Abstract
One of the core goals of the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) is to monitor major meat commodities for antimicrobial resistance. Targeted studies with methodologies based on core surveillance protocols are used to examine other foods, e.g., seafood, for antimicrobial resistance to detect resistances of concern to public health. Here we report the discovery of a novel Ambler class A carbapenemase that was identified in a nontoxigenic strain of Vibrio cholerae (N14-02106) isolated from shrimp that was sold for human consumption in Canada. V. cholerae N14-02106 was resistant to penicillins, carbapenems, and monobactam antibiotics; however, PCR did not detect common β-lactamases. Bioinformatic analysis of the whole-genome sequence of V. cholerae N14-02106 revealed on the large chromosome a novel carbapenemase (referred to here as VCC-1, for Vibrio cholerae carbapenemase 1) with sequence similarity to class A enzymes. Two copies of blaVCC-1 separated and flanked by ISVch9 (i.e., 3 copies of ISVch9) were found in an acquired 8.5-kb region inserted into a VrgG family protein gene. Cloned blaVCC-1 conferred a β-lactam resistance profile similar to that in V. cholerae N14-02106 when it was transformed into a susceptible laboratory strain of Escherichia coli. Purified VCC-1 was found to hydrolyze penicillins, 1st-generation cephalosporins, aztreonam, and carbapenems, whereas 2nd- and 3rd-generation cephalosporins were poor substrates. Using nitrocefin as a reporter substrate, VCC-1 was moderately inhibited by clavulanic acid and tazobactam but not EDTA. In this report, we present the discovery of a novel class A carbapenemase from the food supply.
INTRODUCTION
Carbapenems are potent wide-spectrum antibiotics that are routinely used as last-line drugs to treat resistant bacterial infections. Their continued use is threatened by carbapenemases, which are β-lactamases with an extended spectrum of activity against carbapenems. This family of enzymes is divided into two groups on the basis of their enzymatic mechanism; class A and D enzymes utilize an active-site serine where catalysis proceeds through an acyl-enzyme intermediate, whereas class B enzymes are metalloenzymes that promote the direct hydrolysis of the β-lactam via a metal cofactor (1). Carbapenemases are a major determinant of carbapenem resistance, and public health surveillance efforts have found that their spread has accelerated worldwide over the last decade (2). The incidence of carbapenem-resistant Enterobacteriaceae (CRE) is increasing in health care settings, and useful drugs to treat them are dwindling (3, 4). CRE possess resistance elements that are often highly mobile and can transfer between species and asymptomatic carriers (5, 6), making CRE outbreaks both logistically and epidemiologically difficult to control.
Antibiotic resistance surveillance efforts have largely focused on pathogens isolated from human specimens to identify and track emerging resistance determinants. The environment has tremendous microbial diversity and is a major reservoir for antimicrobial resistance (7, 8); however, it is not subjected to the same level of surveillance as human clinical isolates, and the risk of antimicrobial-resistant infections from such reservoirs is unclear (9, 10). Antibiotic-resistant organisms have been reported from diverse food sources, including raw and processed foods and various plant and animal sources (11–14). A survey of bacterial isolates from seafood and meat collected from Canadian retail sources found carbapenem-resistant organisms with known mechanisms of action, particularly in seafood (12). Recently, Bier and colleagues identified a nontoxigenic Vibrio cholerae isolate that harbored a carbapenemase that could not be identified by standard PCR typing (15). In this work, we present the discovery and characterization of a novel Ambler class A carbapenemase found in a nontoxigenic V. cholerae strain isolated from a shrimp intended for human consumption in Canada.
MATERIALS AND METHODS
Source of isolate.
V. cholerae N14-02106 was collected as part of a targeted study of the carbapenem resistance of Enterobacteriaceae found in imported seafood collected through the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) retail food sampling framework in 2014 and described in detail elsewhere (N. Janecko, S. Martz, B. P. Avery, D. Daignault, A. Desruisseau, D. Boyd, R. J. Irwin, M. R. Mulvey, and R. J. Reid-Smith, submitted for publication). The study used selective media to assess the distribution of carbapenemase-producing Enterobacteriaceae in seafood. The bacterium was isolated from frozen farmed black tiger shrimp imported from India and purchased in Ontario, Canada. Briefly, the shrimp sample was incubated overnight in buffered peptone water. V. cholerae N14-02106 was isolated by directly plating the incubated sample onto chromID Carba medium (bioMérieux, Saint-Laurent, QC, Canada).
Using a disk diffusion test, the isolate had an ertapenem disk (10 μg) diameter of ≤25 mm on a Mueller-Hinton plate, which was the cutoff value for a putative carbapenemase producer in the pilot study. V. cholerae N14-02106 was also positive for carbapenemase production using the Carba NP test (16). The isolate was identified as V. cholerae by the Vitek 2 Compact system, and its identity was confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (Bruker Daltonics, Wissembourg, France). Serotyping was performed using antisera against the O1 and O139 serogroups produced by the National Microbiology Laboratory in a standard slide agglutination assay.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was carried out using a Vitek 2 system and the Etest (bioMérieux Canada Inc., St. Laurent, QC, Canada) and a Sensititre automated microbiology system (Trek Diagnostic Systems Ltd., Thermo Fisher Scientific, Oakwood Village, OH, USA) according to the manufacturers' instructions. The antimicrobial susceptibilities of the isolates were interpreted according to the latest CLSI breakpoints.
Whole-genome sequencing and assembly.
V. cholerae N14-02106 genomic DNA was purified using a MasterPure Complete RNA and DNA purification kit (Epicentre, Madison, WI, USA) according to the manufacturer's instructions. DNA was sequenced by use of both the MiSeq (Illumina, San Diego, CA, USA) and single-molecule real-time sequencing (RSII; Pacific Biosciences, Menlo Park, CA, USA) platforms. The Illumina DNA libraries were prepared with a TruSeq DNA PCR sample preparation kit, and adapter-ligated libraries were size selected for a 500- to 800-bp insert using a Sage Science Blue Pippin instrument (Beverley, MA, USA). Paired-end reads were produced with a MiSeq reagent kit (v3; 600 cycles). Raw reads were preprocessed with FLASH software (17), and de novo assembly was performed by use of the SPAdes algorithm (18). Pacific Biosciences DNA libraries with a 20-kb fragment size were prepared, and sequencing was performed by P6-C4 polymerase/chemistry with one single-molecule real-time (SMRT) cell run for 240 min. A draft assembly was produced using the HGAP (v3.0) protocol (19) via the SMRT portal software (v2.3.0).
Bioinformatic analysis.
Contigs from the Illumina assembly were searched, using the Comprehensive Antibiotic Resistance Database (http://arpcard.mcmaster.ca), to identify putative β-lactamase genes (20). Annotation was carried out by manual NCBI BLAST analysis (21).
PCR and cloning of blaVCC-1.
A multiplex PCR was used to detect the KPC, NDM, GES, VIM, IMP, SME, and OXA-48 β-lactamase genes (22). Primers VibCACF (5′-TTAGCTTATGTGTCCAACCA) and VibCACR (5′-TCTATACAGTTTGTACGACC) were used to amplify blaVCC-1, and the product was cloned into plasmid pCR-XL-TOPO to produce pVCC-1. For VCC-1 production, the blaVCC-1 gene was amplified with primers VCCstart (5′-CCCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACCATGAAACGTATTGCTATG) and VCCstop (5′-TCTCAAGCTTTCACTTTACATTTTCTATTGCAAT), and the product was cloned with XbaI and HindIII into pET-28b(+) to produce pET-VCC-1.
DNA transfer by conjugation.
Escherichia coli J53AZR, resistant to sodium azide, was used as the recipient in mating experiments with V. cholerae N14-02106. Mating was done in LB broth at 25°C and 30°C with selection for transconjugants on LB agar containing 0.125 μg/ml meropenem and 150 μg/ml sodium azide.
VCC-1 protein purification.
Plasmid pET-VCC-1in E. coli BL21(DE3) was grown aerobically at 37°C in Terrific Broth supplemented with 25 μg/ml kanamycin to an A600 of ≈0.6, induced with 100 μM IPTG (isopropyl-β-d-thiogalactopyranoside), and outgrown for 22 h at 18°C. Cells were harvested by centrifugation at 3,000 × g for 10 min, and the periplasm was extracted by resuspending cell pellets in a 1/40 volume of TSE buffer (500 mM sucrose, 200 mM Tris, pH 8, 1 mM EDTA) as described previously (23). Periplasmic extracts were diluted 5-fold with 50 mM sodium acetate buffer (pH 5), loaded onto a 1-ml HiTrap SP XL cation-exchange column (GE Healthcare, Piscataway, NJ, USA) that had been preequilibrated with 50 mM MES (morpholineethanesulfonic acid; pH 6), and eluted with a linear gradient of 0 to 300 mM NaCl. Fractions containing VCC-1 were identified by SDS-PAGE, pooled, buffer exchanged using a centrifugal filter into 50 mM sodium phosphate (pH 7) containing 10% glycerol, and stored at −80°C. The intrinsic molar absorptivity of the mature VCC-1 polypeptide was used to calculate the concentration of protein preparations according to the method of Pace and Schmid (24). The purity of VCC-1 was determined by SDS-PAGE, and the VCC-1 was judged to be greater than 90% pure. The signal peptide cleavage site was determined using the SignalP (v4.1) server (http://www.cbs.dtu.dk/services/SignalP/) (25).
Enzyme kinetics.
Imipenem, meropenem, ertapenem, and aztreonam were purchased from Cedarlane Laboratories (Burlington, ON, Canada), and all other antibiotics were purchased from Sigma-Aldrich (St. Louis, MO, USA). VCC-1 activity was measured by monitoring the change in UV absorbance caused by β-lactam ring opening following hydrolysis using an Agilent Cary 50 spectrophotometer (Santa Clara, CA, USA). Reactions were carried out at 22°C in 50 mM sodium phosphate (pH 7). The concentration VCC-1 varied from 0.5 nM to 20 nM, and substrates were tested at 6 to 8 concentrations typically ranging from 0.5× Km to 5 to 10× Km. The means of duplicate reactions were reported, and the ranges over the means for both Km and Vmax were less than 10%. Km and Vmax were determined by fitting the initial reaction rates by nonlinear regression to a rectangular hyperbola using SigmaPlot (v11.0) software. The following extinction coefficients were used for rate measurements: for benzylpenicillin, Δε232 was −1,230 M−1 cm−1; for oxacillin, Δε260 was 280 M−1 cm−1; for cephalothin, Δε262 was −6,250 M−1 cm−1; for cefoxitin, Δε262 was −6,860 M−1 cm−1; for cefotaxime, Δε265 was −6,730 M−1 cm−1; for ceftazidime, Δε260 was −10,400 M−1 cm−1; for aztreonam, Δε318 was −640 M−1 cm−1; for imipenem, Δε299 was −10,550 M−1 cm−1; and for meropenem, Δε298 was −11,500 M−1 cm−1. Inhibitor potency was measured using nitrocefin as a reporter substrate at 490 nm on a Molecular Devices SpectraMax 384 plate reader (Sunnyvale, CA, USA). VCC-1 (0.5 nM) and inhibitor (0.1 to 1000 μM) were preincubated for 10 min, and reactions were initiated by the addition of substrate. Fifty percent inhibitory concentrations were determined by fitting initial rates by nonlinear regression to a four-parameter logistic equation. Kis were determined using the Cheng-Prusoff equation with correction using a competitive model of inhibition; the means and standard deviations of three reactions are reported (26).
Nucleotide sequence accession number.
The region harboring blaVCC-1, as described here, was deposited in GenBank (accession number KT818596).
RESULTS
Phenotypic and molecular testing of V. cholerae N14-02106.
V. cholerae N14-02106 grew on the selective medium chromID Carba and was subsequently identified to be a carbapenemase producer by Carba NP testing. In antimicrobial susceptibility tests, the isolate was resistant to ampicillin, amoxicillin-clavulanic acid, cefazolin, cefoxitin, and meropenem but susceptible to ceftiofur, cefpodoxime, cefotaxime, ceftazidime, ceftriaxone, amikacin, gentamicin, tobramycin, ciprofloxacin, tigecycline, chloramphenicol, nitrofurantoin, sulfisoxazole, and trimethoprim-sulfamethoxazole. A PCR screen for KPC, NDM, OXA-48, VIM, IMP, and GES-type β-lactamase genes was negative. Limited serotyping showed V. cholerae N14-02106 to be non-O1 and non-O139.
Identification of blaVCC-1 from whole-genome sequence.
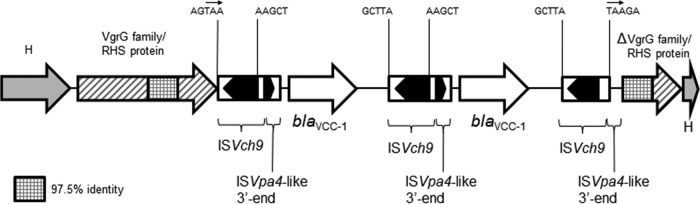
In order to identify the mechanism of carbapenem resistance, V. cholerae N14-02106 was subjected to whole-genome sequencing. An assembly of reads from the Illumina sequencing produced 100 contigs with an average depth of coverage of 164 times and an N50 contig length of 126,070 bp. Analysis of these contigs using the Resistance Gene Identifier tool on the CARD website identified a β-lactamase with 58% identity to the IMI-1 carbapenemase located on a 2,727-bp contig. An 855-bp open reading frame (encoding 284 amino acids) was identified and labeled blaVCC-1 (where VCC-1 represents Vibrio cholerae carbapenemase 1). Sequencing of the N14-02106 genome using the Pacific Biosciences platform produced 952 Mbp of reads with an N50 read length of 17,372 bp. De novo assembly produced two chromosomes of 2,933,987 bp and 1,228,771 bp with an average depth of coverage of 184 times. No plasmids were detected by sequencing. The blaVCC-1 gene was found in an 8.5-kb region on the large chromosome and was inserted into the 3′ end of the virulence-associated vgrG gene. Two identical copies of blaVCC-1 were found in a head-to-tail arrangement, with each copy being surrounded by a novel insertion sequence designated ISVch9 in the ISfinder database (27) (Fig. 1). Beginning 14 bp upstream of the blaVCC-1 start codon is a 534-bp remnant of the 3′ end of an ISVpa4-like element (27), which presumably contains sequences that drive the expression of blaVCC-1. The 8.5-kb region described above is flanked by 3-bp direct repeats (Fig. 1) and resembles a novel composite transposon; thus, this region may have been acquired by a transposition mechanism. Attempts to transfer chromosomal blaVCC-1 to E. coli by conjugation were unsuccessful.
FIG 1.
Schematic map of the region containing blaVCC-1 on the chromosome of V. cholerae N14-02106 (GenBank accession number KT818596). Block arrows, the direction of transcription of the genes; H, hypothetical protein. The sequences of the 5 bp immediately flanking each terminal inverted repeat of the three copies of ISVch9 are shown. The putative direct repeats of TAA flanking the acquired region are indicated by small arrows within the 5-bp sequences.
Comparison of the VCC-1 sequence with the sequences of other β-lactamases.
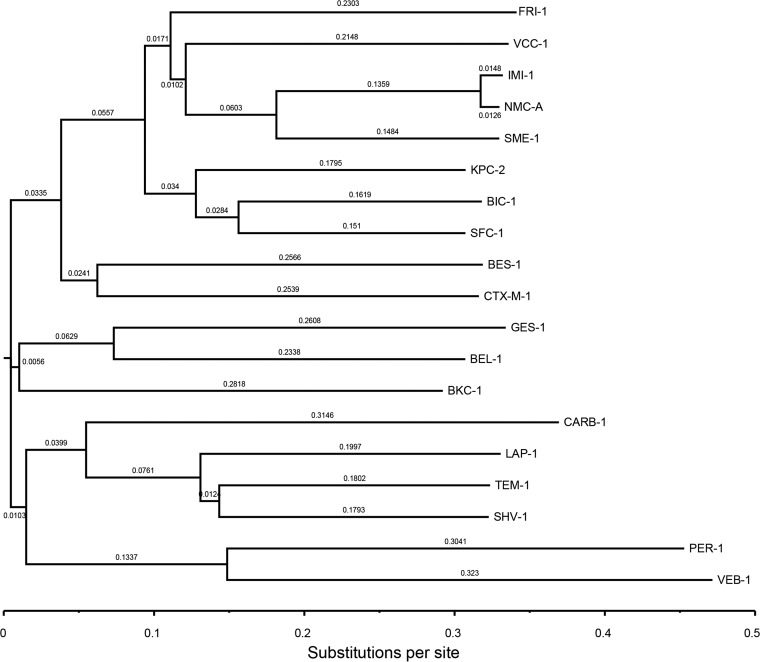
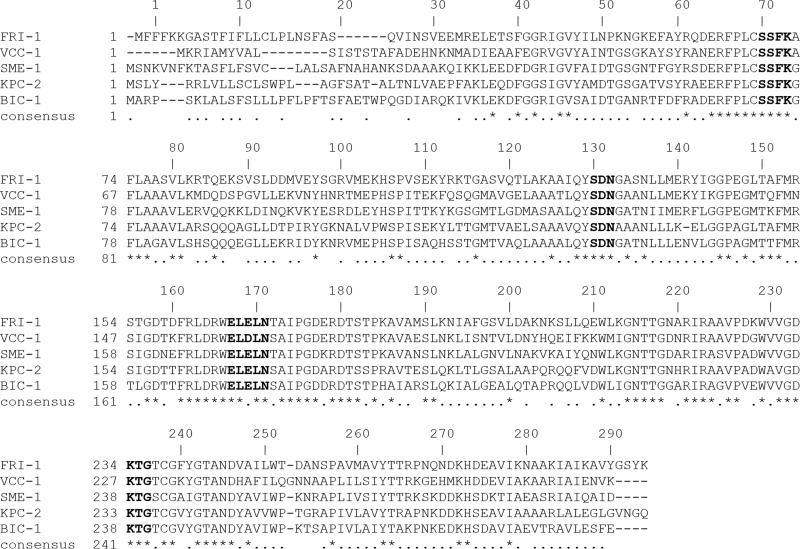
A dendrogram using the VCC-1 amino acid sequence aligned with the sequences of select Ambler class A β-lactamases revealed that it is most closely related to the IMI, NMC-A, and SFC-1 carbapenemases, sharing 59.1%, 59.1%, and 58.3% amino acid sequence identity, respectively (Fig. 2). An alignment of the VCC-1 amino acid sequence with the amino acid sequences of KPC-2, NMC-A, SME-1, and IMI-1 is presented in Fig. 3, where the sequence numbering follows the Ambler scheme for class A β-lactamases (28). VCC-1 contains four conserved motifs of class A β-lactamases: 70S(S/T)FK73 130SDN132, 166EXXXN170, and 234KTG236. It also contains in other positions residues that are characteristic of carbapenemases, namely, 69C, 238C, 105H, 168L, 237T, and 241Y (29–31).
FIG 2.
Dendrogram of 18 representative class A β-lactamases using precursor amino acid sequences. The alignment and neighbor-joining tree were produced using the Clustal Omega program from EMBL-EBI bioinformatics services (http://www.ebi.ac.uk/Tools/msa/clustalo/), and the tree was visualized using the FigTree (v1.4.2) viewer (http://tree.bio.ed.ac.uk/software/figtree/). Branch lengths are scaled to the number of amino acid changes per site. The β-lactamases (GenBank accession numbers) are as follows: BEL-1 (DQ089809), BES-1 (AF234999), BIC-1 (GQ260093), BKC-1 (KP689347), CARB-1 (D86225), CTX-M-1 (X92506), FRI-1 (KT192551) GES-1 (AF156486), IMI-1 (U50278), KPC-2 (AY034847), LAP-1 (EF026092), NMC-A (Z21956), PER-1 (Z21957), SFC-1 (AY354402), SHV-1 (AF148850), SME-1 (Z28968), TEM-1 (AY458016), VEB-1 (AF010416), and VCC-1 (KT818596).
FIG 3.
Amino acid sequence alignment of VCC-1 with the sequences of previously described carbapenemases. Signature motifs of β-lactamases are indicated in bold. The sequence numbering follows the Ambler scheme for class A β-lactamases. GenBank accession numbers are listed in the legend to Fig. 2.
VCC-1 is a carbapenemase.
Etest results showed that V. cholerae N14-02106 was resistant to aztreonam (MIC, >256 μg/ml) and all four carbapenems tested (imipenem, meropenem, ertapenem, and doripenem; MICs, >32 μg/ml) (Table 1). Further, the isolate exhibited intermediate susceptibility to amoxicillin-clavulanic acid, piperacillin-tazobactam, and cefoxitin but was susceptible to 3rd- and 4th-generation cephalosporins. Similar results were obtained by testing with the Vitek 2 and Sensititre systems. Importantly, E. coli TOP10 harboring pVCC-1 exhibited resistance to imipenem and ertapenem and intermediate susceptibility to meropenem and doripenem, indicating that VCC-1 is a carbapenemase. Like the clinical isolate, the pVCC-1 transformant was resistant to amoxicillin-clavulanic acid and aztreonam but susceptible to the 3rd- and 4th-generation cephalosporins; however, MICs were generally lower (except for ceftazidime), as is common for laboratory strains of E. coli harboring cloned β-lactamase genes (32–36).
TABLE 1.
MICs of β-lactams for V. cholerae N14-02106, E. coli TOP10 harboring recombinant plasmid pVCC-1, and the E. coli TOP10 parent strain
| β-Lactam | MIC (μg/ml)a |
||
|---|---|---|---|
| V. cholerae N14-02106 | E. coli TOP10(pVCC-1)b | E. coli TOP10 | |
| Ceftriaxone | 0.39 | 1.5 | 0.064 |
| Cefotaxime | 0.064 | 0.38 | 0.094 |
| Ceftazidime | 0.75 | 3 | 0.75 |
| Cefoxitin | 16 | 12 | 6 |
| Cefepime | 1.5 | 0.5 | 0.094 |
| Aztreonam | >256 | 48 | 0.094 |
| Amoxicillin + CLAc | 16 | 64 | 4 |
| Piperacillin + TZBd | 48 | 32 | 1.5 |
| Imipenem | >32 | 12 | 0.5 |
| Meropenem | >32 | 2 | 0.047 |
| Ertapenem | >32 | 1.5 | 0.004 |
| Doripenem | >32 | 2 | 0.032 |
MICs were determined with Etest gradient strips.
pVCC-1 contains blaVCC-1 cloned into pCR-XL-TOPO.
CLA, clavulanic acid at a fixed concentration of 4 μg/ml.
TZB, tazobactam at a fixed concentration of 4 μg/ml.
Biochemical characterization of VCC-1.
Purified VCC-1 exhibited broad and varied hydrolytic activity toward penicillin, monobactam, and carbapenem antibiotics (Table 2). The highest catalytic turnover (kcat) was observed with oxacillin, cephalothin, and imipenem at 153.3 s−1, 320.1 s−1, and 190.2 s−1, respectively. VCC-1 showed reduced activity toward 2nd- and 3rd-generation cephalosporins, and Kms were too large to be measured. The Km for meropenem was the lowest at 70.1 μM; however, its turnover rate was a modest 11.2 s−1. VCC-1 showed the highest catalytic efficiency (kcat/Km) against oxacillin. Inhibitor studies showed that VCC-1 was inhibited by tazobactam and clavulanic acid, with Kis being 2.0 ± 0.4 μM and 13 ± 3 μM, respectively. No evidence of inhibition by EDTA was observed up to a concentration of 1 mM.
TABLE 2.
Kinetic parameters of VCC-1
| Substrate | kcat (s−1)a | Km (μM)a | kcat/Km (μM−1 s−1) |
|---|---|---|---|
| Benzylpenicillin | 37.8 | 136.0 | 0.278 |
| Oxacillin | 153.3 | 170.2 | 0.901 |
| Cephalothin | 320.1 | 593.0 | 0.540 |
| Cefoxitin | >0.8b | >1,000 | NDc |
| Cefotaxime | >4.5b | >1,000 | ND |
| Ceftazidime | >0.2b | >1,000 | ND |
| Aztreonam | >44.6b | >1,000 | ND |
| Imipenem | 190.2 | 876.8 | 0.217 |
| Meropenem | 11.2 | 70.1 | 0.160 |
kcat and Km were estimated by fitting initial rates to the Michaelis-Menten equation using nonlinear regression. Values represent the averages from duplicates, where the range/mean was <10% in all cases.
kcat not determinable due to high Km; activity at 1,000 µM is reported.
ND, not determinable due to a high Km.
DISCUSSION
The novel carbapenemase VCC-1 was identified in a non-O1, non-O139 strain of V. cholerae with high-level resistance to carbapenems, and this activity was transferable to a susceptible E. coli strain. An alignment of the VCC-1 amino acid sequence with the amino acid sequences of previously characterized carbapenemases revealed signature features of Ambler class A β-lactamases and residues that are indicative of carbapenemase activity. Finally, the VCC-1 protein was able to hydrolyze both meropenem and imipenem in vitro, showing kinetic parameters similar to those of the related enzymes IMI-1 and SFC-1, where hydrolysis of meropenem was approximately 10-fold lower than that of imipenem (36). Additionally, the pattern of hydrolytic activity of this enzyme mirrored that of related class A enzymes, where 2nd- and 3rd-generation cephalosporins are generally poorer substrates than 1st-generation cephalosporins, penicillins, monobactams, and carbapenems (37). In summation, the biochemical and genetic data were consistent with a carbapenemase function for VCC-1.
The blaVCC-1 sequence was present in two copies on the chromosome and was associated with three copies of the insertion element ISVch9 in a novel 8.5-kb region that may have been acquired by transposition from an unknown source (Fig. 1). Direct transfer of blaVCC-1 to E. coli could not be demonstrated in vitro, but it remains to be seen if dissemination in the environment or a clinical setting could occur. The blaVCC-1 region was inserted into the end of the vrgG gene, which encodes an effector protein of the type VI secretion system (38). Thus, in V. cholerae N14-02106, the VgrG protein would be a hybrid, with the last 20 amino acids being encoded by ISVch9. If the VrgG protein is produced, it would be interesting to know if the virulence of V. cholerae N14-02106 is negatively affected by chimeric VrgG. The presence of VCC-1 in a V. cholerae isolate found in frozen shrimp could result in its distribution through the food chain across wide geographical areas and population groups. V. cholerae N14-02106 is not multidrug resistant, so there are options for the treatment of an infection with this organism, possibly with 3rd- or 4th-generation cephalosporins. The isolate was from a farmed shrimp from India, where limited control of antimicrobial use encourages the emergence of multidrug-resistant organisms both in the environment and in health care settings (39).
Expression of VCC in E. coli TOP10 cells via pVCC-1 did not impart the same high-level carbapenem resistance seen with N14-02106; similar MIC differences were seen in the initial characterization of blaFRI, blaBIC, blaBKC, blaSFC, blaIMI, and blaKPC (32–36, 40). In the case of blaKPC, cells of the clinical isolate contained a permeability defect that contributed to the elevated MICs of cell wall-active antibiotics. It is possible that V. cholerae N14-02106 cells have lower permeability than E. coli TOP10 cells, resulting in a higher carbapenem MIC; however, the cephalosporins MICs were lower in N14-02106, indicating that cell permeability was at least not the sole determinant of the differences in the MICs. A number of factors could have contributed to the change in MIC when a VCC was moved from a member of the Vibrionaceae to a member of the Enterobacteriaceae, including differences in gene expression and engagement of secretory machinery. Furthermore, previous work has shown that the complement of penicillin binding proteins and their specific affinities for cell wall-active antibiotics and β-lactamase inhibitors contributed to differences in β-lactam susceptibility (41–44).
In conclusion, this is the first characterization of a class A carbapenemase found in a member of the Vibrionaceae and the first report of a novel class A carbapenemase discovered in food. Since the 1990s, we have seen the emergence of Ambler class A carbapenemases, which are usually first isolated from clinical samples. In this report, we describe one of the few instances where a new carbapenemase was discovered from a nonclinical and nonenteric source, the other example being BIC-1, which was found in a pseudomonad isolated from the Seine River (33). The discovery of VCC-1 was directly facilitated by the use of whole-genome sequencing, which is the first use of this technique to discover an enzyme of this class. Finally, this work highlights the need for inclusion of food and, possibly, environmental sources in antimicrobial resistance surveillance, as we will likely see an increase in the diversity of carbapenemases. Understanding and tracking of the potential spread from the environment into the clinic will be important for future infection control measures.
ACKNOWLEDGMENTS
We thank Amarbeer Bhandari and Mark Miller-Williams (Richardson Centre for Functional Foods and Nutraceuticals, University of Manitoba) for use of their facilities. We thank Shaun Tyler (DNA Core, National Microbiology Laboratory) and David Moraga (Interdisciplinary Center for Biotechnology Research, University of Florida) for whole-genome sequencing services. We thank Ken Fakharuddin (National Microbiology Laboratory) for laboratory technical assistance. We also thank Celine Nadon (Bacteriology and Enterics, National Microbiology Laboratory) for serotyping services.
REFERENCES
- 1.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordmann P, Poirel L. 2014. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 3.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 4.Bradley JS, Guidos R, Baragona S, Bartlett JG, Rubinstein E, Zhanel GG, Tino MD, Pompliano DL, Tally F, Tipirneni P, Tillotson GS, Powers JH, Tillotson GS. 2007. Anti-infective research and development—problems, challenges, and solutions. Lancet Infect Dis 7:68–78. doi: 10.1016/S1473-3099(06)70689-2. [DOI] [PubMed] [Google Scholar]
- 5.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, NISC Comparative Sequencing Program Group, Henderson DK, Palmore TN, Segre JA. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schechner V, Kotlovsky T, Kazma M, Mishali H, Schwartz D, Navon-Venezia S, Schwaber MJ, Carmeli Y. 2013. Asymptomatic rectal carriage of blaKPC producing carbapenem-resistant Enterobacteriaceae: who is prone to become clinically infected? Clin Microbiol Infect 19:451–456. doi: 10.1111/j.1469-0691.2012.03888.x. [DOI] [PubMed] [Google Scholar]
- 7.D'Costa VM, McGrann KM, Hughes DW, Wright GD. 2006. Sampling the antibiotic resistome. Science 311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 8.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 9.Woodford N, Wareham DW, Guerra B, Teale C. 2014. Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: an emerging public health risk of our own making? J Antimicrob Chemother 69:287–291. doi: 10.1093/jac/dkt392. [DOI] [PubMed] [Google Scholar]
- 10.Wright GD. 2010. Antibiotic resistance in the environment: a link to the clinic? Curr Opin Microbiol 13:589–594. doi: 10.1016/j.mib.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Rubin JE, Ekanayake S, Fernando C. 2014. Carbapenemase-producing organism in food, 2014. Emerg Infect Dis 20:1264–1265. doi: 10.3201/eid2007.140534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrison BJ, Rubin JE. 2015. Carbapenemase producing bacteria in the food supply escaping detection. PLoS One 10:e0126717. doi: 10.1371/journal.pone.0126717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerra B, Fischer J, Helmuth R. 2014. An emerging public health problem: acquired carbapenemase-producing microorganisms are present in food-producing animals, their environment, companion animals and wild birds. Vet Microbiol 171:290–297. doi: 10.1016/j.vetmic.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Zurfluh K, Poirel L, Nordmann P, Klumpp J, Stephan R. 2015. First detection of Klebsiella variicola producing OXA-181 carbapenemase in fresh vegetable imported from Asia to Switzerland. Antimicrob Resist Infect Control 4:38. doi: 10.1186/s13756-015-0080-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bier N, Schwartz K, Guerra B, Strauch E. 2015. Survey on antimicrobial resistance patterns in Vibrio vulnificus and Vibrio cholerae non-O1/non-O139 in Germany reveals carbapenemase-producing Vibrio cholerae in coastal waters. Front Microbiol 6:1179. doi: 10.3389/fmicb.2015.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 20.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJV, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Mataseje LF, Bryce E, Roscoe D, Boyd DA, Embree J, Gravel D, Katz K, Kibsey P, Kuhn M, Mounchili A, Simor A, Taylor G, Thomas E, Turgeon N, Mulvey MR, Canadian Nosocomial Infection Surveillance Program. 2012. Carbapenem-resistant Gram-negative bacilli in Canada 2009-10: results from the Canadian Nosocomial Infection Surveillance Program (CNISP). J Antimicrob Chemother 67:1359–1367. doi: 10.1093/jac/dks046. [DOI] [PubMed] [Google Scholar]
- 23.Hiniker A, Bardwell JCA. 2004. In vivo substrate specificity of periplasmic disulfide oxidoreductases. J Biol Chem 279:12967–12973. doi: 10.1074/jbc.M311391200. [DOI] [PubMed] [Google Scholar]
- 24.Pace CN, Schmid FX. 1997. How to determine the molar absorbance coefficient of a protein, p 253–259. In Creighton TE. (ed), Protein structure: a practical approach, 2nd ed. Oxford University Press, New York, New York. [Google Scholar]
- 25.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 26.Cer RZ, Mudunuri U, Stephens R, Lebeda FJ. 2009. IC50-to-Ki: a Web-based tool for converting IC50 to Ki values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res 37:W441–W445. doi: 10.1093/nar/gkp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ambler RP, Coulson AF, Frère JM, Ghuysen JM, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG. 1991. A standard numbering scheme for the class A beta-lactamases. Biochem J 276(Pt 1):269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majiduddin FK, Palzkill T. 2005. Amino acid residues that contribute to substrate specificity of class A beta-lactamase SME-1. Antimicrob Agents Chemother 49:3421–3427. doi: 10.1128/AAC.49.8.3421-3427.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majiduddin FK, Palzkill T. 2003. Amino acid sequence requirements at residues 69 and 238 for the SME-1 beta-lactamase to confer resistance to beta-lactam antibiotics. Antimicrob Agents Chemother 47:1062–1067. doi: 10.1128/AAC.47.3.1062-1067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papp-Wallace KM, Taracila M, Hornick JM, Hujer AM, Hujer KM, Distler AM, Endimiani A, Bonomo RA. 2010. Substrate selectivity and a novel role in inhibitor discrimination by residue 237 in the KPC-2 beta-lactamase. Antimicrob Agents Chemother 54:2867–2877. doi: 10.1128/AAC.00197-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dortet L, Poirel L, Abbas S, Oueslati S, Nordmann P. 2015. Genetic and biochemical characterization of FRI-1, a carbapenem-hydrolyzing class A β-lactamase from Enterobacter cloacae. Antimicrob Agents Chemother 59:7420–7425. doi: 10.1128/AAC.01636-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girlich D, Poirel L, Nordmann P. 2010. Novel Ambler class A carbapenem-hydrolyzing beta-lactamase from a Pseudomonas fluorescens isolate from the Seine River, Paris, France. Antimicrob Agents Chemother 54:328–332. doi: 10.1128/AAC.00961-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henriques I, Moura A, Alves A, Saavedra MJ, Correia A. 2004. Molecular characterization of a carbapenem-hydrolyzing class A beta-lactamase, SFC-1, from Serratia fonticola UTAD54. Antimicrob Agents Chemother 48:2321–2324. doi: 10.1128/AAC.48.6.2321-2324.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasmussen BA, Bush K, Keeney D, Yang Y, Hare R, O'Gara C, Medeiros AA. 1996. Characterization of IMI-1 beta-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob Agents Chemother 40:2080–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walther-Rasmussen J, Høiby N. 2007. Class A carbapenemases. J Antimicrob Chemother 60:470–482. doi: 10.1093/jac/dkm226. [DOI] [PubMed] [Google Scholar]
- 38.Unterweger D, Miyata ST, Bachmann V, Brooks TM, Mullins T, Kostiuk B, Provenzano D, Pukatzki S. 2014. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun 5:3549. doi: 10.1038/ncomms4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdul Ghafur K. 2010. An obituary—on the death of antibiotics! J Assoc Physicians India 58:143–144. [PubMed] [Google Scholar]
- 40.Nicoletti AG, Marcondes MFM, Martins WMBS, Almeida LGP, Nicolás MF, Vasconcelos ATR, Oliveira V, Gales AC. 2015. Characterization of BKC-1 class A carbapenemase from Klebsiella pneumoniae clinical isolates in Brazil. Antimicrob Agents Chemother 59:5159–5164. doi: 10.1128/AAC.00158-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bharat A, Demczuk W, Martin I, Mulvey MR. 2015. Effect of variants of penicillin-binding protein 2 on cephalosporin and carbapenem susceptibilities in Neisseria gonorrhoeae. Antimicrob Agents Chemother 59:5003–5006. doi: 10.1128/AAC.05143-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asli A, Brouillette E, Krause KM, Nichols WW, Malouin F. 2016. Distinctive binding of avibactam to penicillin-binding proteins of Gram-negative and Gram-positive bacteria. Antimicrob Agents Chemother 60:752–756. doi: 10.1128/AAC.02102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farha MA, Leung A, Sewell EW, D'Elia MA, Allison SE, Ejim L, Pereira PM, Pinho MG, Wright GD, Brown ED. 2013. Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to β-lactams. ACS Chem Biol 8:226–233. doi: 10.1021/cb300413m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakoulas G, Kumaraswamy M, Nonejuie P, Werth BJ, Rybak MJ, Pogliano J, Rice LB, Nizet V. 2015. Differential effects of penicillin binding protein deletion on the susceptibility of Enterococcus faecium to cationic peptide antibiotics. Antimicrob Agents Chemother 59:6132–6139. doi: 10.1128/AAC.00486-15. [DOI] [PMC free article] [PubMed] [Google Scholar]