. Neural Circuits. 2013:7. doi: 10.3389/fncir.2013.00201. http://dx.doi.org/10.3389/fncir.2013.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz C, Otis TS, DeSars V, Charpak S, DiGregorio DA, Emiliani V. Holographic photolysis of caged neurotransmitters. Nat. Methods. 2008;5:821–827. doi: 10.1038/nmeth.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski JM. Predictive Control: With Constraints. New York: Prentice Hall; 2002. [Google Scholar]
- MacLean JN, Watson BO, Aaron GB, Yuste R. Internal dynamics determine the cortical response to thalamic stimulation. Neuron. 2005;48:811–823. doi: 10.1016/j.neuron.2005.09.035. [DOI] [PubMed] [Google Scholar]
- Mairal J, Bach F, Ponce J, Sapiro G. Online learning for matrix factorization and sparse coding. J. Mach. Learn. Res. 2010;11:19–60. [Google Scholar]
- Mante V, Sussillo D, Shenoy KV, Newsome WT. Context-dependent computation by recurrent dynamics in prefrontal cortex. Nature. 2013;503:78–84. doi: 10.1038/nature12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus EJ, Qin YL, Leonard B, Skaggs WE, McNaughton BL, Barnes CA. Interactions between location and task affect the spatial and directional firing of hippocampal neurons. J. Neurosci. 1995;15:7079–7094. doi: 10.1523/JNEUROSCI.15-11-07079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshel JH, Mori T, Nielsen KJ, Callaway EM. Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron. 2010;67:562–574. doi: 10.1016/j.neuron.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshel JH, Garrett ME, Nauhaus I, Callaway EM. Functional specialization of seven mouse visual cortical areas. Neuron. 2011;72:1040–1054. doi: 10.1016/j.neuron.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama R, Maeda K, Moroda H, Kato I, Inoue M, Miyakawa H, Aonishi T. Detecting cells using non-negative matrix factorization on calcium imaging data. Neural Netw. 2014;55:11–19. doi: 10.1016/j.neunet.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Mattis J, Tye KM, Ferenczi EA, Ramakrishnan C, O’Shea DJ, Prakash R, Gunaydin LA, Hyun M, Fenno LE, Gradinaru V, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat. Methods. 2012;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurovic I, Baotic M, Petrovic I. Explicit Model Predictive Control for trajectory tracking with mobile robots. 2011 IEEE/ASME International Conference on Advanced Intelligent Mechatronics (AIM); 2011. pp. 712–717. [Google Scholar]
- Maxwell JC. The Scientific Papers of James Clerk Maxwell. Mineola, NY: Dover Publications, Inc; 2003. [Google Scholar]
- Mayblum T, Schejter A, Dana H, Shoham S. New insights and system designs for temporally-focused multiphoton optogenetics. In: Periasamy A, Si PTC, König K, editors. Multiphoton Microscopy in the Biomedical Sciences XV, Proc. of SPIE Vol. 9329; 2015. pp. 932928-2–932928-6. [Google Scholar]
- Mazumder R, Hastie T. The graphical lasso: New insights and alternatives. Electron. J. Stat. 2012;6:2125–2149. doi: 10.1214/12-EJS740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesbah A, Streif S. A probabilistic approach to robust optimal experiment design with chance constraints. arXiv. 2014;1411:2683. http://arxiv.org/abs/1411.2683. [Google Scholar]
- Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minev IR, Musienko P, Hirsch A, Barraud Q, Wenger N, Moraud EM, Gandar J, Capogrosso M, Milekovic T, Asboth L, et al. Biomaterials. Electronic dura mater for long-term multimodal neural interfaces. Science. 2015;347:159–163. doi: 10.1126/science.1260318. [DOI] [PubMed] [Google Scholar]
- Mishchenko Y, Vogelstein JT, Paninski L. A Bayesian approach for inferring neuronal connectivity from calcium fluorescent imaging data. Ann. Appl. Stat. 2011;5:1229–1261. [Google Scholar]
- Mitra P, Bokil H. Observed Brain Dynamics. New York: Oxford University Press; 2007. [Google Scholar]
- Moser M-B, Rowland DC, Moser EI. Place cells, grid cells, and memory. Cold Spring Harb. Perspect. Biol. 2015;7:a021808. doi: 10.1101/cshperspect.a021808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, Schnitzer MJ. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron. 2009;63:747–760. doi: 10.1016/j.neuron.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller RU, Kubie JL. The effects of changes in the environment on the spatial firing of hippocampal complex-spike cells. J. Neurosci. 1987;7:1951–1968. doi: 10.1523/JNEUROSCI.07-07-01951.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh H, Perron A, Akemann W, Iwamoto Y, Knöpfel T. Optogenetic monitoring of membrane potentials. Exp. Physiol. 2011;96:13–18. doi: 10.1113/expphysiol.2010.053942. [DOI] [PubMed] [Google Scholar]
- Nabavi S, Fox R, Proulx CD, Lin JY, Tsien RY, Malinow R. Engineering a memory with LTD and LTP. Nature. 2014;511:348–352. doi: 10.1038/nature13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassi JJ, Callaway EM. Parallel processing strategies of the primate visual system. Nat. Rev. Neurosci. 2009;10:360–372. doi: 10.1038/nrn2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson JL, Yanagawa Y, Obata K, Callaway EM. Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. NeuroScience. 2009;161:441–450. doi: 10.1016/j.neuroscience.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TKT, Navratilova Z, Cabral H, Wang L, Gielen G, Battaglia FP, Bartic C. Closed-loop optical neural stimulation based on a 32-channel low-noise recording system with online spike sorting. J. Neural Eng. 2014;11:046005. doi: 10.1088/1741-2560/11/4/046005. [DOI] [PubMed] [Google Scholar]
- Nikolenko V, Poskanzer KE, Yuste R. Two-photon photostimulation and imaging of neural circuits. Nat. Methods. 2007;4:943–950. doi: 10.1038/nmeth1105. [DOI] [PubMed] [Google Scholar]
- Nikolenko V, Watson BO, Araya R, Woodruff A, Peterka DS, Yuste R. SLM microscopy: scanless two-photon imaging and photostimulation with spatial light modulators. Front Neural Circuits. 2008;2:5. doi: 10.3389/neuro.04.005.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic K, Grossman N, Grubb MS, Burrone J, Toumazou C, Degenaar P. Photocycles of channelrhodopsin-2. Photochem. Photobiol. 2009;85:400–411. doi: 10.1111/j.1751-1097.2008.00460.x. [DOI] [PubMed] [Google Scholar]
- O’Callaghan L, Meyerson A, Motwani R, Mishra N, Guha S. Streaming-data algorithms for high-quality clustering. 2013 IEEE 29th International Conference on Data Engineering (ICDE); 2002. pp. 685–694. [Google Scholar]
- O’Connor DH, Huber D, Svoboda K. Reverse engineering the mouse brain. Nature. 2009;461:923–929. doi: 10.1038/nature08539. [DOI] [PubMed] [Google Scholar]
- O’Connor DH, Hires SA, Guo ZV, Li N, Yu J, Sun Q-Q, Huber D, Svoboda K. Neural coding during active somatosensation revealed using illusory touch. Nat. Neurosci. 2013;16:958–965. doi: 10.1038/nn.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe J, Recce ML. Phase relationship between hippocampal place units and the EEG theta rhythm. Hippocampus. 1993;3:317–330. doi: 10.1002/hipo.450030307. [DOI] [PubMed] [Google Scholar]
- Ogata K. Modern Control Engineering. New York: Prentice Hall; 2010. [Google Scholar]
- Ogunfunmi T. Adaptive Nonlinear System Identification: The Volterra and Wiener Model Approaches. New York: Springer Science & Business Media; 2007. [Google Scholar]
- Ohlsson H, Ljung L. Identification of switched linear regression models using sum-of-norms regularization. Automatica. 2013;49:1045–1050. [Google Scholar]
- Ohlsson H, Ljung L, Boyd S. Segmentation of ARX-models using sum-of-norms regularization. Automatica. 2010;46:1107–1111. [Google Scholar]
- Okun M, Lampl I. Instantaneous correlation of excitation and inhibition during ongoing and sensory-evoked activities. Nat. Neurosci. 2008;11:535–537. doi: 10.1038/nn.2105. [DOI] [PubMed] [Google Scholar]
- Oñativia J, Schultz SR, Dragotti PL. A finite rate of innovation algorithm for fast and accurate spike detection from two-photon calcium imaging. J. Neural Eng. 2013;10:046017. doi: 10.1088/1741-2560/10/4/046017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oron D, Papagiakoumou E, Anselmi F, Emiliani V. Two-photon optogenetics. Prog. Brain Res. 2012;196:119–143. doi: 10.1016/B978-0-444-59426-6.00007-0. [DOI] [PubMed] [Google Scholar]
- Osakada F, Mori T, Cetin AH, Marshel JH, Virgen B, Callaway EM. New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron. 2011;71:617–631. doi: 10.1016/j.neuron.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu Y, Bormuth V, Wong J, Mathieu B, Dugue´ GP, Feltz A, Dieudonné S. Optical monitoring of neuronal activity at high frame rate with a digital random-access multiphoton (RAMP) microscope. J. Neurosci. Methods. 2008;173:259–270. doi: 10.1016/j.jneumeth.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Pachitariu M, Packer AM, Pettit N, Dalgleish H, Hausser M, Sahani M. Extracting regions of interest from biological images with convolutional sparse block coding. In: Burges CJC, Bottou L, Welling M, Ghahramani Z, Weinberger KQ, editors. Advances in Neural Information Processing Systems 26. Red Hook, NY: Curran Associates, Inc; 2013. pp. 1745–1753. [Google Scholar]
- Packer AM, Peterka DS, Hirtz JJ, Prakash R, Deisseroth K, Yuste R. Two-photon optogenetics of dendritic spines and neural circuits. Nat. Methods. 2012;9:1202–1205. doi: 10.1038/nmeth.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Roska B, Häusser M. Targeting neurons and photons for optogenetics. Nat. Neurosci. 2013;16:805–815. doi: 10.1038/nn.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Russell LE, Dalgleish HWP, Häusser M. Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat. Methods. 2015;12:140–146. doi: 10.1038/nmeth.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakman A, Huggins J, Smith C, Paninski L. Fast state-space methods for inferring dendritic synaptic connectivity. J. Comput. Neurosci. 2014;36:415–443. doi: 10.1007/s10827-013-0478-0. [DOI] [PubMed] [Google Scholar]
- Pala A, Petersen CCH. In vivo measurement of cell-type-specific synaptic connectivity and synaptic transmission in layer 2/3 mouse barrel cortex. Neuron. 2015;85:68–75. doi: 10.1016/j.neuron.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paninski L. Asymptotic theory of information-theoretic experimental design. Neural Comput. 2005;17:1480–1507. doi: 10.1162/0899766053723032. [DOI] [PubMed] [Google Scholar]
- Paninski L. Fast Kalman filtering on quasilinear dendritic trees. J. Comput. Neurosci. 2010;28:211–228. doi: 10.1007/s10827-009-0200-4. [DOI] [PubMed] [Google Scholar]
- Paninski L, Ahmadian Y, Ferreira DG, Koyama S, Rahnama Rad K, Vidne M, Vogelstein J, Wu W. Anew look at state-space models for neural data. J. Comput. Neurosci. 2010;29:107–126. doi: 10.1007/s10827-009-0179-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou M, Cassenaer S, Nowotny T, Laurent G. Normalization for sparse encoding of odors by a wide-field interneuron. Science. 2011;332:721–725. doi: 10.1126/science.1201835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiakoumou E. Optical developments for optogenetics. Biol. Cell. 2013;105:443–464. doi: 10.1111/boc.201200087. [DOI] [PubMed] [Google Scholar]
- Papagiakoumou E, de Sars V, Oron D, Emiliani V. Patterned two-photon illumination by spatiotemporal shaping of ultrashort pulses. Opt. Express. 2008;16:22039–22047. doi: 10.1364/oe.16.022039. [DOI] [PubMed] [Google Scholar]
- Pashaie R, Anikeeva P, Lee JH, Prakash R, Yizhar O, Prigge M, Chander D, Richner TJ, Williams J. Optogenetic brain interfaces. IEEE Rev Biomed Eng. 2014;7:3–30. doi: 10.1109/RBME.2013.2294796. [DOI] [PubMed] [Google Scholar]
- Paulson JA, Raimondo DM, Findeisen R, Braatz RD, Streif S. Guaranteed active fault diagnosis for uncertain nonlinear systems. Control Conference (ECC); 2014. pp. 926–931. 2014 European. [Google Scholar]
- Paz JT, Davidson TJ, Frechette ES, Delord B, Parada I, Peng K, Deisseroth K, Huguenard JR. Closed-loop optogenetic control of thalamus as a tool for interrupting seizures after cortical injury. Nat. Neurosci. 2013;16:64–70. doi: 10.1038/nn.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron A, Mutoh H, Akemann W, Gautam SG, Dimitrov D, Iwamoto Y, Knopfel T. Second and third generation voltage-sensitive fluorescent proteins for monitoring membrane potential. Front. Mol. Neurosci. 2009a;2:5. doi: 10.3389/neuro.02.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron A, Mutoh H, Launey T, Knopfel T. Red-shifted voltage-sensitive fluorescent proteins. Chem. Biol. 2009b;16:1268–1277. doi: 10.1016/j.chembiol.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterka DS, Takahashi H, Yuste R. Imaging voltage in neurons. Neuron. 2011;69:9–21. doi: 10.1016/j.neuron.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodop-sin-2-assisted circuit mapping of long-range callosal projections. Nat. Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- Pfau D, Pnevmatikakis EA, Paninski L. Robust learning of low-dimensional dynamics from large neural ensembles. In: Burges CJC, Bottou L, Welling M, Ghahramani Z, Weinberger KQ, editors. Advances in Neural Information Processing Systems 26. Red Hook, NY: Curran Associates, Inc; 2013. pp. 2391–2399. [Google Scholar]
- Pillow JW, Ahmadian Y, Paninski L. Model-based decoding, information estimation, and change-point detection techniques for multineuron spike trains. Neural Comput. 2011;23:1–45. doi: 10.1162/NECO_a_00058. [DOI] [PubMed] [Google Scholar]
- Pisanello F, Sileo L, Oldenburg IA, Pisanello M, Martiradonna L, Assad JA, Sabatini BL, De Vittorio M. Multipoint-emitting optical fibers for spatially addressable in vivo optogenetics. Neuron. 2014;82:1245–1254. doi: 10.1016/j.neuron.2014.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Paninski L. Sparse nonnegative deconvolution for compressive calcium imaging: algorithms and phase transitions. In: Burges CJC, Bottou L, Welling M, Ghahramani Z, Weinberger KQ, editors. Advances in Neural Information Processing Systems 26. Red Hook, NY: Curran Associates, Inc; 2013. pp. 1250–1258. [Google Scholar]
- Pnevmatikakis EA, Kelleher K, Chen R, Saggau P, Josic K, Paninski L. Fast spatiotemporal smoothing of calcium measurements in dendritic trees. PLoS Comput. Biol. 2012;8:e1002569. doi: 10.1371/journal.pcbi.1002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouille F, Scanziani M. Enforcement of temporal fidelity in pyramidal cells by somatic feed-forward inhibition. Science. 2001;293:1159–1163. doi: 10.1126/science.1060342. [DOI] [PubMed] [Google Scholar]
- Pouille F, Marin-Burgin A, Adesnik H, Atallah BV, Scanziani M. Input normalization by global feedforward inhibition expands cortical dynamic range. Nat. Neurosci. 2009;12:1577–1585. doi: 10.1038/nn.2441. [DOI] [PubMed] [Google Scholar]
- Prakash R, Yizhar O, Grewe B, Ramakrishnan C, Wang N, Goshen I, Packer AM, Peterka DS, Yuste R, Schnitzer MJ, Deisseroth K. Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat. Methods. 2012;9:1171–1179. doi: 10.1038/nmeth.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevedel R, Yoon Y-G, Hoffmann M, Pak N, Wetzstein G, Kato S, Schrödel T, Raskar R, Zimmer M, Boyden ES, Vaziri A. Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy. Nat. Methods. 2014;11:727–730. doi: 10.1038/nmeth.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz AA, Abbott LF, Marder E. The dynamic clamp comes of age. Trends Neurosci. 2004;27:218–224. doi: 10.1016/j.tins.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Qin SJ, Badgwell TA. A survey of industrial model predictive control technology. Control Eng. Pract. 2003;11:733–764. [Google Scholar]
- Quirin S, Peterka DS, Yuste R. Instantaneous three-dimensional sensing using spatial light modulator illumination with extended depth of field imaging. Opt. Express. 2013;21:16007–16021. doi: 10.1364/OE.21.016007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancz EA, Franks KM, Schwarz MK, Pichler B, Schaefer AT, Margrie TW. Transfection via whole-cell recording in vivo: bridging single-cell physiology, genetics and connectomics. Nat. Neurosci. 2011;14:527–532. doi: 10.1038/nn.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauov I, Kvasnica M, Cirka L, Fikar M. Real-time model predictive control of a laboratory liquid tanks system. 17th international conference on process control; 2009. ISBN 978-80-227-3081-5. [Google Scholar]
- Rawlings JB. Tutorial overview of model predictive control. IEEE Control Syst. 2000;20:38–52. [Google Scholar]
- Richner TJ, Thongpang S, Brodnick SK, Schendel AA, Falk RW, Krugner-Higby LA, Pashaie R, Williams JC. Optogenetic micro-electrocorticography for modulating and localizing cerebral cortex activity. J. Neural Eng. 2014;11:016010. doi: 10.1088/1741-2560/11/1/016010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer JP, Tank DW. Two-photon excitation of channelrhodopsin-2 at saturation. Proc. Natl. Acad. Sci. USA. 2009;106:15025–15030. doi: 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer JP, Deisseroth K, Tank DW. Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields. Nat. Neurosci. 2014;17:1816–1824. doi: 10.1038/nn.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer S, Zemelman BV, Barbic M, Losonczy A, Buzsa´ki G, Magee JC. Multi-array silicon probes with integrated optical fibers: light-assisted perturbation and recording of local neural circuits in the behaving animal. Eur. J. Neurosci. 2010;31:2279–2291. doi: 10.1111/j.1460-9568.2010.07250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahani M. PhD thesis. Pasadena, CA: California Institute of Technology; 1999. Latent variable models for neural data analysis. [Google Scholar]
- Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- Sakai R, Repunte-Canonigo V, Raj CD, Knöpfel T. Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur. J. Neurosci. 2001;13:2314–2318. doi: 10.1046/j.0953-816x.2001.01617.x. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Takahashi N, Matsuki N, Ikegaya Y. Fast and accurate detection of action potentials from somatic calcium fluctuations. J. Neurophysiol. 2008;100:1668–1676. doi: 10.1152/jn.00084.2008. [DOI] [PubMed] [Google Scholar]
- Sato TR, Svoboda K. The functional properties of barrel cortex neurons projecting to the primary motor cortex. J. Neurosci. 2010;30:4256–4260. doi: 10.1523/JNEUROSCI.3774-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff SJ. Neural Control Engineering: The Emerging Intersection Between Control Theory and NeuroScience. Boston: MIT Press; 2012. [Google Scholar]
- Schiff SJ, Sauer T. Kalman filter control of a model of spatiotemporal cortical dynamics. J. Neural Eng. 2008;5:1–8. doi: 10.1088/1741-2560/5/1/001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DM, Nelson A, Mooney R. A synaptic and circuit basis for corollary discharge in the auditory cortex. Nature. 2014;513:189–194. doi: 10.1038/nature13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheis C, Liewald JF, Bamberg E, Nagel G, Gottschalk A. Optogenetic long-term manipulation of behavior and animal development. PLoS One. 2011;6(4):e18766. doi: 10.1371/journal.pone.0018766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz W. Multiple dopamine functions at different time courses. Annu. Rev. Neurosci. 2007;30:259–288. doi: 10.1146/annurev.neuro.28.061604.135722. [DOI] [PubMed] [Google Scholar]
- Schulz K, Sydekum E, Krueppel R, Engelbrecht CJ, Schlegel F, Schröter A, Rudin M, Helmchen F. Simultaneous BOLD fMRI and fiber-optic calcium recording in rat neocortex. Nat. Methods. 2012;9:597–602. doi: 10.1038/nmeth.2013. [DOI] [PubMed] [Google Scholar]
- Shababo B, Paige B, Pakman A, Paninski L. Bayesian Inference and Online Experimental Design for Mapping Neural Microcircuits. In: Burges CJC, Bottou L, Welling M, Ghahramani Z, Weinberger KQ, editors. Advances in Neural Information Processing Systems (NIPS) 26. Red Hook, NY: Curran Associates, Inc; 2013. pp. 1304–1312. [Google Scholar]
- Sharp AA, O’Neil MB, Abbott LF, Marder E. Dynamic clamp: computer-generated conductances in real neurons. J. Neurophysiol. 1993;69:992–995. doi: 10.1152/jn.1993.69.3.992. [DOI] [PubMed] [Google Scholar]
- Shenoy KV, Carmena JM. Combining decoder design and neural adaptation in brain-machine interfaces. Neuron. 2014;84:665–680. doi: 10.1016/j.neuron.2014.08.038. [DOI] [PubMed] [Google Scholar]
- Shenoy KV, Sahani M, Churchland MM. Cortical control of arm movements: a dynamical systems perspective. Annu. Rev. Neurosci. 2013;36:337–359. doi: 10.1146/annurev-neuro-062111-150509. [DOI] [PubMed] [Google Scholar]
- Shu Y, Hasenstaub A, McCormick DA. Turning on and off recurrent balanced cortical activity. Nature. 2003;423:288–293. doi: 10.1038/nature01616. [DOI] [PubMed] [Google Scholar]
- Siegel MS, Isacoff EY. A genetically encoded optical probe of membrane voltage. Neuron. 1997;19:735–741. doi: 10.1016/s0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- Siegle JH, Wilson MA. Enhancement of encoding and retrieval functions through theta phase-specific manipulation of hippocampus. eLife. 2014;3:e03061. doi: 10.7554/eLife.03061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjulson L, Miesenböck G. Optical recording of action potentials and other discrete physiological events: a perspective from signal detection theory. Physiology (Bethesda) 2007;22:47–55. doi: 10.1152/physiol.00036.2006. [DOI] [PubMed] [Google Scholar]
- Skaggs WE, McNaughton BL. Replay of neuronal firing sequences in rat hippocampus during sleep following spatial experience. Science. 1996;271:1870–1873. doi: 10.1126/science.271.5257.1870. [DOI] [PubMed] [Google Scholar]
- Smith SL, Häusser M. Parallel processing of visual space by neighboring neurons in mouse visual cortex. Nat. Neurosci. 2010;13:1144–1149. doi: 10.1038/nn.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudry D, Keshri S, Stinson P, Oh M, Iyengar G, Paninski L. arXiv, ArXiv13093724 Q-Biol. Stat. 2013. A shotgun sampling solution for the common input problem in neural connectivity inference. http://arxiv.org/abs/1309.3724. [Google Scholar]
- St-Pierre F, Marshall JD, Yang Y, Gong Y, Schnitzer MJ, Lin MZ. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci. 2014;17:884–889. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark E, Koos T, Buzsaki G. Diode probes for spatiotemporal optical control of multiple neurons in freely-moving animals. J. Neurophysiol. 2012;108:349–363. doi: 10.1152/jn.00153.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark E, Roux L, Eichler R, Senzai Y, Royer S, Buzsáki G. Pyramidal cell-interneuron interactions underlie hippocampal ripple oscillations. Neuron. 2014;83:467–480. doi: 10.1016/j.neuron.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetter O, Orlandi J, Soriano J, Battaglia D, Geisel T. Network reconstruction from calcium imaging data of spontaneously bursting neuronal activity. BMC Neurosci. 2013;14:139. [Google Scholar]
- Stuber GD, Sparta DR, Stamatakis AM, van Leeuwen WA, Hardjoprajitno JE, Cho S, Tye KM, Kempadoo KA, Zhang F, Deisseroth K, Bonci A. Excitatory transmission from the amygdala to nucleus accumbens facilitates reward seeking. Nature. 2011;475:377–380. doi: 10.1038/nature10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer V, Bobin J, Chahid M, Mousavi HS, Candes E, Dahan M. Compressive fluorescence microscopy for biological and hyperspectral imaging. Proc. Natl. Acad. Sci. USA. 2012;109:E1679–E1687. doi: 10.1073/pnas.1119511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers T, Lygeros J. Edward B, editor. Optimal sensor and actuator placement in complex dynamical networks. 2014 arXiv, arXiv:1306.2491, http://arxiv.org/abs/1306.2491v2.
- Svoboda K, Denk W, Kleinfeld D, Tank DW. In vivo dendritic calcium dynamics in neocortical pyramidal neurons. Nature. 1997;385:161–165. doi: 10.1038/385161a0. [DOI] [PubMed] [Google Scholar]
- Szabo V, Ventalon C, De Sars V, Bradley J, Emiliani V. Spatially selective holographic photoactivation and functional fluorescence imaging in freely behaving mice with a fiberscope. Neuron. 2014;84:1157–1169. doi: 10.1016/j.neuron.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Tanaka KF, Matsui K, Sasaki T, Sano H, Sugio S, Fan K, Hen R, Nakai J, Yanagawa Y, Hasuwa H, et al. Expanding the repertoire of optogenetically targeted cells with an enhanced gene expression system. Cell Rep. 2012;2:397–406. doi: 10.1016/j.celrep.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis L, Berens P, Froudarakis E, Reimer J, Roman-Roson M, Baden T, Euler T, Tolias AS, Bethge M. Supervised learning sets benchmark for robust spike detection from calcium imaging signals. bioRxiv. 2014:010777. [Google Scholar]
- Thestrup T, Litzlbauer J, Bartholomäus I, Mues M, Russo L, Dana H, Kovalchuk Y, Liang Y, Kalamakis G, Laukat Y, et al. Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat. Methods. 2014;11:175–182. doi: 10.1038/nmeth.2773. [DOI] [PubMed] [Google Scholar]
- Tischer D, Weiner OD. Illuminating cell signalling with optogenetic tools. Nat. Rev. Mol. Cell Biol. 2014;15:551–558. doi: 10.1038/nrm3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towne C, Montgomery KL, Iyer SM, Deisseroth K, Delp SL. Optogenetic control of targeted peripheral axons in freely moving animals. PLoS ONE. 2013;8:e72691. doi: 10.1371/journal.pone.0072691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H-C, Zhang F, Adamantidis A, Stuber GD, Bonci A, de Lecea L, Deisseroth K. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324:1080–1084. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turaga S, Buesing L, Packer AM, Dalgleish H, Pettit N, Hausser M, Macke J. Inferring neural population dynamics from multiple partial recordings of the same neural circuit. In: Burges CJC, Bottou L, Welling M, Ghahramani Z, Weinberger KQ, editors. Advances in Neural Information Processing Systems 26. Red Hook, NY: Curran Associates, Inc; 2013. pp. 539–547. [Google Scholar]
- Tye KM, Prakash R, Kim S-Y, Fenno LE, Grosenick L, Zarabi H, Thompson KR, Gradinaru V, Ramakrishnan C, Deisseroth K. Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 2011;471:358–362. doi: 10.1038/nature09820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlhaas PJ, Singer W. Neural synchrony in brain disorders: relevance for cognitive dysfunctions and pathophysiology. Neuron. 2006;52:155–168. doi: 10.1016/j.neuron.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Vallbo ÅB, Olsson KÅ, Westberg K-G, Clark FJ. Microstimulation of single tactile afferents from the human hand. Sensory attributes related to unit type and properties of receptive fields. Brain. 1984;107:727–749. doi: 10.1093/brain/107.3.727. [DOI] [PubMed] [Google Scholar]
- Vaziri A, Emiliani V. Reshaping the optical dimension in optogenetics. Curr. Opin. Neurobiol. 2012;22:128–137. doi: 10.1016/j.conb.2011.11.011. [DOI] [PubMed] [Google Scholar]
- Vellekoop IM, van Putten EG, Lagendijk A, Mosk AP. Demixing light paths inside disordered metamaterials. Opt. Express. 2008;16:67–80. doi: 10.1364/oe.16.000067. [DOI] [PubMed] [Google Scholar]
- Vidyasagar M. Nonlinear Systems Analysis: Second Edition. Philadelphia: Society for Industrial and Applied Mathematics; 2002. [Google Scholar]
- Vogelstein JT, Watson BO, Packer AM, Yuste R, Jedynak B, Paninski L. Spike inference from calcium imaging using sequential Monte Carlo methods. Biophys. J. 2009;97:636–655. doi: 10.1016/j.bpj.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein JT, Packer AM, Machado TA, Sippy T, Babadi B, Yuste R, Paninski L. Fast nonnegative deconvolution for spike train inference from population calcium imaging. J. Neurophysiol. 2010;104:3691–3704. doi: 10.1152/jn.01073.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorman A, Shojaie A, Witten D. Graph estimation with joint additive models. Biometrika. 2013;101:85–101. doi: 10.1093/biomet/ast053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall NR, Wickersham IR, Cetin A, De La Parra M, Callaway EM. Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc. Natl. Acad. Sci. USA. 2010;107:21848–21853. doi: 10.1073/pnas.1011756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Boyd S. Approximate dynamic programming via iterated Bellman inequalities. Int. J. Robust Nonlinear Control. 2010;00:1–25. [Google Scholar]
- Wang Y, Boyd S. Performance bounds and suboptimal policies for linear stochastic control via LMIs. Int. J. Robust Nonlinear Control. 2011a;21:1710–1728. [Google Scholar]
- Wang Y, Boyd S. Fast evaluation of quadratic control-lyapunov policy. IEEE Trans. Contr. Syst. Technol. 2011b;19:939–946. [Google Scholar]
- Warden MR, Selimbeyoglu A, Mirzabekov JJ, Lo M, Thompson KR, Kim S-Y, Adhikari A, Tye KM, Frank LM, Deisseroth K. A prefrontal cortex-brainstem neuronal projection that controls response to behavioural challenge. Nature. 2012;492:428–432. doi: 10.1038/nature11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden MR, Cardin JA, Deisseroth K. Optical neural interfaces. Annu. Rev. Biomed. Eng. 2014;16:103–129. doi: 10.1146/annurev-bioeng-071813-104733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kessler D, Scott C, Angstadt M, Sripada C. Disease prediction based on functional connectomes using a scalable and spatially-informed support vector machine. Neuroimage. 2014;96:183–202. doi: 10.1016/j.neuroimage.2014.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehr M, Zador AM. Balanced inhibition underlies tuning and sharpens spike timing in auditory cortex. Nature. 2003;426:442–446. doi: 10.1038/nature02116. [DOI] [PubMed] [Google Scholar]
- Wenger N, Moraud EM, Raspopovic S, Bonizzato M, DiGiovanna J, Musienko P, Morari M, Micera S, Courtine G. Closed-loop neuromodulation of spinal sensorimotor circuits controls refined locomotion after complete spinal cord injury. Sci. Transl. Med. 2014;6:255ra133. doi: 10.1126/scitranslmed.3008325. [DOI] [PubMed] [Google Scholar]
- Wentz CT, Bernstein JG, Monahan P, Guerra A, Rodriguez A, Boy-den ES. A wirelessly powered and controlled device for optical neural control of freely-behaving animals. J. Neural Eng. 2011;8:046021. doi: 10.1088/1741-2560/8/4/046021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann K-K, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat. Methods. 2007a;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJO, Mori T, Finke S, Conzelmann K-K, Young JAT, and Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007b;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wietek J, Wiegert JS, Adeishvili N, Schneider F, Watanabe H, Tsunoda SP, Vogt A, Elstner M, Oertner TG, Hegemann P. Conversion of channelrhodopsin into a light-gated chloride channel. Science. 2014;344:409–412. doi: 10.1126/science.1249375. [DOI] [PubMed] [Google Scholar]
- Wilms CD, Häusser M. Twitching towards the ideal calcium sensor. Nat. Methods. 2014;11:139–140. doi: 10.1038/nmeth.2814. [DOI] [PubMed] [Google Scholar]
- Wilson MA, McNaughton BL. Reactivation of hippocampal ensemble memories during sleep. Science. 1994;265:676–679. doi: 10.1126/science.8036517. [DOI] [PubMed] [Google Scholar]
- Wilson NR, Runyan CA, Wang FL, Sur M. Division and subtraction by distinct cortical inhibitory networks in vivo. Nature. 2012;488:343–348. doi: 10.1038/nature11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilt BA, Fitzgerald JE, Schnitzer MJ. Photon shot noise limits on optical detection of neuronal spikes and estimation of spike timing. Biophys. J. 2013;104:51–62. doi: 10.1016/j.bpj.2012.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsching L, Ferreau HJ, Bock HG, Diehl M. An online set strategy for fast adjoint based nonlinear model predictive control. In: Xia X, Camisani-Calzolari F, editors. Nonlinear Control Systems 7. Pretoria: International Federation of Automatic Control; 2007. pp. 234–239. [Google Scholar]
- Witt A, Palmigiano A, Neef A, El Hady A, Wolf F, Battaglia D. Controlling the oscillation phase through precisely timed closed-loop optogenetic stimulation: a computational study. Front. Neural Circuits. 2013;7:49. doi: 10.3389/fncir.2013.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten DM, Friedman JH, Simon N. New insights and faster computations for the graphical lasso. J. Comput. Graph. Statist. 2011;20:892–900. [Google Scholar]
- Wu J, Liu L, Matsuda T, Zhao Y, Rebane A, Drobizhev M, Chang Y-F, Araki S, Arai Y, March K, et al. Improved orange and red Ca2± indicators and photophysical considerations for optogenetic applications. ACS Chem. Neurosci. 2013;4:963–972. doi: 10.1021/cn400012b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Abdelfattah AS, Miraucourt LS, Kutsarova E, Ruangkittisakul A, Zhou H, Ballanyi K, Wicks G, Drobizhev M, Rebane A, et al. A long Stokes shift red fluorescent Ca2+ indicator protein for two-photon and ratiometric imaging. Nat. Commun. 2014:5. doi: 10.1038/ncomms6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Antsaklis PJ. Optimal control of switched autonomous systems. Proceedings of the 41st IEEE Conference on Decision and Control; 2002. pp. 4401–4406. [Google Scholar]
- Xu W, Südhof TC. A neural circuit for memory specificity and generalization. Science. 2013;339:1290–1295. doi: 10.1126/science.1229534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Atallah BV, Scanziani M. Equalizing excitation-inhibition ratios across visual cortical neurons. Nature. 2014;511:596–600. doi: 10.1038/nature13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaksi E, Friedrich RW. Reconstruction of firing rate changes across neuronal populations by temporally deconvolved Ca2+ imaging. Nat. Methods. 2006;3:377–383. doi: 10.1038/nmeth874. [DOI] [PubMed] [Google Scholar]
- Yang S, Papagiakoumou E, Guillon M, de Sars V, Tang C-M, Emiliani V. Three-dimensional holographic photostimulation of the dendritic arbor. J. Neural Eng. 2011;8:046002. doi: 10.1088/1741-2560/8/4/046002. [DOI] [PubMed] [Google Scholar]
- Yeh AJ, Ho JS, Poon ASY. Optical probe for input-impedance measurement of in vivo power-receiving microstructure. 2014 IEEE Antennas and Propagation Society International Symposium (APSURSI) 2014:1409–1410. [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011a;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O’Shea DJ, Sohal VS, Goshen I, Finkelstein J, Paz JT, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011b;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder RM, Pang KCH. Involvement of GABAergic and cholinergic medial septal neurons in hippocampal theta rhythm. Hippocampus. 2005;15:381–392. doi: 10.1002/hipo.20062. [DOI] [PubMed] [Google Scholar]
- Yu BM, Afshar A, Santhanam G, Ryu SI, Shenoy KV, Sahani M. Advances in Neural Information Processing Systems 18. Boston: MIT Press; 2006. Extracting dynamical structure embedded in neural activity. [Google Scholar]
- Yuste R. Dendritic Spines. Boston: MIT Press; 2010. [Google Scholar]
- Zadeh LA. On the Identification Problem. IRE Trans. Circuit Theory. 1956;3:277–281. [Google Scholar]
- Zalocusky K, Deisseroth K. Optogenetics in the behaving rat: integration of diverse new technologies in a vital animal model. Optogenetics. 2013;1:1–17. [Google Scholar]
- Zappavigna A, Colaneri P, Geromel JC, Middleton R. Stabilization of continuous-time switched linear positive systems. In American Control Conference (ACC) 2010;2010:3275–3280. [Google Scholar]
- Zeilinger MN, Jones CN, Morari M. Real-time suboptimal model predictive control using a combination of explicit mpc and online optimization. IEEE Trans. Autom. Control. 2011;56:1524–1534. [Google Scholar]
- Zhang F, Wang L-P, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- Zhang J, Laiwalla F, Kim JA, Urabe H, Van Wagenen R, Song Y-K, Connors BW, Zhang F, Deisseroth K, Nurmikko AV. Integrated device for optical stimulation and spatiotemporal electrical recording of neural activity in light-sensitized brain tissue. J. Neural Eng. 2009;6:055007. doi: 10.1088/1741-2560/6/5/055007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Araki S, Wu J, Teramoto T, Chang Y-F, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, Campbell RE. An expanded palette of genetically encoded Ca2+ indicators. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, El Gamal A, Schnitzer MJ. Long-term dynamics of CA1 hippocampal place codes. Nat. Neurosci. 2013;16:264–266. doi: 10.1038/nn.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Znamenskiy P, Zador AM. Corticostriatal neurons in auditory cortex drive decisions during auditory discrimination. Nature. 2013;497:482–485. doi: 10.1038/nature12077. [DOI] [PMC free article] [PubMed] [Google Scholar]