Abstract
Chikungunya virus (CHIKV) is an arthropod-borne virus, which is known to cause severe disease only in humans. To investigate its potential zoonotic host range and evaluate reservoir competence among these hosts, experimental infections were performed on individuals from nine avian and 12 mammalian species representing both domestic and wild animals common to North America. Hamsters and inbred mice have previously been shown to develop viremia after inoculation with CHIKV and were used as positive controls for infection. Aside from big brown bats (Eptesicus fuscus), none of the mammals or birds developed detectable viremia or overt clinical disease. However, most mammals and a smaller proportion of birds developed neutralizing antibody responses to CHIKV. On the basis of these results, it seems unlikely that CHIKV poses a significant health threat to most domestic animals or wildlife and that the species examined do not likely contribute to natural transmission cycles. Additional studies should further evaluate bats and wild rodents as potential reservoir hosts for CHIKV transmission during human epidemics.
Chikungunya virus (CHIKV; Togaviridae: Alphavirus) is a mosquito-borne virus of medical importance that is distributed throughout much of Africa and southeast Asia, and more recently has expanded to regions in Europe and the Western Hemisphere.1 In the past decade, CHIKV has caused numerous large-scale epidemics, estimated to have involved millions of people.2–4 During this time frame, CHIKV underwent a mutation that subsequently permitted Aedes albopictus to serve as a competent vector, permanently altering the epidemiology of this virus.5,6 The virus is thought to be maintained in nature through a human–mosquito–human cycle involving Aedes aegypti and Ae. albopictus mosquitoes, although other species may be involved as reservoirs.7–9 The recent emergence of CHIKV, including introductions to the New World via viremic travelers and subsequent autochthonous infections, has led to a heightened interest in better understanding the ecology and epidemiology of this virus.10–12 For example, little is known about the sylvatic cycle of CHIKV and whether nonhuman vertebrates are involved in transmission, although forest-dwelling primates have been implicated as potential hosts in both Africa and Asia.9,13–15 Here, we report on a series of experimental trials designed to identify potentially competent host species representative of domestic and wild animals common to North America.
Animal species were chosen for inoculation with CHIKV based on their abundance, availability, and to serve as representatives of broader taxonomic groups. Two strains of CHIKV were used for inoculation: the SAH2123 strain was isolated in 1976 from a human in South Africa and the COM2005 strain was isolated from mosquitoes collected during an outbreak in the Comoros Islands in 2005. Individuals of nine avian and 12 mammalian species were prescreened as negative for neutralizing anti-CHIKV antibodies and inoculated with 104–105 plaque forming units (PFU) of CHIKV by subcutaneous injection. All animals were observed twice daily to assess clinical signs of disease, which in some cases, included recording rectal temperature. Blood samples were collected daily (except for big brown bats) for up to 7 days postinoculation (DPI) and also at the time of euthanasia on 14 DPI. Because of their small size, groups of bats were terminally bled and euthanized on 1–5, 7, and 14 DPI. Sera were tested for CHIKV by plaque assay on Vero cells; the threshold for detection varied depending on volume of serum available and is depicted in Table 1. Antibody titers were determined by plaque reduction neutralization tests (PRNT, 90% reduction of ∼100 PFU SAH2123 virus) using procedures previously described.16 This study was performed in accordance with regulations established by the Institutional Animal Care and Use Committee at Colorado State University and all experimentation was carried out in biosafety level-3 laboratory conditions.
Table 1.
Viremia titers and antibody responses in birds and mammals experimentally inoculated with chikungunya virus
| Species | Virus strain | Number tested | Number viremic (range of peak viremia*) | Percentage PRNT positive at 14 DPI | PRNT90 antibody titer range |
|---|---|---|---|---|---|
| Hamster | SAH2123 | 5 | 3 (2.3–5.5) | 0 | < 10 |
| Mesocricetus auratus | COM2005 | 5 | 4 (2.3–2.9) | 0 | < 10 |
| C57BL/6 mouse | SAH2123 | 8 | 2 (2–2.6) | 0 | < 10 |
| Mus musculus | COM2005 | 8 | 1 (2.7) | 0 | < 10 |
| Big brown bat† | SAH2123 | 7 | 4 (1.5–5.5) | 50 | 20 |
| Eptesicus fuscus | COM2005 | 7 | 3 (2.6–3.7) | 0 | < 10 |
| Horse | SAH2123 | 2 | 0 (< 1.0) | 100 | 20–80 |
| Equus caballus | COM2005 | 2 | 0 (< 1.0) | 50 | 40 |
| Calf | SAH2123 | 2 | 0 (< 1.0) | 100 | 10–20 |
| Bos taurus | COM2005 | 2 | 0 (< 1.0) | 100 | 20 |
| Goat | SAH2123 | 2 | 0 (< 1.0) | 100 | 20 |
| Capra aegagrus hircus | |||||
| Pig | SAH2123 | 2 | 0 (< 1.0) | 100 | 40 |
| Sus scrofa domesticus | COM2005 | 2 | 0 (< 1.0) | 100 | 10–40 |
| Dog | SAH2123 | 2 | 0 (< 1.0) | 50 | 10–20 |
| Canis lupus familiaris | |||||
| Rabbit | SAH2123 | 2 | 0 (< 1.0) | 100 | 40–80 |
| Oryctolagus cuniculus | COM2005 | 2 | 0 (< 1.0) | 100 | 40–160 |
| Mink | SAH2123 | 3 | 0 (< 1.0) | 0 | < 10 |
| Neovison vison | |||||
| Armadillo | SAH2123 | 3 | 0 (< 1.0) | 100 | 10–40 |
| Dasypus novemcinctus | |||||
| Raccoon | SAH2123 | 2 | 0 (< 1.0) | 100 | 20–40 |
| Procyon lotor | COM2005 | 2 | 0 (< 1.0) | 100 | 20–40 |
| Chicken | SAH2123 | 3 | 0 (< 2.0) | 67 | 10–20 |
| Gallus gallus domesticus | COM2005 | 3 | 0 (< 2.0) | 67 | 10 |
| Mallard | SAH2123 | 2 | 0 (< 2.0) | 50 | 10 |
| Anas platyrhynchos | COM2005 | 2 | 0 (< 2.0) | 0 | < 10 |
| Red-winged blackbird | SAH2123 | 2 | 0 (< 2.0) | 0 | < 10 |
| Agelaius phoeniceus | COM2005 | 2 | 0 (< 2.0) | 0 | < 10 |
| Double-crested cormorant | SAH2123 | 2 | 0 (< 2.0) | 0 | < 10 |
| Phalacrocorax auritus | COM2005 | 3 | 0 (< 2.0) | 0 | < 10 |
| American white pelican | SAH2123 | 2 | 0 (< 2.0) | 50 | 20 |
| Pelecanus erythrorhynchus | COM2005 | 2 | 0 (< 2.0) | 50 | 10 |
| House sparrow | SAH2123 | 4 | 0 (< 2.0) | 0 | < 10 |
| Passer domesticus | COM2005 | 4 | 0 (< 2.0) | 0 | < 10 |
| Rock pigeon | SAH2123 | 3 | 0 (< 2.0) | 0 | < 10 |
| Columba livia | COM2005 | 2 | 0 (< 2.0) | 0 | < 10 |
| Ring-billed gull | SAH2123 | 2 | 0 (< 2.0) | 50 | 10 |
| Larus delawarensis | |||||
| Cattle egret | SAH2123 | 3 | 0 (< 2.0) | 33 | 10 |
| Bubulcus ibis | COM2005 | 2 | 0 (< 2.0) | 0 | < 10 |
DPI = days postinoculation; PRNT = plaque reduction neutralization tests.
Peak viremia titer is in log10 plaque-forming units/mL serum.
Five bats per virus strain were used to assess viremia; seroconversion was assayed in two additional bats per virus strain.
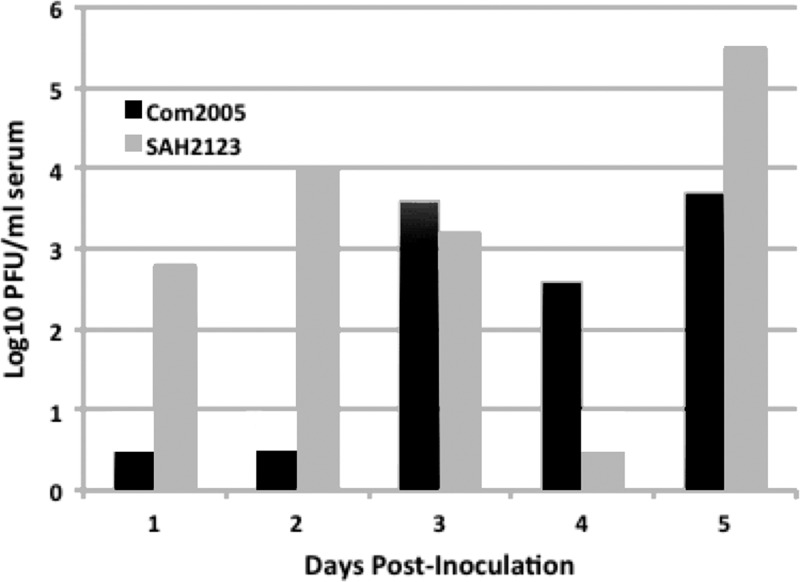
Individuals of nine avian species were inoculated, none of which developed detectable viremia. However, individuals of five different species seroconverted by 14 DPI (Table 1). Domestic mammals failed to develop detectable viremia, but at least one individual from each species seroconverted by 14 DPI. Among the wild mammals inoculated, only big brown bats developed detectable viremia, while one or more individuals of all species (i.e., bats, raccoons, armadillos, and rabbits) except mink seroconverted. Seven of 10 bats euthanized from 1–5 DPI were viremic during that time (Figure 1 ). Hamsters and C57BL/6 mice were used as positive controls for infection with CHIKV.16–18 Three of 16 mice and seven of 10 hamsters developed detectable viremia, but neither mice nor hamsters seroconverted by 14 DPI. Clinical signs of disease were not evident in any animal, and an increase above preinoculation body temperatures was not observed in any individuals in which rectal temperatures were recorded through the course of the study.
Figure 1.
Viremia in 10 Eptesicus fuscus bats inoculated with two strains of chikungunya virus. Each bar depicts an individual bat and values of 0.5 log10 plaque-forming units (PFU)/mL indicate that viremia was not detected (< 10 PFU/mL).
Many arboviruses have complex transmission cycles and use multiple reservoir host species for viral maintenance and replication, whereas others, such as dengue viruses, appear to be exclusively primate pathogens, which can include multiple species.8 Serological evidence of CHIKV infection from Africa and Asia suggests that nonhuman primates and possibly rodents may serve as reservoir hosts.9,10,19,20 However, details of these reservoir host associations have not yet been well elucidated and humans are the only known natural host of CHIKV. In addition, humans develop a viremia of sufficient magnitude to transmit the virus back to mosquitoes.10,21,22 Thus, the involvement of alternative reservoir hosts in CHIKV epidemiology is currently unknown. Results from this study help to narrow the search among animal hosts by lessening the likelihood that numerous bird and mammal species are competent reservoir hosts. The results of the experimental infections reported suggest that some bats and rodents may potentially serve as virus-amplifying hosts of CHIKV. Bats and rodents encompass enormous and diverse taxa with a worldwide distribution, and based on this initial assessment into reservoir host competence, it appears that these groups may justify further study in terms of understanding the ecology and epidemiology of endemic CHIKV. In contrast to bats and rodents, results from this study indicate that various domestic and wild animal species, including birds, mammalian mesocarnivores, lagomorphs, and members of the family Dasypodidae (i.e., armadillos), are unlikely to be competent hosts of CHIKV in nature. However, this study included small numbers of adult animals from each species, and was limited to two strains of CHIKV, thus warranting more thorough evaluations of the range of potential vertebrate amplifying hosts. However, based on these results, in conjunction with a lack of field evidence, it seems unlikely that CHIKV poses a significant threat to most New World livestock and some wild avian and mammalian species. Elucidation of the transmission epidemiology of CHIKV will improve the understanding of how the virus is currently maintained and established in new geographic regions, and can help develop strategies for predicting the source of outbreaks and targeting control and prevention efforts.
ACKNOWLEDGMENTS
We are grateful to Ann Powers from the U.S. Centers for Disease Control and Prevention for providing virus isolates used in this study. We thank United States Department of Agriculture (USDA)/Animal and Plant Health Inspection Service/Wildlife Services biologists Patrick Whitley, Scott Stopak, Paul Wolf, Ryan Powers, Kerri Pedersen, and USDA volunteer Jennifer Watson Marlow for assistance in procuring animals.
Footnotes
Financial support: This work was funded through a cooperative agreement 11-7100-0331-CA from the USDA.
Authors' addresses: Angela M. Bosco-Lauth and Richard A. Bowen, Department of Biomedical Sciences, Colorado State University, Fort Collins, CO, E-mails: mopargal@rams.colostate.edu and rbowen@colostate.edu. Nicole M. Nemeth, Department of Pathobiology, University of Guelph, Guelph, ON, Canada, E-mail: nnemeth@uoguelph.ca. Dennis J. Kohler, United States Department of Agriculture/Animal and Plant Health Inspection Service, Wildlife Services, Fort Collins, CO, E-mail: dennis.kohler@aphis.usda.gov.
References
- 1.Weaver SC, Lecuit M. Chikungunya virus and the global spread of mosquito-borne disease. N Engl J Med. 2015;372:1231–1239. doi: 10.1056/NEJMra1406035. [DOI] [PubMed] [Google Scholar]
- 2.Yergolkar PN, Tandale BV, Arankalle VA, Sathe PS, Sudeep AB, Gandhe SS, Gokhle MD, Jacob GP, Hundekar SL, Mishra AC. Chikungunya outbreaks caused by African genotype, India. Emerg Infect Dis. 2006;12:1580–1583. doi: 10.3201/eid1210.060529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgs S. The 2005–2006 chikungunya epidemic in the Indian Ocean. Vector Borne Zoonotic Dis. 2006;6:115–116. doi: 10.1089/vbz.2006.6.115. [DOI] [PubMed] [Google Scholar]
- 4.Charrel RN, de Lamballerie X, Raoult D. N Engl J Med. 2007;356:769–771. doi: 10.1056/NEJMp078013. [DOI] [PubMed] [Google Scholar]
- 5.Tsetsarkin KA, Vanlandingham DL, McGee GE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vega-Rúa A, Zouache K, Girod R, Failloux AB, Lourenco-de-Oliviera R. High vector competence of Aedes aegypti and Aedes albopictus from ten American countries as a critical factor in the spread of chikungunya virus. J Virol. 2014;88:6294–6306. doi: 10.1128/JVI.00370-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tesh RB. Arthritides caused by mosquito-borne viruses. Annu Rev Med. 1982;33:31–40. doi: 10.1146/annurev.me.33.020182.000335. [DOI] [PubMed] [Google Scholar]
- 8.Turell MJ, Beaman JR, Tammariello RF. Susceptibility of selected strains of Aedes aegypti and Aedes albopictus (Dipteral: Culicidae) to chikungunya virus. J Med Entomol. 1992;29:49–53. doi: 10.1093/jmedent/29.1.49. [DOI] [PubMed] [Google Scholar]
- 9.Diallo M, Thonnon J, Traore-Lamizana M, Fontenille D. Vectors of chikungunya virus in Senegal: current data and transmission cycles. Am J Trop Med Hyg. 1999;60:281–286. doi: 10.4269/ajtmh.1999.60.281. [DOI] [PubMed] [Google Scholar]
- 10.MacKenzie JS, Jeggo M. Reservoirs and vectors of emerging viruses. Curr Opin Virol. 2013;3:170–179. doi: 10.1016/j.coviro.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuehn BM. Chikungunya virus transmission found in the United States: US health authorities brace for wider spread. JAMA. 2014;312:776–777. doi: 10.1001/jama.2014.9916. [DOI] [PubMed] [Google Scholar]
- 12.Morrison TE. Reemergence of chikungunya virus. J Virol. 2014;88:11644–11647. doi: 10.1128/JVI.01432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chevillon C, Briant L, Renaud F, Devauz C. The chikungunya threat: an ecological and evolutionary perspective. Trends Microbiol. 2008;16:80–88. doi: 10.1016/j.tim.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Diallo D, Sall AA, Buenemann M, Chen R, Faye R, Diagne CT, Faye O, Ba Y, Dia I, Watts D, Weaver SC, Hanley KA, Diallo M. Landscape ecology of sylvatic chikungunya virus and mosquito vectors in southeastern Senegal. PLoS Negl Trop Dis. 2012;6:e1649. doi: 10.1371/journal.pntd.0001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sam I, Chua CL, Rovie-Ryan JJ, Fu JYL, Tong C, Sitam FT, Chan YF. Chikungunya virus in macaques, Malaysia. Emerg Infect Dis. 2015;21:1683–1685. doi: 10.3201/eid2109.150439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosco-Lauth AM, Han S, Hartwig A, Bowen RA. Development of a hamster model for chikungunya virus infection and pathogenesis. PLoS One. 2015;10:e0130150. doi: 10.1371/journal.pone.0130150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziegler SA, Lu L, da Rosa AP, Xiao SY, Tesh RB. An animal model for studying the pathogenesis of chikungunya virus infection. Am J Trop Med Hyg. 2008;79:133–139. [PubMed] [Google Scholar]
- 18.Teo TH, Lum FM, Lee WW, Ng LF. Mouse models for chikungunya virus: deciphering immune mechanisms responsible for disease and pathology. Immunol Res. 2012;53:136–147. doi: 10.1007/s12026-012-8266-x. [DOI] [PubMed] [Google Scholar]
- 19.Inoue S, Morita K, Matias RR, Tuplano JV, Resuello RR, Candelario JR, Cruz DJ, Mapua CA, Hasebe F, Igarashi A, Natividad FF. Distribution of three arbovirus antibodies among monkeys (Macaca fascicularis) in the Philippines. J Med Primatol. 2003;32:89–94. doi: 10.1034/j.1600-0684.2003.00015.x. [DOI] [PubMed] [Google Scholar]
- 20.Vourc'h G, Halos L, Desvars A, Boué F, Pascal M, Lecollinet S, Zientara S, Duval T, Nzonza A, Brémont M. Chikungunya antibodies detected in non-human primates and rats in three Indian Ocean islands after the 2006 ChikV outbreak. Vet Res. 2014;45:52–57. doi: 10.1186/1297-9716-45-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M, Cordioli P, Fortuna C, Boros S, Magurano F, Silvi G, Angelini P, Dottori M, Ciufolini MG, Majori GC, Cassone A. CHIKV Study Group Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370:1840–1846. doi: 10.1016/S0140-6736(07)61779-6. [DOI] [PubMed] [Google Scholar]
- 22.Thiberville SD, Boisson V, Gaudart J, Simon F, Flahault A, de Lamballerie X. Chikungunya fever: a clinical and virological investigation of outpatients on Reunion Island, South-West Indian Ocean. PLoS Negl Trop Dis. 2013;7:e2004. doi: 10.1371/journal.pntd.0002004. [DOI] [PMC free article] [PubMed] [Google Scholar]