Abstract
In a search for new antifungal compounds, we screened a library of 4,454 chemicals for toxicity against the human fungal pathogen Aspergillus fumigatus. We identified sr7575, a molecule that inhibits growth of the evolutionary distant fungi A. fumigatus, Cryptococcus neoformans, Candida albicans, and Saccharomyces cerevisiae but lacks acute toxicity for mammalian cells. To gain insight into the mode of inhibition, sr7575 was screened against 4,885 S. cerevisiae mutants from the systematic collection of haploid deletion strains and 977 barcoded haploid DAmP (decreased abundance by mRNA perturbation) strains in which the function of essential genes was perturbed by the introduction of a drug resistance cassette downstream of the coding sequence region. Comparisons with previously published chemogenomic screens revealed that the set of mutants conferring sensitivity to sr7575 was strikingly narrow, affecting components of the endoplasmic reticulum-associated protein degradation (ERAD) stress response and the ER membrane protein complex (EMC). ERAD-deficient mutants were hypersensitive to sr7575 in both S. cerevisiae and A. fumigatus, indicating a conserved mechanism of growth inhibition between yeast and filamentous fungi. Although the unfolded protein response (UPR) is linked to ERAD regulation, sr7575 did not trigger the UPR in A. fumigatus and UPR mutants showed no enhanced sensitivity to the compound. The data from this chemogenomic analysis demonstrate that sr7575 exerts its antifungal activity by disrupting ER protein quality control in a manner that requires ERAD intervention but bypasses the need for the canonical UPR. ER protein quality control is thus a specific vulnerability of fungal organisms that might be exploited for antifungal drug development.
INTRODUCTION
The burden of fungal infections in the human population is very high, with an estimated 1.5 million annual deaths worldwide, despite antifungal prophylaxis (1–3). The evolutionary proximity between mammalian and fungal cells creates a challenge for the identification of selective drug targets. Consequently, there are only a few mechanistically distinct classes of antifungal agents. The major antifungal drugs in clinical use disrupt membrane homeostasis by targeting ergosterol (4), impair cell wall integrity by inhibiting β-1,3-glucan synthase (5), or perturb nucleic acid synthesis via a fluorinated nucleotide analogue (6). The limited number of therapeutic options impedes effective management of invasive fungal infections, particularly when resistance to a drug is either emerging or an intrinsic characteristic of the fungal pathogen.
The identification of novel drugs and their targets can follow several strategies, ranging from the inhibition of a known protein target with a panel of inhibitors to the analysis of mutant strain sensitivity to toxic compounds (7). Chemical genomic screens analyze large collections of genetically defined mutant strains for their sensitivity to chemical libraries in a systematic manner. Data from these screens can provide insight into candidate targets for a given drug, as well as the cellular pathways required to buffer drug toxicity (8–11). The interpretation of chemogenomic screens depends on the type of mutant collection utilized for the analysis. For example, the absence of a general dosage compensation mechanism in yeast (12) allows heterozygous deletion strains to be used as tools to determine how a reduction in the level of a gene product impacts drug sensitivity. However, since heterozygous deletion strains retain some level of gene function, compensatory mechanisms could mask changes in drug sensitivity. Haploid deletion strains can circumvent this problem and increase the sensitivity of the screen. The resulting chemogenomic profile, or pattern of mutants that are affected by a given compound, is predictive of the mechanism of action and has been successfully applied to drug lead identification and target classification in yeast (13). Further insight into pathways that are modulated by a compound can be obtained by developing in silico comparison tools that link the results of a chemogenomic analysis to the data of other published large-scale chemogenomic or genetic interaction screens (for examples, see references 14 and 15).
Most of the large chemogenomic data sets currently available have investigated nonessential gene deletions in haploid strains (10, 16). Essential genes, representing about one-sixth of all Saccharomyces cerevisiae genes, are more difficult to study in haploid or heterozygous deletion strains, so an alternative approach is the use of DAmP (decreased abundance by mRNA perturbation) strains. DAmP strains contain a drug resistance marker inserted into the 3′ untranslated region (UTR) of a gene, resulting in defects in mRNA stability that can create hypomorphic alleles for phenotypic analysis of essential gene function (17). These strains have provided important insights into gene function as well as the response of cells to stress (for example, see reference 18).
In this study, we report the identification of sr7575, a small molecule with fungistatic activity against S. cerevisiae and three genera of human fungal pathogens: Aspergillus fumigatus, Cryptococcus neoformans, and Candida albicans. We employed a genome-wide approach to characterize the mode of action of sr7575, using a systematic determination of S. cerevisiae deletion and DAmP mutant sensitivity to the drug, combined with extensive in silico comparisons with large-scale data sets from published chemogenomic and genetic interaction screens. The strategy led to the conclusion that S. cerevisiae and A. fumigatus mutants that are deficient in endoplasmic reticulum-associated degradation (ERAD), a degradative pathway that disposes of misfolded proteins that arise in the ER membrane or lumen (19), are hypersensitive to sr7575. Collectively, these data implicate ER protein quality control as the target of sr7575 toxicity in evolutionarily distant fungi and suggest that further analysis of compounds that disrupt ER homeostasis may provide novel avenues for antifungal drug development.
MATERIALS AND METHODS
Screening procedure of the CERMN chemical library.
All robotic steps were performed on a Tecan Freedom EVO platform. Compounds were transferred from mother plates into clear, flat-bottom, barcoded tissue culture 96-well plates (Greiner Bio One): 1 μl of a dimethyl sulfoxide (DMSO) solution containing 3.3 mg/ml of each compound was spiked into dry wells of daughter plates (80 compounds per plate). For each plate, columns 1 and 12 served as controls: 8 positive controls spiked with DMSO alone provided the reference as 100% growth, and 8 negative controls contained the antifungal drug amphotericin B at 15 μg/ml to kill all cells. A mixture (130 μl) containing 10 volumes of a conidial suspension of 105 conidia/ml (in RPMI with 0.1% Tween 20) and 3 volumes of 0.01% resazurin was added to each well. After 48 h of incubation at 37°C, the absorbances at 570 nm (measurement wavelength) and 604 nm (reference wavelength) were measured on a Safire2 (Tecan) microplate reader. The data were normalized using the following formula: % viability = 100 × (sample value − average value of negative controls)/(average of positive controls − average of negative controls).
For analysis of toxicity to human cells, compounds were added to HeLa cells at a concentration of 10 μM (2.8 μg/ml for sr7575) and the release of cytoplasmic lactate dehydrogenase was measured using the enzyme-linked immunosorbent assay (ELISA)-based cytotoxicity detection kit (Roche) according to the manufacturer's recommendations. Mutagenic activity was tested in the bacterial reverse mutation test, either in the presence or in the absence of a rat metabolizing system (performed by CiToxLAB Safety and Health Research Laboratories). To determine acute mouse toxicity, groups of four NMRI mice were given a single dose of sr7575 (100 mg/kg of body weight) by intraperitoneal (i.p.) injection, and mortality was monitored for 3 days.
Yeast strains, growth, and media.
All strains used in this study are described in Table S4 in the supplemental material. The pooled haploid deletion library (MATa) contained deletions in 4,885 nonessential genes along with DAmP modifications of 977 essential genes (20, 21). Wild-type (WT) S. cerevisiae strain BY4741 was routinely maintained on YPD agar (YPDA; consisting of 1% yeast extract, 2% peptone, 2% dextrose, and 2% Bacto agar).
For S. cerevisiae serial dilution spot assays, fresh colonies from plates were used to inoculate overnight cultures in YPD. The next morning, cultures were washed once and diluted to an optical density at 600 nm (OD600) of 1 in phosphate-buffered saline (PBS), and serial 10-fold dilutions were carried out in a 96-well plate. Ten microliters of each dilution was spotted onto SC medium (containing 6.7 g/liter yeast nitrogen base with ammonium sulfate [BD Difco], with all amino acids, 2% dextrose, and 2% Bacto agar) lacking or supplemented with 0.25 μg/ml sr7575. Plates were incubated at 30°C, and growth was monitored every 24 h over 3 days. Spot assays on SC supplemented with the analog sr7576 were conducted in a similar fashion.
The same serial dilution spot assay used to assess sensitivity in S. cerevisiae was used for C. albicans and C. neoformans, with the exception that the plates were supplemented with sr7575 between 1 and 8 μg/ml and were incubated at 37°C.
Verification of S. cerevisiae mutant strains.
The following mutants were extracted from the gene deletion library maintained in the 96-well format (8): YBR283C (SSH1), YCL045C (EMC1), YDL020C (RPN4), YDL226C (GCS1), YER019C-A (SBH2), YER090W (TRP2), YKL126W (YPK1), YKL207W (EMC3), YML105C (SEC65; DAmP strain), YMR022W (UBC7), YMR264W (CUE1), YOL013C (HRD1), YOR008C (SLG1), YOR153W (PDR5), YPR060C (ARO7), YHR079C (IRE1), YFL031W (HAC1), YBR201W (DER1), YLR207W (HRD3), YIL030C (SSM4/DOA10), YDL190C (UFD2), YGL013C (PDR1), YNL181W (DAmP strain), and YML125C (PGA3; DAmP strain). Genomic DNA was extracted using phenol-chloroform followed by ethanol precipitation. Mutants were verified by PCR amplification using a common forward primer annealing to the KanMX cassette (KaniF) and gene-specific reverse primers (oligonucleotides are listed in Table S5 in the supplemental material).
Complementation tests in S. cerevisiae.
Complementation tests were performed with plasmids from the Molecular Barcoded Yeast ORF collection (MoBY-ORF) (22). URA3 plasmids carrying open reading frame (ORFs) corresponding to genes ARO7, CUE1, EMC1, EMC3, HRD1, RPN4, SSH1, and UBC7 (see Table S4 in the supplemental material) were recovered from Escherichia coli grown in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl, 1.5% Bacto agar) supplemented with chloramphenicol (60 μg/ml) and kanamycin (50 μg/ml) (22). Five hundred nanograms of each plasmid was transformed into the appropriate yeast deletion parent following the lithium acetate protocol (23), and URA3-expressing transformants were selected on SC medium lacking uracil (−URA). The resulting transformants were purified by passaging onto fresh SC (−URA) medium, and four clones of each transformant set were screened by colony PCR using a gene-specific primer pair (see Table S5 in the supplemental material), generating product sizes ranging between 500 and 1,000 bp. The deletion parent was always included as a negative control. Complemented strains were screened in parallel with the parental deletion strains in spot assays.
Overexpression tests in S. cerevisiae.
2μm-based LEU2 plasmids from the systematic overexpression library (24) corresponding to regions of the yeast genome that contain the ORFs PDR1, PDR5, and PDR12 and a control lacking intact genes (see Table S4 in the supplemental material) were recovered from E. coli DH10B cultures grown in LB medium supplemented with kanamycin (50 μg/ml) and transformed into wild-type BY4741, and transformants were selected on SC lacking leucine (−LEU) plates. Transformants were purified by passaging onto fresh SC (−LEU) plates. Overexpressing strains were screened by serial dilution spot assays on SC medium supplemented with increasing concentrations of sr7575.
Chemogenomic profiling.
Concentrations of sr7575 that inhibit WT growth by 10 to 20% in liquid culture were determined using the haploid strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). Single colonies from fresh YPDA plates were inoculated into 10 ml YPD and incubated at 30°C for 14 h. Cultures were diluted to an OD600 of 0.01 and grown to an OD600 of 0.05 prior to the addition of increasing concentrations of sr7575 (0.0625 μg/ml to 0.5 μg/ml). DMSO was used as a vehicle control, but there was no observable difference in growth rate between the no-vehicle and DMSO-treated cultures. Growth was monitored by measuring the OD600 every hour, starting from 0 h until 10 h (see Fig. S2A in the supplemental material).
Pooled 400-ml cultures of the haploid deletion library were grown for 12 generations in the presence of sr7575 at 0.125 μg/ml or DMSO vehicle control. Amplified TAG products from the pooled cultures were hybridized to Agilent barcode-specific microarrays (GEO platform GPL18088) as previously described (14). Images obtained with a GenePix 4200AL scanner were annotated by using GenePix Pro 7 (Molecular Devices, CA, USA). Gpr files were normalized separately for the UP and DOWN barcodes and aggregated for each mutant. Raw and normalized data were deposited in the GEO database (see below). Only results for which data were obtained in two independent biological replicates were further considered for analysis. A total of 4,909 mutants, including both deletion and DAmP-modified strains, showed consistent growth measurements (see Table S6 in the supplemental material), with a Pearson correlation coefficient between the two series of log-transformed values of 0.78.
Gene set enrichment analysis and correlations.
Overrepresentation of gene ontology (GO) terms in the chemogenomic screen results was analyzed using the web interface at http://go.princeton.edu/cgi-bin/GOTermFinder to the GO Term finder program (25). This program identifies enriched GO terms by calculating the frequency with which one expects to encounter a number of genes having the same annotation in a subset of genes (hypergeometric distribution). Correlations with published large-scale data sets were computed using the R project (https://cran.r-project.org/) function “cor.test,” using either “pearson” or “spearman” as comparison methods. Treatments or gene deletion perturbations were ranked in decreasing order of calculated correlation coefficients.
A. fumigatus strains, growth, and media.
WT A. fumigatus strain kuA and deletion mutant derAΔ, hacAΔ, hrdAΔ, and hrdAΔ derAΔ strains were maintained on malt slants (2% malt extract, 2% Bacto agar) while ireAΔ and hacAΔ derAΔ strains were maintained on Aspergillus minimal medium (MM) with 5 mM ammonium tartrate as the nitrogen source and osmotically stabilized with 1.2 M sorbitol (26). G418 was obtained from Invitrogen, and Sigma-Aldrich was the source for ampicillin, kanamycin, and chloramphenicol.
Conidia from ireAΔ and hacAΔ hrdAΔ strains were recovered from Aspergillus MM plus 1.2 M sorbitol slants, while those of the parental strain kuA and the remaining deletion strains were recovered from 10-day-old malt slants in 0.05% Tween. Conidia were diluted to 107 conidia/ml, and serial 10-fold dilutions were carried out in a 96-well plate prior to spotting 10 μl of each dilution onto MOPS (morpholinepropanesulfonic acid)-buffered RPMI 1640, pH 7.0, plates in the presence or absence of 5 μg/ml sr7575. The analog sr7576 precipitated out of solution in RPMI 1640 medium and was therefore not included in the analysis. Plates were incubated at 37°C, and growth was monitored over 4 days.
MIC determination.
Determination of the MIC for yeast strains was carried out by the CLSI M27-A3 broth microdilution method (27). Growth inhibition of Aspergillus strains was monitored using a colorimetric test described earlier (28). The MIC of the A. fumigatus strain that constitutively expresses DsRed fluorescent protein (29) was determined following growth for 24 h at 37°C by measuring fluorescence using a Biotek Synergy fluorescent microplate reader with an excitation wavelength of 254 nm and emission filter set at 291 nm. The relative fluorescence units were plotted against the compound concentrations to determine the MIC.
Measurement of fungistatic or fungicidal activity.
For yeast, freshly growing YPD cultures were diluted to an OD600 of 0.001. sr7575 was added at 0.625 μg/ml, amphotericin B was added at 0.5 μg/ml, and DMSO was used as a vehicle control. One hundred-microliter aliquots were recovered for plating on YPDA. Cultures were grown for 16 h, cells were washed once in 1× PBS, and 100-μl volumes of serially diluted samples were plated. Colonies were counted following 48 h and normalized to the OD600.
For A. fumigatus, 50-ml RPMI cultures with a starting cell number of 1 × 105 conidia/ml were set up in the presence or absence of 5 μg/ml sr7575 (in duplicate). One hundred-microliter aliquots were recovered for enumeration of CFU. Following 16 h of growth, mycelia from one pair of flasks were filtered and mycelial dry weight was estimated. From the second pair, 100 μl from the drug-treated flask was serially diluted and plated to assess viability.
qRT-PCR.
A. fumigatus conidia were inoculated into YG medium (0.5% yeast extract, 2% glucose) and incubated overnight at 37°C, 200 rpm. The mycelium was treated with the indicated concentrations of sr7575 or dithiothreitol (DTT), along with appropriate vehicle controls, for 1 h. The mycelia were harvested by filtration and lysed by crushing in liquid nitrogen. RNA was isolated using the TRIzol reagent, treated with DNase to remove traces of DNA, and reverse transcribed using Moloney murine leukemia virus (M-MuLV) reverse transcriptase (NEB) together with an oligo(dT) primer. Quantitation of bipA and tigA mRNA expression was performed by reverse transcriptase quantitative PCR (qRT-PCR), as previously described (30).
Microarray data accession number.
Raw and normalized data obtained in this study were deposited in the GEO database under identifier GSE60934.
RESULTS
Identification of a new inhibitor of fungal growth.
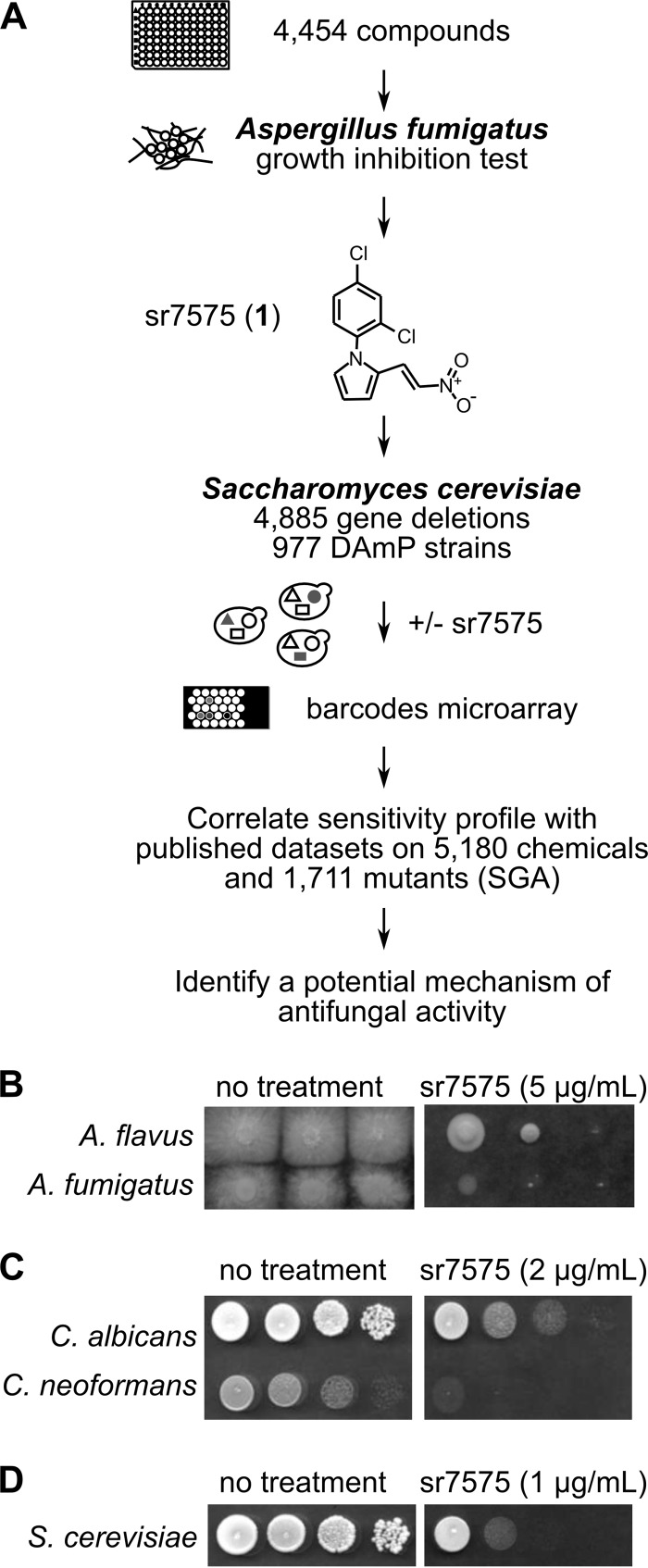
In a search for new antifungals, we tested the toxicity of 4,454 chemicals from the CERMN compound library against A. fumigatus using the strategy outlined in Fig. 1A. The CERMN library is part of the French national collection of chemicals (31) and has been developed since 1998 to be used in the framework of partnerships with public research laboratories. The dynamic range and degree of separation between positive and negative controls in the screen were evaluated by computing the Z' score (32). The average Z' value was 0.92 ± 0.03, indicating a robust and reliable assay. Data analysis identified 76 hits showing greater than 90% fungal growth inhibition, which were clustered into 7 chemical families and 29 singletons (see Table S1 in the supplemental material). Compounds with known effects on human physiology (33) or that showed cytotoxicity for HeLa cells in a lactate dehydrogenase release assay were eliminated from further consideration. The compound sr1810 was active against A. fumigatus and was selected for further analysis. Since sr1810 consisted of a mixture of two isomers, 75% of sr7575 (1) (Fig. 1A) and 25% of sr7576 (2), we synthesized each isomer (see Fig. S1A and B in the supplemental material) and found that it was only sr7575 that was responsible for the antifungal activity. The sr7575 compound showed no mutagenic activity in the bacterial reverse mutation test, and no acute toxicity was observed in mice at a dose of 100 mg/kg.
FIG 1.
Identification of a compound with broad antifungal activity. (A) Selection of a new antifungal, sr7575, through a chemical library screen of A. fumigatus growth inhibition was followed by chemogenomic profiling in S. cerevisiae to identify a potential mechanism of action. (B to D) sr7575 inhibited growth of various fungi, including A. flavus (48 h, 37°C, RPMI medium, 5 μg/ml) (B), C. albicans and C. neoformans (24 h, 37°C, SC medium, 2 μg/ml) (C), and S. cerevisiae (48 h, 30°C, SC medium, 1 μg/ml) (D).
To gain insight into the structural basis for sr7575 antifungal activity, we prepared 30 analogues using aniline derivatives with different substitutions in the first reaction (compounds 3 to 32; see Tables S2 and S3 in the supplemental material and the details of synthesis in the supplemental text). Growth inhibition tests with these compounds showed that at least two features of sr7575 were required for its antifungal potency: the chlorine at position 4 of the phenyl group and the positioning of the nitro group in relation to the pyrrole moiety (see Table S2 in the supplemental material).
In addition to its effects on A. fumigatus, sr7575 was active against Aspergillus flavus (Fig. 1B), C. neoformans, C. albicans (Fig. 1C), and S. cerevisiae (Fig. 1D) on plates and in liquid medium at inhibitory concentrations ranging from 0.6 to 10 μg/ml (see Fig. S2A, B, C, and D in the supplemental material). More than 90% of either S. cerevisiae or A. fumigatus cells were able to resume growth after 16 h of incubation in the presence of sr7575 (0.625 μg/ml and 5 μg/ml, respectively) indicating that the compound exerts a fungistatic effect.
ERAD-deficient mutants of S. cerevisiae are hypersensitive to sr7575.
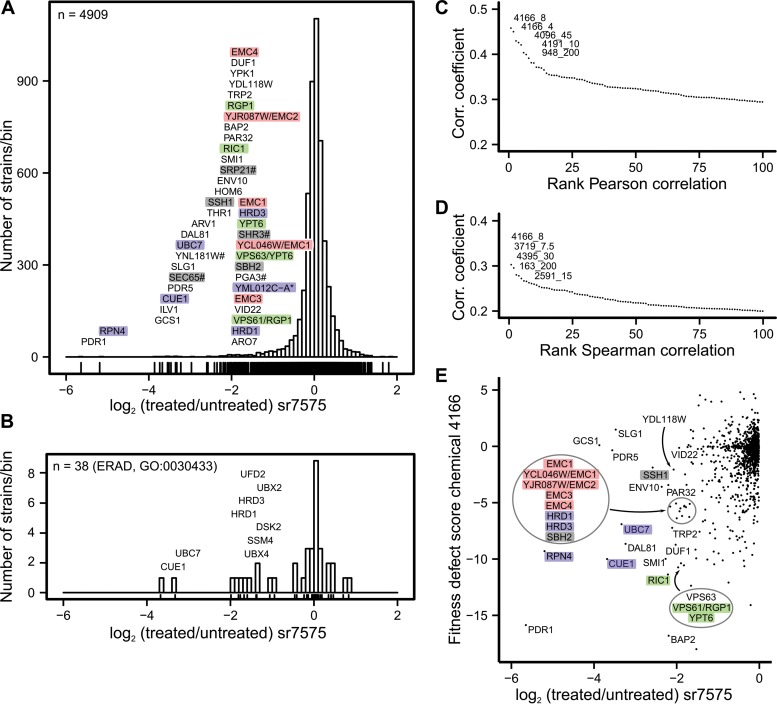
To gain insight into the mechanism by which sr7575 perturbs fungal physiology, the effect of sr7575 was tested on the growth rate of each of 4,885 haploid yeast deletion strains in the systemic deletion collection (20). In addition, sr7575 activity was measured on 977 locus-tagged barcoded DAmP (17) mutants of essential genes that were previously generated in our laboratory (21). This collection of gene knockout and DAmP strains contains molecular barcodes to facilitate detection and quantitation of DNA by custom Agilent microarrays (34, 35). Following the strategy outlined in Fig. 1A, the normalized ratio of the hybridization signal in the presence or absence of treatment was used as an estimate of relative growth rates in pools of mutants. Only a fraction of mutant strains showed hypersensitivity to sr7575, as indicated by the left tail of the distribution for sensitivity values (Fig. 2A). The strain that showed the most dramatic increase in sr7575 sensitivity harbors a deletion of the PDR1 gene, encoding the main regulator of multidrug resistance in yeast (36). Pdr1 is a transcriptional activator for xenobiotic efflux transporter genes, thereby governing resistance to numerous toxic compounds. It is likely that the effect of PDR1 deletion on sensitivity to sr7575 is mediated through the plasma membrane ATP-binding cassette (ABC) transporter Pdr5, since PDR5 is a known target of Pdr1 (37) and the pdr5 mutant was ranked 6th among deletion strains that were most affected by sr7575.
FIG 2.
Chemogenomic profiling reveals an ERAD-enriched signature for sr7575 toxicity. (A) Distribution of relative growth values for S. cerevisiae mutant strains grown in the presence of sr7575. Colors indicate functional categories from the pooled library with genes annotated as ERAD (violet), protein translocation (gray), ER membrane complex (EMC; pink), and vesicular traffic (green) showing the most sensitivity to sr7575 when mutated. Note that YML012C-A* overlaps UBX2; #, DAmP strain. (B) Distribution of sensitivity values for deletion strains affected for genes annotated with the GO term 0030433, ERAD. (C) Pearson correlation coefficients between results obtained with sr7575 and a published large-scale chemogenomics data set identifies chemical 4166 as having a profile that is most similar to sr7575. Only the scores for the top 100 correlated treatments are displayed. (D) Same as panel C, but with the Spearman rank correlation. (E) Comparison of the fitness defect scores between sr7575 and chemical 4166; gene names are color coded as described for panel A.
To identify cellular pathways or protein complexes that allow cells to counteract sr7575 effects, we used a gene set enrichment analysis on 89 mutant strains that showed an average increase in generation time of at least 10% relative to the WT in the presence of sr7575. The most overrepresented pathway in the data set was ER-associated protein degradation (ERAD), specifically the GO term “ER-associated ubiquitin-dependent protein catabolic process” (GO:0030433), with a P value corrected for multiple-hypothesis testing of 1.5 × 10−6. This set included CUE1, UBC7, HRD1, HRD3, UFD2, UBX4, SSM4 (DOA10), DSK2, and UBX2, encompassing one-fifth of the total number of genes annotated to this term (Fig. 2B). The second most overrepresented GO term was “aromatic amino acid family biosynthetic process” (GO:0009073). However, strains deficient in this pathway are known to exhibit a multidrug response signature (9), so the study of the corresponding strains was not pursued further.
Cellular component enrichment analysis was used to determine whether any of the 89 proteins selected in the screen were linked to the same protein complex or intracellular location. The ER membrane protein complex (EMC) was the most overrepresented group by this analysis, with a P value of 2 × 10−7. In addition to gene deletions directly affecting EMC1, EMC3, EMC4, and EMC5, deletions affecting dubious ORFs that overlap EMC2 (YJR087W) and EMC1 (YCL046W), which are independent mutants of these genes, were also present in this data set. Members of the EMC complex are required for efficient protein folding in the ER (38), potentially through roles in phospholipid metabolism at the ER membrane (39). Other components of the ER membrane showed enrichment, including 10 of 58 genes annotated as “intrinsic components of the ER membrane” (GO:0031227) and two DAmP-modified essential genes of uncharacterized function (YNL181W and PGA3). In addition, several genes encoding components of the signal recognition particle (SRP) involved in cotranslational targeting of proteins into the ER showed enrichment: sec65-DAmP, srp21-DAmP, shr3-DAmP, ssh1Δ, and sbh2Δ. Taken together, these findings indicate that sensitivity to the inhibitory effects of sr7575 is exacerbated by defects in the ERAD stress response, as well as by alterations in ER membrane composition that affect optimal ER protein translocation and folding.
The sr7575 sensitivity profile suggests an unfolded protein response-independent stress response.
The effects of sr7575 on haploid yeast deletion strains were compared to profiles obtained from 1,824 different chemicals in a recently published large-scale chemogenomic screen (16). The compound CMB4166 had the highest Spearman correlation coefficient in this comparison (r = 0.44) (Fig. 2C) and showed a profile remarkably similar to that of sr7575 (Fig. 2D). Most of the strains showing sensitivity to sr7575 were also sensitive to CMB4166 (Fig. 2E), suggesting that the two compounds trigger similar cellular responses. However, CMB4166 is a macrolide (D. Hoepfner, personal communication) and shares no structural homology to sr7575.
To acquire insights into the specificity of the response to sr7575, we compared its sensitivity profile to published results on 3,356 other chemical compounds (10). The pattern of sr7575 sensitive mutants revealed little to no similarity to profiles obtained from the other compounds in this comparison. For example, the maximum computed Pearson correlation coefficient was 0.27 for the compound k048-0007 (screen SGTC_352; see Fig. S3A in the supplemental material). However, this correlation was due to strains with deletions in PDR1, RPN4, or GCS1, which confer sensitivity to multiple stresses (see Fig. S3C in the supplemental material). To avoid the typically large effect of outliers on the Pearson correlation, we also tested correlation via the Spearman nonparametric test that uses ranks rather than values. The maximum correlation by this approach was also low (0.18), identifying the compound 4245-1575 used in screen SGTC_513 (see Fig. S3B). Most of the correlation in this case could be attributed to the hypersensitivity of the ubc7Δ and cue1Δ ERAD mutants (see Fig. S3D). Since this second compound was annotated as having an unfolded protein response signature (10), we also tested the correlation between the profile of sr7575 and that of tunicamycin, a well-known and widely used inducer of the UPR (for reviews, see references 40 and 41). However, no similarity was found (see Fig. S3E). Collectively, these comparisons suggest that while many sensitivity profiles are related and indicate the most frequent types of cellular responses to chemical toxicity (10), the profile obtained for sr7575 was specific, with similarity to only one compound of over 5,000 chemicals analyzed.
Large-scale chemical toxicity screens are complementary to synthetic genetic array analyses (SGA), in which double deletion mutant strains are used to determine functional interactions between genes. We compared the sensitivity profile of sr7575 with the results from 1711 SGA screens (42). The closest hit was the profile shown by a strain harboring a DAmP modification of the essential gene PGA3 (17) (see Fig. S4A in the supplemental material). The correlation between the sr7575 and PGA3 profiles was robust, since it also ranked 5th when estimated using the Spearman correlation (see Fig. S4B). Despite the low value of the correlation coefficient (0.23), several strains containing gene deletions were affected by both sr7575 treatment and replacement of PGA3 with the pga3-DAmP allele, including the ERAD-associated genes cue1Δ, ubc7Δ, ufd2Δ, ssm4Δ, and EMC complex components (see Fig. S4C). A role in newly synthesized protein trafficking has been proposed for Pga3 (43), raising the possibility that its presence in our data set is due to a function that impacts ER homeostasis.
The gene deletion that ranked second in terms of correlation with the sr7575 profile involved CHO2, encoding a phosphatidyl N-methyltransferase required for phosphatidylcholine synthesis. The absence of CHO2 renders yeast cells dependent on the UPR for survival (42, 44), suggesting that CHO2 contributes to ER homeostasis. Consistent with this, CHO2 deletion shows an aggravating interaction with the loss of EMC genes in terms of yeast growth (39).
A summary of the correlations between sr7575 sensitivity profiles and those derived from published chemogenomic screens is shown in Fig. 3. Since the numerical values reported for genetic and chemogenomic screens are not readily comparable, the ranking of the different mutants in each screen was used to generate a meaningful graphical display. The resulting heatmap (Fig. 3A) highlights the unique ERAD signature of sr7575 relative to currently published screens. A schematic illustrating the ER membrane proteins involved in the ERAD pathway is shown for perspective (Fig. 3B). Since ERAD is known to work in concert with the UPR to relieve ER stress, it is interesting that hypersensitivity to sr7575 was observed for ERAD mutants but not for the UPR-inactivated ire1Δ and hac1Δ strains. Taken together, these data are consistent with a model in which sr7575 toxicity is counteracted by a functional ERAD machinery, independent of signaling through the UPR pathway.
FIG 3.
Summary of mutations conferring increased susceptibility to sr7575. (A) Heatmap showing the unique ERAD signature of sr7575 compared with published chemogenomic and SGA growth defect profiles. (B) Model showing the two main pathways responsible for ERAD in fungi: the Doa10 pathway (green) for clearing misfolded proteins with cytosolic lesions and the Hrd1 pathway (violet), which degrades misfolded proteins with lumenal or transmembrane lesions. Shared components (Ubx2, Ubc7, Cue1, and the Cdc48 complex) are denoted in gray or black.
Specific ERAD deficiencies enhance sr7575 toxicity in S. cerevisiae.
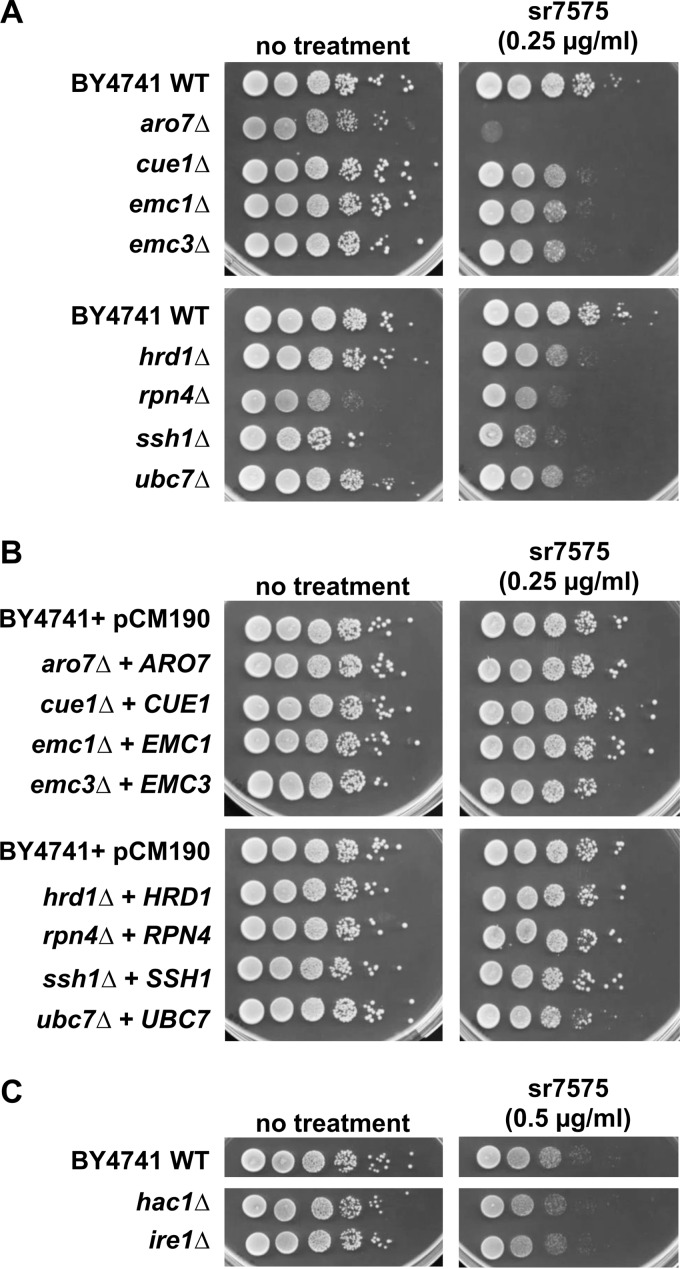
Seventeen S. cerevisiae mutant strains were selected to validate the results of the chemogenomic screen, encompassing strains with deletions in components of the ERAD and proteasome pathways, the EMC, the Ssh1 cotranslocase, the PDR network, aromatic acid biosynthesis, and DAmP modifications of YNL181W and PGA3. Each strain was analyzed individually for sr7575 susceptibility, using a subinhibitory concentration for the WT (Fig. 4A; see also Fig. S5 in the supplemental material). A strain in which the ARO7 gene for aromatic amino acid biosynthesis was deleted showed increased sr7575 sensitivity (Fig. 4A), consistent with the pleiotropic effects of this mutation on stress response.
FIG 4.
Sensitivity to sr7575 depends on EMC and ERAD components. (A) Serial 10-fold dilutions of the WT and selected haploid deletion mutants were grown on SC plates in the absence or presence of 0.25 μg/ml sr7575 for 48 h at 30°C. (B) Complementation of sr7575 sensitivity for the strains shown in panel A was tested by using single-copy plasmids carrying the corresponding genes. (C) Strains lacking core UPR components HAC1 and IRE1 were tested for sensitivity against sr7575 at 0.5 μg/ml.
As predicted by the chemogenomic screen, mutants in the PDR5 multidrug transporter and its transcriptional activator PDR1 were hypersensitive to sr7575 (see Fig. S5 in the supplemental material). Conversely, overexpression of PDR1 and PDR5, but not PDR12, rendered S. cerevisiae cells tolerant to high concentrations of sr7575 (see Fig. S6A). This phenotype was conserved across fungal species, since clinical isolates and laboratory C. albicans strains that overexpress CDR1, the ortholog of S. cerevisiae PDR5, were also tolerant to sr7575 (see Fig. S6B).
Deletions of genes coding for EMC members, EMC1 and EMC3, and the cotranslational translocase gene SSH1 conferred increased sensitivity to sr7575, as suggested by the chemogenomic screen. However, a mutant in SBH2, which functions in the Ssh1 translocase complex (45), did not show increased sensitivity, at least at this concentration (see Fig. S5 in the supplemental material). Hypersensitivity to sr7575 was confirmed for components of the ERAD complex, including the Hrd1 E3 ubiquitin ligase, the E2 ubiquitin conjugating enzyme Ubc7, and the ER membrane resident recruiter Cue1 (46–48) (Fig. 4A). Although the ERAD component Der1 (49) was not identified in our chemogenomic screen, a mild increase in sr7575 sensitivity was observed for this mutant (see Fig. S5), consistent with ERAD involvement in sr7575 effects. sr7575 hypersensitivity was also validated for a strain lacking RPN4, which encodes a transcriptional activator of proteasome genes (Fig. 4A). Since proteasomal degradation is the final step in the disposal of misfolded proteins by ERAD, this finding is consistent with the notion that sr7575 affects protein quality control in the ER. In conclusion, these findings demonstrate that components of the ERAD pathway are necessary to protect yeast cells from the toxic effects of sr7575 in S. cerevisiae, suggesting a mechanism of action that involves perturbation of ER protein quality control.
ERAD protects against sr7575 toxicity in A. fumigatus but is UPR independent.
The UPR is a stress response pathway that communicates information on ER homeostasis to the nucleus (40, 41). The pathway is triggered by misfolded proteins, which accumulate in the ER when the demand for secretion exceeds ER folding capacity or when the cell encounters adverse environmental conditions. Unfolded proteins are sensed by the ER transmembrane sensor Ire1, which triggers the synthesis of Hac1, a transcription factor. Hac1 translocates to the nucleus and upregulates the expression of chaperones, folding enzymes, and other proteins that support ER function (44, 50). Since ERAD mutants are hypersensitive to sr7575 and ERAD capacity can be regulated by the UPR, we were surprised to find that neither HAC1 nor IRE1 was identified in the sr7575 chemogenomic screen. The UPR independence of this response was confirmed by susceptibility testing: yeast ire1Δ and hac1Δ mutants were not affected by sr7575 at concentrations of up to 0.5 μg/ml (Fig. 4C). These findings suggest that ERAD protects against sr7575 toxicity through a mechanism that is independent of the UPR in S. cerevisiae.
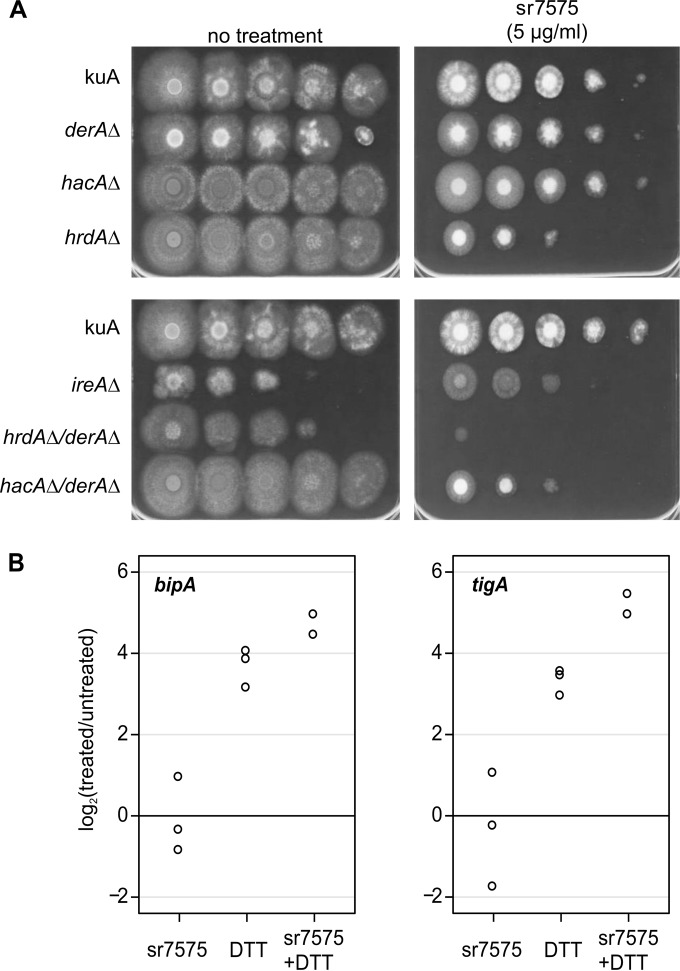
Consistent with the results obtained in S. cerevisiae, UPR mutants of A. fumigatus that lack either the ER sensor IreA or the transcription factor HacA showed no hypersensitivity to sr7575 (Fig. 5A). As in yeast, the hrdAΔ mutant, which lacks the ortholog of S. cerevisiae HRD1, showed increased sensitivity to sr7575. A derAΔ mutant that is deficient in the DerA component of the HrdA ERAD complex showed no increase in sr7575 sensitivity. However, a double deletion mutant lacking both DerA and HrdA showed greater sensitivity to sr7575 than a mutant lacking HrdA alone, underscoring the importance of the Hrd1 complex in the response to sr7575 toxicity. We conclude that sr7575 action involves the inhibition of an evolutionarily conserved target, necessitating the intervention of the ERAD complex Hrd1/HrdA in both S. cerevisiae and A. fumigatus.
FIG 5.
The hypersensitivity of ERAD mutants to sr7575 is conserved in A. fumigatus. (A) Conidia from A. fumigatus WT and deletion mutants were recovered in 0.05% Tween–water, and serial dilutions were spotted onto sr7575-containing RPMI 1640, pH 7.0. Plates were incubated at 37°C for 72 h. (B) Analysis of UPR target gene expression (bipA and tigA) by qRT-PCR. Cultures were treated with sr7575, DTT, or sr7575 for 1 h followed by DTT. RNA was extracted and analyzed by qRT-PCR, using tubA mRNA for normalization. The results of treated versus untreated samples from three independent experiments are shown.
The ERAD-enriched signature for sr7575 suggested that some aspect of ER protein quality control is adversely affected by this compound. However, since UPR-deficient strains of S. cerevisiae or A. fumigatus showed no increase in sr7575 sensitivity, the results suggest that a UPR-independent mechanism of ERAD activity is involved in the sr7575 response. To confirm UPR independence, qRT-PCR was used to measure mRNA levels for two well-known UPR target genes: the ER chaperone bipA and the protein disulfide isomerase tigA. As expected, the expression of both genes was strongly induced by treatment with a subinhibitory concentration of dithiothreitol (1 mM), a well-known inducer of the UPR (Fig. 5B). In contrast, no increase in expression was observed following treatment with a subinhibitory concentration of sr7575 (0.1 μg/ml). In addition, pretreatment with sr7575 for 1 h prior to DTT exposure failed to block UPR activation. These results indicate that while exposure to sr7575 does not trigger the UPR, it was also unable to prevent UPR activation by DTT. We conclude that sr7575 is unlikely to target UPR signaling for its toxic effects in A. fumigatus or S. cerevisiae, consistent with the UPR-independent response suggested by the chemogenomic screen.
DISCUSSION
In this study, we describe the identification of a novel antifungal compound, sr7575, that was active against species from four fungal genera. Chemogenomic profiling in S. cerevisiae demonstrated that the set of genes required for protection against sr7575 was markedly narrow, involving components of the ERAD stress response and other components of the ER membrane. The function of the ERAD pathway is to maintain protein quality control in the ER by eliminating toxic unfolded proteins that may accumulate in the fungus during periods of high secretory activity or when the organism encounters adverse environmental conditions. This disposal mechanism centers on a multiprotein complex in the ER membrane that selectively identifies misfolded proteins in the ER lumen or membrane and transports them back into the cytoplasm for degradation by the proteasome. The results from our chemogenomic screen demonstrate that mutants of this complex, either in S. cerevisiae or A. fumigatus, are hypersensitive to sr7575 inhibition, suggesting that the antifungal effects of this compound involves a disruption of ER protein quality control.
ER protein quality control is also affected by the UPR, a signaling pathway that counters the accumulation of unfolded proteins in the ER by increasing the expression of chaperones and other proteins involved in protein folding when the demand for secretion exceeds the folding capacity of the organelle. A tight coordination between the UPR and ERAD pathways was demonstrated in yeast, where UPR mutants have decreased ERAD activity whereas ERAD mutants exhibit constitutive UPR upregulation (50). In addition, although ERAD is sufficient to eliminate misfolded proteins that continually arise during normal growth, it requires the UPR for optimal degradative capacity under conditions of severe ER stress (51). Basal ERAD activity is thus sufficient to handle low levels of unfolded proteins and is UPR independent. However, upregulation of ERAD activity by the UPR is needed when the level of unfolded proteins reaches a critical threshold of toxicity. The connection between these pathways is also evident in A. fumigatus, in which mutants deficient in both the UPR and ERAD are less fit than those lacking the UPR or ERAD alone (52).
In view of the link between the UPR and ERAD pathways, we were surprised to find that neither ire1Δ nor hac1Δ was among the strains most affected by sr7575 in our chemogenomic screen and that these strains showed no increase in sr7575 sensitivity when tested individually. In addition, our experiments revealed that sr7575 did not trigger the UPR, nor did it prevent the UPR from being activated by DTT, a strong inducer of unfolded proteins. These observations suggest that sr7575 does not cause the widespread protein unfolding that is typical of strong ER stress aggravators such as DTT and tunicamycin. Specific ER stress can be induced, for example, by expressing topologically abnormal ERAD-targeted integral membrane proteins without inducing the canonical UPR pathway in yeast (53).
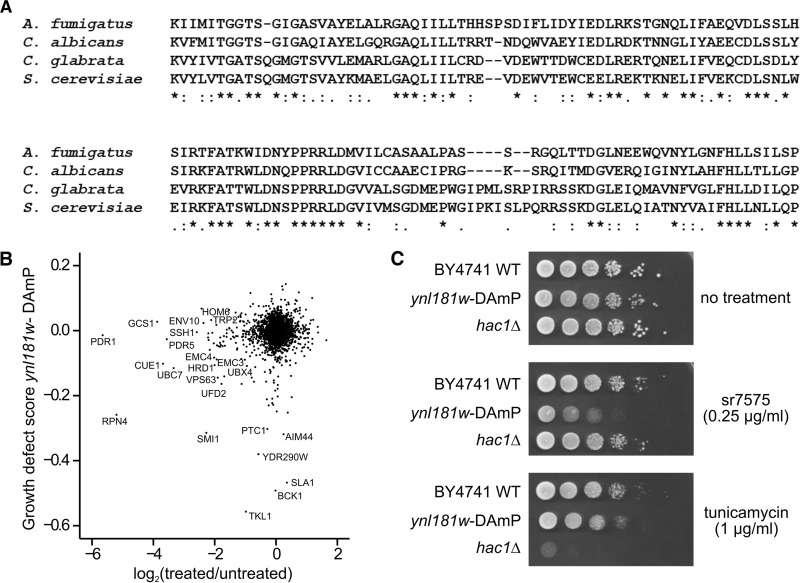
The ability of sr7575 to inhibit the growth of fungi but not human cells raises the possibility that it targets a fungus-specific process. Our chemogenomic screen identified ynl181w-DAmP as one of the top 10 strains most affected by sr7575 toxicity. YNL181W encodes an essential ER membrane protein, the function of which is currently unknown but is speculated to involve an oxidoreductase activity (54, 55). The Ynl181w protein is conserved among fungi (Fig. 6A) and has no metazoan ortholog, as defined in the OrthoMCL database (56). Since the protein is essential, a heterozygous YNL181W/ynl181wΔ deletion strain was previously used to study its function in chemogenomic investigations (10, 16). We were especially interested in the effects of chemical CMB4166 in these studies because our data revealed that the strain sensitivity profile for that compound (16) most closely resembled that of sr7575 (Fig. 2C, D, and E). A striking finding from this comparison was that the heterozygous deletion strain of YNL181W was among the strains most affected by CMB4166 (Fig. 3A), indicating that the absence of Ynl181w sensitizes yeast to both sr7575 and CMB4166.
FIG 6.
Ynl181w is an ER protein conserved in fungi and involved in adaptation to sr7575. (A) T-Coffee alignment of the conserved short-chain dehydrogenase region within Sc Ynl181w (PFAM 54-187) and its orthologs in pathogenic fungi. Gene annotations with a number range indicate the position of the PFAM domain: A. fumigatus (Afu5g10790; positions 54 to 205), C. albicans (orf19.6233; positions 58 to 204), C. glabrata (XP_448202; positions 54 to 193). (B) Scatter plot showing the correlation between sr7575 sensitivity values and the previously published SGA scores for ynl181w-DAmP. (C) Spot assays showing the difference in sensitivity to sr7575 and UPR inducer tunicamycin (TM) for ynl181w-DAmP compared with a strain defective for UPR (hac1Δ).
A DAmP modification of YNL181W has been previously combined with a large-scale genetic screen to identify mutations that synergize with loss of YNL181W function (42). A comparison of the sr7575 sensitivity profile with the results of this genetic screen identified several ERAD mutants that were affected both by DAmP modification of YNL181W and by treatment with sr7575, including UBC7, CUE1, and RPN4 (Fig. 6B). The connection to UBC7 was particularly remarkable because a similar synergistic growth defect associated with ynl181w-DAmP and the ubc7Δ mutation was observed in two other large-scale studies (17, 57). The correlation between the Ynl181w mutation and sr7575 sensitivity obtained by chemogenomics was confirmed on plates by showing that a ynl181w-DAmP mutant was hypersensitive to sr7575 (Fig. 6C). We speculate that Ynl181w could be involved in processes that are targeted by sr7575.
In conclusion, we report the identification of a novel compound that has activity against both S. cerevisiae and A. fumigatus. The data are consistent with a model for s7575 action in which the compound disrupts the structure of one or more proteins in the ER lumen or membrane, resulting in a situation that necessitates ERAD intervention to eliminate the abnormal protein(s) but does not require UPR activation. These findings underscore the importance of ER homeostasis to the growth fungi and suggest the presence of fungus-specific ER processes that could represent new opportunities for antifungal intervention.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michel Boulouard (Groupe Mémoire et Plasticité Comportementale, GMPc, EA4259, Caen) for the in vivo toxicity tests on mice, Dominique Sanglard (CHUV Lausanne, Switzerland) for the C. albicans strains, Françoise Dromer (CNRMA, Institut Pasteur) for the A. fumigatus clinical isolates, and Alain Jacquier (Unité de Génétique des Interactions Macromoléculaires, Institut Pasteur) for support and critical reading of the manuscript.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02239-15.
REFERENCES
- 1.Brown GD, Denning DW, Levitz SM. 2012. Tackling human fungal infections. Science 336:647. doi: 10.1126/science.1222236. [DOI] [PubMed] [Google Scholar]
- 2.Lai C-C, Tan C-K, Huang Y-T, Shao P-L, Hsueh P-R. 2008. Current challenges in the management of invasive fungal infections. J Infect Chemother 14:77–85. doi: 10.1007/s10156-007-0595-7. [DOI] [PubMed] [Google Scholar]
- 3.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 4.Odds FC, Brown AJP, Gow NAR. 2003. Antifungal agents: mechanisms of action. Trends Microbiol 11:272–279. doi: 10.1016/S0966-842X(03)00117-3. [DOI] [PubMed] [Google Scholar]
- 5.Perlin DS. 2011. Current perspectives on echinocandin class drugs. Future Microbiol 6:441–457. doi: 10.2217/fmb.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hope WW, Tabernero L, Denning DW, Anderson MJ. 2004. Molecular mechanisms of primary resistance to flucytosine in Candida albicans. Antimicrob Agents Chemother 48:4377–4386. doi: 10.1128/AAC.48.11.4377-4386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schenone M, Dančík V, Wagner BK, Clemons PA. 2013. Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol 9:232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giaever G, Nislow C. 2014. The yeast deletion collection: a decade of functional genomics. Genetics 197:451–465. doi: 10.1534/genetics.114.161620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, Giaever G. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee AY, St Onge RP, Proctor MJ, Wallace IM, Nile AH, Spagnuolo PA, Jitkova Y, Gronda M, Wu Y, Kim MK, Cheung-Ong K, Torres NP, Spear ED, Han MKL, Schlecht U, Suresh S, Duby G, Heisler LE, Surendra A, Fung E, Urbanus ML, Gebbia M, Lissina E, Miranda M, Chiang JH, Aparicio AM, Zeghouf M, Davis RW, Cherfils J, Boutry M, Kaiser CA, Cummins CL, Trimble WS, Brown GW, Schimmer AD, Bankaitis VA, Nislow C, Bader GD, Giaever G. 2014. Mapping the cellular response to small molecules using chemogenomic fitness signatures. Science 344:208–211. doi: 10.1126/science.1250217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho C-H, Wang J, Ketela T, Brenner C, Brill JA, Fernandez GE, Lorenz TC, Payne GS, Ishihara S, Ohya Y, Andrews B, Hughes TR, Frey BJ, Graham TR, Andersen RJ, Boone C. 2006. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 12.Springer M, Weissman JS, Kirschner MW. 2010. A general lack of compensation for gene dosage in yeast. Mol Syst Biol 6:368. doi: 10.1038/msb.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hillenmeyer ME, Ericson E, Davis RW, Nislow C, Koller D, Giaever G. 2010. Systematic analysis of genome-wide fitness data in yeast reveals novel gene function and drug action. Genome Biol 11:R30. doi: 10.1186/gb-2010-11-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decourty L, Saveanu C, Zemam K, Hantraye F, Frachon E, Rousselle J-C, Fromont-Racine M, Jacquier A. 2008. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc Natl Acad Sci U S A 105:5821–5826. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong AHY, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Ménard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu A-M, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. 2004. Global mapping of the yeast genetic interaction network. Science 303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 16.Hoepfner D, Helliwell SB, Sadlish H, Schuierer S, Filipuzzi I, Brachat S, Bhullar B, Plikat U, Abraham Y, Altorfer M, Aust T, Baeriswyl L, Cerino R, Chang L, Estoppey D, Eichenberger J, Frederiksen M, Hartmann N, Hohendahl A, Knapp B, Krastel P, Melin N, Nigsch F, Oakeley EJ, Petitjean V, Petersen F, Riedl R, Schmitt EK, Staedtler F, Studer C, Tallarico JA, Wetzel S, Fishman MC, Porter JA, Movva NR. 2014. High-resolution chemical dissection of a model eukaryote reveals targets, pathways and gene functions. Microbiol Res 169:107–120. doi: 10.1016/j.micres.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, Krogan NJ. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 18.Berry DB, Guan Q, Hose J, Haroon S, Gebbia M, Heisler LE, Nislow C, Giaever G, Gasch AP. 2011. Multiple means to the same end: the genetic basis of acquired stress resistance in yeast. PLoS Genet 7:e1002353. doi: 10.1371/journal.pgen.1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thibault G, Ng DTW. 2012. The endoplasmic reticulum-associated degradation pathways of budding yeast. Cold Spring Harb Perspect Biol 4(12):pii:a013193. doi: 10.1101/cshperspect.a013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian K-D, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Güldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kötter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang C, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 21.Decourty L, Doyen A, Malabat C, Frachon E, Rispal D, Séraphin B, Feuerbach F, Jacquier A, Saveanu C. 2014. Long open reading frame transcripts escape nonsense-mediated mRNA decay in yeast. Cell Rep 6:593–598. doi: 10.1016/j.celrep.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 22.Ho CH, Magtanong L, Barker SL, Gresham D, Nishimura S, Natarajan P, Koh JLY, Porter J, Gray CA, Andersen RJ, Giaever G, Nislow C, Andrews B, Botstein D, Graham TR, Yoshida M, Boone C. 2009. A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat Biotechnol 27:369–377. doi: 10.1038/nbt.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiestl RH, Gietz RD. 1989. High efficiency transformation of intact yeast cells using single-stranded nucleic acids as a carrier. Curr Genet 16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 24.Jones GM, Stalker J, Humphray S, West A, Cox T, Rogers J, Dunham I, Prelich G. 2008. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat Methods 5:239–241. doi: 10.1038/nmeth.1181. [DOI] [PubMed] [Google Scholar]
- 25.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. 2004. GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richie DL, Hartl L, Aimanianda V, Winters MS, Fuller KK, Miley MD, White S, McCarthy JW, Latgé J-P, Feldmesser M, Rhodes JC, Askew DS. 2009. A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus. PLoS Pathog 5:e1000258. doi: 10.1371/journal.ppat.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard, 3rd ed CLSI document M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 28.Clavaud C, Beauvais A, Barbin L, Munier-Lehmann H, Latgé J-P. 2012. The composition of the culture medium influences the β-1,3-glucan metabolism of Aspergillus fumigatus and the antifungal activity of inhibitors of β-1,3-glucan synthesis. Antimicrob Agents Chemother 56:3428–3431. doi: 10.1128/AAC.05661-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jhingran A, Mar KB, Kumasaka DK, Knoblaugh SE, Ngo LY, Segal BH, Iwakura Y, Lowell CA, Hamerman JA, Lin X, Hohl TM. 2012. Tracing conidial fate and measuring host cell antifungal activity using a reporter of microbial viability in the lung. Cell Rep 2:1762–1773. doi: 10.1016/j.celrep.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng X, Krishnan K, Richie DL, Aimanianda V, Hartl L, Grahl N, Powers-Fletcher MV, Zhang M, Fuller KK, Nierman WC, Lu LJ, Latgé J-P, Woollett L, Newman SL, Cramer RA, Rhodes JC, Askew DS. 2011. HacA-independent functions of the ER stress sensor IreA synergize with the canonical UPR to influence virulence traits in Aspergillus fumigatus. PLoS Pathog 7(10):e1002330. doi: 10.1371/journal.ppat.1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hibert MF. 2009. French/European academic compound library initiative. Drug Discov Today 14:723–725. doi: 10.1016/j.drudis.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J-H, Chung TDY, Oldenburg KR. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 33.Prunier H, Rault S, Lancelot JC, Robba M, Renard P, Delagrange P, Pfeiffer B, Caignard DH, Misslin R, Guardiola-Lemaitre B, Hamon M. 1997. Novel and selective partial agonists of 5-HT3 receptors. 2. Synthesis and biological evaluation of piperazinopyridopyrrolopyrazines, piperazinopyrroloquinoxalines, and piperazinopyridopyrroloquinoxalines. J Med Chem 40:1808–1819. [DOI] [PubMed] [Google Scholar]
- 34.Smith AM, Heisler LE, Mellor J, Kaper F, Thompson MJ, Chee M, Roth FP, Giaever G, Nislow C. 2009. Quantitative phenotyping via deep barcode sequencing. Genome Res 19:1836–1842. doi: 10.1101/gr.093955.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eason RG, Pourmand N, Tongprasit W, Herman ZS, Anthony K, Jejelowo O, Davis RW, Stolc V. 2004. Characterization of synthetic DNA bar codes in Saccharomyces cerevisiae gene-deletion strains. Proc Natl Acad Sci U S A 101:11046–11051. doi: 10.1073/pnas.0403672101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, Keniya MV, Tanabe K, Niimi M, Goffeau A, Monk BC. 2009. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev 22:291–321. doi: 10.1128/CMR.00051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyers S, Schauer W, Balzi E, Wagner M, Goffeau A, Golin J. 1992. Interaction of the yeast pleiotropic drug resistance genes PDR1 and PDR5. Curr Genet 21:431–436. doi: 10.1007/BF00351651. [DOI] [PubMed] [Google Scholar]
- 38.Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, Schuldiner M. 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lahiri S, Chao JT, Tavassoli S, Wong AKO, Choudhary V, Young BP, Loewen CJR, Prinz WA. 2014. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol 12:e1001969. doi: 10.1371/journal.pbio.1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hampton RY. 2000. ER stress response: getting the UPR hand on misfolded proteins. Curr Biol 10:R518–R521. doi: 10.1016/S0960-9822(00)00583-2. [DOI] [PubMed] [Google Scholar]
- 41.Schröder M. 2008. Endoplasmic reticulum stress responses. Cell Mol Life Sci 65:862–894. doi: 10.1007/s00018-007-7383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin Z-Y, Liang W, Marback M, Paw J, San Luis B-J, Shuteriqi E, Tong AHY, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pál C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras A-C, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C. 2010. The genetic landscape of a cell. Science 327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu L, Peña Castillo L, Mnaimneh S, Hughes TR, Brown GW. 2006. A survey of essential gene function in the yeast cell division cycle. Mol Biol Cell 17:4736–4747. doi: 10.1091/mbc.E06-04-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thibault G, Shui G, Kim W, McAlister GC, Ismail N, Gygi SP, Wenk MR, Ng DTW. 2012. The membrane stress response buffers lethal effects of lipid disequilibrium by reprogramming the protein homeostasis network. Mol Cell 48:16–27. doi: 10.1016/j.molcel.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finke K, Plath K, Panzner S, Prehn S, Rapoport TA, Hartmann E, Sommer T. 1996. A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae. EMBO J 15:1482–1494. [PMC free article] [PubMed] [Google Scholar]
- 46.Biederer T, Volkwein C, Sommer T. 1997. Role of Cue1p in ubiquitination and degradation at the ER surface. Science 278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 47.Bordallo J, Plemper RK, Finger A, Wolf DH. 1998. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell 9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hampton RY, Gardner RG, Rine J. 1996. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell 7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mehnert M, Sommer T, Jarosch E. 2014. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat Cell Biol 16:77–86. doi: 10.1038/ncb2882. [DOI] [PubMed] [Google Scholar]
- 50.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249–258. doi: 10.1016/S0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 51.Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. 2000. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol 2:379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- 52.Richie DL, Feng X, Hartl L, Aimanianda V, Krishnan K, Powers-Fletcher MV, Watson DS, Galande AK, White SM, Willett T, Latgé J-P, Rhodes JC, Askew DS. 2011. The virulence of the opportunistic fungal pathogen Aspergillus fumigatus requires cooperation between the endoplasmic reticulum-associated degradation pathway (ERAD) and the unfolded protein response (UPR). Virulence 2:12–21. doi: 10.4161/viru.2.1.13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buck TM, Jordan R, Lyons-Weiler J, Adelman JL, Needham PG, Kleyman TR, Brodsky JL. 2015. Expression of three topologically distinct membrane proteins elicits unique stress response pathways in the yeast Saccharomyces cerevisiae. Physiol Genomics 47:198–214. doi: 10.1152/physiolgenomics.00101.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hazbun TR, Malmström L, Anderson S, Graczyk BJ, Fox B, Riffle M, Sundin BA, Aranda JD, McDonald WH, Chiu C-H, Snydsman BE, Bradley P, Muller EGD, Fields S, Baker D, Yates JR, Davis TN. 2003. Assigning function to yeast proteins by integration of technologies. Mol Cell 12:1353–1365. doi: 10.1016/S1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- 55.Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 56.Chen F, Mackey AJ, Stoeckert CJ, Roos DS. 2006. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res 34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. 2011. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol 195:323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.