Abstract
SXT/R391 integrative and conjugative elements (ICEs) were detected in 8 out of 125 Proteus mirabilis isolates from food-producing animals in China. Whole-genome sequencing revealed that seven ICEs were identical to ICEPmiJpn1, carrying the cephalosporinase gene blaCMY-2. Another one, designated ICEPmiChn1, carried five resistance genes. All eight ICEs could be transferred to Escherichia coli via conjugation. The results highlight the idea that animal farms are important reservoir of the SXT/R391 ICE-containing P. mirabilis.
TEXT
Integrative and conjugative elements (ICEs) are self-transmissible chromosomal mobile genetic elements (MGEs) that can bestow new traits on bacteria, such as increased virulence, biofilm formation, and resistance to antimicrobials and heavy metals (1), which have an important impact on bacterial evolution and can mediate the dissemination of antimicrobial resistance (1–4). SXT/R391 ICEs are chromosomal MGEs sharing a conserved integrase that could mediate site-specific integration into the 5′ end of prfC (5). This family of ICEs contains 52 nearly identical core genes, many of which are involved in integration/excision, conjugative transfer, and regulation of the ICEs (6, 7). In addition, five hot spots (HS1 to -5) and four variable regions (VRI to -IV) have been identified which contain variable genes conferring element-specific properties and providing their hosts with fitness functions (6, 8–10).
SXT/R391 ICEs have been found in several genera of Gammaproteobacteria (http://db-mml.sjtu.edu.cn/ICEberg/) (11), such as Vibrio, Providencia, Proteus, and some marine bacteria (3, 6, 9, 10). Proteus mirabilis, a member of the family Enterobacteriaceae, has become one of the most important opportunistic pathogens in nosocomial infections in China (12). Four SXT/R391 ICEs, R997 (India), ICEPmiUSA1 (United States), ICEPmiJpn1 (Japan and Spain), and ICEPmiSpn1 (Spain), have been detected in P. mirabilis (3, 13–16). It is worth noting that the cephalosporinase gene blaCMY-2 is inserted into ICEPmiJpn1 and ICEPmiSpn1 via IS10-mediated transposition (15, 16), indicating that SXT/R391 ICEs are the important mobile platforms to capture new resistance genes. However, data on the SXT/R391 ICEs in P. mirabilis isolates from China are still lacking.
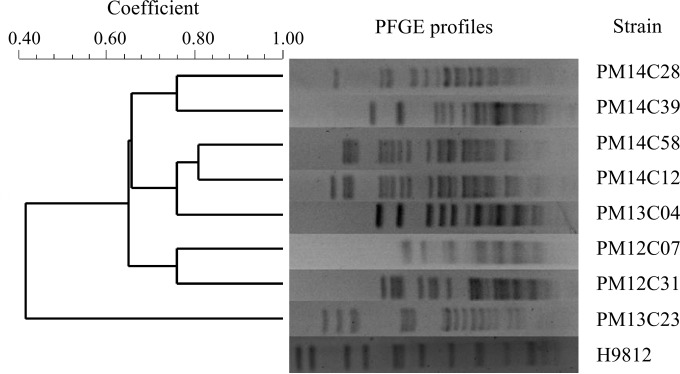
In this study, a total of 125 P. mirabilis strains (64 from chicken and 61 from swine), which were isolated from the intestinal tracts of animals among 11 poultry and 35 swine farms in 16 provinces in China between March 2012 and December 2014, were screened for the presence of the integrase gene int of SXT/R391 ICEs and blaCMY-2 by PCR with primers listed in Table S1 in the supplemental material (9, 15, 17, 18). SXT/R391 ICEs and blaCMY-2 were detected in eight and 16 strains, respectively. The eight strains carrying SXT/R391 ICEs displayed multidrug resistance profiles, as determined by the disk diffusion method according to the CLSI guidelines (19) (Table 1), and seven of these also contained blaCMY-2. The clonal relationships of the ICE-harboring strains were determined by pulsed-field gel electrophoresis (PFGE) analysis with genomic DNA digested by SmaI (Fig. 1). Two strains, Pm14C12 and Pm14C58, from different chicken farms in Sichuan Province had similar PFGE profiles, indicating the presence of clonal spread of ICE-harboring strains. The origin and antimicrobial resistance profiles of the eight ICE-harboring strains are listed in Table 1. To the best of our knowledge, this is the first report of SXT/R391 ICEs in P. mirabilis from China.
TABLE 1.
Origin, antimicrobial resistance profiles, and transconjugative frequencies of SXT/R391 ICEs of the eight P. mirabilis strains
| Strain | Province of isolation | Date of isolation | Host | Antimicrobial resistance profilea | ICE type | Transconjugative frequency |
|---|---|---|---|---|---|---|
| PM12C07 | Shandong | 29 October 2012 | Chicken | AMX, AMC, FOX, CTX, NAL, NOR, CIP, DOX, SXT | ICEPmiJpn1 | 5.5 × 10−5 |
| PM12C31 | Chongqing | 3 November 2012 | Swine | AMX, AMC, FOX, CTX, DOX, STR, SPT, SXT | ICEPmiJpn1 | 2.3 × 10−5 |
| PM13C04 | Hubei | 16 April 2013 | Chicken | AMX, NAL, NOR, DOX, STR, SPT, SXT, FFC | ICEPmiChn1 | 2.8 × 10−6 |
| PM13C23 | Henan | 27 November 2013 | Chicken | AMX, AMC, FOX, CTX, NAL, NOR, CIP, DOX, SXT | ICEPmiJpn1 | 1.5 × 10−4 |
| PM14C12 | Sichuan | 5 March 2014 | Chicken | AMX, AMC, FOX, CTX, NAL, NOR, DOX, STR, SPT, SXT | ICEPmiJpn1 | 2.5 × 10−5 |
| PM14C28 | Heibei | 22 April 2014 | Chicken | AMX, AMC, FOX, CTX, NAL, NOR, CIP, DOX, SXT, FFC | ICEPmiJpn1 | 3.2 × 10−5 |
| PM14C39 | Sichuan | 29 April 2014 | Swine | AMX, AMC, FOX, CTX, NAL, NOR, DOX, SXT, FFC | ICEPmiJpn1 | 1.9 × 10−7 |
| PM14C58 | Sichuan | 12 December 2014 | Chicken | AMX, AMC, FOX, CTX, NAL, NOR, DOX, STR, SPT, SXT | ICEPmiJpn1 | 3.6 × 10−6 |
AMX, amoxicillin; AMC, amoxicillin-clavulanate; FOX, cefoxitin; CTX, cefotaxime; NAL, nalidixic acid; NOR, norfloxacin; CIP, ciprofloxacin; DOX, doxycycline; STR, streptomycin; SPT, spectinomycin; SXT, trimethoprim-sulfamethoxazole; FFC, florfenicol. Resistance to the antimicrobials shown in bold is conferred by genes present on SXT/R391 ICE.
FIG 1.
PFGE analysis of the eight P. mirabilis strains carrying SXT/R391 ICEs. PFGE analysis with SmaI digestion was carried out on CHEF Mapper under the following conditions: Auto Algorithm; 30 kb, low MW; 400 kb, high MW; 16.5 h. Salmonella enterica serovar Braenderup strain H9812 was used as a reference.
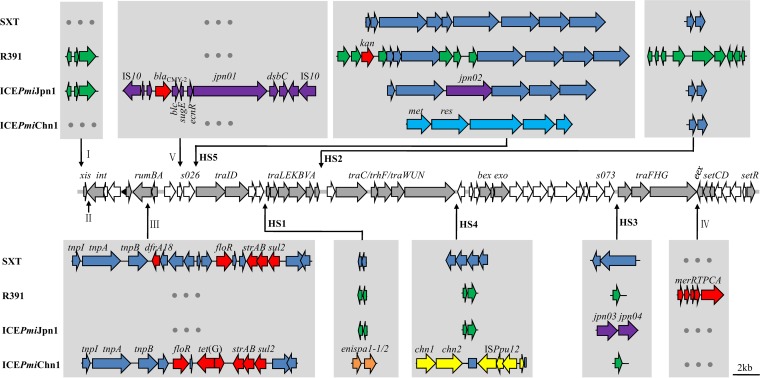
Whole-genome sequencing was performed on the Illumina MiSeq (Majorbio, Shanghai) using a 400-bp paired-end library with ∼200-fold average coverage. The paired-end reads were assembled de novo using the SOAPdenovo v2.04 and GapCloser v1.12. The gaps among contigs were filled in by PCR linkage. The complete nucleotide sequences of SXT/R391 ICEs were analyzed by using the BLAST programs (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequence analysis showed that two different SXT/R391 ICEs were present in the eight strains. Seven R391-like ICEs (91,091 bp) were identical to the ICEPmiJpn1, which carried the blaCMY-2 gene and was previously reported in Japan and Spain (15, 16). Given that the IS10-mediated transposition region carrying the blaCMY-2 gene was inserted into S025 in ICEPmiJpn1 and ICEPmiSpn1, we renamed this region as variable region V (Fig. 2). The nucleotide sequences of ICEPmiJpn1 reported in this study were almost 100% identical to the corresponding incomplete nucleotide sequences of ICEPmiJpn1 in Japan (15), with only a 2-bp difference in HS3. Our data showed that 43.75% (7 of 16) blaCMY-2 genes in P. mirabilis were carried by ICEPmiJpn1. Mata et al. also reported that 36.84% (7 of 19) blaCMY-2 genes in P. mirabilis clinical isolates from Spain were associated with SXT/R391 ICEs (16). The results indicated that SXT/R391 ICEs are the important MGEs for the dissemination of blaCMY-2 genes in P. mirabilis.
FIG 2.
Schematic view of the two SXT/R391 ICEs, ICEPmiJpn1 and ICEPmiChn1, and comparison with SXT and R391. The line of arrows in the middle represents the 52 conserved core genes of the SXT/R391 family of ICEs. HS1 to HS5 represent hot spots 1 to 5, and I to V represent the variable regions I to V. Variable genes from originally described ICEs are shown with different colors: SXT (21), blue; R391 (22), green; ICEPmiJpn1, purple; ICEPmiChn1, yellow; ICEValSpaI (10), light blue; ICEEniSpaI (9), orange. Resistance genes are in red.
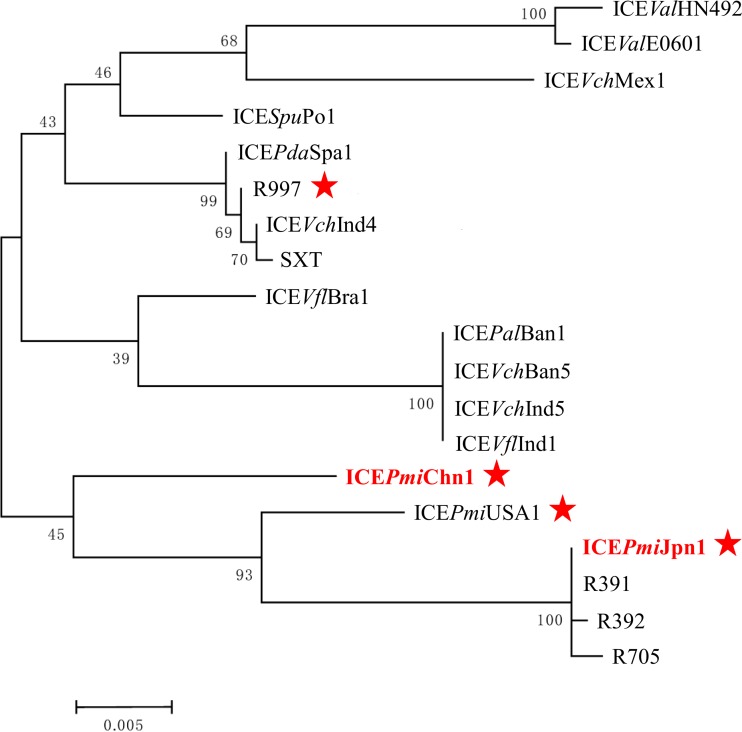
Strain PM13C04 contained a novel multidrug-resistant SXT-like ICE that was designated ICEPmiChn1 (92,751 bp), according to the nomenclature proposed by Burrus et al. (5). The ICEPmiChn1 belonged to the S exclusion group determined by the amino acid sequences of TraG and Eex proteins (20). The nucleotide sequences of int (1242 bp) in ICEPmiChn1 showed 97% identity to that of SXT and 96% identity to that of R391 (21, 22), which also varied from those of previously reported SXT/R391 ICEs according to the maximum likelihood tree obtained by using MEGA 6 software (23) (Fig. 3). ICEPmiChn1 harbored a multidrug resistance region in VRIII, containing five resistance genes, floR, tet(G), strA, strB, and sul2. ICEPmiChn1 shared HS1, HS2, and HS3 gene contents with ICEEniSpaI from Enterovibrio nigricans (9), SXT (21) and R391 (22), respectively, indicating that ICEPmiChn1 might come from homologous recombination among different SXT/R391 ICEs (4, 6, 24). It was noticeable that two open reading frames (ORFs) (chn1 and chn2) and the insertion sequence ISPpu12 in HS4 were reported here for the first time in a SXT/R391-related ICE (Fig. 2). chn1 and chn2 showed 93% and 92% nucleotide identity to the corresponding genes in Vibrio sp. EJY3 (CP003241). A protein BLAST search showed that chn1 encodes the predicted SIR2 superfamily of protein (472 amino acids [aa]), including the silent information regulator 2 (Sir2) enzyme, which catalyzes NAD+-dependent protein/histone deacetylation, and chn2 encodes a predicted ATPase (679 aa). The insertion sequence ISPpu12 (3,372 bp), which was first characterized in Pseudomonas putida (25), was inserted into s062, leading to an 8-bp target site duplication (TAAAGAAA). ISPpu12 consists of four ORFs, tnpA, lspA, orf1, and tnpR. orf1 encodes a possible divalent heavy metal/H+ antiporter protein (26), which might provide fitness in the event of exposure to heavy metal. In HS5 five genes showed 97 to 98% nucleotide identity to the corresponding genes that encoded the type III restriction-modification system conferring resistance to phage infection in ICEValSpaI from Vibrio alginolyticus (10), with the exception of the met gene, which showed only 71% nucleotide identity.
FIG 3.
Maximum likelihood phylogenetic tree based on 19 int gene sequences from SXT/R391 ICEs by using MEGA 6. Bootstrap analysis was performed with 1,000 replications. The SXT/R391 ICEs from P. mirabilis are indicated by a red star. The int gene sequences reported in this study are in red, and others are in black and were downloaded from GenBank with the accession numbers listed in Table S2 in the supplemental material. Bar, 0.05 substitution per nucleotide position.
Most of the SXT/R391 ICEs could be excised from the chromosome and transferred to new hosts via conjugation (5). We detected the circular form of ICEs using the primers LE4 and RE4 (17). A 550-bp target band was detected in all eight ICE-harboring strains (see Fig. S1 in the supplemental material), indicating that the ICEs could form the circular intermediates. Conjugation experiments were performed by using the rifampin-resistant strain Escherichia coli EC600 as the recipient strain, with selection on agar plates containing 300 μg/ml rifampin and 4 μg/ml cefotaxime (or 8 μg/ml florfenicol and 16 μg/ml tetracycline together). The transconjugants were further determined by the mobilization of antimicrobial resistances and by PCR targeting the ICE int gene. The results confirmed that all eight ICEs could be transferred to E. coli, with transfer rates ranging from 1.5 × 10−4 to 1.9 × 10−7 transconjugants per recipient cell (Table 1), suggesting that the horizontal transfer might be an important mechanism for the dissemination of SXT/R391 ICEs.
In conclusion, this is the first report of SXT/R391 ICEs in P. mirabilis in China. The results highlight the idea that animal farms are important reservoirs of the P. mirabilis strains carrying SXT/R391 ICEs. A novel multidrug-resistant ICE, ICEPmiChn1, was characterized in P. mirabilis for the first time. SXT/R391 ICEs could mediate the dissemination of antimicrobial resistance in P. mirabilis and be transferred to other members of the Enterobacteriaceae, which should receive more attention.
Nucleotide sequence accession numbers.
The nucleotide sequences of the complete ICEPmiJpn1 and ICEPmiChn1 characterized in this study were submitted to GenBank and assigned accession numbers KT894734 and KT962845, respectively.
Supplementary Material
ACKNOWLEDGMENT
We are very grateful to Wang Ming-Gui (Institute of Antibiotics, Huashan Hospital, Fudan University) for providing E. coli EC600.
Funding Statement
This work was supported by “973” National Basic Research Program of China (project 2013CB127200), Earmarked Fund for Modern Agroindustry Technology Research System (grant CARS-41-K09), Special Fund for Agro-scientific Research in the Public Interest of China (grants 201303044 and 201403054), Science & Technology Pillar Program in Sichuan Province (grant 2013NZ0025), and the general program of National Natural Science Foundation of China (grants 31572547 and 31572548).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02852-15.
REFERENCES
- 1.Wozniak RA, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 2.Burrus V, Waldor MK. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol 155:376–386. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 3.Toleman MA, Walsh TR. 2011. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol Rev 35:912–935. doi: 10.1111/j.1574-6976.2011.00294.x. [DOI] [PubMed] [Google Scholar]
- 4.Garriss G, Waldor MK, Burrus V. 2009. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet 5:e1000775. doi: 10.1371/journal.pgen.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burrus V, Marrero J, Waldor MK. 2006. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55:173–183. doi: 10.1016/j.plasmid.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Wozniak RA, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, Dery C, Burrus V, Waldor MK. 2009. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet 5:e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carraro N, Poulin D, Burrus V. 2015. Replication and active partition of integrative and conjugative elements (ICEs) of the SXT/R391 family: the line between ICEs and conjugative plasmids is getting thinner. PLoS Genet 11:e1005298. doi: 10.1371/journal.pgen.1005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bordeleau E, Brouillette E, Robichaud N, Burrus V. 2010. Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ Microbiol 12:510–523. doi: 10.1111/j.1462-2920.2009.02094.x. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez-Blanco A, Lemos ML, Osorio CR. 2012. Integrating conjugative elements as vectors of antibiotic, mercury, and quaternary ammonium compound resistance in marine aquaculture environments. Antimicrob Agents Chemother 56:2619–2626. doi: 10.1128/AAC.05997-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balado M, Lemos ML, Osorio CR. 2013. Integrating conjugative elements of the SXT/R391 family from fish-isolated Vibrios encode restriction-modification systems that confer resistance to bacteriophages. FEMS Microbiol Ecol 83:457–467. doi: 10.1111/1574-6941.12007. [DOI] [PubMed] [Google Scholar]
- 11.Bi D, Xu Z, Harrison EM, Tai C, Wei Y, He X, Jia S, Deng Z, Rajakumar K, Ou HY. 2012. ICEberg: a Web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res 40:D621–626. doi: 10.1093/nar/gkr846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu YY, Cai JC, Zhang R, Zhou HW, Sun Q, Chen GX. 2012. Emergence of Proteus mirabilis harboring blaKPC-2 and qnrD in a Chinese hospital. Antimicrob Agents Chemother 56:2278–2282. doi: 10.1128/AAC.05519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy DB, Pembroke JT. 1999. Monitoring of chromosomal insertions of the IncJ elements R391 and R997 in Escherichia coli K-12. FEMS Microbiol Lett 174:355–361. doi: 10.1111/j.1574-6968.1999.tb13590.x. [DOI] [PubMed] [Google Scholar]
- 14.Pearson MM, Sebaihia M, Churcher C, Quail MA, Seshasayee AS, Luscombe NM, Abdellah Z, Arrosmith C, Atkin B, Chillingworth T, Hauser H, Jagels K, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Walker D, Whithead S, Thomson NR, Rather PN, Parkhill J, Mobley HL. 2008. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J Bacteriol 190:4027–4037. doi: 10.1128/JB.01981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada S, Ishii Y, Saga T, Tateda K, Yamaguchi K. 2010. Chromosomally encoded blaCMY-2 located on a novel SXT/R391-related integrating conjugative element in a Proteus mirabilis clinical isolate. Antimicrob Agents Chemother 54:3545–3550. doi: 10.1128/AAC.00111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mata C, Navarro F, Miro E, Walsh TR, Mirelis B, Toleman M. 2011. Prevalence of SXT/R391-like integrative and conjugative elements carrying blaCMY-2 in Proteus mirabilis. J Antimicrob Chemother 66:2266–2270. doi: 10.1093/jac/dkr286. [DOI] [PubMed] [Google Scholar]
- 17.McGrath BM, O'Halloran JA, Piterina AV, Pembroke JT. 2006. Molecular tools to detect the IncJ elements: a family of integrating, antibiotic resistant mobile genetic elements. J Microbiol Methods 66:32–42. doi: 10.1016/j.mimet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Koeck JL, Arlet G, Philippon A, Basmaciogullari S, Thien HV, Buisson Y, Cavallo JD. 1997. A plasmid-mediated CMY-2 beta-lactamase from an Algerian clinical isolate of Salmonella senftenberg. FEMS Microbiol Lett 152:255–260. doi: 10.1016/S0378-1097(97)00206-1. [DOI] [PubMed] [Google Scholar]
- 19.CLSI. 2012. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement CLSI document M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA, USA. [Google Scholar]
- 20.Marrero J, Waldor MK. 2007. The SXT/R391 family of integrative conjugative elements is composed of two exclusion groups. J Bacteriol 189:3302–3305. doi: 10.1128/JB.01902-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beaber JW, Hochhut B, Waldor MK. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol 184:4259–4269. doi: 10.1128/JB.184.15.4259-4269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boltner D, MacMahon C, Pembroke JT, Strike P, Osborn AM. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J Bacteriol 184:5158–5169. doi: 10.1128/JB.184.18.5158-5169.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burrus V, Waldor MK. 2004. Formation of SXT tandem arrays and SXT-R391 hybrids. J Bacteriol 186:2636–2645. doi: 10.1128/JB.186.9.2636-2645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams PA, Jones RM, Shaw LE. 2002. A third transposable element, IS Ppu12, from the toluene-xylene catabolic plasmid pWW0 of Pseudomonas putida mt-2. J Bacteriol 184:6572–6580. doi: 10.1128/JB.184.23.6572-6580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christie-Oleza JA, Nogales B, Lalucat J, Bosch R. 2010. TnpR encoded by an ISPpu12 isoform regulates transposition of two different ISL3-like insertion sequences in Pseudomonas stutzeri after conjugative interaction. J Bacteriol 192:1423–1432. doi: 10.1128/JB.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.