Abstract
The evolution of antibiotic resistance in bacteria has become one of the defining problems in modern biology. Bacterial resistance to antimicrobial therapy threatens to eliminate one of the pillars of the practice of modern medicine. Yet, in spite of the importance of this problem, only recently have the dynamics of the shift from antibiotic sensitivity to resistance in a bacterial population been studied. In this study, a novel chemostat method was used to observe the evolution of resistance to streptomycin in a sensitive population of Escherichia coli, which grew while the concentration of antibiotic was constantly increasing. The results indicate that resistant mutants remain at a low frequency for longer than expected and do not begin to rise to a high frequency until the antibiotic concentrations are above the measured MIC, creating a “lull period” in which there were few bacterial cells growing in the chemostats. Overall, mutants resistant to streptomycin were found in >60% of the experimental trial replicates. All of the mutants detected were found to have MICs far above the maximum levels of streptomycin to which they were exposed and reached a high frequency within 96 h.
INTRODUCTION
Incidents of resistance of bacterial populations to antibiotic treatments are on the rise worldwide (1, 2). In human pathogens, resistant strains represent a growing threat to public health and foretell of a loss in medical and technological capabilities that is unprecedented in the history of humanity. The loss of antibiotic treatments would all but cripple not only the field of infectious disease medicine but all aspects of medicine, including surgery (postsurgical wound management) and oncology (chemotherapeutic immune suppression). Given that antibiotic resistance is an acknowledged and growing problem, and given the innumerable negative impacts that the loss of anti-infection treatments would bring, the question remains: how does antibiotic resistance arise within populations?
We know that resistance can arise de novo in susceptible populations through mutations that inhibit the action of the antibiotic or that increase the efflux of the antibiotic or through horizontal gene transfer from resistant strains to susceptible strains (3, 4). However, while the mechanisms of resistance are well understood, the population-level dynamics inherent in frequency shifts from sensitivity to resistance are not well understood. There has been some excellent work showing that resistance can be selected for in populations that are in very low-concentration (≪MIC) antibiotic environments (5), and some clever approaches to investigating intermediate mutational steps in the evolution of resistance have been developed (6–8). For example, using the morbidostat (7), a culturing device that keeps population growth below the maximal growth rate (μmax) by the addition of sublethal doses of an antibiotic, Toprak et al. (6) showed that trimethoprim resistance in Escherichia coli increases in a stepwise fashion, with a preponderance of mutations occurring in the folic acid synthesis genes, while doxycycline and chloramphenicol resistance increases more continuously, and mutations promoting resistance are found in membrane proteins and in proteins involved in transcription and translation. Thus, the target of selection will be important in the dynamic of selection for resistance. Also, Girgis et al. (9) used transposon-induced mutations (mostly amorphs) to characterize many genes that increase antibiotic tolerance. Of particular note, reduced flagellar synthesis increased tolerance to β-lactams, and the disruption of electron transport and oxidative respiration increased tolerance to aminoglycosides.
In another example, Miller and colleagues (8), using turbidostats (which maintain populations at the μmax), identified conserved mutational paths to daptomycin resistance in a clinically relevant strain of Enterococcus faecalis. The paths were the result of mutation and selection interacting on a genomic scale to explore the phenotypic landscape available to extant populations as the concentration of daptomycin was increased in a stepwise fashion. Their observation of conserved multistep mutational paths in the relevant cellular pathways recalls the pleiotropic interactions identified at the protein level by Weinreich et al. (10) in β-lactamase-mediated resistance to a cephalosporin.
Changes such as these are not always likely to be those that are selected to lead to resistance in clinical practice. However, they all help highlight the importance of the questions at hand, namely, when and how does resistance rise in frequency within a population. These questions are basic to our understanding of the nature of antibiotic resistance, particularly in situations in which there are suspected environmental reservoirs of resistant phenotypes, such as water treatment facilities or livestock waste pools in agriculture. Here, we attempt to look more deeply at how and when a mutation granting resistance to the antibiotic streptomycin comes into existence and rises in frequency within a population that is initially streptomycin sensitive.
MATERIALS AND METHODS
Strain.
The ancestral strain used is the DD1953 strain of E. coli, an MG1655 derivative of K-12 lab strains used widely. DD1953 is rpoS-, having a mutation causing a premature stop codon early in the coding region of the rpoS gene, which codes for the σS protein. The MIC of streptomycin for DD1953 was measured as being 3 (±1) μg/ml. DD1953 has been used extensively in our lab, both in serial transfer and especially in chemostat experiments. For the purposes of a molecular marker, resistance to the bacteriophage T5 was used. T5 resistance is caused by a naturally occurring and fairly common mutation (fhu) in this strain and has been shown to be selectively neutral with respect to fitness (11) in chemostats under glucose-limiting conditions. The ancestral strain stock used for all experiments was grown from a single plated colony of DD1953 in order to ensure that the ancestral strain utilized was genetically homogeneous at the start of experimentation. That initial stock was plated with T5 to screen for a T5-resistant mutant. Once a T5-resistant mutant colony was found, a stock of T5-resistant DD1953 was grown from this single colony, ensuring a genetically homogeneous stock. The two stocks were independent in their array of new mutations.
Chemostats.
All experimental trials were conducted in chemostats using a protocol modified from that previously described elsewhere (12–14). Basically, a chemostat is a continuous culture device that supplies a measured amount of fresh medium to an exponentially growing population while it removes an equal volume of spent medium and cells, always allowing the population size (N) to remain constant. For these experiments, the chemostats were all 30-ml experimental volumes with a population size (carrying capacity) of 108 cells/ml (N = 3.0 × 109). The experimental temperature was maintained at 37°C at all times.
The medium used was Davis salts minimal medium with 0.01% glucose added. The medium was metered via a Wizco pump set to deliver 5.625 ml of medium per hour to the chemostat chamber (D = 0.1875). This rate of dilution results in a generation time of 3.69 h per generation (15). This generation time was held constant throughout all replicate trials. Of note, in control experiments that utilized the same or similar starting strains with higher dilution rates (D = 0.35 and 0.46) in chemostats, no differing results were observed in terms of gene targets outside those observed here.
In addition to the usual single-medium jar, a second medium jar containing the same Davis salts minimal medium and glucose, supplemented with streptomycin at a concentration of 100 μg/ml, was connected to the primary medium jar via a siphon line. This siphon line replenished half of the volume of medium metered out of the primary jar on a continuous basis. A magnetic stir bar ensured that the medium in the primary jar was well mixed. This means that at T0, the concentration of streptomycin in the medium delivered to the chemostat, was 0. As the experiment progressed and more of the medium used was replenished with streptomycin-laden medium, the concentration of the medium in the primary jar, and hence, the chemostat, increased. The concentration of streptomycin in the medium delivered to the chemostat can be calculated, to a close approximation, for any time point by using the equation
Where h is the time point in hours, X is the volume of medium in the primary medium jar, Y is the concentration of streptomycin, and c is the constant delivered total mass of streptomycin to primary medium jar from h − 1 to h; here, that rate is 281.25 μg/h.
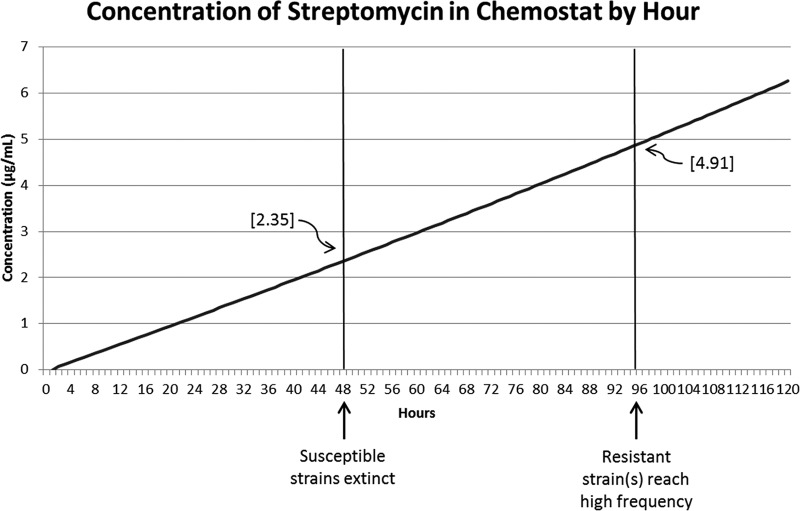
The concentration of streptomycin in the chemostat will be less than that in the medium bottle. By the iteration of adding 5.625 ml from the medium bottle each hour, we estimated that the concentration of streptomycin in the chemostat would be one-third of the concentration in the medium bottle (Fig. 2).
FIG 2.
Concentration of streptomycin over time. As the concentration of antibiotic increased, susceptible strains died off, but resistant mutants did not increase in frequency until later, even though streptomycin continued to increase. In almost all cases for which resistance was observed, the resistant strain(s) reached high frequency by the 96th hour.
Plating and population tracking.
A sample was taken from all chemostats at roughly 24-h intervals. This sample was diluted 100,000-fold and replicate plated on LB agar in petri dishes. The diluted samples were plated 4 times on LB agar and 4 times on LB agar with T5 bacteriophage as a screen for T5-resistant (T5R) clones. The samples were pour plated using soft agar (LB agar with a reduced concentration of Difco agar) and then topped with top agar (Difco agar and water). All plates were incubated at 37°C overnight and counted the next day using a ProtoCOL automatic plate reader and ProtoCOL software version 3.15, both by Synbiosis (Cambridge, United Kingdom). The number of T5-sensitive (T5S) colonies was calculated as the mean number of T5R colonies subtracted from the mean number of total colonies. Populations of T5S and T5R were then able to be tracked over the time course of the experiment.
Sequencing and bioinformatics.
Resistance mutations to streptomycin in E. coli strains are most often linked to single nucleotide polymorphisms (SNPs) in the rpsL gene. This gene codes for the ribosomal protein S12, which makes up part of the small (30S) subunit of the E. coli ribosome. In particular, this protein interacts with rRNA and mRNA at the codon-anticodon recognition site (A site) of the ribosome. This implies that S12 plays a role in codon-anticodon recognition, aminoacyl-tRNA selection, and/or the translational proofreading process (16–19).
The rpsL gene was PCR amplified using forward and reverse primers and then Sanger sequenced at the University of Arizona Genetics Core, Tucson, AZ. These sequence reads were then vetted for quality and aligned to the ancestral rpsL gene from the DD1953 strain and the reference K-12 MG1655 genome (GenBank accession no. NC_000913.3) sequence for rpsL. SNPs were identified only when forward and reverse reads of high quality agreed.
Along with rpsL, compensatory mutations in the rpsD and rpsE genes have been reported in the literature (17, 20, 21). These genes were sequenced using a similar protocol for comparative reasons. All alignments and assemblies were completed using Geneious bioinformatics software tools (Biomatters Ltd., Auckland, New Zealand), version 7 or later.
MICs.
The MICs for all strains were found experimentally using optical density (OD) measures in Luria-Bertani (LB) liquid medium. Basically, the strain of E. coli to be tested was grown overnight in a test tube in LB at 37°C and shaken at 200 rpm. After the population had entered stationary phase, 100 μl of the overnight growth was put into fresh LB medium and allowed to enter exponential-growth phase. At this point, 100 μl of the growing population was put into an array of fresh test tubes containing 1.9 ml of fresh LB along with a spectrum of streptomycin concentrations ranging from 0 to 1,000 μg/ml. Growth in these tubes was tracked by measuring optical density at 600 nm at regular intervals with a WPA Biowave model CO8000 cell density meter.
RESULTS
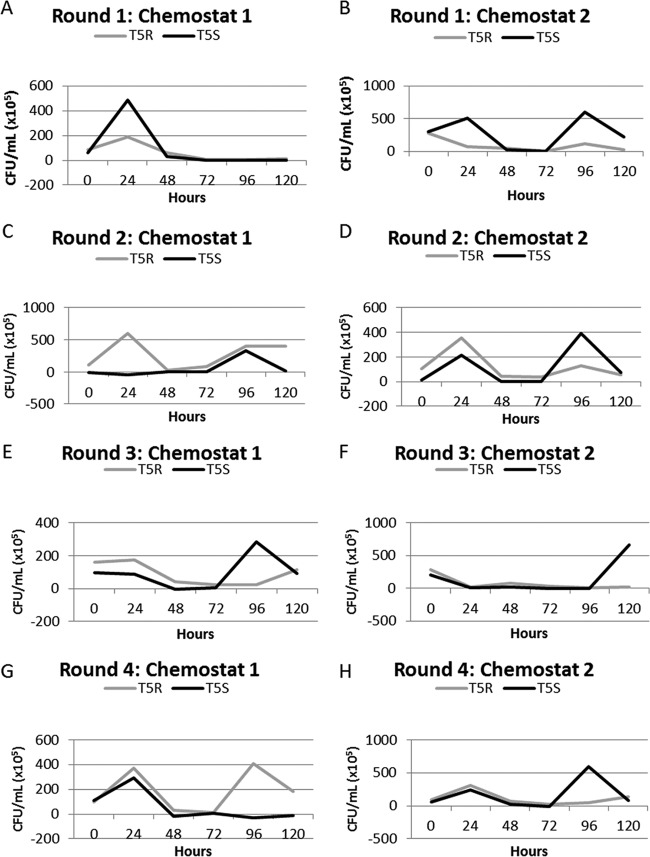
Four rounds of experiments were conducted. Each round consisted of two chemostats each inoculated with approximately equal amounts of the two E. coli strains, DD1953 T5R and DD1953 T5S. This gave a total of four experimental replicates per round of two chemostats, resulting in 16 replicate experimental runs over those 4 rounds (2 strains per chemostat, 2 chemostats per round, for 4 total replicate rounds). Each chemostat was continued for a minimum of 120 h, although the results were usually clear by 96 h. In 5 of the 16 (31.25%) replicate trials, the populations of E. coli went extinct within 48 h as the concentration of streptomycin in the chemostat approached 2.5 μg/ml (see Fig. 2). In 10 replicates, the population was observed to decline to low levels and then rapidly recover within the next 48 h (Fig. 1, colony counts for experiments). Of the 8 chemostats run (2 chemostats per round), only 1 had no observed streptomycin mutant come into the population at a detectable level within 120 h. The remaining 7 chemostats had at least 1 streptomycin-resistant mutant rise to a high frequency within that time frame.
FIG 1.
(A to H) CFU/ml counts within experimental chemostats. Within 48 h, all chemostats indicated that sensitive strains were significantly depleted to or near 0. In all but one case, the resistant mutant had reached a high frequency (when a mutant was present) by 96 h. The two strains within each chemostat were labeled with or without resistance to T5 bacteriophage. Note that a Bacillus contaminant was found in chemostat 2 of round 4 at the 120-h mark, which was not present in the 72-h sample.
In one replicate, a contaminant was observed at 120 h that was later identified as belonging to the genus Bacillus (Fig. 1H, T5R). This contaminant was found at a low frequency (approximately 6.7% of CFU) and was not present at the 96-h sample, at which point the E. coli strain was extinct. This contamination is not wholly unexpected when we consider that the chemostat was an available niche with all the resources needed for microbial growth. The streptomycin-resistant mutant in this chemostat, K42N in the rpsL gene, was increasing in frequency but did not completely fill the niche space available during this time.
Sequences from the populations that recovered showed that in all cases, one of three SNPs had swept to fixation (Table 1). The three SNPs observed all occurred at the same codon position within the rpsL gene, codon 42, which codes for the amino acid lysine (K) (codon AAA), in the ancestor, DD1953 (both strains, T5R and T5S). The observed SNPs were either AAA→ACA (lysine to threonine), AAA→AAC (lysine to asparagine), and AAA→ATA (lysine to isoleucine). The threonine mutant was observed to fix in 7 of the 11 (63.6%) replicates in which a streptomycin-resistant mutant was observed. In three cases (round 2, chemostat 1, T5R; round 4, chemostat 1, T5R; and round 4, chemostat 2, T5S), the K→N change was observed. One case (round 3, chemostat 2, T5S) was found to be the isoleucine mutant K42I. The MICs for all mutants were measured to be >1,000 μg/ml for streptomycin.
TABLE 1.
Chemostat resultsa
| Round | Results of chemostat trials |
||
|---|---|---|---|
| Chemostat | T5S | T5R | |
| 1 | 1 | Extinct | Extinct |
| 2 | K42T | K42T | |
| 2 | 1 | K42T | K42N |
| 2 | K42T | K42T | |
| 3 | 1 | K42T | K42T |
| 2 | K42I | Extinct | |
| 4 | 1 | Extinct | K42N |
| 2 | K42N | Extinct | |
All mutations were found in the rpsL gene. In cases in which no mutant was observed, the populations both went extinct. Note that in round 2, chemostat 1, both the T5R and T5S strains were observed to have a K42T mutation (these have to be independent mutations) and that in round 4, chemostat 2, a Bacillus contaminant was found in place of a T5R strain at 120 h.
In order to test whether the discrepancy between the number of trials that resulted in the K42T mutant fixing as opposed to the K42N mutant fixing was an experimental condition or a result of mutant presence/absence in the initial ancestor stocks, a control experiment was conducted. The ancestor strains, DD1953 T5R and DD1953 T5S, were grown overnight in LB. Twenty milliliters of this overnight growth (OD at 600 nm [OD600] at time of inoculation, 1.96, for density of 1.96 × 109 and a total population size of 3.92 × 1010 cells) was added to 180 ml of soft agar supplemented with streptomycin at a concentration of 100 μg/ml. The E. coli and soft agar mixtures were then plated in 250-ml glass petri dishes and incubated for 7 days at 37°C. This control was performed in two replicates.
After the 7-day incubation period, a total of 59 streptomycin-resistant colonies were observed (from 4 plates; see Tables 2 and 3). These colonies were collected, and their rpsL, rpsD, and rpsE genes were PCR amplified and Sanger sequenced. As in the experimental condition, no SNPs were found in rpsD or rpsE; once again, all SNPs found were in the rpsL gene. However, not all of the SNPs found in the control experiment were in the codon 42 position, although most were. The complete list of SNPs from the control experiments is given in Table 3. Of the 59 streptomycin-resistant mutants from the control experiments, 41 (69.5%) mutants were exact matches of the threonine mutant (K42T) observed in the chemostat experiments. This represents the largest portion of the control mutant list and is a close match in terms of percentage to the chemostat experiments (63.6% in the experiment versus 69.5% in the control). The second most frequent mutant observed from the control experiments was a match to the asparagine mutant (K42N) (11.8% [7/59 mutants] versus 27.3% in the experiment). Of course, other streptomycin mutants were also found in the controls (5 others) that were capable of growing in streptomycin concentrations of at least 100 μg/ml.
TABLE 2.
Complete list of SNPs found in the control experimenta
| rpsL SNP | Replicate 1 |
Replicate 2 |
||
|---|---|---|---|---|
| T5S | T5R | T5S | T5R | |
| K42T | 11 | 11 | 11 | 8 |
| K42N | 1 | 2 | 1 | 3 |
| K42R | 0 | 0 | 1 | 0 |
| K87R | 1 | 2 | 2 | 0 |
| P90Q | 0 | 1 | 0 | 1 |
| P90L | 0 | 0 | 0 | 1 |
| G91D | 0 | 1 | 0 | 1 |
| Total | 13 | 17 | 15 | 14 |
All SNPs were identified in the rpsL gene, which is known to play a role in streptomycin resistance.
TABLE 3.
Control experiment results: 59 streptomycin mutants found in rpsL in the control experiment with MICs for streptomycin of >100 μg/ml
| rpsL SNP | Codon | Sequence | Amino acid | Quantity | % |
|---|---|---|---|---|---|
| WTa | 42 | AAA | Lysine | NAb | |
| K42T | 42 | ACA | Threonine | 41 | 69.5 |
| K42N | 42 | AAC | Asparagine | 7 | 11.8 |
| K42R | 42 | AGA | Arginine | 1 | 1.7 |
| WT | 87 | AAA | Lysine | NA | |
| K87R | 87 | AGA | Arginine | 5 | 8.5 |
| WT | 90 | CCG | Proline | NA | |
| P90L | 90 | CTG | Leucine | 1 | 1.7 |
| P90Q | 90 | CAG | Glutamine | 2 | 3.4 |
| WT | 91 | GGT | Glycine | NA | |
| G91D | 91 | GAT | Aspartic acid | 2 | 3.4 |
WT, wild type.
NA, not available.
DISCUSSION
This series of experiments highlights the incredible speed with which a streptomycin-resistant mutant can sweep to fixation within a population exposed to low levels of streptomycin. The sensitive clones within the population die off quickly, and the resistant mutants, with MICs of >1,000 μg/ml, grow to fill the emptied environmental space of the chemostat. We should note here that the MIC of the resistant populations (>1,000 μg/ml) was over 400 times higher than the streptomycin concentration in the medium at the time when the susceptible population had already completely died off (2.35 μg/ml streptomycin at 48 h). The rise was so rapid, in fact, that the overall time to fixation for the streptomycin-resistant mutants observed in these experiments was far shorter than the typical 10- to 14-day treatment cycle for many antibiotics, including streptomycin. Further, toxic levels of streptomycin are reached at about 430 mg/kg of body weight (as per the material safety data sheet [MSDS] for streptomycin sulfate: 50% lethal dose [LD50] in rats, oral ingestion [http://www.pfizer.com/files/products/material_safety_data/284.pdf]), far below the 1,000 μg/ml level of resistance and still at least 160 times higher than the highest concentration of the antibiotic to which the populations were exposed (Fig. 2).
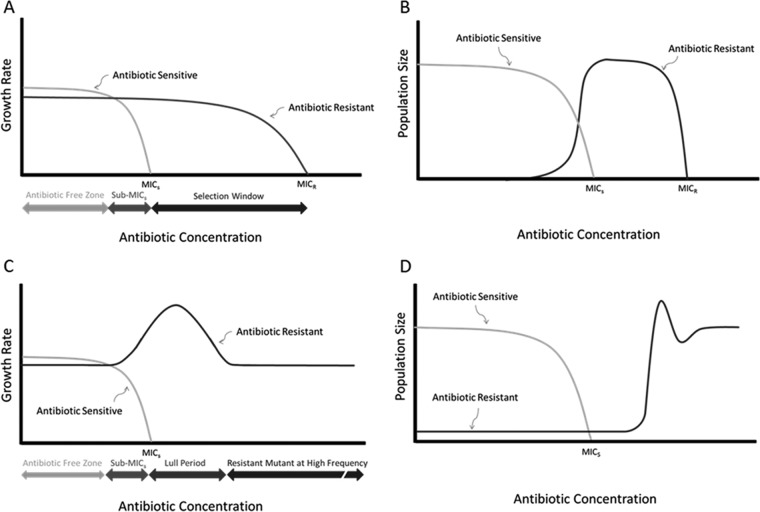
Additionally, while the overall shift of the populations from streptomycin sensitivity to resistance occurred rapidly, in each individual case, the dynamics of the population shift differed from the expectation provided by current theory. In each case, there was a short but definitive period in which the sensitive clones within the population were at or near extinction and the resistant clones were not yet occurring at a high frequency within the chemostats. This lull period, which was present in every case in which a resistant mutant was later recovered from a chemostat trial, indicates that resistant mutants do not increase in frequency as the environment approaches the MIC of the susceptible population, as expected. This is so even though the chemostat systems are designed to maintain population size over time. Instead, the streptomycin-resistant mutant was found to be at low frequency until well after the sensitive population declined to extinction or near extinction (Fig. 3). This suggests that the resistant mutant remains at a low frequency until the susceptible clones decline.
FIG 3.
Theory versus observation. (A) Expected graphs of relative growth rates for a population of bacteria in an antibiotic gradient. The expectation was that the susceptible population would go extinct when the antibiotic concentration reached the MIC of the susceptible bacteria (MICS). Another expectation was that the growth rate of any of the resistant mutants in the population would continue unabated until the antibiotic concentration reached a threshold capable of killing these resistant cells (MICR). The underlying assumption was that both strains grew at the maximum growth rate (μmax), with the antibiotic-resistant clone having a slightly lower maximum growth rate. (B) Same scenario as in panel A but in terms of population size (N) instead of growth rate (μ). The resistant clone filled the available niche space and increased in frequency as the growth rate of the susceptible clone declined. Overall, N remained constant. (C) Results of chemostat experiments are graphed in panel A. Contrary to expectation, the resistant mutant underwent an apparent bump in growth rate (in chemostats, bacteria experience submaximal μ at equilibrium) and then a return to baseline after a lull period in which the total population size was greatly diminished. The resistant strain increased in growth rate to μmax as the growth rate of the susceptible strain declined. (D) Results of the current experiments in terms of N. The low starting frequency of the resistant strain did not take up niche space as quickly as it was yielded by the susceptible strain even though the resistant mutant grew as quickly as physiologically possible. This resulted in the observed lull period. Whether similar dynamics would be observed in hosts with an active immune system is unknown.
The results from the control experiment indicate that by the time of inoculation, the streptomycin-resistant mutants were already present in the population, and the shift in environment was into one in which the antibiotic selects for the mutants, while also selecting against the sensitive strain. This is evident by the number of mutants that were recovered from the control experiments. If the streptomycin-resistant mutants were present in the 20 ml of inoculum that went into each control petri dish containing agar and streptomycin, the colonies would grow and they would be recoverable. This is indeed what happened. If the streptomycin-resistant mutants had not been present in the inoculum, we would expect a dynamic similar to that observed in the chemostat trials in which no resistant strains were found (see Fig. 1A), namely, that the sensitive cells would very rapidly be killed by the antibiotic in the agar, and no mutant colonies would have been expected. Additionally, the distributions of SNPs found in the control experiment were consistent across the two experimental conditions (Tables 2 and 3), and these distributions also correlate well with the distribution of SNPs observed in the chemostat experiments (Table 1).
The total volume of inoculum for the control experiments was 20 ml of E. coli at an OD600 of about 1.96. This means that the petri dishes with agar and streptomycin were each seeded with 3.92 × 1010 live growing cells. The average number of streptomycin-resistant colonies recovered from the 4 trial petri dishes of the control experiment was 14.75 (P = 0.5238 for the four trials, G-test of independence) (22). Put another way, we can calculate a conditional mutation rate for the rpsL gene for streptomycin resistance. Taking the average of results from the four control plates, the mutation frequency for rpsL is calculated to be 2.69 × 10−9, meaning that this is the rate at which we can expect to find a streptomycin-resistant mutant with an MIC of >1,000 μg/ml.
Since the mutation frequency and distribution are about the same for both the T5S and the T5R control cultures, we can assume that all the mutations happened in the last three generations. Assuming a mutation rate of 10−10 (lower than measured) and a population size of 4 × 1010, there is an expectation of 4 new mutations per generation. When the population size is 2 × 1010, there is an expectation of 2 new mutations per generation, which will double to give 4 mutations in the final generation. Likewise, when the population size is 1 × 1010, the expectation is one new mutation, which will increase to four mutations in the final generation. The expectation in the previous generation is less than one that there will be a new mutation, and if there is, it will give the appearance of unevenness in the distribution of mutation frequency across populations. This will give an expectation of 16 mutations for each petri plate, compared to the observed 14.75 mutations. Thus, we can conclude that the mutation rate of streptomycin resistance is just a little more than 10−10, and that the last two generations contributed most of the observed mutations.
In the chemostat experiments, each chemostat was inoculated with 1 ml of T5S and 1 ml of T5R, both at an OD600 of about 0.8. This means that each milliliter of inoculant contained about 8 × 108 cells, for a total of 1.6 × 109 cells per chemostat inoculation. Given the calculated frequency of streptomycin-resistant strains, we would expect 59.3% of chemostat replicate trials to result in an observed streptomycin-resistant mutant in the rpsL gene (1.6 × 109 total E. coli cells/2.7 × 10−9 expected gene frequency). At equilibrium, these chemostats contain about 3 × 109 cells. Across 8 chemostats, this would be 2.4 × 1010 cells. With a mutation rate of 10−10, this gives an expectation of an additional 2.5 new mutations in the chemostats. Since about half the populations already carry at least one cell that is resistant to streptomycin, only half of these new mutations in the chemostat will be in populations consisting of sensitive cells only. Thus, we expect 10.75 (9.5 + 1.25) populations to contain a strain that is resistant to streptomycin. This agrees closely with the observed 11/16 populations that contained streptomycin-resistant cells.
The chemostat trials support the conclusion of the control experiment that most of the mutants come into the population during the exponential-growth phase prior to exposure to the antibiotic in the chemostat. If the SNPs that are responsible for resistance were to occur after inoculation of the chemostats, we would have to expect that the time of mutation would again be before the level of streptomycin reaches a concentration capable of killing most of the bacterial cells. This must be the case, because once streptomycin concentrations reach a level that results in bacterial killing, population sizes drop very quickly (Fig. 1), representing a decreasing probability of an SNP occurring, not an increasing one. In fact, keeping pathogen population sizes small is a basic tenet of controlling many infectious diseases, including HIV (23), tuberculosis (24), and malaria (25). This is because host immune systems function well when infection size is controlled, as opposed to the uncontrolled growth of pathogens that warrants antibiotic treatments. By keeping the overall population size of the pathogen in check, the immune system can most probably eliminate the antibiotic-resistant mutants that occur through random SNPs before they are selected for and become the dominant strains in an infection.
Extrapolating the results observed here to general infections treated with streptomycin beginning in the 1940s would mean that streptomycin should almost immediately have been rendered effectively useless as an antimicrobial therapy. In fact, this was a concern for researchers investigating streptomycin efficacy in tuberculosis as early as 1948 (26, 27). The Marshall et al. (27) study found resistance to streptomycin after treatment with streptomycin in samples collected from tuberculosis patients in 35 of 41 cases (85.4%). Initially, all the populations were sensitive to streptomycin. After treatment, no bacteria were isolated from sputum samples for a few days to months, and then the populations were resistant to killing by streptomycin (27). Putting aside the large number of differences between the two studies, the in vivo outcome of the 1948 study did not differ drastically from the 63.6% rate of high-level resistance in vitro observed here, suggesting that the streptomycin-resistant mutations that arose were preexisting in the population. This is particularly clear when you consider that the present study looked at a window of only 5 days, as opposed to up to 5.5 months in the 1948 study.
However, if many infections can reach pathogen population sizes of 1010 (28), like our control trial population sizes, we should expect that most cases of streptomycin treatment would have provided for the selection of resistant mutants present in the patients at rates similar to those seen in the present series of experiments. One reasonable explanation for why this did not occur would be the major component of host-pathogen dynamics not present in the chemostat, the host immune system. The immune system would have acted on the relatively small number of resistant mutants, along with any persister cells that escaped chemotherapy, thus ending the line of streptomycin-resistant E. coli organisms before they had a chance to grow under positive selective conditions and spread beyond the host. This would be particularly effective during the lull period observed here, when overall bacterial population size is low, thus preserving streptomycin effectiveness. While antibiotics helped the immune system, the implication is that the immune system also helped control the spread of antibiotic-resistant mutants.
ACKNOWLEDGMENTS
This work was supported in part by the Stony Brook Dean's Fund (DED) and by Lawrence B. Slobodkin and George C. Williams Fellowships (to F.S.).
We thank Walt Eanes, John True, Peter Tonge, and Omar Warsi for many helpful discussions. This work was greatly improved by the comments of two anonymous reviewers.
Footnotes
This article is contribution 1,236 from the Ecology & Evolution Program of Stony Brook University.
REFERENCES
- 1.Bush K, Courvalin P, Dantas G, Davies J, Eisenstein B, Huovinen P, Jacoby GA, Kishony R, Kreiswirth BN, Kutter E, Lerner SA, Levy S, Lewis K, Lomovskaya O, Miller JH, Mobashery S, Piddock LJV, Projan S, Thomas CM, Tomasz A, Tulkens PM, Walsh TR, Watson JD, Witkowski J, Witte W, Wright G, Yeh P, Zgurskaya HI. 2011. Tackling antibiotic resistance. Nat Rev Microbiol 9:894–896. doi: 10.1038/nrmicro2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies SC. 2011. Annual report of the chief medical officer, vol 2. Infections and the rise of antimicrobial resistance. Department of Health, London, United Kingdom: https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/138331/CMO_Annual_Report_Volume_2_2011.pdf. [DOI] [PubMed] [Google Scholar]
- 3.MacLean RC, Hall AR, Perron GG, Buckling A. 2010. The population genetics of antibiotic resistance: integrating molecular mechanisms and treatment contexts. Nat Rev Genet 11:405–414. doi: 10.1038/nrg2778. [DOI] [PubMed] [Google Scholar]
- 4.Hall BG. 2004. Predicting the evolution of antibiotic resistance genes. Nat Rev Microbiol 2:430–435. doi: 10.1038/nrmicro888. [DOI] [PubMed] [Google Scholar]
- 5.Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI. 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 7:e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toprak E, Veres A, Michel J-B, Chait R, Hartl DL, Kishony R. 2011. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet 44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toprak E, Veres A, Yildiz S, Pedraza JM, Chait R, Paulsson J, Kishony R. 2013. Building a morbidostat: an automated continuous-culture device for studying bacterial drug resistance under dynamically sustained drug inhibition. Nat Protoc 8:555–567. doi: 10.1038/nprot.2013.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller C, Kong J, Tran TT, Arias CA, Saxer G, Shamoo Y. 2013. Adaptation of Enterococcus faecalis to daptomycin reveals an ordered progression to resistance. Antimicrob Agents Chemother 57:5373–5383. doi: 10.1128/AAC.01473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girgis HS, Hottes AK, Tavazoie S. 2009. Genetic architecture of intrinsic antibiotic susceptibility. PLoS One 4:e5629. doi: 10.1371/journal.pone.0005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 11.Moser H. 1958. The dynamics of bacterial populations maintained in the chemostat. Carnegie Institute publ. no. 614. Carnegie Institution for Science, Washington, DC. [Google Scholar]
- 12.Dykhuizen D, Dean AM, Hartl DL. 1987. Metabolic flux and fitness. Genetics 115:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dykhuizen DE. 1993. Chemostats used for studying natural selection and adaptive evolution. Methods Enzymol 224:613–631. doi: 10.1016/0076-6879(93)24046-W. [DOI] [PubMed] [Google Scholar]
- 14.Hartl DL, Dykhuizen DE. 1984. The population genetics of Escherichia coli. Annu Rev Genet 18:31–68. doi: 10.1146/annurev.ge.18.120184.000335. [DOI] [PubMed] [Google Scholar]
- 15.Dykhuizen DE, Hartl DL. 1983. Selection in chemostats. Microbiology 47:150–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurland CG. 1992. Translational accuracy and the fitness of bacteria. Annu Rev Genet 26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 17.Kurland CG, Hughes D, Ehrenberg M. 1996. Limitations of translational accuracy, p 979–1004. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin EC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington DC. [Google Scholar]
- 18.Kurland CG, Jorgensen F, Richter A, Ehrenberg M, Bilgin N, Rojas A-M. 1990. Through the accuracy window, p 513–526. In Hill WE, Dahlberg A, Garrett RA, Moore PB, Schlessinger D, Wagner JR (ed), The ribosome: structure, function, and evolution. ASM Press, Washington, DC. [Google Scholar]
- 19.Noller HF, Nomura M. 1996. Ribosomes, p 167–186. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin EC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington DC. [Google Scholar]
- 20.Björkman J, Nagaev I, Berg OG, Hughes D, Andersson DI. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287:1479–1482. doi: 10.1126/science.287.5457.1479. [DOI] [PubMed] [Google Scholar]
- 21.Björkman J, Andersson DI. 2000. The cost of antibiotic resistance from a bacterial perspective. Drug Resist Updat 3:237–245. doi: 10.1054/drup.2000.0147. [DOI] [PubMed] [Google Scholar]
- 22.Sokal RR, Rohlf FJ. 1995. Biometry, 3rd ed W.H. Freeman & Co, New York, NY. [Google Scholar]
- 23.Coffin JM. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 24.Dorman SE, Chaisson RE. 2007. From magic bullets back to the magic mountain: the rise of extensively drug-resistant tuberculosis. Nat Med 13:295–298. doi: 10.1038/nm0307-295. [DOI] [PubMed] [Google Scholar]
- 25.Hartl DL. 2004. The origin of malaria: mixed messages from genetic diversity. Nat Rev Microbiol 2:15–22. doi: 10.1038/nrmicro795. [DOI] [PubMed] [Google Scholar]
- 26.Crofton J, Mitchison DA. 1948. Streptomycin resistance in pulmonary tuberculosis. Br Med J 2:1009–1015. doi: 10.1136/bmj.2.4588.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall G, Blacklock JWS, Cameron C, Capon NB, Cruickshank R, Gaddum JH, Heaf FRG, Hill AB, Houghton LE, Hoyle JC, Raistrick H, Scadding JG, Tytler WH, Wilson GS, Hart PD. 1948. Streptomycin treatment of pulmonary tuberculosis: a medical research council investigation. Br Med J 2:769–782. doi: 10.1136/bmj.2.4582.769.18890300 [DOI] [Google Scholar]
- 28.Drlica K. 2003. The mutant selection window and antimicrobial resistance. J Antimicrob Chemother 52:11–17. doi: 10.1093/jac/dkg269. [DOI] [PubMed] [Google Scholar]