SUMMARY
In recently-developed approaches for high-resolution imaging within intact tissue, molecular characterization over large volumes has been largely restricted to labeling of proteins. But volumetric nucleic acid labeling may represent a far greater scientific and clinical opportunity, enabling detection of not only diverse coding RNA variants but also non-coding RNAs. Moreover, scaling immunohistochemical detection to large tissue volumes has limitations, due to high cost, limited renewability/availability, and restricted multiplexing capability of antibody labels. With the goal of versatile, high-content, and scalable molecular phenotyping of intact tissues, we developed a method using carbodiimide-based chemistry to stably retain RNAs in clarified tissue, coupled with amplification tools for multiplexed detection. The resulting technology enables robust measurement of activity-dependent transcriptional signatures, cell-identity markers, and diverse non-coding RNAs in rodent and human tissue volumes. The growing set of validated probes is deposited in an online resource for nucleating related developments from across the scientific community.
INTRODUCTION
An exciting theme in modern biology is moving toward joint maximization of the content and context of molecular-level observations—that is, obtaining high-resolution and content-rich information about the biological system, while also maintaining this system largely or fully intact to preserve crucial contextual information. Historically these two goals of content and context have been in opposition, since higher-resolution analyses have tended to require disassembling the system or accepting a limited field of view. But the value of obtaining and integrating information about the identity, function and connectivity of cells in intact 3D volumes has been increasingly appreciated.
For example, one of the current challenges in neuroscience is to query molecular identity, activity level, and circuit wiring of individual cells within intact brain networks, which would require linkage of information spanning several orders of magnitude in spatial scale. Until recently, investigating the structure of neural networks in this way required sectioning for optical access and molecular labeling, followed by computer-assisted alignment and 3D reconstruction (Denk and Horstmann, 2004; Micheva and Smith, 2007; Oh et al., 2014). Such reconstructions have been valuable, but are often laborious, limited to small volumes, and susceptible to loss of information at section boundaries, making tract-tracing and circuit-mapping particularly difficult (Wanner et al., 2015). However, tissue-clearing techniques have emerged that, to various degrees, enable the visualization of cell morphology (and in some cases molecular phenotype, as well as local and long-range wiring) embedded within intact neural circuits (Chung et al., 2013; Tomer et al., 2014; Yang et al., 2014; Dodt et al., 2007; Ertürk et al., 2012; Hama et al., 2011; Kuwajima et al., 2013; Renier et al., 2014; Richardson and Lichtman, 2015; Staudt et al., 2007; Susaki et al., 2014; Tainaka et al., 2014).
To date these technologies have chiefly focused on interrogating proteins, whether transgenically-expressed or immunohistochemically-detected (with the exception of single probes tested in CLARITY-based hydrogel experiments in sectioned tissue; Chung et al., 2013; Yang et al., 2014), and many such approaches may not be compatible with accessing the wealth of biological information contained in the RNA of large intact volumes. This untapped opportunity spans untranslated species, including microRNAs (which, among other reasons for investigation, are particularly relevant to human genetically-determined diseases; Esteller, 2011), the majority of splice variants, many immediate early gene (IEG) RNAs used to infer activity of particular regions or cells during behavior (Guzowski et al., 1999; Loebrich and Nedivi, 2009), and even the vast majority of translated gene products, due to limited antibody specificity and availability. We sought to address this challenge by developing generalizable methods for versatile and robust RNA preservation and access within transparent, intact tissue volumes.
RESULTS
Advancing clarified tissue chemistry with carbodiimide-based RNA retention
Many existing clearing methods rely on incubation of tissue for prolonged periods of time at temperatures of 37°C or greater (Chung et al., 2013 ; Tomer et al., 2014; Yang et al., 2014; Renier et al., 2014; Susaki et al., 2014; Tainaka et al., 2014); however, formalin is known to revert its crosslinks at elevated temperatures, and the bonds made to nucleic acids are particularly vulnerable (Masuda et al., 1999; Srinivasan et al., 2002). Therefore, to improve retention of RNA during high-temperature tissue clearing, we sought to introduce temperature- resistant covalent linkages to RNA molecules prior to clearing, by targeting functional groups on the RNA molecule for fixation to surrounding proteins or the hydrogel matrix.
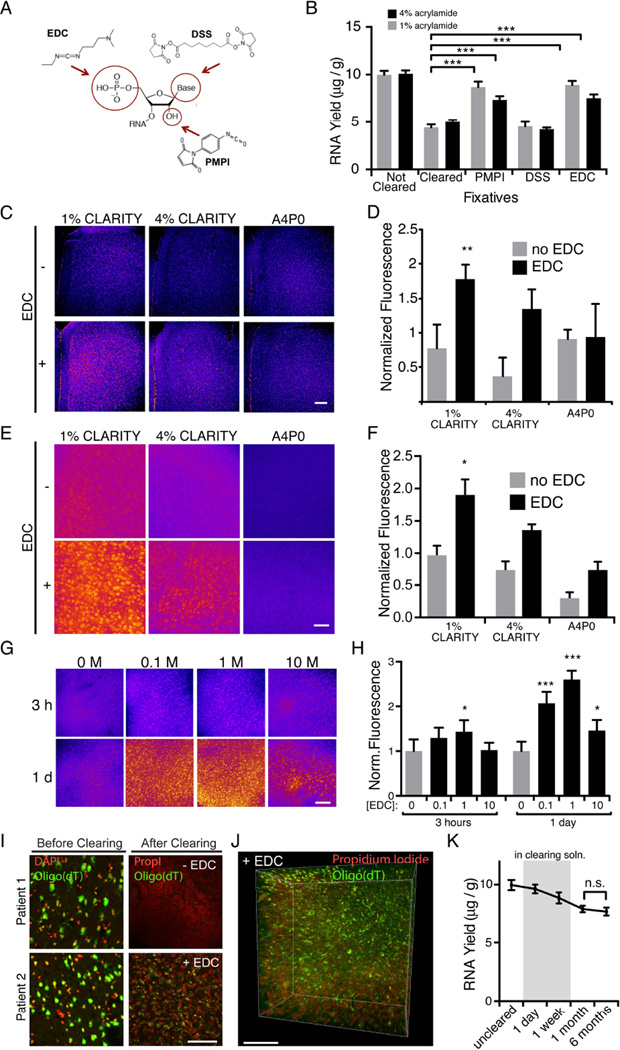
We explored three tissue-chemistry strategies: EDC (1-Ethyl-3-3-dimethyl-aminopropyl carbodiimide) for linkage of the 5’-phosphate group to surrounding amine-containing proteins (Pena et al., 2009; Tymianski et al., 1997); PMPI (p-maleimidophenyl isocyanate) for linkage of the 2’ hydroxyl group to surrounding sulfydryl-containing proteins (Shen et al., 2004); and DSS (disuccinimidyl suberate) for linkage of amine-containing side chains in RNA to surrounding amine-containing proteins (Mattson et al., 1993) (Figure 1A). These crosslinks were introduced after hydrogel embedding (Chung et al., 2013). After fixation, samples were fully cleared and RNA was extracted from each preparation. We observed that although DSS provided no significant increase in RNA yield (potentially due to overfixation of RNA through multiple amine groups on each RNA molecule), there was markedly improved retention of RNA in EDC and PMPI-fixed samples compared with control for both 1% and 4% acrylamide hydrogel compositions (Figure 1B). However, since PMPI doubled tissue-clearing time, while EDC only marginally increased clearing time (1–2 extra days in 1mm tissue blocks), we proceeded with EDC as an RNA-fixation agent for CLARITY.
Figure 1. Fixation in EDC significantly improves RNA retention in CLARITY volumes.
(A) Chemical compounds targeting functional groups on RNA (red circles) were characterized and assessed for RNA fixation and retention.
(B) 1mm mouse brain blocks were embedded in CLARITY hydrogel containing either 1% or 4% acrylamide, then either immediately processed for RNA extraction (uncleared), or post-fixed overnight in PMPI, DSS, EDC, or no-fix, then cleared until visually transparent, and processed for RNA extraction. There was a significant increase in RNA yield in PMPI and EDC treated groups relative to cleared no-fix controls (*** P < 0.001, one-way ANOVA, with Sidak’s post-hoc multiple comparisons test n = 6 slices per group). (C) 1mm blocks embedded in hydrogel (containing 1% or 4% acrylamide, or 4% acrylamide with no PFA) were post-fixed with EDC (+) or no fix (−), then cleared and stained with acridine orange to visualize total RNA levels (false colored; RNA signal in pink). Scale bar, 200 µm. Relative intensities are quantified in (D).
(D) 1% hydrogel embedded slices post-fixed in EDC showed significantly more RNA than all other conditions tested. Fluorescence intensities are normalized to mean intensity for all conditions (P<0.01, one-way ANOVA, with Sidak’s post-hoc multiple comparisons test. n = 5 slices per group).
(E) 1mm blocks prepared as in (C), hybridized with an oligo(dT) probe to detect mRNA (false colored). Scale bar, 50 µm. Relative intensities are quantified in (F).
(F) 1% hydrogel embedded slices post-fixed in EDC showed more mRNA than all other conditions tested. Fluorescence intensities are normalized to mean intensity for all conditions for each experiment (P<0.01 One way ANOVA, Tukey’s post-hoc test for multiple comparisons. n = 4 slices per condition). (G) 1mm blocks of tissues were embedded in a 1% CLARITY hydrogel and post fixed with 0, 0.1, 1, or 10M EDC, either for 3 hours or 1 day at 37°C. Oligo(dT) was performed as in (E). Relative intensities are quantified in (H).
(H) Fixation with 0.1M or 1M EDC for 1 day produced optimal RNA hybridization in 1% CLARITY tissue. Fluorescence intensities from oligo(dT) are normalized to mean intensity of the no EDC condition. Asterisks indicate statistical significance compared to 0M EDC, 3 hour condition (***P<0.001, *P<0.05, One way ANOVA, Tukey’s post-hoc test for multiple comparisons). n=4 slices per condition. (I) In situ hybridization in human tissue from temporal lobe resection. Left, small samples of resection from each patient were PFA fixed and oligo(dT) hybridization was performed to confirm that mRNA was intact before clearing. The remaining tissue was immersion fixed in 1% CLARITY hydrogel (2 days), embedded, then cleared immediately (-EDC), or fixed in EDC overnight at 37°C prior to clearing (+EDC). Right, mRNA was detected by oligo(dT) and DNA was stained with propidium iodide (PropI). Scale bar, 100 µm.
(J) 3D rendering of EDC fixed human temporal lobe volume (same patient as in (G)). Scale bar, 100 µm.
(K) 1 mm tissue blocks (1% hydrogel, EDC postfix) were processed for RNA extraction at various time points: uncleared (immediately after post-fixation); 1 day and 1 week (while in clearing solution); 1 month and 6 months (after clearing and stored in PBST). There is no significant loss of RNA during storage even up to 6 months at 4°C (n = 6 slices per group, n.s. paired t-test). All data are means ± S.D. See also Figure S1.
To complement these quantitative total-RNA biochemical measures with direct visualization of retained RNA within tissue, we stained tissue samples of different hydrogel compositions with acridine orange, an intercalating RNA dye. We found significantly increased RNA staining in EDC fixed samples, with EDC-treated 1% CLARITY tissue showing the best RNA labeling (Figure 1C,D). While promising, these total RNA measures did not specifically address mRNA, the population most relevant to molecular phenotyping and activity-dependent gene expression (in contrast to the more abundant rRNA, which by virtue of tight association with proteins could contribute disproportionately to the improvement seen with EDC). To determine if EDC improved mRNA preservation, we performed in situ hybridization with a 50 base deoxy-thymine oligonucleotide (oligo(dT)) to target the polyA tail of mature mRNA. Again, we found that 1% CLARITY with EDC samples exhibited the highest RNA signal (Figure 1E, F). Surprisingly, the 4% acrylamide hydrogel composition with EDC exhibited significantly reduced RNA detection with both acridine orange staining and oligo(dT) in situ hybridization (as well as weaker staining in target-specific in situ hybridization; Figure S1A). This consistent picture may reveal that the dense hydrogel network in 4% CLARITY makes mRNA targets less accessible for probe hybridization. In support of this notion, we find high concentrations (10M) of EDC also reduced mRNA staining, whereas more modest fixation (0.1M-1.0M EDC) provided the most effective labeling of RNAs (Figure 1G,H).
A major motivation for RNA detection includes broad application to clinical tissue, but human samples are particularly prone to RNA degradation, since pre-fixation post-mortem intervals vary, immersion-fixation crosslinks tissue more slowly than transcardial perfusion, and clinical samples are often banked for extended periods of time. We have also found that human tissue clears more slowly and, in some cases, demands higher clearing temperatures. To test if EDC could improve RNA retention in human tissue, we compared two human samples collected during temporal lobe resection, one treated with EDC and one an untreated control (1% CLARITY hydrogel). Although both samples showed comparably strong mRNA signal prior to clearing, we found that only the EDC-treated sample exhibited detectable mRNA after clearing (Figure 1I,J). We reasoned that EDC might not only be critical for the immediate processing of CLARITY samples, but might enable long-term storage with little RNA loss. To test this idea, we extracted and measured total RNA from rodent tissue during each stage of the clearing process. After a small loss of RNA during clearing, there was no significant loss during subsequent storage at 4 °C for up to 6 months (Figure 1K), demo nstrating a surprising level of stability (also reflected in target-specific in situ hybridization; Figure S1B). Together, these data identify and validate a nucleic acid-tuned CLARITY chemistry with EDC.
Quantifying diffusion of in situ hybridization components into clarified tissue
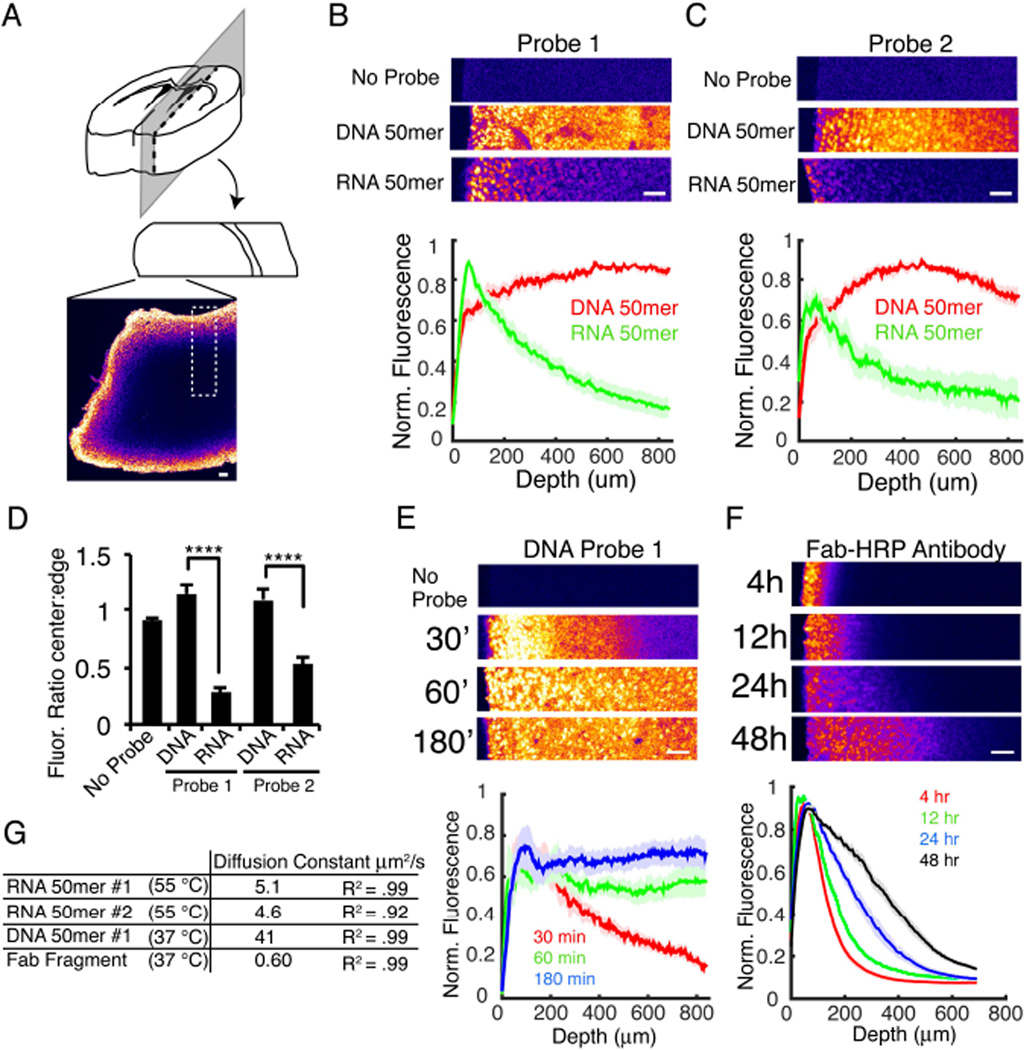
After ensuring stable retention of RNAs, we next focused on access to target RNAs for specific labeling in transparent tissue volumes. Traditional in situ hybridization (ISH) uses labeled DNA or RNA probes, which are detected by enzyme-conjugated antibodies that catalyze the deposition of chromophores or fluorophores at the target location. Interrogation of RNA by these methods requires the penetration of each component to the target location. Since prior work had only shown detection of RNA in small volumes (100–500µm thick; Chung et al., 2013; Yang et al., 2014), we sought to test the ability of ISH components to diffuse into intact EDC-CLARITY tissue.
We began by characterizing the diffusion of nucleic acid probes into EDC-CLARITY tissue. We incubated tissue blocks with 50-base DIG-labeled DNA or RNA probes, and visualized the diffusion profile of these probes by cutting cross-sections through the center of the tissue blocks and quantifying probe density on the newly exposed surface via antibody-based enzymatic amplification (tyramide signal amplification; TSA) (Figure 2A). We found that DNA probes diffused significantly faster into EDC-CLARITY tissue than corresponding RNA probes (Figure 2B–D); this important effect may be due to greater nonspecific tissue binding of RNA at this temperature, hindering penetration. Strikingly (and with substantial implications for nucleic acid labeling as the potential approach of choice for transparent tissue molecular phenotyping), we consistently observed DNA probes reaching the center of 2 mm tissue blocks within 3 hours. It should be noted that this detection method (TSA) may saturate at higher concentrations and obscure more subtle underlying concentration gradients expected to be present at 1–3 hour time points, but these diffusion rates are still considerably faster than observed for antibodies (Chung et al., 2013; Tomer et al., 2014).
Figure 2. DNA diffuses into CLARITY tissue more quickly than antibodies.
(A) Tissue configuration for B,C,E, and F. 2 mm EDC-CLARITY blocks are incubated in nucleic acid probes or antibody for time indicated and fixed in 4% PFA. 200 µm cross-sections are cut, probe diffusion is detected by TSA on the newly exposed tissue surface, and ROIs are selected as indicated by the dotted box and quantified in B,C,E, and F.
(B, C) 3h incubation with DIG-labeled riboprobes or DNA oligonucleotides (50 bases) targeting two different mRNAs in 50% formamide, 5x SSC at 55°C. Top, example ROIs of tissue as shown in (A), pseudocolored. Cross-section is incubated in anti-DIG Fab fragment antibody conjugated to HRP and detected with TSA using FITC. Bottom, quantification of signal intensity as a function of depth for 10-15 ROIs from 3 experiments. For each ROI, no probe control is subtracted, and signal is normalized to peak intensity.
(D) Quantification of ratio of signal intensity at tissue edge to center, calculated as maximum intensity over first 100 µm to average intensity of last 100 µm. (****P<0.0001, One way ANOVA, Tukey’s post hoc test for multiple comparisons).
(E) Diffusion of 50 base DNA oligonucleotide at shorter incubation times with hybridization conditions optimized for in situ hybridization with DNA probes (30, 60 and 180 minutes; 2x SSC, 40% formamide, 37°C). n= 6-12 ROIs.
(F) Antibody diffusion. CLARITY tissue is incubated in 50 base oligonucleotide probes overnight, washed, and transferred to anti-DIG antibody conjugated to HRP for time indicated. Tissue is sectioned as in (A), and antibody diffusion is detected by TSA. For 4 hours, n=25 ROIs; 12 hours, n=8; 24 hours, n=17; 48 hours, n=24.
(G) Diffusion constants and R2 values for nucleic acid and antibody diffusion in CLARITY tissue. Constants calculated by fitting average curves to Fick’s Law: y=n0*erfc(x/(2*√(D*t))) for one dimensional diffusion in a uniform medium with constant boundary condition. Diffusion rate is slower than reported previously in CLARITY tissue (Li et al., 2015), which may arise from additional crosslinking during EDC fixation, or changes in tissue properties during in situ hybridization. Curves used for fitting: RNA, 3 hours incubation; DNA, 30 min incubation; Antibody, 4 hours incubation. All error bars indicate SEM. All scale bars = 100 µm.
At 37°C (optimized for DNA-RNA hybridization), DNA probes reached the center of a 2 mm-thick block in <1 hour (Figure 2E). In contrast, enzyme-linked Fab antibody fragments penetrated only ~500µm into tissue even after 2 days (Figure 2F). Importantly, the rate of diffusion for the Fab fragment was almost two orders of magnitude slower than that of the DNA oligonucleotide (Figure 2G) under the EDC-CLARITY-ISH condition. Taken together, these experiments reveal that short DNA probes rapidly diffuse throughout large volumes of EDC-CLARITY tissue and suggest that an optimal approach to labeling native RNA species in large intact volumes could leverage the speed and specificity of short DNA probes in addition to EDC tissue chemistry.
In situ hybridization in EDC-CLARITY
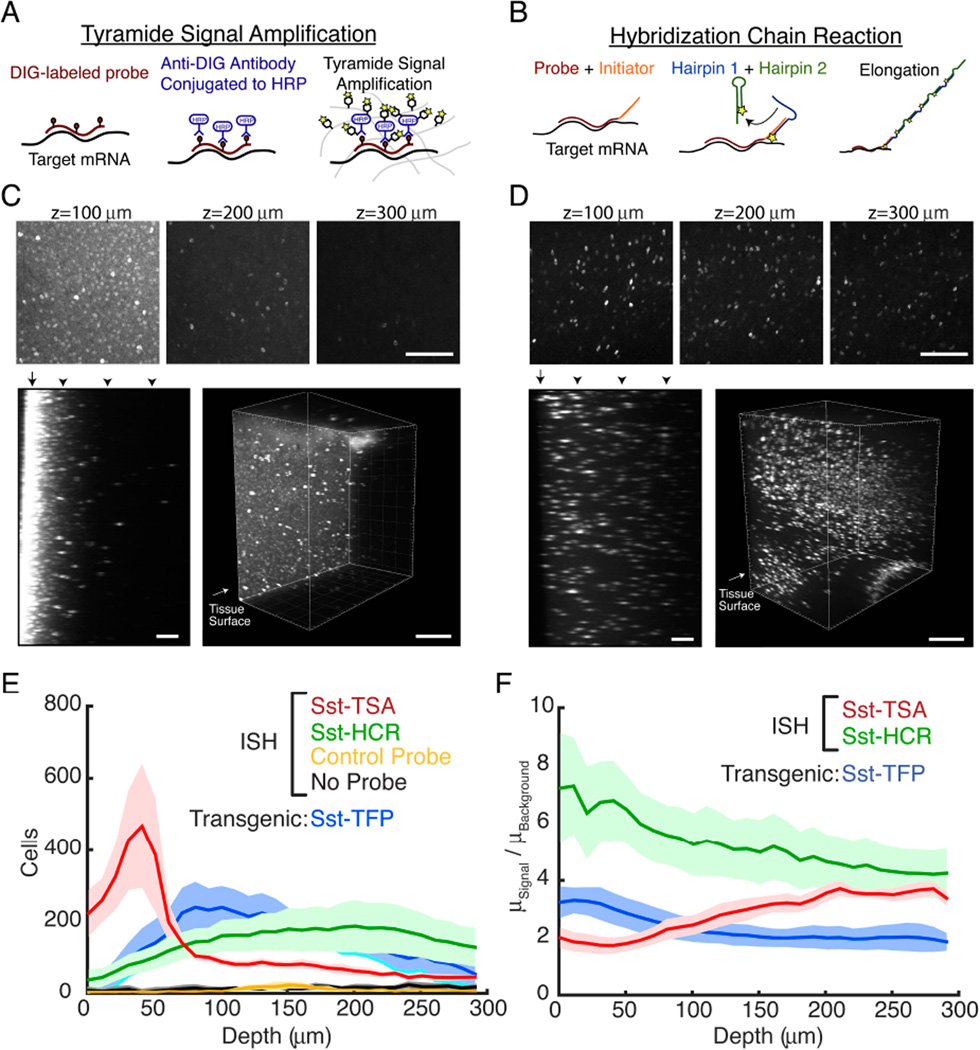
Based on these findings that demonstrate stable retention of RNA with EDC-CLARITY and rapid penetration with short DNA probes, we next sought to develop a panel of oligonucleotide-based ISH techniques for application to large transparent tissue volumes. We began with digoxigenin (DIG)-labeled DNA oligonucleotide probes targeting somatostatin mRNA (3 probes) and amplified with anti-DIG HRP-conjugated antibody and TSA (Figure 3A). In initial tests, we were readily able to resolve individual cells expressing somatostatin mRNA, demonstrating that specific mRNA species within the EDC-CLARITY hydrogel can be retained and are accessible to ISH probes (Figure 3C).
Figure 3. Comparison of antibody-based and DNA-based amplification.
(A) Workflow for TSA reaction. DIG-labeled probes are hybridized to target mRNA. HRP conjugated anti-DIG antibodies bind to hybridized probes and are detected by TSA.
(B) Workflow for HCR reaction. Initiator-labeled probes are hybridized to target mRNA. In a second step, initiator sequences hybridize to toehold of fluorophore bearing hairpins, starting a chain reaction of hairpin assembly.
(C-D) In situ hybridization for somatostatin mRNA in CLARITY tissue. Above, z = 100 µm, 200 µm, and 300 µm into CLARITY tissue volume of mouse cortex using traditional ISH (C) or hybridization chain reaction (D). Scale bars, 100 µm. Below, yz-subsections of CLARITY volume and 3D rendering of 1mm sections. Arrowheads indicate the z-location of the sections above. Arrows indicate the tissue surface. Due to high surface background, the top 130 µm of tissue are not shown for the 3D rendering in (C). Scale bars, 100 µm (left), 300 µm (right).
(C) DIG-labeled oligonucleotide probes detected with anti-DIG antibody (2 days) and TSA. Somatostatin expressing cells can be detected, but surface has high background and signal diminishes deeper in the tissue.
(D) Initiator-labeled oligonucleotide is detected with HCR reaction (1 day), resulting in more uniform staining.
(E) Number of cells as a function of tissue depth after local thresholding and cell segmentation on each imaging plane, 10 µm z-interval. High surface background in TSA reaction yields a large number of putative false positives 0-75 µm into the tissue section. Detection with HCR amplification shows a more uniform labeling of cells, comparable to the distribution of somatostatin cells in a genetically encoded reporter mouse (Sst-TFP).
(F) Ratio of signal to background as a function of depth in tissue, calculated from ratio of mean signal intensities segmented in (F) to the mean background intensity. For (E-F), No Probe, n=3; Scrambled Control, n=3; TSA, n=5; HCR, n=9; Sst-TFP, n=4. P<0.05, Kruskal-Wallis test on mean ratio over entire depth. All error bars indicate SEM. See also Figure S2
However, using this technique in larger volumes revealed two major limitations: (1) the surface of the tissue sections showed non-specific staining that could result in false positives during cell detection, and (2) the signal was visible only to a depth of <300 µm (Figure 3C). A similar pattern was seen in parallel experiments with a probe set targeting YFP mRNA in a Thy1-YFP transgenic mouse, confirming that under these conditions TSA signal at the tissue surface lacks specificity (Figure S2C). We hypothesized that the main sources of surface staining and signal heterogeneity resulted from a concentration gradient of antibody penetrating the EDC-CLARITY hydrogel and, consequently greater surface deposition of fluorophore during enzymatic amplification. We and others have found that probes can be labeled directly with fluorophore when RNA copy-number is high and little amplification needed (Yang et al., 2014), though with limitations on sensitivity and volume size (up to 1 mm blocks, still far greater than the 20–40µm queried with traditional techniques). Nevertheless, this restriction in volume, the need for exclusion of superficial tissue, and the severe limitation to highly-expressed transcripts together pointed to the need for further innovation to exploit the speed of DNA penetration into EDC-CLARITY tissue (Fig. 2B,E).
DNA-based ISH signal amplification
We hypothesized that an all-DNA based amplification system rather than the traditional antibody approach might be an ideal solution. Recent work has capitalized upon the programmable base-pairing of DNA molecules to design DNA structures that amplify signal by several orders of magnitude (Battich et al., 2013; Choi et al., 2010). We explored integrating this approach with EDC-CLARITY tissue chemistry, selecting the hairpin chain reaction amplification system (HCR; Choi et al., 2010) for further development since HCR (a) involves only small DNA oligonucleotides (<150 bases) which self-assemble at the target mRNA, and (b) requires only two hybridization steps (Fig 3B).
In the first hybridization step, an oligonucleotide probe containing a 36-base initiator sequence binds to target mRNA. In a second step, two fluorophore-tagged oligonucleotides are added, which are kinetically trapped in a hairpin conformation in the absence of the initiator sequence. As they diffuse into the tissue and encounter initiator sequences on hybridized probes, base pairing between the initiator sequences and the single-stranded toehold on Hairpin 1 open the hairpin, revealing a new initiator sequence capable of opening Hairpin 2. In turn, Hairpin 2 opens to reveal the original initiator sequence, starting the cycle anew. As the chain self-assembles, fluorophores accumulate at the target location. It is estimated that the hairpin chain reaction can amplify the signal approximately 200 fold (Choi et al., 2014), and we expected that this degree of amplification might be sufficient to detect RNA in EDC-CLARITY.
To test this approach, we appended initiator sequences to the 3’ and 5’ ends of the three somatostatin oligonucleotide probes used above, hybridized the probes to EDC-CLARITY tissue, and amplified with HCR hairpins. We found that the combination of EDC-CLARITY and HCR amplification exhibited excellent signal, low background, produced no non-specific surface staining and significantly improved the depth at which we could identify individual cells (Figure 3D). The signal-to-background ratio was significantly higher than in TSA-based amplification (Figure 3F) with the characteristic sparse pattern of somatostatin mRNA expression clearly distinguishable from background (Figure 3D). Moreover, the distribution and cell density detected with HCR amplification mirrors somatostatin expression in transgenic reporter mice, underscoring the specificity of this method (Sst-TFP, Figure 3E).
In situ hybridization in intact tissue
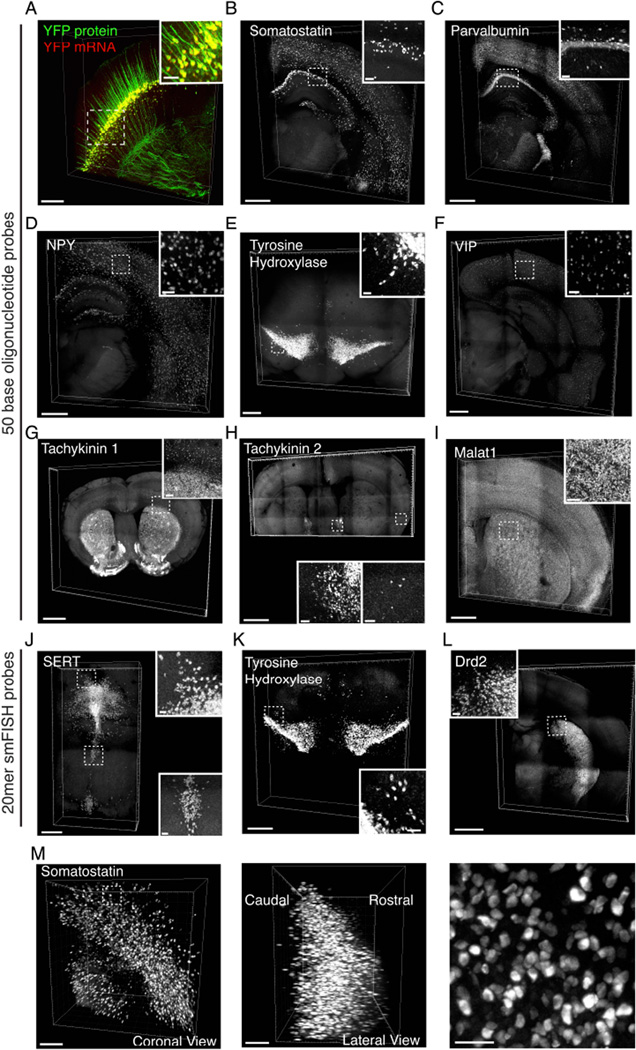
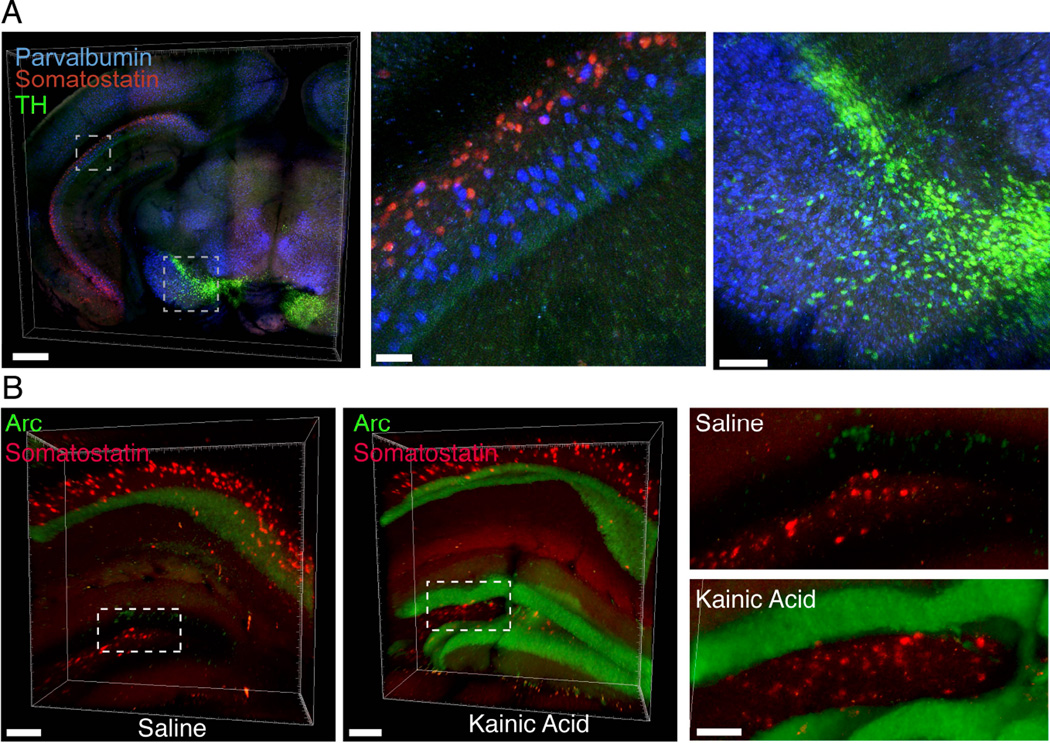
Linking information on cellular morphology, connectivity, and activity to information on RNA expression will be of substantial value; accordingly, we sought conditions for in situ hybridization in EDC-CLARITY that maintained fluorescence of transgenically expressed proteins. As a proof of concept, we performed in situ hybridization for YFP mRNA on Thy1-YFP transgenic mouse tissue and formulated a hybridization buffer that allowed reduction of hybridization temperature from 45°C to 37°C, which improved fluorescence in d endrites and axons while maintaining ISH specificity (Figures 4A, S2; Supplemental Video 1). To provide a generalizable framework for HCR-based RNA detection in EDC-CLARITY, we used these hybridization conditions to design, test, and refine sets of 50mer DNA probes for several representative and broadly-useful target RNAs for molecular phenotyping in nervous system tissue: somatostatin, parvalbumin, neuropeptide Y (NPY), vasoactive intestinal peptide (VIP), tachykinin1, tachykinin2, tyrosine hydroxylase, and Malat1. These targets showed reliable signal in EDC-CLARITY tissue and corresponded to known anatomical distributions in both neural and non-neural tissue (Figure 4B–I, Figure S3).
Figure 4. Cell-type phenotyping in CLARITY tissue using DNA probes and HCR amplification.
(A) 3D rendering of 1-mm-thick coronal section from Thy1-YFP mouse, in situ hybridization for YFP mRNA in red, endogenous YFP fluorescence in green. Scale bar, 200 µm. Inset, 3D rendering of boxed section in cortex. Scale bar, 50 µm.
(B-L) 3D rendering of in situ hybridization performed in 0.5mm coronal CLARITY sections using 50mer DNA oligonucleotide probes. Scale bars, 500 µm; insets, 50 µm, unless otherwise noted.
(B) Somatostatin mRNA (4 probes).
(C) Parvalbumin mRNA (4 probes). Inset scale bar, 70 µm.
(D) Neuropeptide Y mRNA (5 Probes).
(E) Tyrosine hydroxylase mRNA (10 probes).
(F) Vasoactive Intestinal Peptide (VIP) mRNA (10 probes).
(G) Tachykinin1 mRNA (5 probes). Scale bar, 1 mm; inset 100 µm.
(H) Tachykinin2 mRNA (4 probes). Scale bar, 1000 µm; inset of BNST and cortex, 50 µm.
(I) Malat1 mRNA (4 probes).
(J-L) 3D rendering of in situ hybridization performed in 0.5mm CLARITY sections using 20mer DNA oligonucleotides.
(J) SERT mRNA (47 probes).
(K) Tyrosine hydroxylase mRNA (39 probes).
(L) Drd2 mRNA (39 probes). Scale bar, 1500 µm, inset; 50 µm.
(M) Left, 3D rendering of 2mm block of mouse cortex, processed with EDC-CLARITY with in situ hybridization for somatostatin using HCR amplification. (Middle) orthogonal view of volume at left, showing signal throughout tissue depth. Scale bars, 200 µm. (Right) magnified view of somatostatin expressing cells in cortex from volume at left. Scale bar, 50 µm. See also Figure S3,4.
In refining these probe sets, we typically performed initial testing on pools of 5–10 probes; in cases where we observed non-specific staining, we then tested probes individually to identify and discard probes contributing significantly to background, which improved image quality (Figure S4A). Under these conditions, we estimate that HCR in CLARITY tissue results in ~50 fold amplification per double-initiator-labeled probe (Figure S4D–F). In agreement with previous results, increasing the number of initiators, whether by adding initiators to both 5’ and 3’ ends or by adding more probes, enhances signal substantially. This effect may eventually saturate if limited by slightly sublinear amplification (Figure S4C), but low copy number transcripts may still benefit from a larger set of probes. In comparing these results to published data from single-cell transcriptomics (Zeisel et al., 2015), we find that our data capture relative differences among gene expression levels across 2 orders of magnitude (Figure S4G–H); indeed, with 4 probes per target, this approach allows detection of mRNAs present at as low as ~50 copies/cell (Figure S4H). Although not as sensitive as RNAseq, probe sets can be expanded as shown below, and the large volumes processed in a single CLARITY experiment enable inclusion of spatial information and sampling from many more cells than would be achieved with RNAseq (particularly important if genes are expressed in sparse subsets within a tissue).
Since low copy-number transcripts may benefit from additional probes, and since it was important to determine if our methodology could be readily adapted to diverse probe design strategies, we tested the feasibility of using a larger set of shorter probes by attaching initiators to the 5’ end of probe sets originally designed for single-molecule fluorescent in situ hybridization (smFISH), which typically uses 20mer oligonucleotides (30–50 probes) that tile the mRNA target sequence. As with directly fluorophore-labeled 20mer probes, we expected that with many HCR-labeled 20mers, the on-target signal would accumulate in cells in which many probes bind and amplify (whereas off-target binding would be uniform across the sample); we did not, however, expect that HCR with these probes would provide single-molecule capability. Using this strategy, we were able to detect tyrosine hydroxylase, SERT, and Drd2 mRNA in EDC-CLARITY tissue, demonstrating that the HCR approach is adaptable to other probe types in CLARITY and compatible with larger pools of short probes (Figure 4J–L, Supplemental Video 2–3).
Because longer nucleotides are more expensive to synthesize and purify, the strategy of using short probes would reduce overall cost and may enable significantly greater signal amplification. Likely owing to the quick and uniform diffusion of DNA probes and hairpins, we find that tissue blocks up to at least 3 mm thick could be reliably used for intact in situ hybridization (Figure 4M, Supplemental Video 4,5). Another unique advantage of nucleic acid detection (relative to antibody-based detection) is that once the target sequence is known, it is possible to design probes for the target which are highly specific, permanently renewable, and cost-effective. We therefore anticipate that this methodology for RNA detection in EDC-CLARITY may be versatile for probing a variety of transcriptional products across many tissue-types and species.
Detection of activity-dependent genes and non-coding RNAs in intact volumes
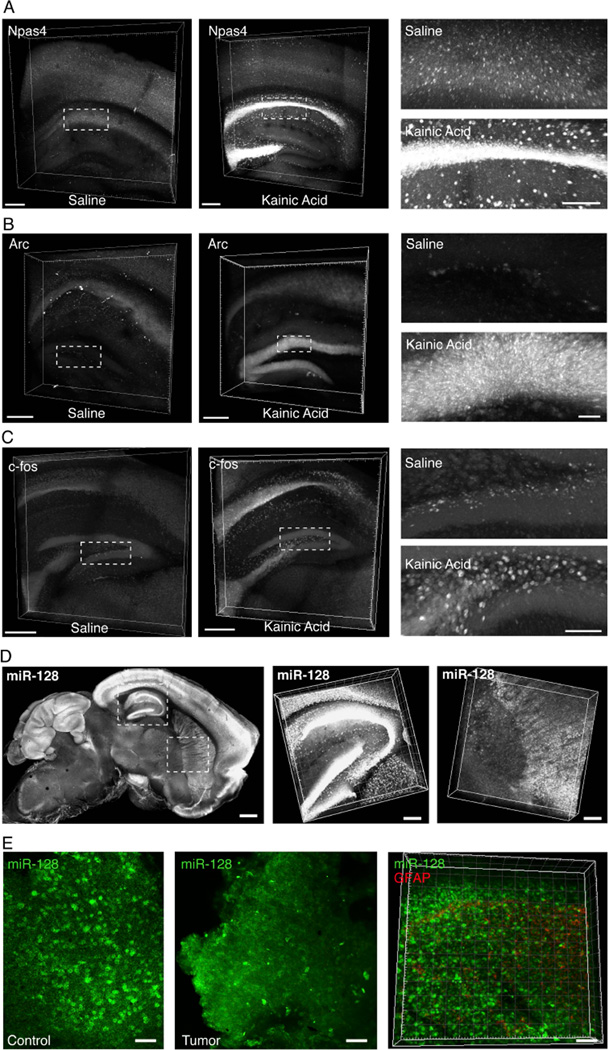
Many mRNAs are transiently up-regulated by activity, a fact that has been instrumental in identifying cells and circuits recruited during particular behaviors (e.g. Loebrich and Nedivi, 2009). Using such immediate early genes (IEGs), it has been possible to identify neurons involved in complex behaviors (even multiple behaviors separated in time; Guzowski et al., 1999; Reijmers et al., 2007), to visualize behaviorally relevant neurons in transgenic mice, in some cases long after the behavior itself (Barth et al., 2004; Guenthner et al., 2013; Smeyne et al., 1992), and to manipulate these IEG-expressing neurons to modify or recapitulate the observed behavior (Garner et al., 2012; Liu et al., 2012; Ramirez et al., 2013). Yet a major unmet goal is linking form and function: to align these transcriptional activity changes with molecular phenotype and connectivity information in large intact volumes. We therefore next designed HCR probe sets against several canonical activity-regulated transcripts: Arc (Lyford et al., 1995), c-fos (Sheng et al., 1990), and Npas4 (Bloodgood et al., 2014), and tested these probes in a kainic acid seizure model (known to induce robust hippocampal transcription of many activity-regulated genes; Nedivi et al., 1993)). We found that we were able to reliably track changes in expression of all of these activity-regulated genes in EDC-CLARITY. For example, Npas4 is normally expressed in scattered cells in cortex but robustly transcribed in both hippocampal pyramidal cells and interneurons after seizure activity (Figure 5A). In parallel experiments, increases in c-Fos transcription in hilar neurons and Arc transcription in dentate granule cells were readily detectable (Figure 5B–C), as described previously in hippocampal seizure models (Lyford et al., 1995).
Figure 5. Detecting activity-induced transcripts and non-coding RNAs in CLARITY volumes.
(A-C) 3D rendering of 0.5mm CLARITY section, HCR in situ hybridization in control saline injected (left) and kainic acid injected (right) animals. Kainic acid, 12 mg/kg, i.p., 2 hours prior to perfusion.
(A) Npas4 mRNA (4 probes). Scale bar, 200µm. Right, magnified view of indicated boxes. Scale bar, 100µm.
(B) Arc mRNA (5 probes). Right, magnified view of indicated boxes. Scale bar, 50µm.
(C) c-fos mRNA (45 probes). Scale bars 500µm; Right, magnified view of dentate gyrus as indicated by dotted box. Scale bar, 100 µm.
(D) Left, projection image of 1 mm mouse brain sagittal section, cleared, and hybridized with DIG-labeled LNA probes for mature miR-128 sequence. Scale bar, 500 µm. Middle, right 10x zoom of hippocampal and striatal volumes respectively. Scale bar, 150 µm.
(E) Left, projection images of human brain control (left) and tumor (GBM) (middle) samples, cleared and in situ hybridized for miR-128 (green). Scale: 50 µm (Right). Volume reconstruction of human GBM tumor biopsy sample (200 µm thick; scale: 50 µm) also stained with antibody to GFAP (red). miR-128 and GFAP have orthogonal signal gradients within the tumor preparation. See also Figure S5.
Lastly, we assessed detection of small-noncoding RNAs-- a major motivation for this entire approach since these are undetectable by antibodies yet also are 1) critical for the modulation of post-transcriptional gene expression; 2) play key roles in human genetic diseases (Esteller, 2011); and 3) represent a wealth of biological information not yet approached by any tissue clearing technique. Indeed, due to small size, microRNAs have fewer amines to react with paraformaldehyde or acrylamide and are easily lost from fixed tissues (Pena et al., 2009; Renwick et al., 2013).
Consistent with this expected challenge, we found that post-treatment with EDC was critical for the retention of miRNAs in EDC-CLARITY (Figure S5A). Using DIG-labeled locked nucleic acid probes, we targeted several miRNAs with known function in the mammalian brain and in neuropsychiatric disease (miR-10, miR-124, miR-128), as well as a miRNA known to exist only outside the mammalian brain (miR-21) to serve as a negative control (Landgraf et al., 2007). We detected robust expression of these miRNAs in volumes of mouse brain, in a pattern largely limited to areas with anticipated expression (as inferred from miRNA deep sequencing experiments). miR-10 signal (for instance) was almost exclusively recovered in the thalamus, miR-124 was observed to be more ubiquitously present throughout the brain, and miR-128 showed characteristic forebrain and cerebellar enrichment (Figure 5D, S5B–C). We observed minimal signal for miR-21 under the same detection and amplification conditions, as expected, highlighting the specificity of the miRNA signals observed.
miR-128 is particularly well-studied in the context of its known disease-relevance for oncogenic suppression (Pang et al., 2009) and predisposition to mood disorders (Zhou et al., 2009) but until now, miR-128 expression has not been visualized volumetrically in the mammalian brain at single cell resolution, which we were readily able to achieve here (Figure 5D). To further test potential utility of this approach in the study of brain disease, we endeavored to detect miR-128 in human clinical samples to determine if differences in expression might be associated with human glioblastomas (suggested but not directly observed; Ciafrè et al., 2005). We indeed were able to detect miR-128 in human GBM samples processed in EDC-CLARITY hydrogel; moreover, by integrating antibody staining (in this case, GFAP to mark tumor location) with ISH in EDC-CLARITY, we could track the crucial relative relationships of GFAP and miR-128 expression across the tissue volume at cellular resolution (Figure 5E). Such an approach designed to provide 3D volumetric access to miRNAs in biopsied or post-mortem human brain samples may be valuable in the search for tissue-level disease insights, biomarkers, and therapeutic targets for neurological and psychiatric disease.
Multiplexed molecular phenotyping
Finally, we sought to develop methods for multiplexed detection of RNA in EDC-CLARITY to address the critical and rapidly-growing need for multiple overlaid markers of cell identity or activity in the native anatomical context. Using multiplexed hybridization and amplification with orthogonal hairpin sets (Choi et al, 2014), we were able to simultaneously label multiple mRNAs in EDC-CLARITY. Of note, although orthogonal hairpins were equivalent in amplification, individual fluorophores varied in fluorescence signal, as may be expected by differences in tissue autofluorescence, fluorophore efficiency, and light transmittance at different wavelengths (Figure S6). Nevertheless, somatostatin, parvalbumin, and tyrosine hydroxylase could be simultaneously hybridized and amplified with sets of orthogonal hairpins carrying Alexa514, Alexa647 and Alexa546 fluorophores, respectively (Figure 6A; Supplemental Video 5). We were also able to combine in situ hybridization for cell-type markers with in situ hybridization for activity markers (Figure 6B). Taken together, these data demonstrate key steps toward integrated investigation of cellular structure and typology, microRNA expression, and activity-regulated gene transcription within intact tissue volumes.
Figure 6. Multiplexed detection of mRNAs in CLARITY.
(A) Left, multiplexed in situ hybridization of 0.5mm coronal CLARITY section treated with kainic acid, using somatostatin (red), parvalbumin (blue) and tyrosine hydroxylase (green) probe sets. Scale bar, 500 µm. Middle, inset of caudal hippocampus showing parvalbumin and somatostatin interneurons in CA1 region. Scale bar, 50 µm. Right, parvalbumin and tyrosine hydroxylase positive cells in midbrain. Scale bar, 100 µm.
(B) 3D rendering of 1mm CLARITY block, HCR in situ hybridization for somatostatin (red) and Arc mRNA (green) in control saline injected (left) and kainic acid injected (right) animals. Right, magnified view of indicated boxes. Scale bar, 100 µm. See also Figure S6
DISCUSSION
While whole-mount in situ hybridization has long been a standard technique to query RNA populations in small transparent or embryonic tissue, the scattering properties of adult tissues have prevented extension to most adult samples. Moreover, while tissue clearing methods to meet the challenge of scattering have been described for over one hundred years (Spalteholz, 1914), along with a resurgence in interest and innovation over the past decade, visualization of RNA in large blocks of intact tissue has remained largely unaddressed (Richardson and Lichtman, 2015) despite significant advantages over protein labeling for reasons discussed above (briefly: scaling, cost, renewability, availability for all transcripts, stability across preparations, and inclusivity of nontranslated mRNAs). Here we report development of general methodology, tools, and resources for cellular-resolution transcriptional profiling of large and intact transparent mammalian tissue volumes, with reliable detection of diverse markers for non-coding transcripts, cell identity, and activity history.
Critically, while hydrophobic solvent- and immersion-based techniques have emerged as methods for clearing and immunostaining tissue, none have demonstrated successful RNA detection. Although these techniques might be optimized in this direction, key components could be incompatible with RNA detection. For example, in ClearT2 (an immersion-based method) the use of formamide during clearing (which is also used during hybridization to destabilize RNA duplexes) may pose challenges for probe hybridization (Kuwajima et al., 2013). Likewise, ScaleA2, ScaleU2, ScaleS, and CUBIC use urea (Hama et al., 2011; 2015; Susaki et al., 2014; Tainaka et al., 2014), also a destabilizing agent for hybridization (Simard et al., 2001). Finally, hydrophobic solvent-based clearing methods (such as the 3DISCO and iDISCO techniques (Ertürk et al., 2012; Renier et al., 2014)) crosslink tissue with methanol prior to solvent clearing, which may limit access to RNA for hybridization in large volumes, and indeed RNA labeling has not yet been shown in any of these volumetric methods. Since the specificity of nucleic acid probe hybridization requires maintenance of specific concentrations of salt and organic solvent (such as formamide) in a window that favors probe hybridization to target but not off-target RNA, aqueous-based CLARITY/hydrogel approaches appear to provide unique RNA detection advantages for compatibility with commonly-used hybridization reagents.
Almost all in situ hybridization techniques for fixed tissue require permeabilization prior to hybridization to enable access of probes to target RNA (Wilkinson, 1999). Intrinsic to the CLARITY process is permeabilization of tissue via removal of lipid membranes and denaturation of proteins, enabling RNA access without necessity of proteases (though proteases may be used as well in some forms of CLARITY). Without stabilization via cross-linking, RNA is susceptible to rapid degradation by endogenous RNAses; in EDC-CLARITY RNA is stabilized by both PFA cross-linking (which fixes RNA and renders RNAses immobile) and EDC. Together, these properties may make EDC-CLARITY particularly suitable for RNA detection; nevertheless, the methodologies we describe might also be suitable for hybrid approaches to volumetric RNA detection in other clearing methods, given sufficient optimization of fixation and permeabilization parameters.
We have presented validation for select microRNAs, cell-type markers, and immediate-early genes, but with the RNA landscape preserved via EDC fixation, we anticipate that this method could be extended to additional RNA species. Oligonucleotide or LNA probes could be designed for splice junctions to visualize brainwide expression of particular mRNA variants. Because CLARITY tissue maintains subcellular structures, this method also dovetails with catFISH approaches, where intronic or exonic targeting probes could be used to temporally differentiate two activity-triggered transcriptional events (Guzowski et al., 1999; Lin et al., 2011). The ability in EDC-CLARITY to use a variety of different probe types (50mer oligos, 20mer oligos, LNAs) and amplification methods (HCR for multiplexing and larger volumes, TSA for smaller volumes) suggests versatility, but as always, in situ hybridization must be further tailored to RNA target type, volume size, copy number, transcript length, and species.
The ability to monitor many transcripts simultaneously will be of growing relevance as the field moves toward increasingly rich and detailed molecular phenotyping. For HCR amplification there are currently five validated hairpin sets allowing up to five targets to be labeled simultaneously (Choi et al., 2014), but spectral separation will be a limiting factor. The sharp emission spectra of quantum dots could address this problem, though the effect of steric hindrance on hairpin assembly and diffusion of oligonucleotides will require testing. Opportunities associated with multi-feature typology extend beyond multiple molecular features, and including fusing molecular and anatomical datasets. Whereas typical in situ protocols degrade fluorescence, the approach shown here is compatible with maintained fluorescence of genetically encoded cell-filling proteins for visualization of three-dimensional neuronal morphology and wiring (and thus allows integration of information about molecular identity and functional history with information on local and global connectivity across intact nervous systems).
The goal of identifying low-copy number transcripts can be approached from several angles. During amplification, initiators could be concatenated, or combined with a branched DNA approach (Battich et al., 2013), to incorporate more initiator sequences onto each probe. Additional fluorophores, or photobleach-resistant quantum dots, could be conjugated to hairpins to improve signal (Resch-Genger et al., 2008). During hybridization, we have demonstrated that 20mer probes could be used to tile the entire sequence with initiators, which would be particularly effective in improving detection for rare, but long transcripts. Alternatively, one could directly reverse-transcribe mRNA and amplify the resulting cDNA in situ (successful for in situ sequencing strategies), though hydrogel properties may require optimization for best enzymatic activity and diffusion of key components (Ke et al., 2013; Lee et al., 2014).
Having characterized in EDC-CLARITY an array of probe sets for canonical markers of cell identity and activity, to facilitate the community’s further benefit from (and engagement with) this growing effort of probe testing and publication, we have established a database of validated probes, with sequences and hybridization conditions available as an open online resource (clarityresourcecenter.org, wiki.claritytechniques.org/index.php/ISH). Requiring little specialized equipment, low startup costs, and moderate operational costs, this approach could provide an alternative to immunohistochemistry when specific antibodies are lacking, or to traditional in situ hybridization and serial sectioning when streamlined data acquisition is needed, as with large cohorts or large tissue volumes. While each step will benefit from further innovation and optimization, flexible design of EDC-CLARITY methodologies may favor adoption across a broad application domain, from clinical samples to integrative basic analysis of structure-function properties spanning cell typology, activity in behavior, and local and global tissue relationships.
EXPERIMENTAL PROCEDURES
CLARITY Tissue Preparation
CLARITY tissue was prepared as described in Tomer et al. (2014). For A4P0 samples, tissues were prepared as described (Yang et al., 2014). Tissue was incubated with RNA fixatives after acrylamide polymerization (EDC, 0.1M; PMPI, 0.1M; or DSS, 0.1M, overnight at 37°C). Tissue was cleared passiv ely in a 4%SDS/ 0.2M Boric acid (pH=8.5) clearing solution at 37°C until transparent and sto red in 1x PBS with 0.3% TX-100 (PBST) at 4°C.
Total RNA Isolation and Acridine Orange Staining
Cleared tissue was homogenized in 20 µg/ml proteinase K, extracted with Trizol and then acidic phenol:chloroform:isoamyl alcohol before precipitation with ethanol. For acridine orange staining, sections were rinsed in sodium citrate (SC) buffer for 10 minutes, incubated in acridine orange solution (100 µg/mL) for 3h, then rinsed in SC buffer, then PBS, and transferred to refractive index matching in FocusClear.
Probe Design
Riboprobes were generated from cDNA templates, reverse transcribed with DIG-labeled dNTPs (Roche), and purified. smFISH probes were designed and synthesized by BioSearch (Petaluma, CA). DNA 50mer oligonucleotide probes were purchased from Molecular Instruments (Caltech) or designed using OligoWiz software (Wernersson et al., 2007) and synthesized by Integrated DNA Technologies. LNA probes were synthesized by Exiqon.
Probe and Antibody Diffusion
For RNA and DNA probe diffusion, cleared tissue (2mm) was incubated in hybridization solution for the time indicated, then cooled to 4°C, fixed with PFA, and re-sectioned (200µm). Cross-sections of the center of tissue were selected for staining with anti-DIG antibody conjugated to HRP and detected with TSA.
For antibody diffusion, tissue was incubated in 50mer DIG-labeled oligonucleotides overnight in 40% formamide and 2xSSC, cooled to 4°C, fixed in 4% PFA for one hour at RT. The tissue was then incubated with anti-DIG Fab fragment antibody coupled to HRP (1:1000) in PBST for the corresponding time and further processed as above for re-sectioning and TSA amplification.
In Situ Hybridization
For all in situ hybridizations, cleared tissue was equilibrated in hybridization solution for 1h, hybridized in the same solution overnight at 37°C unless otherwise noted, then stringency washes were performed at the hybridization temperature to remove excess or non-specifically bound probe. Solutions and temperatures varied for each probe type and are as follows. Oligo(dT): hybridization with 15% formamide, 2xSSC, 10% dextran sulfate, 50nM probe; stringency 3×1 hour in 15% formamide, 2 × SSC then 2×1 hour in 2xSSC. DIG-labeled 50mers: hybridization with 50% formamide, 5xSSC, 0.5mg/ml yeast tRNA; stringency 3×1hour in 50% formamide, 5xSSC plus 2×1 hour in 2xSSC and then transferred to PBST. Initiator-labeled 50mers: hybridization with 40% formamide, 2xSSC, 10% dextran sulfate, 0.5mg/ml yeast tRNA; stringency 3×1hour in 40% formamide, 2xSSC plus 2×1hour in 2xSSC. DNA 20mers (smFISH sets): hybridization with 10% formamide, 2xSSC, 10% dextran sulfate; stringency 3×1 hour in 10% formamide, 2xSSC plus 2×1 hour in 2xSSC. LNA probes: hybridization with 50% formamide, 5xSSC, 0.5mg/ml yeast tRNA, 12.5nM DIG labeled probe at 20°C below Tm; stringency 2×1 hour in 5xSSC plus 1 hour in 2xSSC at the same temperature.
For DIG labeled probes, tissue was washed in PBST after stringency. Tissue was incubated overnight in anti-DIG antibody conjugated to HRP (1:500) for 2 days per mm tissue thickness, washed overnight in PBST, developed with tyramide signal amplification (1:50 dilution, 30 minutes), washed 3x in PBST, and transferred to FocusClear for imaging. For initiator probes, tissue was equilibrated in amplification buffer (5xSSC, 0.1% Tween20, 10% dextran sulfate). DNA hairpins were separately heated to 90°C, cooled to RT, and added to amplification buffer. Tissue was incubated in hairpins overnight at RT, then washed 5×1 hour with 5xSSC plus 0.1% Tween20, and transferred to FocusClear for imaging.
Propidium iodine staining, where applicable, was performed using a PropI/RNase solution after stringency washes. Sections were transferred to FocusClear for 4 hours prior to imaging. Tissue shrinks once equilibrated to FocusClear for imaging; all scale bars represent the imaged volume, which is approximately 50% of original tissue volume
Human Tissue
Human tissue is putative healthy tissue obtained from temporal lobe resections from two patients (46 y.o. female, 18 y.o. male). Tissue was equilibrated in 1% hydrogel solution for 2 days at 4°C, polymerized for 5 hours at 37°C, and cleared for 5 weeks in 4% SDS at 37°C.
Confocal Microscopy
All images were taken on a Leica SP5 confocal microscope with a 10x/0.4 objective (WD: 2.2 mm) or 20x/0.75 objective (WD: 0.66 mm) at 488 nm (FITC), 514 nm, 543 nm, or 647 nm excitation.
Experimental subjects
Animal husbandry and all aspects of animal care and euthanasia as described were in accordance with guidelines from the National Institutes of Health and have been approved by members of the Stanford Institutional Animal Care and Use Committee. Use of surgical and post-mortem human tissue was in accordance with guidelines from the National Institutes of Health and approved by the Stanford Institutional Review Board.
Supplementary Material
Highlights.
Carbodiimide-based chemistry in CLARITY hydrogels for RNA preservation and detection.
Rapid diffusion of DNA oligonucleotides for volumetric in situ hybridization.
Detection of microRNAs and mRNAs in clarified mouse and human tissues.
DNA-based amplification for multiplexed in situ hybridization in CLARITY.
Acknowledgments
We thank Niles Pierce and Harry Choi for generously making HCR reagents freely available and for advice during protocol troubleshooting. We thank Michelle Monje for providing human glioblastoma tumor samples and the entire Deisseroth lab for helpful discussions. This work is supported by an Ellison LSRF fellowship (PR), NIH T32 Fellowship (MW), and NIMH, NIDA, and the U.S. Army Research Laboratory and Defense Advanced Research Projects Agency (Cooperative Agreement Number W911NF-14-2-0013); nothing in this material represents official views or policies of our funders. The authors have disclosed these findings to the Stanford Office of Technology Licensing, and all tools, protocols and resources are freely distributed and supported (clarityresourcecenter.org, wiki.claritytechniques.org/index.php/ISH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
E.L.S., P.R., M.W., and K.D. designed experiments; P.R. and A.J. developed and characterized fixation conditions, designed and implemented microRNA in situ hybridization experiments and analyzed data; E.L.S. and M.W. characterized diffusion conditions, designed and implemented mRNA in situ hybridization experiments and analyzed data; E.L.S., P.R., M.W., and K.D. wrote the paper; K.D. supervised all aspects of the work.
REFERENCES
- Barth AL, Gerkin RC, Dean KL. Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. J. Neurosci. 2004;24:6466–6475. doi: 10.1523/JNEUROSCI.4737-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battich N, Stoeger T, Pelkmans L. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat Meth. 2013:1–10. doi: 10.1038/nmeth.2657. [DOI] [PubMed] [Google Scholar]
- Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature. 2014;503:121–125. doi: 10.1038/nature12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Beck VA, Pierce NA. Next-Generation in Situ Hybridization Chain Reaction: Higher Gain, Lower Cost, Greater Durability. ACS Nano. 2014;8:4284–4294. doi: 10.1021/nn405717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Chang JY, Le A Trinh, Padilla JE, Fraser SE, Pierce NA. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim S-Y, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciafrè SA, Galardi S, Mangiola A, Ferracin M, Liu C-G, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt H-U, Leischner U, Schierloh A, Jährling N, Mauch CP, Deininger K, Deussing JM, Eder M, Zieglgänsberger W, Becker K. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Meth. 2007;4:331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG, Hellal F, Bradke F, Sheng M, Dodt H-U. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nature Protocols. 2012;7:1983–1995. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Garner AR, Rowland DC, Hwang SY, Baumgaertel K, Roth BL, Kentros C, Mayford M. Generation of a synthetic memory trace. Science. 2012;335:1513–1516. doi: 10.1126/science.1214985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron. 2013;78:773–784. doi: 10.1016/j.neuron.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa H, Ishidate F, Kaneko T, Akagi T, Saito T, Saido T, et al. ScaleS: an optical clearing palette for biological imaging. Nat Neurosci. 2015:1–14. doi: 10.1038/nn.4107. [DOI] [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wählby C, Nilsson M. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Meth. 2013;10:857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- Kuwajima T, Sitko AA, Bhansali P, Jurgens C, Guido W, Mason C. ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development. 2013;140:1364–1368. doi: 10.1242/dev.091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SSF, Li C, Amamoto R, et al. Highly Multiplexed Subcellular RNA Sequencing in Situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Czajkowsky DM, Li X, Shao Z. Fast immuno-labeling by electrophoretically driven infiltration for intact tissue imaging. Sci Rep. 2015;5:10640. doi: 10.1038/srep10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Boyle MP, Dollar P, Lee H, Lein ES, Perona P, Anderson DJ. Functional identification of an aggression locus in the mouse hypothalamus. Nature. 2011;470:221–226. doi: 10.1038/nature09736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, Tonegawa S. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebrich S, Nedivi E. The Function of Activity-Regulated Genes in the Nervous System. Physiological Reviews. 2009;89:1079–1103. doi: 10.1152/physrev.00013.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Research. 1999;27:4436–4443. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson G, Conklin E, Desai S, Nielander G, Savage MD, Morgensen S. A practical approach to crosslinking. Mol. Biol. Rep. 1993;17:167–183. doi: 10.1007/BF00986726. [DOI] [PubMed] [Google Scholar]
- Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature. 1993;363:718–722. doi: 10.1038/363718a0. [DOI] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang JC-S, Kwok WK, Chen Z, Ng H-K. Oncogenic role of microRNAs in brain tumors. Acta Neuropathol. 2009;117:599–611. doi: 10.1007/s00401-009-0525-0. [DOI] [PubMed] [Google Scholar]
- Pena JTG, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Meth. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez S, Liu X, Lin P-A, Suh J, Pignatelli M, Redondo RL, Ryan TJ, Tonegawa S. Creating a false memory in the hippocampus. Science. 2013;341:387–391. doi: 10.1126/science.1239073. [DOI] [PubMed] [Google Scholar]
- Reijmers LG, Perkins BL, Matsuo N, Mayford M. Localization of a stable neural correlate of associative memory. Science. 2007;317:1230–1233. doi: 10.1126/science.1143839. [DOI] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Renwick N, Cekan P, Masry PA, McGeary SE, Miller JB, Hafner M, Li Z, Mihailovic A, Morozov P, Brown M, et al. Multicolor microRNA FISH effectively differentiates tumor types. J. Clin. Invest. 2013;123:2694–2702. doi: 10.1172/JCI68760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum dots versus organic dyes as fluorescent labels. Nat Meth. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- Richardson DS, Lichtman JW. Clarifying Tissue Clearing. Cell. 2015;162:246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G, Anand MFG, Levicky R. X-ray photoelectron spectroscopy and infrared spectroscopy study of maleimide-activated supports for immobilization of oligodeoxyribonucleotides. Nucleic Acids Research. 2004;32:5973–5980. doi: 10.1093/nar/gkh932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, McFadden G, Greenberg ME. Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron. 1990;4:571–582. doi: 10.1016/0896-6273(90)90115-v. [DOI] [PubMed] [Google Scholar]
- Simard C, Lemieux R, Côté S. Urea substitutes toxic formamide as destabilizing agent in nucleic acid hybridizations with RNA probes. Electrophoresis. 2001;22:2679–2683. doi: 10.1002/1522-2683(200108)22:13<2679::AID-ELPS2679>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Smeyne RJ, Schilling K, Robertson L, Luk D, Oberdick J, Curran T, Morgan JI. fos-lacZ transgenic mice: mapping sites of gene induction in the central nervous system. Neuron. 1992;8:13–23. doi: 10.1016/0896-6273(92)90105-m. [DOI] [PubMed] [Google Scholar]
- Song W, Zhu K, Cao Z, Lau C, Lu J. Hybridization chain reaction-based aptameric system for the highly selective and sensitive detection of protein. Analyst. 2012;137:1396–1396. doi: 10.1039/c2an16232f. [DOI] [PubMed] [Google Scholar]
- Spalteholz W. Uber das Durchsichtigmachen von menschlichen und tierischen Präparaten und seine theoretischen Bedingungen (Leipzig: S. Hirzel) 1911. [Google Scholar]
- Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. The American Journal of Pathology. 2002;161:1961–1971. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt T, Lang MC, Medda R, Engelhardt J, Hell SW. 2,2’-thiodiethanol: a new water soluble mounting medium for high resolution optical microscopy. Microsc. Res. Tech. 2007;70:1–9. doi: 10.1002/jemt.20396. [DOI] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Tainaka K, Kubota SI, Suyama TQ, Susaki EA, Perrin D, Ukai-Tadenuma M, Ukai H, Ueda HR. Whole-body imaging with single-cell resolution by tissue decolorization. Cell. 2014;159:911–924. doi: 10.1016/j.cell.2014.10.034. [DOI] [PubMed] [Google Scholar]
- Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nature Protocols. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymianski M, Bernstein GM, Abdel-Hamid KM, Sattler R, Velumian A, Carlen PL, Razavi H, Jones OT. A novel use for a carbodiimide compound for the fixation of fluorescent and non-fluorescent calcium indicators in situ following physiological experiments. Cell Calcium. 1997;21:175–183. doi: 10.1016/s0143-4160(97)90042-7. [DOI] [PubMed] [Google Scholar]
- Wanner AA, Kirschmann MA, Genoud C. Challenges of microtome-based serial block-face scanning electron microscopy in neuroscience. J Microsc. 2015;259:137–142. doi: 10.1111/jmi.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernersson R, Juncker AS, Nielsen HB. Probe selection for DNA microarrays using OligoWiz. Nature Protocols. 2007;2:2677–2691. doi: 10.1038/nprot.2007.370. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG. In Situ Hybridization: A Practical Approach. Oxford: University Press; 1999. [Google Scholar]
- Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen C-K, Lubeck E, Shah S, Cai L, Gradinaru V. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zheng H, Rinaman L. Simplified CLARITY for visualizing immunofluorescence labeling in the developing rat brain. Brain Struct Funct. 2015:1–9. doi: 10.1007/s00429-015-1020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yuan P, Wang Y, Hunsberger JG, Elkahloun A, Wei Y, Damschroder-Williams P, Du J, Chen G, Manji HK. Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology. 2009;34:1395–1405. doi: 10.1038/npp.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.