Abstract
Pseudomonas aeruginosa plays an important role in chronic lung infections among patients with cystic fibrosis (CF) through its ability to form antibiotic-resistant biofilms. In P. aeruginosa, biofilm development and the production of several virulence factors are mainly regulated by the rhl and las quorum-sensing (QS) systems, which are controlled by two N-acyl-homoserine lactone signal molecules. In a previous study, we discovered an original QS inhibitor, N-(2-pyrimidyl)butanamide, called C11, based on the structure of C4-homoserine lactone, and found that it is able to significantly inhibit P. aeruginosa biofilm formation. However, recent data indicate that P. aeruginosa grows under anaerobic conditions and forms biofilms in the lungs of CF patients that are denser and more robust than those formed under aerobic conditions. Our confocal microscopy observations of P. aeruginosa biofilms developed under aerobic and anaerobic conditions confirmed that the biofilms formed under these two conditions have radically different architectures. C11 showed significant dose-dependent antibiofilm activity on biofilms grown under both aerobic and anaerobic conditions, with a greater inhibitory effect being seen under conditions of anaerobiosis. Gene expression analyses performed by quantitative reverse transcriptase PCR showed that C11 led to the significant downregulation of rhl QS regulatory genes but also to the downregulation of both las QS regulatory genes and QS system-regulated virulence genes, rhlA and lasB. Furthermore, the activity of C11 in combination with antibiotics against P. aeruginosa biofilms was tested, and synergistic antibiofilm activity between C11 and ciprofloxacin, tobramycin, and colistin was obtained under both aerobic and anaerobic conditions. This study demonstrates that C11 may increase the efficacy of treatments for P. aeruginosa infections by increasing the susceptibility of biofilms to antibiotics and by attenuating the pathogenicity of the bacterium.
INTRODUCTION
Pseudomonas aeruginosa is the most common opportunistic Gram-negative bacterium found in nosocomial and life-threatening infections, such as pneumonia and septicemia, in immunocompromised patients (1). More notoriously, P. aeruginosa is also the main pathogen in patients with cystic fibrosis (CF), playing an important role in progressive lung destruction and the subsequent respiratory failure, and is thus responsible for high rates of morbidity and mortality (2).
The pathogenesis of chronic P. aeruginosa infections is mainly due to the capacity of P. aeruginosa to form biofilms, which are structured communities of bacteria encased in a self-produced polymeric matrix, during the infection process (3). This adds to its resistance to many antibiotics through a naturally lower outer membrane permeability and many adaptive mechanisms when it is growing as a biofilm (4, 5), making it difficult to eradicate infections by conventional antibiotherapy since bacterial cells living as biofilms are more tolerant toward antibiotics than their planktonic counterparts (6). Hence, novel targets are needed to combat biofilm infections. One of them could be the bacterial quorum-sensing (QS) communication system. It has been shown that the QS system plays a key role in orchestrating the expression of many P. aeruginosa virulence factors (7), such as exoproteases, siderophores, exotoxins, and several secondary metabolites, and participates in the development of biofilms (8).
QS is a regulatory mechanism that enables bacteria to make collective decisions with respect to the expression of a specific set of genes. P. aeruginosa possesses three QS systems, las, rhl and 2-heptyl-3-hydroxy-4-quinolone, the last of which is referred to as the Pseudomonas quinolone signal (PQS). Each of these QS systems consists of one transcriptional regulator, LasR, RhlR, or PqsR, respectively, and their associated signaling molecules, N-(3-oxo-dodecanoyl)-l-homoserine lactone (3-oxo-C12-HSL), N-butanoyl-l-homoserine lactone (C4-HSL), and PQS, respectively (9). The N-acyl-homoserine lactone (HSL) signaling molecules have been well studied and are synthesized by the autoinducer enzymes LasI and RhlI for the las and rhl QS systems, respectively (10). PQS synthesis requires the expression of multiple operons, one of which is pqsABCDE (11). When a certain threshold concentration of signaling molecules is reached, binding to receptor molecules is promoted and the activated complex forms dimers or multimers, which in turn act as transcriptional regulators on target virulence genes. The las and rhl QS systems are hierarchically arranged, with the las system being at the top of the signaling cascade and positively regulating expression of the rhl system. Both are required for all virulence determinants (12), and together, these systems control 6 to 11% of the P. aeruginosa genome (13). The PQS system also plays a significant role in the transcription of multiple P. aeruginosa virulence genes (14) and is intertwined in the P. aeruginosa QS hierarchy, with its production and bioactivity requiring the las and rhl QS systems, respectively (15).
P. aeruginosa extracellular quorum-sensing signal molecules have been detected in the sputum and lungs of infected CF patients (16, 17), and many studies have shown that P. aeruginosa mutants lacking functional QS systems form biofilms with a drastically altered structure and exhibit significantly reduced virulence in comparison with the biofilm structures and virulence of P. aeruginosa strains with functional QS systems (8, 18). Therefore, the QS system constitutes a highly attractive target for novel therapeutics that may be developed. There are basically three different points of attack in the QS systems of Gram-negative bacteria: the signal generator, the signal molecule, and the receptor protein. The last one has been the most intensively investigated, which has been done with HSL analogs able to block the QS receptor site by antagonism. In recent years, a large variety of synthetic HSL analogs and natural product (i.e., furanone) libraries have been proposed, and a number of QS inhibitors (QSIs) have shown promising results in studies conducted in vitro and sometimes in experimental animal models of infection (19–21).
However, no QSI has been introduced on the market, and therefore, we looked into new approaches by considering information presented in the literature. The first concerned the predominance of C4-HSL in the lungs of CF patients and led to our previous discovery of an original QSI, N-(2-pyrimidyl)butanamide, called C11, which is based on the structure of C4-HSL and which is able to significantly inhibit P. aeruginosa biofilm formation in a dose-dependent manner in the absence of cytotoxicity for lung cells (22). Second, many studies have considered that P. aeruginosa grows in an anaerobic environment in the lungs of infected CF patients (23–25). In contrast to aerobic in vitro biofilms, P. aeruginosa biofilms in CF patient lungs grow in stagnant mucus, and recent data have revealed that this environment is anaerobic and favors the production of the viscous exopolysaccharide alginate (24). Meanwhile, it has been shown that P. aeruginosa forms biofilms that are denser and more robust under anaerobic than aerobic conditions, thus calling into question many previous works that were performed on biofilms grown under aerobic conditions (23).
Actually, intensive antipseudomonal treatment has greatly improved the prospects of patients with CF. However, high doses of antibiotics, most commonly used in combination, lead to significant side effects for the patients, as well as the emergence of bacterial strains resistant to these antibiotics. Therefore, the use of molecules like QSIs to attenuate bacterial pathogenicity rather than planktonic bacterial growth is attractive. However, little is known about the relationship between the antibiofilm effect of QSIs and the susceptibility of biofilms to antibiotics.
In order to expand our preliminary results (22), the present study investigated the effects of the new QSI C11 on P. aeruginosa biofilms placed under both aerobic and anaerobic conditions and evaluated the relationship between its antibiofilm activity and the expression of QS regulatory and QS-regulated genes. Finally, the susceptibility of biofilms grown under aerobic and anaerobic conditions to C11 in combination with conventional systemic (ciprofloxacin, ceftazidime), topical (colistin), or systemic/topical (tobramycin) antibiotics used in P. aeruginosa lung infections was analyzed.
MATERIALS AND METHODS
Chemicals.
Ciprofloxacin, tobramycin, ceftazidime, colistin, and all chemicals used for C11 and C4-HSL synthesis were obtained from Sigma-Aldrich (Saint-Quentin-Fallavier, France).
C11 [N-(2-pyrimidyl)butanamide] was prepared by condensation of butyryl chloride with 2-aminopyrimidine (in pyridine). The reaction mixture was stirred under reflux for 30 min. After the mixture was cooled at room temperature, it was filtered and the resulting solution was evaporated under reduced pressure. The crude product was washed with water and recrystallized from ethanol to obtain colorless needles with a melting point of 103°C at 60% yield. The compound was characterized by its 1H nuclear magnetic resonance (NMR) and infrared (IR) spectra.
N-(2-Pyrimidyl)butanamide: molecular weight, 165 g/mol; melting point, 103°C; 1H NMR (CDCl3), δ (parts per million) 8.63, 2H, d (J = 4.9 Hz) H4′, H6′; 8.40, 1H br, NH; 7.01 1H, t (J = 4.85 Hz) H5′; 2.75, 2H, t (J = 7.40 Hz), CH2—CO; 1.81, 2H, m, CH2; 1.05, 3H, t, CH3; IR (KBr) ν (centimeters−1): 3,400, 3,218, 3,074, 3,001, 2,961, 2,902, 2,873, 1,685, 1,583, 1,525, 1,446.
C4-HSL was synthesized following the procedure previously described (26) and was characterized by its 1H NMR and IR spectra.
All compounds tested were dissolved at the appropriate concentrations in sterile distilled water.
Bacterial strain, culture media, and growth conditions.
P. aeruginosa PAO1 was obtained from the Institute Pasteur collection (Paris, France). The strain was frozen and kept at −80°C. Before each experiment, 2 subcultures were prepared on Trypticase soy agar (bioMérieux, Craponne, France) and incubated under aerobic conditions at 37°C for 24 h. Bacterial suspensions were freshly prepared in sterile distilled water before each experiment.
The bacterial enumerations were done on Trypticase soy agar. The antimicrobial susceptibility tests were performed in Trypticase soy broth. The basic medium used for biofilm formation (BBM), modified from the medium described by Campanac et al. (27), contained MgSO4·2H2O (0.2 g/liter), FeSO4·7H2O (0.0005 g/liter), anhydrous Na2HPO4 (1.25 g/liter), KH2PO4 (0.5 g/liter), (NH4)2SO4 (0.1 g/liter), and glucose (0.05 g/liter) (Sigma-Aldrich, Saint-Quentin-Fallavier, France).
Control of planktonic growth.
Liquid cultures of P. aeruginosa PAO1, inoculated at about 102 CFU/ml, were performed in BBM or in Trypticase soy broth in a neutral glass tube. The tubes were incubated at 37°C and 200 rpm using an orbital shaker (New Brunswick Scientific, Edison, NJ, USA) for 48 h. Growth was monitored by measuring the optical density (OD) at 640 nm (OD640) at 6, 24, and 48 h.
Susceptibility testing of planktonic cells.
The MIC and minimum bactericidal concentration (MBC) of C11 and each antibiotic against P. aeruginosa were determined using the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (28). Briefly, 100 μl of C11 or antibiotics was loaded into the wells of a sterile 96-well microtiter plate and subjected to 2-fold serial dilutions with Trypticase soy broth (bioMérieux, Craponne, France) to achieve final concentrations ranging from 128 to 0.125 μg/ml. A bacterial suspension was prepared in sterile distilled water and adjusted to an OD640 of 0.150, corresponding to a concentration of 108 CFU/ml. The microtiter plates were inoculated using a multipoint inoculator (Denley; Mast Diagnostics, Bootle, United Kingdom) to obtain a final concentration of 106 CFU/ml. Cell suspensions were added to all wells except the outer wells, which were used as sterility controls. Growth wells (with bacteria and without test solution) were also included. The plates were prepared in triplicate and then incubated at 37°C for 24 h. The MIC was determined as the lowest concentration of test solution that inhibited the visible growth of the tested microorganism.
The MBC was determined by removing 100-μl aliquots from each well without visible growth and then spread plating the aliquots onto Trypticase soy agar. The plates were incubated at 37°C for 24 h. The MBC was defined as the lowest concentration of test compound that did not permit any visible growth on the appropriate agar plate during the incubation period (99.9% of the bacteria were killed). Each concentration of the tested compounds was tested in triplicate.
C11 antibiofilm activity under aerobic and anaerobic conditions.
The biofilm was assayed by use of the model developed by Khalilzadeh et al. (22) with few modifications. Briefly, biofilms were developed in a sterile 24-well cell culture plate, each well of which contained 1 ml of BBM (2× solution) and 1 ml of sterile distilled water (control) or test compound solution (C11) prepared at different concentrations in sterile distilled water (final concentrations, 8 to 1 μg/ml). The highest concentration of C11 tested (8 μg/ml) corresponded to 50 μM. The bacterial inoculum was prepared by dilution in sterile distilled water of an overnight culture of PAO1 to an OD640 of 0.15, corresponding to a bacterial density of 108 CFU/ml. The inoculum then underwent 10-fold serial dilutions (from 10−1 to 10−6), and 100 μl of the 10−6 dilution (equivalent to 102 CFU/ml) was added to each well. The plates were then incubated at 37°C. After 2, 4, 6, 20, and 24 h of incubation, the wells were rinsed twice with sterile distilled water, and the medium (BBM with or without C11) was replaced.
For the biofilms developed under anaerobic conditions, BBM was supplemented with 1% KNO3 (Sigma-Aldrich, Saint-Quentin-Fallavier, France) as previously recommended (23). Between each medium renewal, the plates were placed in a Mart anaerobic jar (Mart Microbiology B.V., Netherlands) and then flushed with a gas mixture (80% N2, 10% CO2, 10% H2) using a Mart Anoxomat system (Mart Microbiology B.V., Netherlands) before incubation at 37°C.
After 48 h of incubation, biofilms were observed by confocal laser scanning microscopy (CLSM). For the quantification of adherent cells, the wells were rinsed twice with sterile distilled water, and the cells at the bottoms of the wells were scraped with a sterile spatula and placed into 1 ml of sterile distilled water. Harvested biofilm samples were disaggregated by vortexing vigorously for 2 min and by sonication (VibraCell 72441; Bioblock Scientific) at 45 W for 5 s and then analyzed by cell culture (CFU method) and by real-time quantitative PCR (qPCR).
CLSM.
Noninvasive confocal imaging of the biofilms that had been incubated for 48 h was accomplished with a confocal visible Leica DMR upright TCS SP2 AOBS microscope (Leica Microsystems, Wetzlar, Germany) fitted with water-immersion dipping lenses (×40 HCX Apo objective; numerical aperture, 0.8). Specimens were stained using the stain from a LIVE/DEAD BacLight bacterial viability kit (Invitrogen, Cergy-Pontoise, France) for microscopy according to the manufacturer's instructions. An excitation wavelength of 488 nm was used, and the fluorescence emitted at between 500 and 540 nm for SYTO 9 and at 543 nm for propidium iodide was recorded. The biofilm structure was analyzed in duplicate by taking a series of horizontal sections, each of which was 0.5 μm thick. Digital images were processed using Leica confocal software Lite (Leica Microsystems).
Quantification of adherent cells by cell culture (CFU method).
When necessary, logarithmic dilutions of harvested biofilm samples were performed in sterile distilled water in order to decrease the numbers of CFU to between 30 and 300 per plate. Aliquots of bacterial suspensions were then plated out on Trypticase soy agar plates. After 24 h of incubation at 37°C, the numbers of viable CFU for triplicate wells were averaged and subjected to logarithmic transformation. Following this protocol, the detection and quantification limits were fixed at, respectively, 1 CFU/ml and 30 CFU/ml (equivalent to 4 CFU/cm2).
Quantification of adherent cells by qPCR.
Bacterial DNA of the harvested biofilm samples was extracted using a QIAamp DNA stool minikit (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions. Extraction was performed with 200 μl of bacterial suspension (control or samples). The extracted DNA was eluted in 200 μl of elution buffer and immediately stored at −20°C. The primers were designed using Primer Express (version 3.0) software (Applied Biosystems, Foster City, CA, USA), and the sequences were as follows: for the rhlR forward primer, 5′-AACGCGAGATCCTGCAATG-3′, and for the rhlR reverse primer, 5′-GCGCGTCGAACTTCTTCTG-3′. The primer sequences were subjected to a BLAST analysis against the P. aeruginosa PAO1 genome sequence to eliminate the possibility of nonspecific binding. qPCR was carried out using the IQ SYBR green supermix (Bio-Rad, Marnes-la-Coquette, France) with a 25-μl reaction mixture containing 12.5 μl of 2× IQ SYBR green supermix, 0.2 μM each forward and reverse primer, and 5 μl of extracted DNA. qPCR was performed using an iCycler IQ5 real-time PCR detection system (Bio-Rad, Marnes-la-Coquette, France) and the following cycling parameters: an initial denaturation at 95°C for 3 min, followed by 50 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s. Depending on the melting temperature (Tm) of a specific amplicon, the melt curve identifies a characteristic Tm that can distinguish between amplicons differing by only a single base. For our study, a melt curve analysis was performed immediately after amplification at 95°C for 1 min and 55°C for 1 min, followed by 81 repeats of heating for 10 s starting at 55°C and increasing in 0.5°C increments. Negative controls (RNase-free water) were added in each assay.
In order to construct standard curves, three control biofilms were scraped and the number of CFU per square centimeter of each biofilm was determined by cell culture. Then, 10-fold dilutions of each biofilm were made with sterile distilled water and bacterial DNA was extracted from each dilution. The threshold cycle (CT) values of each dilution amplified in triplicate were plotted as a function of the logarithm of the starting quantity of P. aeruginosa (number of CFU per square centimeter). Standard curves were then used to determine the starting quantity of PAO1 biofilms in equivalent numbers of CFU per square centimeter. The slope (s) of the standard curve were used to determine the PCR efficiency (E) in conformity with E = 10(−1/s) − 1 (29). Thus, a standard curve with a slope of −3.33 corresponded to a reaction with an efficiency value of 100%. The quantification limit of the qPCR corresponded to 101 equivalent CFU/cm2.
Gene expression.
The effects of C11 and C4-HSL (used as a positive control) on gene expression in biofilms were tested by adding into each well of a sterile 24-well cell culture plate 1 ml of 2× BBM and 1 ml of sterile distilled water (control) or 1 ml of test compound solution (C11 or C4-HSL) prepared in sterile distilled water (final concentration, 4 μg/ml). Then, biofilm formation was performed as described above for the test of the C11 antibiofilm activity under aerobic and anaerobic conditions. After 48 h of incubation, wells were rinsed twice with sterile distilled water, and the cells at the bottoms of the wells were scraped with a sterile spatula and placed into 1 ml of RNAprotect Bacteria reagent (Qiagen, Courtaboeuf, France) for immediate RNA stabilization.
Total RNA of harvested biofilm samples was extracted using the RNeasy minikit (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions, and a supplementary DNase digestion step was performed with an RNase-free DNase set (Qiagen, Courtaboeuf, France). A preliminary phase of destruction of the bacterial envelope was performed by incubation in a solution of 1 mg/ml of lysozyme for 10 min at room temperature. The primers and probes for the genes rhlR (PA3477), rhlI (PA3476), lasR (PA1430), lasI (PA1432), rhlA (PA3479), and lasB (PA3724) were designed with Primer Express software (Applied Biosystems, Foster City, CA, USA) and were specific for P. aeruginosa PAO1 (NCBI GenBank). The rpoD (PA0576) gene was used as a control for standardization. All primers and probes were purchased from Life Technologies (Saint-Aubin, France) (Table 1). The probes were labeled with the reporter dye 6-carboxyfluorescein (6-FAM) at the 5′ end and the quencher 6-carboxytetramethylrhodamine (TAMRA) at the 3′ end. Quantitative reverse transcriptase PCR (qRT-PCR) was carried out using an iTaq universal probes one-step kit (Bio-Rad, Marnes-la-Coquette, France) with a 25-μl reaction mixture containing 12.5 μl of 2× iTaq universal probes reaction mix, 0.6 μl of iScript advanced reverse transcriptase, 200 nM each forward and reverse primer, 400 nM fluorescently labeled probe, and 5 μl of extracted RNA. qRT-PCR was performed using an iCycler IQ5 real-time PCR detection system (Bio-Rad, Marnes-la-Coquette, France) under the following conditions: incubation at 50°C for 10 min (reverse transcription reaction) followed by an activation step at 95°C for 5 min and then 45 cycles of amplification with denaturation at 95°C for 10 s and annealing/extension at 60°C for 30 s. Negative controls (RNase-free water) were added in each assay.
TABLE 1.
Primers and TaqMan probes used to amplify genes by quantitative reverse transcriptase PCR
| Gene | Sequence |
||
|---|---|---|---|
| Forward primer | Reverse primer | Probea | |
| rhlR | AACGCGAGATCCTGCAATG | GCGCGTCGAACTTCTTCTG | TGAGCATCTCCGAGAGCACGGT |
| rhlI | GCAGCTGGCGATGAAGATATTC | CGAACGAAATAGCGCTCCAT | AGCCTGCAATGCGCCTGGTACCT |
| lasR | GACCAGTTGGGAGATATCGGTTA | TCCGCCGAATATTTCCCATA | CAACTGCTCGGAAGCCAATGTGAACTTC |
| lasI | GCCCCTACATGCTGAAGAACA | CGAGCAAGGCGCTTCCT | CTTCCCGGAGCTTCTGCACGGC |
| rhlA | GGCGATCGGCCATCTG | AGCGAAGCCATGTGCTGAT | TCAACGAGACCGTCGGCAAATACCTG |
| lasB | CGACAACGCGTCGCAGTA | AGGTAGAACGCACGGTTGTACA | TCGACGTGCACCACTCCAGCG |
| rpoD | ACAAGATCCGCAAGGTACTGAAG | CGCCCAGGTGCGAATC | CGCCAAAGAGCCGATCTCCATGG |
The probes were labeled at the 5′ end with 6-FAM and at the 3′ end with TAMRA.
To determine the impact of the test compound on gene expression in biofilms, each biofilm condition (C11 treatment, C4-HSL treatment, and control treatment) was tested in triplicate, and the RNA was extracted from each triplicate biofilm. Then, each extracted RNA was amplified in triplicate and the means of the CT values were used for further analysis. For each sample, the mean of the CT values obtained for a given gene (CT gene) was normalized to the mean of the CT values obtained for the rpoD gene (CT rpoD) amplified from the same sample at the same time. To quantify the fold change in gene expression, we used a relative quantification in which the expression levels of genes in treated biofilms were compared to the expression levels of genes in control biofilms using the following formula: 2−ΔΔCT, where ΔCT = CT gene − CT rpoD and ΔΔCT = ΔCT for the treated biofilm − ΔCT for the control biofilm.
Antibiofilm activity of C11 in combination with antibiotics.
Biofilm formation was performed as described above for the test of the C11 antibiofilm activity under aerobic and anaerobic conditions. At time zero (T0), 100 μl of the bacterial inoculum was added to each well of a sterile 24-well cell culture plate. For control biofilms and biofilms treated with C11 alone, 1 ml of 2× BBM with, respectively, 1 ml of sterile distilled water (control) or 1 ml of C11 solution (1 μg/ml, C11 alone) was added to the wells at T0. After 2, 4, 6, 20, and 24 h of incubation at 37°C, the wells were rinsed and the medium (BBM with or without C11) was renewed. For biofilms treated with antibiotics alone, 1 ml of 2× BBM with 1 ml of sterile distilled water was added to the wells at 0, 2, 4, 6, and 20 h, and then at 24 h the medium was replaced with 1 ml of 2× BBM with 1 ml of antibiotic solution. For biofilms treated with a C11-antibiotic combination, 1 ml of 2× BBM with 1 ml of C11 solution was added to the wells at 0, 2, 4, 6, and 20 h, and then at 24 h the medium was replaced by 1 ml of 2× BBM with 1 ml of a solution containing C11 and antibiotics. For all conditions, biofilms were analyzed at 48 h by confocal laser scanning microscopy, and the number of adherent cells was quantified by the CFU method and qPCR.
Statistical analysis.
The Student t test was used to calculate the significance of the difference between the mean effects of a given compound and those for the control group. For each assay, all determinations were carried out in triplicate from three independent experiments. Statistically significant values were defined as a P value of <0.05, <0.01, or <0.001.
RESULTS
Effects of C11 on growth of planktonic cells.
Suspensions of P. aeruginosa cells were made in Trypticase soy broth, as a positive control, and in BBM and incubated at 37°C for 48 h under agitation in order to limit biofilm development. Planktonic growth was monitored after 6, 24, and 48 h (Table 2). As expected, in Trypticase soy broth, the OD640 increased strongly with time up to 2.984 ± 0.010. In contrast, no visible growth occurred in BBM, with the OD640 reaching only 0.034 ± 0.002 after 48 h of incubation.
TABLE 2.
Planktonic growth of P. aeruginosa in Trypticase soy broth or BBM in tubesa
| Medium |
P. aeruginosa OD640 at: |
||
|---|---|---|---|
| 6 h | 24 h | 48 h | |
| Trypticase soy broth | 0.001 ± 0.001 | 0.016 ± 0.004 | 0.034 ± 0.002 |
| BBM | 0.000 ± 0.001 | 2.162 ± 0.066 | 2.984 ± 0.010 |
Growth was monitored by determination of the OD at 640 nm after 6, 24, and 48 h of incubation at 37°C. Values are means ± standard deviations (n = 3).
Determination of the MICs and MBCs of C11 and antibiotics against P. aeruginosa was therefore performed in Trypticase soy broth (Table 3). No bactericidal activity was obtained for C11 at the highest concentration tested of 128 μg/ml, corresponding to a molarity of greater than 500 μM. In contrast, the antibacterial activities of all the antibiotics tested against P. aeruginosa showed low MIC and MBC values.
TABLE 3.
Antimicrobial activities of C11 and antibiotics against P. aeruginosaa
| Compound | MICb (μg/ml) | MBCc (μg/ml) |
|---|---|---|
| C11 | >128 | >128 |
| Ciprofloxacin | <0.125 | <0.125 |
| Tobramycin | 1 | 2 |
| Ceftazidime | 4 | 8 |
| Colistin | 4 | 8 |
Data are from three experiments.
MIC, the concentration at which no bacterial growth was visible after 24 h of incubation at 37°C.
MBC, minimal bactericidal concentration (the concentration at which the number of CFU was reduced by 99.9%).
Antibiofilm activity of C11 under aerobic and anaerobic conditions.
In order to promote the growth of adherent P. aeruginosa bacteria and not simple cell adhesion and to produce a more accurate model with a monospecies structured biofilm for in vitro purposes, the wells of a sterile 24-well cell culture plate were inoculated with a low bacterial inoculum and BBM was renewed after 2, 4, 6, 20, and 24 h of incubation. After 48 h, control biofilms observed by confocal microscopy and developed under aerobic and anaerobic conditions clearly showed a typical biofilm structure (Fig. 1) and consisted of an average of 5.79 ± 0.04 and 6.33 ± 0.12 log CFU/cm2, respectively (Fig. 2). These results and the demonstration that there was no visible planktonic growth in BBM bring out that biofilms formed in microplate wells result only from the growth of the adhered bacteria.
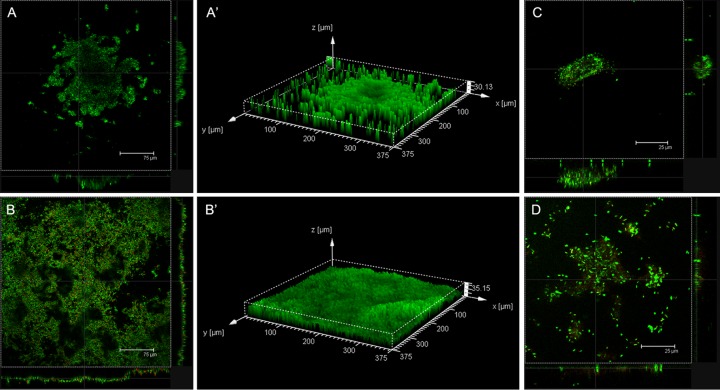
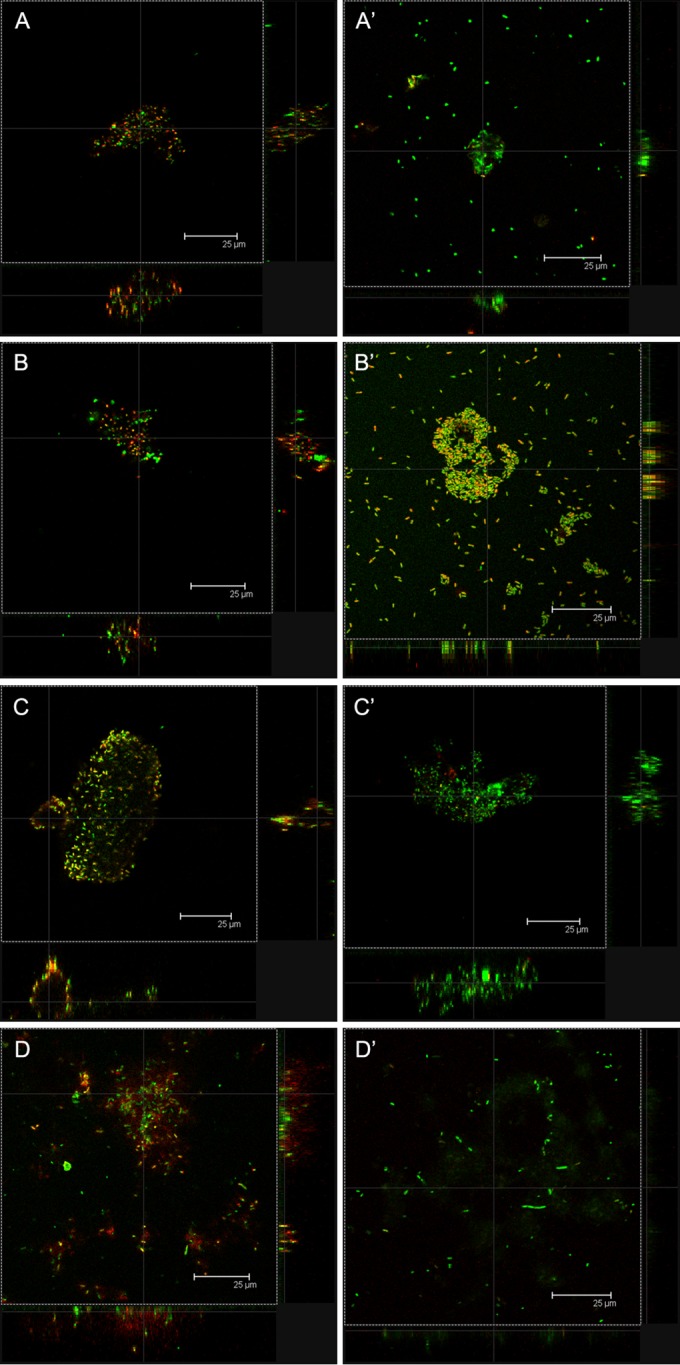
FIG 1.
Confocal laser scanning microscopy images of P. aeruginosa biofilms incubated at 37°C for 48 h under aerobic (A, A′, C) or anaerobic (B, B′, D) conditions and then stained using stain from a LIVE/DEAD BacLight bacterial viability kit. (A, A′, B, B′) Control biofilms; (C, D) biofilms grown in the presence of C11 at 1 μg/ml in the medium; (A′, B′) three-dimensional images of control biofilms.
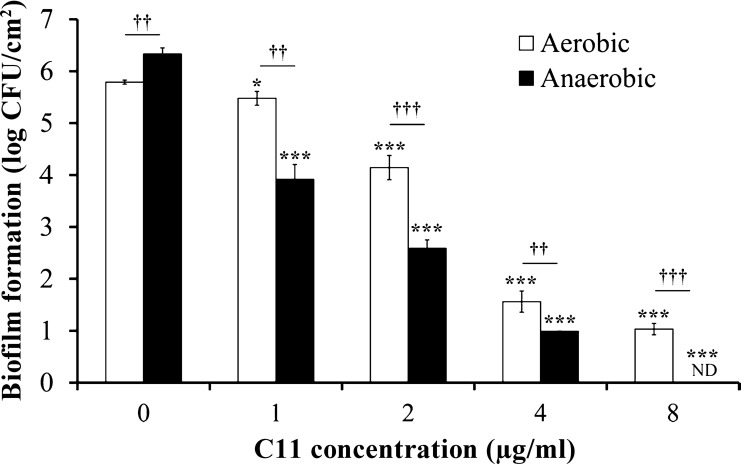
FIG 2.
Effect of C11 on P. aeruginosa biofilm development under aerobic and anaerobic conditions at 37°C for 48 h. Results are expressed as the means and standard deviations from triplicate experiments. Statistically significant differences (*, P < 0.05; ***, P < 0.001) between the test compound and its control under aerobic or anaerobic conditions (n = 3) and statistically significant differences (††, P < 0.01; †††, P < 0.001) between the control or test compound under aerobic conditions and the control or test compound under anaerobic conditions (n = 3) are indicated. ND, not detected.
Confocal microscopy of control biofilms developed for 48 h at 37°C under aerobic or anaerobic conditions showed that they had very different structures (Fig. 1). Biofilms developed under aerobic conditions were characterized by the formation of typical microcolonies with more bacteria at the biofilm base (thickness, 30.13 μm; Fig. 1A and A′). Biofilms grown under anaerobic conditions (Fig. 1B and B′) were thicker (35.15 μm) and more compact than biofilms grown under aerobic conditions, and the bacteria were distributed more uniformly over the well surface. Moreover, the quantification of adherent cells by cell culture of these two control biofilms showed that biofilms grown under anaerobic conditions contained significantly more P. aeruginosa bacteria than ones grown under aerobic conditions (Fig. 2).
The activity of C11 on P. aeruginosa biofilm development was tested under aerobic and anaerobic conditions (Fig. 1C and D and 2). Under both conditions, C11 showed significant inhibition of biofilm formation in a dose-dependent manner (Fig. 2). For all concentrations tested, the antibiofilm activity of C11 was significantly greater on biofilms grown under anaerobic conditions, with no biofilm development at 8 μg/ml, than on biofilms grown under aerobic conditions.
Effects of C11 on QS gene expression.
The effects of C11 and C4-HSL on the expression of QS regulatory genes (rhlR, rhlI, lasR, lasI) and QS-regulated genes (rhlA, lasB) by P. aeruginosa biofilms grown at 37°C for 48 h under aerobic and anaerobic conditions were measured by quantitative RT-PCR (Fig. 3). The levels of QS gene expression were normalized to the levels of rpoD gene expression and then compared with the levels of expression in control biofilms. The compounds were tested at the same concentration of 4 μg/ml because this was the highest concentration that did not result in the complete inhibition of biofilm formation by C11 (Fig. 2). The quantification of adherent cells in biofilms grown in the presence of 4 μg/ml of C4-HSL showed that they contained significantly (P < 0.001, n = 3) more bacteria than their respective untreated biofilms: 6.41 ± 0.08 and 7.19 ± 0.08 log CFU/cm2 under aerobic and anaerobic conditions, respectively (data not shown).
FIG 3.
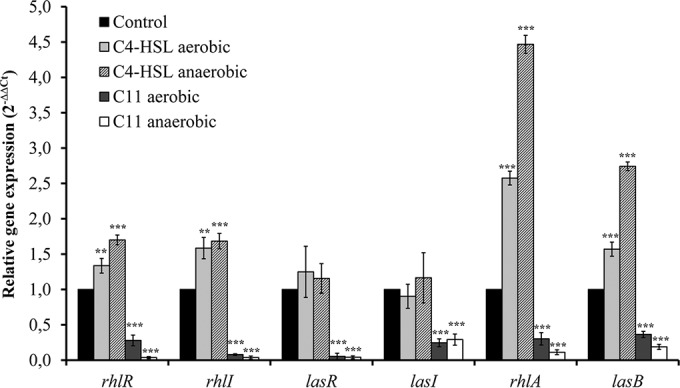
Relative expression of quorum-sensing genes (rhlR, rhlI, lasR, lasI, rhlA, lasB) measured by quantitative reverse transcriptase PCR from P. aeruginosa biofilms untreated (control) or treated with C4-HSL (4 μg/ml) or C11 (4 μg/ml). Biofilms were grown at 37°C for 48 h under aerobic or anaerobic conditions. The levels of QS gene expression were normalized to the level of rpoD gene expression and then compared with the level of expression by control biofilms. Results are expressed as the means and standard deviations from triplicate experiments. Statistically significant differences (**, P < 0.01; ***, P < 0.001) between the test compound and its control are indicated (n = 3).
Under both biofilm development culture conditions, C4-HSL led to the significant upregulation of QS regulatory genes belonging to the rhl system (rhlR and rhlI), but no significant effect was observed on las system genes (lasR and lasI). C4-HSL led to the significant upregulation of the QS-regulated virulence genes rhlA (rhamnolipid synthesis) and lasB (elastase synthesis), which are mainly controlled by the rhl system, with fold changes in expression under aerobic and anaerobic conditions being 2.58 and 4.47, respectively, for rhlA and 1.57 and 2.74, respectively, for lasB. In contrast, C11 showed a significant downregulation of QS regulatory genes belonging to both the rhl and las systems, as well as the QS-regulated virulence genes rhlA and lasB, with fold changes in expression being between −2.75 for lasB and −18.84 for lasR under aerobic conditions and between −3.44 for lasI and −32.96 for rhlR under anaerobic conditions.
Compared to the regulation of gene expression in biofilms grown under aerobic conditions, the upregulation of gene expression induced by C4-HSL on biofilms developed under anaerobic conditions was significantly (P < 0.05) greater for the rhlR, rhlA, and lasB genes and the downregulation induced by C11 was significantly (P < 0.05) higher for the rhlR, rhlI, rhlA, and lasB genes.
Antibiofilm activity of C11 in combination with antibiotics.
The activity of antibiotics on 24-h-old C11-untreated P. aeruginosa biofilms (treated with an antibiotic alone) or C11-pretreated P. aeruginosa biofilms (treated with the C11-antibiotic combination) grown under aerobic (Fig. 4A) and anaerobic (Fig. 4B) conditions at 37°C for 48 h was analyzed. When it was tested, C11 was added to the wells at T0 and at each medium renewal at 1 μg/ml, corresponding to the lowest concentration showing a significant inhibitory effect on biofilms grown under aerobic conditions (P < 0.05; reduction of 0.29 ± 0.13 log CFU/cm2), in an attempt to identify a possible synergy of activity when C11 was combined with antibiotics. All antibiotics were added to the wells alone or in combination with C11 after 24 h of biofilm formation. The highest antibiotic concentrations tested were based on the average peak levels achieved in serum 1 h after administration of a standard single intravenous or oral dose: tobramycin, 8 μg/ml; ceftazidime, 32 μg/ml; and ciprofloxacin, 2 μg/ml. Colistin was tested at 32 μg/ml, which is the median concentration in sputum after 1 h of inhalation of the recommended dose of colistin. However, the recommended concentrations of 8 μg/ml for tobramycin and 32 μg/ml for colistin led to high levels of inhibition of biofilm formation (greater than 3 log units; data not shown), and in order to observe a possible synergy of activity, the concentrations of these two antibiotics were reduced by half: the tobramycin concentration was reduced to 4 μg/ml and the colistin concentration was reduced to 16 μg/ml.
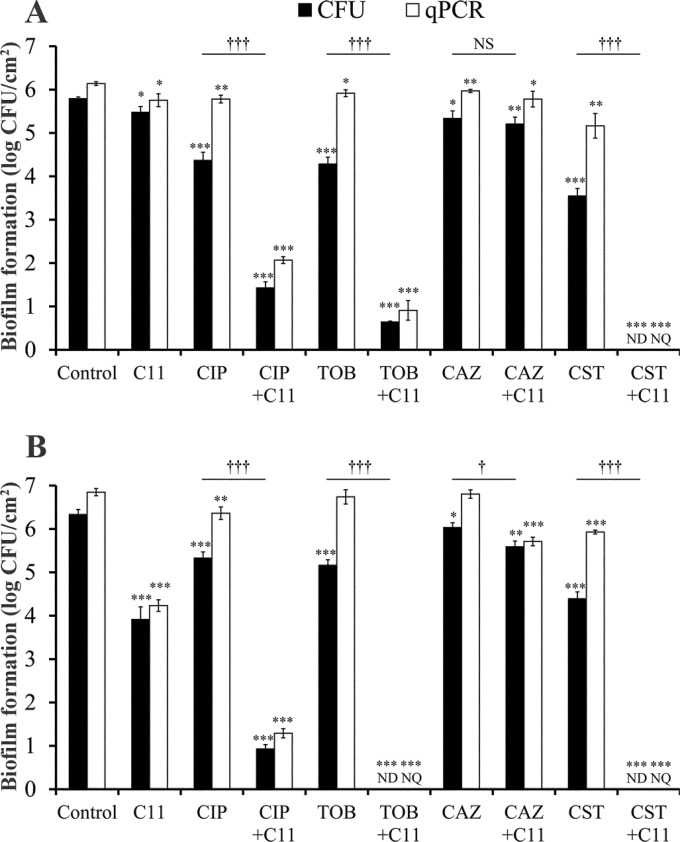
FIG 4.
Effect of C11 and antibiotics, tested alone or in combination, on P. aeruginosa biofilms grown under aerobic (A) and anaerobic (B) conditions at 37°C for 48 h. C11 (1 μg/ml) was added to the wells at T0. Ciprofloxacin (CIP; 2 μg/ml), tobramycin (TOB; 4 μg/ml), ceftazidime (CAZ; 32 μg/ml), and colistin (CST; 16 μg/ml) were tested on 24-h-old C11-untreated biofilms (treated with antibiotics alone) or C11-pretreated biofilms (treated with the C11-antibiotic combination). Quantification of adhered cells was carried out after 48 h of incubation by cell culture (the CFU method; log number of CFU per square centimeter) and by qPCR (equivalent log number of CFU per square centimeter). Results are expressed as the means and standard deviations from triplicate experiments. Statistically significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) between the test compound and its control (n = 3), measured by CFU enumeration or qPCR, and statistically significant differences (†, P < 0.05; †††, P < 0.001) between antibiotics tested alone and in combination with C11, measured by CFU enumeration (n = 3), are indicated. NS, not significant; ND, not detected; NQ, not quantified.
We observed a loss of activity of antibiotics against P. aeruginosa biofilms grown under aerobic conditions in comparison to that against planktonic cells, with reductions of between 0.49 log unit for ceftazidime and 2.27 log units for colistin, even though the concentrations tested were 2- to 16-fold higher than the MBCs. For ciprofloxacin, tobramycin, and colistin, the addition of C11 led to a significant (P < 0.001) increase in the antibiofilm activity of the antibiotics, whereas for ceftazidime, the addition had no significant effect. These two observations were also obtained with biofilms developed under anaerobic conditions. However, the effects of the antibiotics tested alone on biofilms grown under anaerobic conditions were generally less than those on biofilms grown under aerobic conditions, with reductions of between 0.30 log unit for ceftazidime and 2.01 log units for colistin, while the effects of the combination of the antibiotics with C11 were significantly greater on biofilms grown under anaerobic conditions.
On the other hand, when we compared the results obtained by cell culture (number of CFU) and qPCR for antibiotics tested alone on biofilms developed under both conditions, we observed that the number of cells quantified by qPCR was always higher than the number measured by culture, indicating that the activity of the antibiotics was mainly bactericidal or at least led to cell damage with no regrowth. This difference between the results of the two quantification methods was not significant for C11, indicating that its antibiofilm activity may be due to other mechanisms, which was foreseeable, since it was tested at a concentration considerably lower than its MIC/MBC (>128 μg/ml).
This hypothesis was confirmed by confocal microscopy observations, which revealed that P. aeruginosa biofilms treated with 1 μg/ml of C11 were thick and contained very few dead cells (in red) relative to the number of living cells (in green) (Fig. 1C). In comparison, biofilms exposed to antibiotics alone had equivalent thicknesses but their matrices showed many dead cells due to the cell damage caused by the antibiotics (Fig. 5, left). In addition, biofilms treated with the combination of antibiotics and C11 were finer (Fig. 5, right), underlining the combination of actions, and the number of dead cells was less than the number of live cells, suggesting that C11 caused a modification in the structure of the biofilms, causing the bacteria to revert to the planktonic state, which allowed the antibiotics to exercise their activity. Damaged cells were then removed by rinsing just before observations were made by CSLM.
FIG 5.
Confocal laser scanning microscopy images of P. aeruginosa biofilms incubated at 37°C for 48 h under aerobic conditions and treated with antibiotics alone (A to D) or with a combination of antibiotics and C11 (A′ to D′). (A) Ciprofloxacin at 2 μg/ml; (A′) ciprofloxacin at 2 μg/ml combined with 1 μg/ml of C11; (B) tobramycin at 4 μg/ml; (B′) tobramycin at 4 μg/ml combined with 1 μg/ml of C11; (C) ceftazidime at 32 μg/ml; (C′) ceftazidime at 32 μg/ml combined with 1 μg/ml of C11; (D) colistin at 16 μg/ml; (D′) colistin at 16 μg/ml combined with 1 μg/ml of C11. Antibiotics were added in the screening model at 24 h, and C11 was added at T0. Biofilms were stained using stain from a LIVE/DEAD BacLight bacterial viability kit.
DISCUSSION
P. aeruginosa is known to grow in a sessile form in the lungs of cystic fibrosis patients and to undergo a specific transition from colonization to infection as a result of the expression of virulence factors and host responses (4). Biofilm formation also induces a loss of microbial susceptibility, leading to the use of higher concentrations of antibiotics and/or combinations (30, 31). Therefore, we focused our study on the search for new molecules that act upstream to prevent biofilm formation, for molecules that potentiate the action of conventional antibiotics on a structured biofilm, or for molecules having both activities.
The quorum-sensing bacterial communication system in P. aeruginosa plays a key role in the control of the production of several virulence factors and in the regulation of biofilm formation (7, 8, 32, 33), making it a very attractive target in the search for antibiofilm compounds. Many studies have been aimed at affecting the synthesis of 3-oxo-C12-HSL because in vitro studies, performed under aerobic conditions, have shown that this HSL is required for biofilm differentiation and that the lasI gene is expressed in a large number of cells during the initial phase of biofilm development. In contrast, the rhlI gene was considered to be expressed in fewer cells, suggesting that C4-HSL plays no central role in biofilm formation (34). In vivo, both HSLs have been detected in the sputum of patients with cystic fibrosis (16, 17, 35). However, it has been shown more recently that when P. aeruginosa isolates from the sputum of CF patients are grown in biofilms, they produce larger amounts of C4-HSL than 3-oxo-C12-HSL, with the ratio being 3:1 (16). Moreover, it has been observed that the amount of biofilm produced by a lasI mutant (deficient in 3-oxo-C12-HSL) of PAO1 strain PT5 is similar to the amount produced by the wild type, whereas an rhlI mutant (deficient in C4-HSL) produces 70% less biofilm than the wild type (36). On the other hand, C4-HSL controls the production of rhamnolipids, shown to be required for the maintenance of the biofilm architecture (37). All these observations suggest that C4-HSL also plays a significant role in biofilm formation. Therefore, we previously designed and then synthesized HSL analogs on the basis of the structure of C4-HSL (22). Among them, N-(2-pyrimidyl)butanamide (called C11) showed significant inhibition of P. aeruginosa biofilm formation in a dose-dependent manner.
Recent data indicate that the stagnant mucus of the CF airway is anaerobic (24), suggesting that P. aeruginosa infection in CF patients reflects biofilm formation and persistence in an anaerobic environment. Yoon et al. (23) have demonstrated that P. aeruginosa forms more robust biofilms during anaerobic growth, and we confirmed this finding not only by cell culture (the CFU method) and qPCR quantification but also by confocal microscopy observations (Fig. 1 and 2). Moreover, we confirmed the antibiofilm effect of C11, which showed an amazing enhancement of biofilm regulation under anaerobic conditions. This higher antibiofilm activity of C11 under these conditions compared to that under aerobic conditions confirms our preliminary hypothesis (22) that C11 acts by inhibition of the rhl system.
Yoon et al. (23) also considered that the most efficient mode of growth under anaerobic conditions required enzymes/proteins that allowed P. aeruginosa to (i) utilize nitrate (NO3−) and nitrite (NO2−) as terminal electron acceptors, (ii) influence the activity/stability of denitrifying enzymes (OprF protein), and (iii) modulate the production (rhl QS system) and removal (nitric oxide reductase) of toxic nitric oxide (NO), a by-product of anaerobic respiration. Thus, we suggest that these pathways represent candidate targets for a novel suite of antimicrobials, founded on the basis of the proposal that successful treatment of the disease may require the disruption of anaerobic respiration within the anaerobic airway mucus. These data provide one more reason to search for an inhibitor of the rhl system and, thus, to select analogs based on the C4-HSL structure.
Moreover, in order to confirm that C11 exerts its activity on biofilm formation via inhibition of the rhl system, we evaluated its effect on the expression of QS regulatory and QS-regulated genes. As expected, when C4-HSL was added, a significant increase in rhlR and rhlI expression was observed, while no impact on lasR and lasI expression was noted. In contrast, the addition of C11 induced a significant downregulation of regulatory genes belonging to both the rhl and las systems (Fig. 3). So, it is likely that C11 reduced the production of both C4-HSL and 3-oxo-C12-HSL. This result is not surprising, as structure-activity relationship studies have already revealed some interactions between the compound C11 and the receptor protein LasR (22). As 3-oxo-C12-HSL is required for biofilm differentiation (6), it is likely that C11, by reducing the effects of both HSLs, not only leads to a reduction in the total amount of biofilm formed but also affects its differentiation. On the other hand, many of the virulence factors produced by P. aeruginosa are regulated by the QS system (38). Among these are rhamnolipids and elastase, whose synthesis partly depends on the activation of the rhlA and lasB genes, respectively (39, 40). Rhamnolipids and elastase are capable of degrading or inactivating important biological tissues and immune system components (40, 41) and have been found to contribute to the virulence of P. aeruginosa in clinical studies (42, 43). In our study, C11 led to a significant inhibition of the expression of rhlA and lasB and so could certainly contribute to the fight against P. aeruginosa pathogenesis.
Concerning antibiotic activity, our results (Table 3 and Fig. 4) confirm the loss of antibiotic susceptibility when P. aeruginosa is grown as a biofilm. In addition, an important demonstration in our work concerns the higher levels of resistance to antibiotics of P. aeruginosa biofilms growing under anaerobic conditions. This loss of biofilm susceptibility to antimicrobial treatments is well-known and is often the result of a combination of transfer limitation, the consumption/neutralization of the antibiotics by biofilm matrix components, and drastic modifications in gene expression and cell physiology (31). This process must thus be considered ubiquitous, as the various antibiotics tested have different cell targets, i.e., peptidoglycan synthesis for ceftazidime (44), DNA synthesis for quinolones, and protein synthesis for tobramycin (45). For colistin, the target is lipid A of the lipopolysaccharide (LPS) of the outer membrane of Gram-negative bacteria (46). Thanks to its positive charge, colistin interacts electrostatically with these molecules and competitively displaces divalent cations from them, causing disruption of the membrane. This results in an increase in cell wall permeability and, subsequently, cell death (47, 48). Other combined effects are expected to produce lethality leading to rapid bactericidal activity combined with a systemic toxicity. Thus, colistin could be considered a biocide rather than an antibiotic. The loss of activity on biofilms could be linked to its chemical characteristics. Colistin is a cyclic lipodecapeptide, which is strongly cationic and can be neutralized by the polyanionic exopolysaccharides (49). In spite of this, our results underline the fact that neither the bacterial target nor the antibacterial activity (inhibition of growth or bactericidal activity) is discriminating for the selection of antibiotics active against biofilms.
Because of the interest in C11 as a QS inhibitor, we combined it with antibiotics (Fig. 4 and 5). As previously described, C11 had no killing effect on a mature biofilm (22). The activities of antibiotics on 24-h-old C11-untreated or C11-pretreated biofilms were therefore tested in order to demonstrate the effects of combination treatment. A dramatic increase in the antibiofilm activities of the antibiotics was observed when C11 was combined with ciprofloxacin, tobramycin, and, above all, colistin, which resulted in the detection of no adherent cells. Surprisingly, in our study, C11 did not restore the efficiency of ceftazidime against biofilms grown under either aerobic or anaerobic conditions. For the other antibiotics tested, impairment of the QS system (i) led to a significant modification of the biofilm matrix, resulting in poor interactions with active molecules, which were thus able to reach their cell targets, and/or (ii) maintained the intracellular machinery for antibiotic efficiency. For colistin, the first hypothesis is more likely, considering its cationic structure and its membrane target (50). On the other hand, previous important work of Wagner et al. (51) on the shift in gene expression between planktonic and sessile P. aeruginosa cells showed that genes implicated in DNA replication, recombination, modification, and repair could be considered to be repressed in biofilm cells, which could explain the restoration of ciprofloxacin activity when C11 is present. In the same way, transcription and RNA processing and degradation were considered to be downregulated. For genes implicated in cell wall/LPS/capsule production, the results are more difficult to interpret because of the balance between the genes whose expression was promoted and repressed. Our observations by CLSM (Fig. 5) did not reveal any modification of cell morphology, while the cell damage cause by ceftazidime treatment is considered characteristic of the damage induced by beta-lactam molecules, which consisted of the elongation of specific cells (52), and molecules such as tobramycin induced abnormal proteins that were even incorporated into the cell wall, resulting in an apparent loss of integrity or elasticity and a rough appearance, as demonstrated by atomic force microscopy observations (53). So, under our conditions, the inhibition of C4-HSL did not allow ceftazidime to reach its cellular target (periplasmic transpeptidases and carboxypeptidases) and/or to induce significant effects, given that peptidoglycan synthesis is involved in the cell division process.
These effects emphasizing the great therapeutic value of QSIs in rendering biofilms more susceptible to antimicrobial compounds have been previously observed and encouraged us to conduct the same type of experiment with combinations of compounds. Hentzer et al. (54) demonstrated that, unlike untreated biofilms, P. aeruginosa biofilms pretreated with the synthetic halogenated furanone C-30 were easily killed by tobramycin treatment. Similarly, P. aeruginosa biofilms were more sensitive to tobramycin when they were pretreated with either garlic extract or extracts from Penicillium species (patulin and penicillic acid), all of which inhibited the QS system, as judged from the findings of DNA array-based transcriptomics (55, 56). Moreover, Christensen et al. (57) demonstrated that the furanone C-30, ajoene from garlic, and horseradish juice extract exhibited QSI activity in vitro and provided synergistic antimicrobial effects with tobramycin in combination treatments initiated early after infection against in vivo P. aeruginosa biofilms in a mouse model of an intraperitoneal foreign-body infection. Unfortunately, these compounds are unsuitable for human use: halogenated furanones are unstable, fungal compounds are mycotoxins, and the active components from natural sources exist at very low concentrations (21). However, C11 is a stable compound that can be easily synthesized in large quantities and is not cytotoxic to lung cells (22). Thus, these data observed for in vivo biofilms are promising for the development of a prophylactic or early C11-antibiotic combination treatment strategy for patients with P. aeruginosa infections.
To date, only azithromycin (AZM), a macrolide antibiotic known to be a QSI, has been gaining favor for the treatment of chronic lung infections caused by P. aeruginosa due to the improved outcomes seen in CF patients (58). The mechanisms of action of AZM are multiple and may include an anti-inflammatory effect, inhibition of alginate production, inhibition of protein synthesis, and/or inhibition of the QS system and the associated virulence factors that are produced (59). AZM was found to inhibit biofilm formation at a sub-MIC (2 μg/ml), and this activity was specific to the initial stages of biofilm formation, since mature biofilms were not affected by the presence of 8 μg/ml of AZM (60). Moreover, AZM reduced the levels of production of both C4-HSL and 3-oxo-C12-HSL (36). These data are comparable to those observed for C11, suggesting its clinical efficacy. Concerning the combination of AZM with antibiotics, it was shown that the addition of AZM decreased the MICs of many antipseudomonal agents (61), and synergistic activity in eradicating a 24-h-old P. aeruginosa biofilm was observed only with the combination of a high dose of AZM (256 μg/ml) and ciprofloxacin (1 μg/ml) (62).
In conclusion, it appears that C11, by reducing cell-to-cell signal production, interferes with biofilm formation at different levels under both aerobic and anaerobic conditions, resulting in a decrease in the levels of production of extracellular virulence factors, such as rhamnolipids and elastase, and also, in combination with antibiotics, allowing the recovery of their antibacterial activity against P. aeruginosa biofilms. The use of original synthetic molecules capable of interfering with biofilm development and the expression of the associated virulence factors constitutes an interesting alternative to the massive use of antibiotics and promises new approaches for the prevention and/or treatment of P. aeruginosa infections. The clinical application of such molecules should be considered further, as prophylactic or early treatment with the combination of C11 and conventional antibiotics may prevent the formation of highly resistant P. aeruginosa biofilms on medical devices as well as in the lungs of CF patients before a chronic infection is established. Moreover, C11 may improve the outcomes of patients with P. aeruginosa infections by increasing the efficacy of the antibiotics against biofilm-forming bacteria and by attenuating the pathogenicity of the bacterium. Therefore, this study should be continued to determine the best combination of molecules effective against P. aeruginosa biofilms and to define optimal conditions for further investigations in a murine model of chronic lung infection.
ACKNOWLEDGMENTS
We thank the Association Vaincre la Mucoviscidose and the Association Gregory Lemarchal for support and advice with this project.
We also thank the individuals from the FRAIB-TRI imaging platform (Toulouse, France) for advice and technical assistance with confocal imaging.
REFERENCES
- 1.Van Delden C, Iglewski BH. 1998. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis 4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canton R, Cobos N, de Gracia J, Baquero F, Honorato J, Gartner S, Alvarez A, Salcedo A, Oliver A, Garcia-Quetglas E. 2005. Antimicrobial therapy for pulmonary pathogenic colonisation and infection by Pseudomonas aeruginosa in cystic fibrosis patients. Clin Microbiol Infect 11:690–703. doi: 10.1111/j.1469-0691.2005.01217.x. [DOI] [PubMed] [Google Scholar]
- 3.Matsui H, Wagner VE, Hill DB, Schwab UE, Rogers TD, Button B, Taylor RM II, Superfine R, Rubinstein M, Iglewski BH, Boucher RC. 2006. A physical linkage between cystic fibrosis airway surface dehydration and Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci U S A 103:18131–18136. doi: 10.1073/pnas.0606428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strateva T, Yordanov D. 2009. Pseudomonas aeruginosa—a phenomenon of bacterial resistance. J Med Microbiol 58:1133–1148. doi: 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- 5.Breidenstein EB, de la Fuente-Nunez C, Hancock RE. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Hill D, Rose B, Pajkos A, Robinson M, Bye P, Bell S, Elkins M, Thompson B, Macleod C, Aaron SD, Harbour C. 2005. Antibiotic susceptibilities of Pseudomonas aeruginosa isolates derived from patients with cystic fibrosis under aerobic, anaerobic, and biofilm conditions. J Clin Microbiol 43:5085–5090. doi: 10.1128/JCM.43.10.5085-5090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams P, Camara M, Hardman A, Swift S, Milton D, Hope VJ, Winzer K, Middleton B, Pritchard DI, Bycroft BW. 2000. Quorum sensing and the population-dependent control of virulence. Philos Trans R Soc Lond B Biol Sci 355:667–680. doi: 10.1098/rstb.2000.0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 9.Schuster M, Greenberg EP. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol 296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 10.Parsek MR, Greenberg EP. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc Natl Acad Sci U S A 97:8789–8793. doi: 10.1073/pnas.97.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wade DS, Calfee MW, Rocha ER, Ling EA, Engstrom E, Coleman JP, Pesci EC. 2005. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol 187:4372–4380. doi: 10.1128/JB.187.13.4372-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pesci EC, Pearson JP, Seed PC, Iglewski BH. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol 179:3127–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whiteley M, Lee KM, Greenberg EP. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGrath S, Wade DS, Pesci EC. 2004. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol Lett 230:27–34. doi: 10.1016/S0378-1097(03)00849-8. [DOI] [PubMed] [Google Scholar]
- 16.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 17.Favre-Bonte S, Pache JC, Robert J, Blanc D, Pechere JC, van Delden C. 2002. Detection of Pseudomonas aeruginosa cell-to-cell signals in lung tissue of cystic fibrosis patients. Microb Pathog 32:143–147. doi: 10.1006/mpat.2001.0487. [DOI] [PubMed] [Google Scholar]
- 18.Pearson JP, Feldman M, Iglewski BH, Prince A. 2000. Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect Immun 68:4331–4334. doi: 10.1128/IAI.68.7.4331-4334.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith KM, Bu Y, Suga H. 2003. Induction and inhibition of Pseudomonas aeruginosa quorum sensing by synthetic autoinducer analogs. Chem Biol 10:81–89. doi: 10.1016/S1074-5521(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 20.Persson T, Hansen TH, Rasmussen TB, Skinderso ME, Givskov M, Nielsen J. 2005. Rational design and synthesis of new quorum-sensing inhibitors derived from acylated homoserine lactones and natural products from garlic. Org Biomol Chem 3:253–262. doi: 10.1039/b415761c. [DOI] [PubMed] [Google Scholar]
- 21.Rasmussen TB, Givskov M. 2006. Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol 296:149–161. [DOI] [PubMed] [Google Scholar]
- 22.Khalilzadeh P, Lajoie B, El Hage S, Furiga A, Baziard G, Berge M, Roques C. 2010. Growth inhibition of adherent Pseudomonas aeruginosa by an N-butanoyl-l-homoserine lactone analog. Can J Microbiol 56:317–325. doi: 10.1139/W10-013. [DOI] [PubMed] [Google Scholar]
- 23.Yoon SS, Hennigan RF, Hilliard GM, Ochsner UA, Parvatiyar K, Kamani MC, Allen HL, DeKievit TR, Gardner PR, Schwab U, Rowe JJ, Iglewski BH, McDermott TR, Mason RP, Wozniak DJ, Hancock RE, Parsek MR, Noah TL, Boucher RC, Hassett DJ. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev Cell 3:593–603. doi: 10.1016/S1534-5807(02)00295-2. [DOI] [PubMed] [Google Scholar]
- 24.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC, Doring G. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109:317–325. doi: 10.1172/JCI0213870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Son MS, Matthews WJ Jr, Kang Y, Nguyen DT, Hoang TT. 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun 75:5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chhabra SR, Harty C, Hooi DS, Daykin M, Williams P, Telford G, Pritchard DI, Bycroft BW. 2003. Synthetic analogues of the bacterial signal (quorum sensing) molecule N-(3-oxododecanoyl)-l-homoserine lactone as immune modulators. J Med Chem 46:97–104. doi: 10.1021/jm020909n. [DOI] [PubMed] [Google Scholar]
- 27.Campanac C, Pineau L, Payard A, Baziard-Mouysset G, Roques C. 2002. Interactions between biocide cationic agents and bacterial biofilms. Antimicrob Agents Chemother 46:1469–1474. doi: 10.1128/AAC.46.5.1469-1474.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed, document M07-A8, vol 29, no. 2 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 29.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonak J, Lind K, Sindelka R, Sjoback R, Sjogreen B, Strombom L, Stahlberg A, Zoric N. 2006. The real-time polymerase chain reaction. Mol Aspects Med 27:95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Hengzhuang W, Wu H, Ciofu O, Song Z, Hoiby N. 2011. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 55:4469–4474. doi: 10.1128/AAC.00126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Olmos A, Garcia-Castillo M, Maiz L, Lamas A, Baquero F, Canton R. 2012. In vitro prevention of Pseudomonas aeruginosa early biofilm formation with antibiotics used in cystic fibrosis patients. Int J Antimicrob Agents 40:173–176. doi: 10.1016/j.ijantimicag.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Erickson DL, Endersby R, Kirkham A, Stuber K, Vollman DD, Rabin HR, Mitchell I, Storey DG. 2002. Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect Immun 70:1783–1790. doi: 10.1128/IAI.70.4.1783-1790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christensen LD, Moser C, Jensen PO, Rasmussen TB, Christophersen L, Kjelleberg S, Kumar N, Hoiby N, Givskov M, Bjarnsholt T. 2007. Impact of Pseudomonas aeruginosa quorum sensing on biofilm persistence in an in vivo intraperitoneal foreign-body infection model. Microbiology 153:2312–2320. doi: 10.1099/mic.0.2007/006122-0. [DOI] [PubMed] [Google Scholar]
- 34.De Kievit TR, Gillis R, Marx S, Brown C, Iglewski BH. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl Environ Microbiol 67:1865–1873. doi: 10.1128/AEM.67.4.1865-1873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Middleton B, Rodgers HC, Camara M, Knox AJ, Williams P, Hardman A. 2002. Direct detection of N-acylhomoserine lactones in cystic fibrosis sputum. FEMS Microbiol Lett 207:1–7. doi: 10.1111/j.1574-6968.2002.tb11019.x. [DOI] [PubMed] [Google Scholar]
- 36.Favre-Bonte S, Kohler T, Van Delden C. 2003. Biofilm formation by Pseudomonas aeruginosa: role of the C4-HSL cell-to-cell signal and inhibition by azithromycin. J Antimicrob Chemother 52:598–604. doi: 10.1093/jac/dkg397. [DOI] [PubMed] [Google Scholar]
- 37.Davey ME, Caiazza NC, O'Toole GA. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol 185:1027–1036. doi: 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson JP, Pesci EC, Iglewski BH. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol 179:5756–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rust L, Pesci EC, Iglewski BH. 1996. Analysis of the Pseudomonas aeruginosa elastase (lasB) regulatory region. J Bacteriol 178:1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Gennip M, Christensen LD, Alhede M, Phipps R, Jensen PO, Christophersen L, Pamp SJ, Moser C, Mikkelsen PJ, Koh AY, Tolker-Nielsen T, Pier GB, Hoiby N, Givskov M, Bjarnsholt T. 2009. Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS 117:537–546. doi: 10.1111/j.1600-0463.2009.02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultz DR, Miller KD. 1974. Elastase of Pseudomonas aeruginosa: inactivation of complement components and complement-derived chemotactic and phagocytic factors. Infect Immun 10:128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaffar-Bandjee MC, Lazdunski A, Bally M, Carrere J, Chazalette JP, Galabert C. 1995. Production of elastase, exotoxin A, and alkaline protease in sputa during pulmonary exacerbation of cystic fibrosis in patients chronically infected by Pseudomonas aeruginosa. J Clin Microbiol 33:924–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woods DE, Sokol PA, Bryan LE, Storey DG, Mattingly SJ, Vogel HJ, Ceri H. 1991. In vivo regulation of virulence in Pseudomonas aeruginosa associated with genetic rearrangement. J Infect Dis 163:143–149. doi: 10.1093/infdis/163.1.143. [DOI] [PubMed] [Google Scholar]
- 44.Tan JS, File TM Jr. 1995. Antipseudomonal penicillins. Med Clin North Am 79:679–693. [DOI] [PubMed] [Google Scholar]
- 45.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biswas S, Brunel JM, Dubus JC, Reynaud-Gaubert M, Rolain JM. 2012. Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti Infect Ther 10:917–934. doi: 10.1586/eri.12.78. [DOI] [PubMed] [Google Scholar]
- 47.Yahav D, Farbman L, Leibovici L, Paul M. 2012. Colistin: new lessons on an old antibiotic. Clin Microbiol Infect 18:18–29. doi: 10.1111/j.1469-0691.2011.03734.x. [DOI] [PubMed] [Google Scholar]
- 48.Velkov T, Thompson PE, Nation RL, Li J. 2010. Structure-activity relationships of polymyxin antibiotics. J Med Chem 53:1898–1916. doi: 10.1021/jm900999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hale JD, Hancock RE. 2007. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev Anti Infect Ther 5:951–959. doi: 10.1586/14787210.5.6.951. [DOI] [PubMed] [Google Scholar]
- 50.Zhang L, Dhillon P, Yan H, Farmer S, Hancock RE. 2000. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:3317–3321. doi: 10.1128/AAC.44.12.3317-3321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol 185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horii T, Ichiyama S, Ohta M, Kobayashi M. 1999. Relationship between morphological changes and endotoxin release induced by carbapenems in Pseudomonas aeruginosa. J Med Microbiol 48:309–315. doi: 10.1099/00222615-48-3-309. [DOI] [PubMed] [Google Scholar]
- 53.Formosa C, Grare M, Duval RE, Dague E. 2012. Nanoscale effects of antibiotics on P. aeruginosa. Nanomedicine 8:12–16. doi: 10.1016/j.nano.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 54.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, Manefield M, Costerton JW, Molin S, Eberl L, Steinberg P, Kjelleberg S, Hoiby N, Givskov M. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J 22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rasmussen TB, Bjarnsholt T, Skindersoe ME, Hentzer M, Kristoffersen P, Kote M, Nielsen J, Eberl L, Givskov M. 2005. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J Bacteriol 187:1799–1814. doi: 10.1128/JB.187.5.1799-1814.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rasmussen TB, Skindersoe ME, Bjarnsholt T, Phipps RK, Christensen KB, Jensen PO, Andersen JB, Koch B, Larsen TO, Hentzer M, Eberl L, Hoiby N, Givskov M. 2005. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 151:1325–1340. doi: 10.1099/mic.0.27715-0. [DOI] [PubMed] [Google Scholar]
- 57.Christensen LD, van Gennip M, Jakobsen TH, Alhede M, Hougen HP, Hoiby N, Bjarnsholt T, Givskov M. 2012. Synergistic antibacterial efficacy of early combination treatment with tobramycin and quorum-sensing inhibitors against Pseudomonas aeruginosa in an intraperitoneal foreign-body infection mouse model. J Antimicrob Chemother 67:1198–1206. doi: 10.1093/jac/dks002. [DOI] [PubMed] [Google Scholar]
- 58.Equi A, Balfour-Lynn IM, Bush A, Rosenthal M. 2002. Long term azithromycin in children with cystic fibrosis: a randomised, placebo-controlled crossover trial. Lancet 360:978–984. doi: 10.1016/S0140-6736(02)11081-6. [DOI] [PubMed] [Google Scholar]
- 59.Hoffmann N, Lee B, Hentzer M, Rasmussen TB, Song Z, Johansen HK, Givskov M, Hoiby N. 2007. Azithromycin blocks quorum sensing and alginate polymer formation and increases the sensitivity to serum and stationary-growth-phase killing of Pseudomonas aeruginosa and attenuates chronic P. aeruginosa lung infection in Cftr−/− mice. Antimicrob Agents Chemother 51:3677–3687. doi: 10.1128/AAC.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gillis RJ, Iglewski BH. 2004. Azithromycin retards Pseudomonas aeruginosa biofilm formation. J Clin Microbiol 42:5842–5845. doi: 10.1128/JCM.42.12.5842-5845.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lutz L, Pereira DC, Paiva RM, Zavascki AP, Barth AL. 2012. Macrolides decrease the minimal inhibitory concentration of anti-pseudomonal agents against Pseudomonas aeruginosa from cystic fibrosis patients in biofilm. BMC Microbiol 12:196. doi: 10.1186/1471-2180-12-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saini H, Chhibber S, Harjai K. 2015. Azithromycin and ciprofloxacin: a possible synergistic combination against Pseudomonas aeruginosa biofilm-associated urinary tract infections. Int J Antimicrob Agents 45:359–367. doi: 10.1016/j.ijantimicag.2014.11.008. [DOI] [PubMed] [Google Scholar]