Abstract
Enterobacteriaceae with blaNDM-7 are relatively uncommon and had previously been described in Europe, India, the United States, and Japan. This study describes the characteristics of Enterobacteriaceae (Klebsiella pneumoniae [n = 2], Escherichia coli [n = 2], Serratia marcescens [n = 1], and Enterobacter hormaechei [n = 1] isolates) with blaNDM-7 obtained from 4 patients from Calgary, Canada, from 2013 to 2014. The 46,161-bp IncX3 plasmids with blaNDM-7 are highly similar to other blaNDM-harboring IncX3 plasmids and, interestingly, showed identical structures within the different isolates. This finding may indicate horizontal transmission within our health region, or it may indicate contact with individuals from areas of endemicity within the hospital setting. Patients infected or colonized with bacteria containing blaNDM-7 IncX3 plasmids generate infection control challenges. Epidemiological and molecular studies are required to better understand the dynamics of transmission, the risk factors, and the reservoirs for bacteria harboring blaNDM-7. To the best of our knowledge, this is the first report of S. marcescens and E. hormaechei with blaNDM-7.
INTRODUCTION
The metallo-β-lactamase (MBL) NDM-1 was first described in Klebsiella pneumoniae and Escherichia coli recovered from a Swedish patient who was previously hospitalized in New Delhi, India (1). Subsequently, bacteria carrying NDM have been recognized in over 50 countries on every continent, except Antarctica (2). Gram-negative bacteria carrying blaNDM are endemic in South Asia (especially the Indian subcontinent), South-East Asia (3, 4), and certain countries within the Middle East and the Balkans (5). Infections with NDM-producing bacteria in areas where these organisms are nonendemic, such as Europe and North America, have most often been associated with patients who required hospitalization while visiting an area where NDM-producing bacteria are endemic (6).
NDM carbapenemases are commonly reported in K. pneumoniae and E. coli but have also been found in a variety of other members of the Enterobacteriaceae family, including Acinetobacter spp., Pseudomonas spp., and Vibrio cholerae (7, 8). The treatment of infections caused by multidrug-resistant NDM-producing Enterobacteriaceae is causing serious therapeutic challenges for the medical community because isolates are often also resistant to non-β-lactam antibiotics (9). Bacteria with NDMs often remain susceptible only to agents such as colistin (CST), fosfomycin (FOF), and tigecycline (TGC) (10).
During 2013 and 2014, six Enterobacteriaceae with blaNDM-7 were isolated from four different Calgary patients over a period of 18 months. One patient had recently been hospitalized in India, while the remaining three did not have a history of recent travel outside Alberta, Canada. NDM-7 is an infrequent blaNDM allele; therefore, a study was designed to characterize these isolates and their respective plasmids using traditional and next-generation sequencing techniques.
MATERIALS AND METHODS
Patients and isolates.
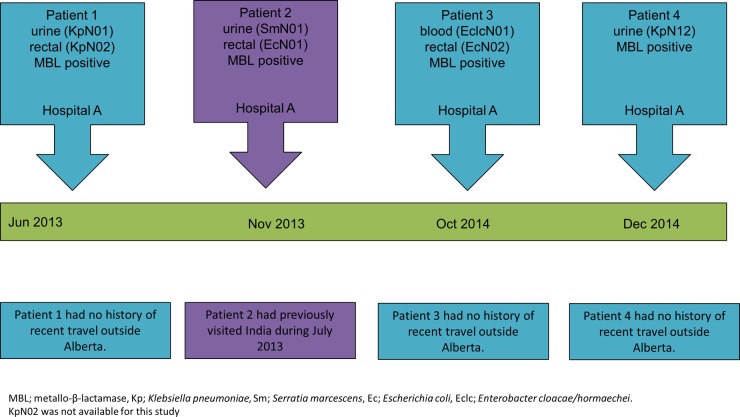
For a summary of the clinical features of patients 1 to 4 and a timeline of events, please see Table 1 and Fig. 1.
TABLE 1.
Characteristics of patients infected with Enterobacteriaceae harboring blaNDM-7 in Calgary, Canada
| Characteristic | Patient 1 | Patient 2 | Patient 3 | Patient 4 |
|---|---|---|---|---|
| Sex | Male | Male | Male | Female |
| Hospital | A | B | A | A |
| Clinical diagnosis | Asymptomatic bacteriuria | Lower UTIa | Septicemia | Lower UTI |
| Date of clinical presentation | June 2013 | November 2013 | October 2014 | December 2014 |
| Treatment | In-out catheterization | Ciprofloxacin | Gentamicin and colistin | Ciprofloxacin |
| Travel history | None | India | None | None |
UTI, urinary tract infection.
FIG 1.
The timeline of events of patients infected with Enterobacteriaceae with blaNDM-7, Calgary, Canada.
The isolates were identified using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Vitek AMS; bioMérieux Vitek Systems Inc., Hazelwood, MO). Further identification of Enterobacter cloacae was performed by partial sequencing of the leuS gene (11).
Antimicrobial susceptibilities.
MICs were determined using the MicroScan NEG 38 panel (Siemens, Burlington, Ontario, Canada) and were interpreted by using CLSI guidelines for broth dilution (12). The following drugs were tested: piperacillin-tazobactam (TZP), cefoxitin (FOX), ceftriaxone (CRO), ceftazidime (CAZ), cefepime (FEP), aztreonam (ATM), meropenem (MEM), ertapenem (ERT), amikacin (AMK), gentamicin (GEN), tobramycin (TOB), ciprofloxacin (CIP), tigecycline (TGC), and trimethoprim-sulfamethoxazole (SXT). Colistin (CST), fosfomycin (FOF), imipenem (IPM), MEM, and ERT MICs were determined using Etests (bioMérieux Inc., Hazelwood, MO, USA) according to the manufacturer's instructions. The IPM, MEM, ERT, and FOF Etests were also performed on the E. coli J53 transconjugants (see below). The European Committee for Antimicrobial Susceptibility Testing (EUCAST) breakpoint was used for CST, and the FDA breakpoint was used for TGC.
Carbapenemase gene identification.
The presence of carbapenemases was detected using the CLSI guidelines for the modified Hodge test (MHT) and Mastdiscs ID inhibitor combination disks (13) (Mast Group Ltd., Merseyside, United Kingdom). PCR amplification and sequencing for blaKPC, blaVIM, blaIMP, blaNDM, and blaOXA-48-like genes were undertaken using primers and conditions as previously described (13).
Plasmid analysis.
Plasmid sizes were determined as previously described (14) and were assigned to plasmid incompatibility (Inc) groups by PCR-based replicon typing (15–17). Conjugation experiments were performed by mating-out assays with nutrient agar containing 1 μg/ml MEM and by using E. coli J53 (azide, 100 μg/ml) as the recipient. Plasmids from the transconjugants were sequenced using the Pacific Biosciences RS II platform (Menlo Park, CA, USA) and the Illumina MiSeq system (San Diego, CA, USA) (see details below).
MLST.
Multilocus sequencing typing (MLST) of the K. pneumoniae (18), E. coli (19), and E. cloacae (20) isolates was performed as previously described.
Complete sequencing of blaNDM-7-harboring plasmids.
The genome of isolate KpN01 from patient 1 (chromosome and plasmids) was sequenced using the Pacific Biosciences RS II platform (Menlo Park, CA, USA). Assembly of the data was performed using the hierarchical genome assembly process (HGAP) compiled specifically for quality trimming, de novo assembly, and polishing of PacBio data. The blaNDM-7-harboring plasmids from isolates SmN01 (Serratia marcescens isolate from patient 2), EcN01 (E. coli isolate from patient 2), EclcN01 (Enterobacter cloacae/hormaechei isolate from patient 3), EcN02 (from patient 3), and KpN12 (K. pneumoniae isolate from patient 4) were sequenced using a previously described method (21). In brief, plasmid DNA from E. coli J53 transconjugants with a single blaNDM-7-harboring plasmid was extracted using a Qiagen plasmid maxikit (Qiagen, Valencia, CA) and was sequenced using the Illumina MiSeq system (San Diego, CA, USA). Sequencing reads were de novo assembled into consensus contigs using Velvet algorithms (22), and sequence gaps were closed by PCR and standard Sanger sequencing. The resultant plasmids were annotated using the Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) available at NCBI (http://www.ncbi.nlm.nih.gov/). The plasmid replicon type (InC) was examined by PlasmidFinder (23), and antibiotic resistance genes were identified with the Comprehensive Antibiotic Resistance Database (CARD) (24). The plasmids were compared to each other and were then compared to publicly available plasmid references using BLAST at GenBank (www.ncbi.nlm.nih.gov/GenBank/). The plasmid comparison and visualization were generated by Easyfig (25) according to the online protocol (http://easyfig.sourceforge.net/).
Nucleotide sequence accession number.
The complete nucleotide sequence of pKpN01-NDM-7 was deposited under GenBank accession number CP012990.
RESULTS AND DISCUSSION
Susceptibilities and carbapenemase genes.
Table 2 shows the susceptibilities of the clinical isolates. EclcN01 was identified as Enterobacter hormaechei. All of the isolates tested positive with the modified Hodge test, and the Mastdiscs ID inhibitor combination disks indicated that they were all MBL producers. All of the isolates were positive for blaNDM-7 according to PCR and sequencing results (Table 2).
TABLE 2.
Characteristics of Enterobacteriaceae with NDM-7 from Calgary, Canada, from 2013 to 2014
| Characteristic | KpN01 | SmN01 | EcN01 | EclcN01 | EcN02 | KpN12 |
|---|---|---|---|---|---|---|
| Patient | 1 | 2 | 2 | 3 | 3 | 4 |
| Hospital | A | B | B | A | A | A |
| Specimen | Urine | Urine | Rectal swab | Blood | Rectal swab | Urine |
| Date of clinical presentation | June 2013 | November 2013 | November 2013 | October 2014 | October 2014 | December 2014 |
| Susceptibilities (MIC, μg/ml) | ||||||
| TZP | >64/4 | >64/4 | >64/4 | >64/4 | >64/4 | >64/4 |
| FOX | >16 | >16 | >16 | >16 | >16 | >16 |
| CRO | >32 | >32 | >32 | >32 | >32 | >32 |
| CAZ | >16 | >16 | >16 | >16 | >16 | >16 |
| FEP | >16 | >16 | >16 | >16 | >16 | >16 |
| ATM | >16 | ≤4 | >16 | >16 | >16 | >16 |
| MEM | >8 | >8 | >8 | >8 | >8 | >8 |
| ERT | >4 | >4 | >4 | >4 | >4 | >4 |
| AMK | ≤4 | ≤4 | 16 | ≤4 | ≤4 | ≤4 |
| GEN | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 |
| TOB | ≤1 | ≤1 | >8 | ≤1 | ≤1 | ≤1 |
| CIP | 1 | ≤0.5 | >2 | >2 | ≤0.5 | ≤0.5 |
| SXT | >2/38 | ≤2/38 | >2/38 | >2/38 | ≤2/38 | ≤2/38 |
| TGC | ≤1 | 2 | ≤1 | 1 | 1 | ≤1 |
| CST | 0.19 | >256 | 0.125 | 0.09 | 0.125 | 0.125 |
| FOF | 16 | >256 | 1 | 32 | 1 | 16 |
| MHT | Positive | Positive | Positive | Positive | Positive | Positive |
| Mastdiscs | MBL positive | MBL positive | MBL positive | MBL positive | MBL positive | MBL positive |
| Carbapenemase | NDM-7 | NDM-7 | NDM-7 | NDM-7 | NDM-7 | NDM-7 |
| MLST | ST654 | ST44 | ST113 | ST91 | ST138 | |
| Plasmid size(s) (kb) | 190, 130, 50 | 50 | 175, 105, 80, 55, 50 | 170, 80, 50 | 135, 50 | 200, 50 |
| Transconjugant (plasmid size, kb) | KpN01T (50) | SmN01T (50) | EcN01T (50) | EclcNT01T (50) | EcN02T (50) | KpN12T (50) |
| ERT MIC, μg/ml | 16 | 10 | 16 | 8 | 16 | 16 |
| MER MIC, μg/ml | 8 | 6 | 4 | 4 | 8 | 4 |
| IPM MIC, μg/ml | >32 | >32 | >32 | >32 | >32 | >32 |
| FOS MIC, μg/ml | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Replicon typing | IncX | IncX | IncX | IncX | IncX | IncX |
Enterobacteriaceae with blaNDM-7 are relatively uncommon, having only been previously described in E. coli from France (26), Germany (27), India (28), the United States (29), and Japan (30) and in K. pneumoniae from the United States (29), Spain (31), and Denmark (32). The single biggest risk factor in all patients was a history of recent travel to India with two exceptions; the patient in the French case had previously traveled to Burma, while no connection with the Indian subcontinent was established among the Spanish patients (26, 31). One of the Calgary patients (patient 2) recently visited India (but he had no contact with the health care system in that country to the best of our knowledge), and the remaining patients (patients 1, 3, and 4) had no recent history of travel outside Alberta. The cases were not linked by time or place, and we were unable to establish any epidemiological linkage between the four patients. The patients from hospital A were admitted to different wards over different time periods and stayed in different parts of Calgary.
Plasmids and MLST.
MLST showed that KpN01 belonged to sequence type 278 (ST278), EcN01 belonged to ST44, EclcN01 belonged to ST113, EcN02 belonged to ST91, and KpN12 belonged to ST138 (Table 1). The isolates contained several plasmids of different sizes, but interestingly, they all harbored a plasmid of approximately 50 kb (Table 1). The transconjugants obtained with the different isolates (i.e., KpN01T, SmN01T, etc.) contained the 50-kb plasmid, which was positive for blaNDM and was typed with the IncX3 replicon (Table 1). The international case reports of E. coli with blaNDM-7 belonged to various sequence types, including ST167 from France (26), ST599 from Germany (27), ST617 from the United States (29), and ST648 from Japan (30). The K. pneumoniae isolates from the United States and Denmark were typed as ST147 (29, 32), while the Spanish isolates belonged to ST437 (31). The conjugative plasmid with blaNDM-7 from the French isolate was untypeable (26), and blaNDM-7 from the German E. coli isolate was flanked upstream by IS5 and ISAba125 and downstream by bleMBL and was located on a 60-kb IncX3 plasmid (27).
Plasmid sequencing and comparative plasmid analysis.
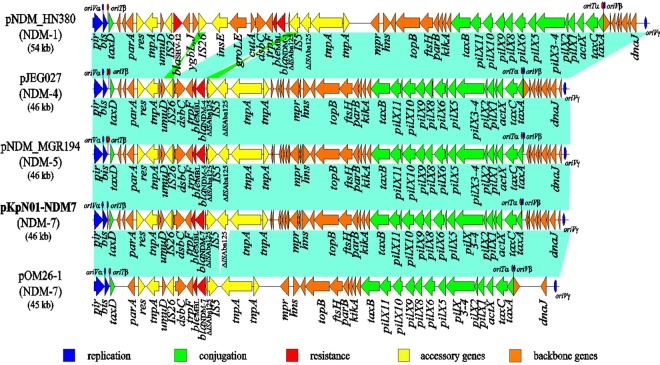
The blaNDM-7-harboring plasmid in KpN01 (namely pKpN01-NDM7) is 46,161 bp in size with an average GC content of 46.6% and contains 54 predicted open reading frames (ORFs) (Fig. 2). Complete sequencing of blaNDM-7-harboring plasmids from the other five transconjugants (i.e., SmN01T, EcN01T, EclcN01T, EcN02T, and KpN12T) interestingly showed 100% identities to that of pKpN01-NDM7. This suggests that the same blaNDM-7-containing plasmid was horizontally transferred to several different Enterobacteriaceae genera that colonized/infected the patients in the Calgary region. Alternatively, there may have been common connections with patients from settings where these organisms are endemic who harbored the same or a closely related plasmid. However, we were unable to establish such a link.
FIG 2.
The comparative structures of IncX3 plasmids harboring blaNDM: pKpN01-NDM7 (blaNDM-7; GenBank accession number CP012990), pNDM-HN380 (blaNDM-1; GenBank accession number JX104760), pNDM_MGR194 (blaNDM-5; GenBank accession number KF220657), pJEG027 (blaNDM-4; GenBank accession number KM400601), and pOM26-1 (blaNDM-7; GenBank accession number KP776609). Light blue shading denotes shared regions of homology, while green shading indicates inversely displayed regions of homology. Open reading frames (ORFs) are portrayed by arrows and colored based on predicted gene function, while the putative oriT and oriV are shown by red and blue ellipses, respectively. The blaNDM-7-harboring plasmid, pKpN01-NDM7, is in bold.
pKpN01-NDM7 is an IncX3 plasmid; IncX3 plasmids are usually self-transferable and have been associated with the spread of several antimicrobial-resistant genes, including carbapenemase genes, such as blaNDM (33–37), blaKPC (38), and blaOXA-181 (39, 40). A BLAST search against the GenBank sequence database revealed that pKpN01-NDM7 is highly similar to other blaNDM-harboring IncX3 plasmids, including pNDM-HN380 (blaNDM-1) (36), p112298-NDM (blaNDM-1; GenBank accession number KP987216), pEc1929 (blaNDM-5; GenBank accession number KT824791), pNDM_MGR194 (blaNDM-5) (34), pJEG027 (blaNDM-4) (35), and pOM26-1 (blaNDM-7; GenBank accession number KP776609) (Fig. 2). pKpN01-NDM7 has a typical backbone of IncX plasmids, including the genes encoding replication (pir and bis), partitioning (par), maintenance (topB and hns), and conjugal transfer (pil and tax) (17, 41). Similar to other IncX3 plasmids, pKpN01-NDM7 carries three putative origins of replication (oriV-α, oriV-β, or oriV-γ), and two origins of conjugal transfer (oriT-α and oriT-β) (41–43) (Fig. 1). The backbone regions among these blaNDM-harboring IncX3 plasmids showed very high identities (>99.7% compared with each other), suggesting they likely evolved from the same ancestor plasmid.
Common to other IncX3 plasmids, the blaNDM-7-containing accessory region in pKpN01-NDM7 was located downstream of the serine resolvase gene res. blaNDM-7 was carried by an IS26-dsbC-trpF-bleMBL-blaNDM-7-ΔISAba125-IS5-ΔISAba125 genetic element; this is the same structure as that of the blaNDM-4- and blaNDM-5-containing elements reported previously (34, 35) and is similar to the blaNDM-7 plasmid described from Germany (27). The overall plasmid genome synteny in pKpN01-NDM7 is also identical to that of blaNDM-5-harboring pNDM_MGR194 (from a ST11 K. pneumoniae isolate in India) (34) and blaNDM-4-harboring pJEG027 (from a K. pneumoniae isolate in Australia) (35). Importantly, IS5 was inserted into ISAba125 located upstream of blaNDM, leading to the interruption of ISAba125 into two segments (1,018 bp and 73 bp) (Fig. 1). Downstream from blaNDM are the genes for bleMBL (encoding bleomycin-resistant protein), trpF, dsbC, and IS26.
There are two main differences between pKpN01-NDM7, pNDM_MGR194, and pJEG027. First, they carry different blaNDM alleles; pKpN01-NDM7 carried blaNDM-7, while pJEG027 and pNDM_MGR194 harbored blaNDM-4 and blaNDM-5, respectively. Of note, blaNDM-7 and blaNDM-5 each differ from blaNDM-4 by a single nucleotide (G388A and G262T, respectively). Second, pKpN01-NDM7 (46,161 bp) is 92 bp shorter than pNDM_MGR194 and pJEG027 (both are 46,253 bp). This is due to an extra 92-bp palindromic sequence in pNDM_MGR194 and pJEG027 that is upstream of taxD and that forms an additional oriT-β site. It is not clear whether this extra oriT-β site will enhance plasmid transfer efficiency, because a previous study indicated that the IncX plasmid can simultaneously cleave multiple nic sites, thereby initiating conjugal transfer (42). In addition, pKpN01-NDM7 also showed high identity to a blaNDM-7-bearing plasmid pOM26-1 (GenBank accession number KP776609), which was isolated from an E. coli isolate in Oman. pOM26-1 differs from pKpN01-NDM7 by a 1,039-bp deletion, encompassing the aforementioned 1,018 bp ΔISAba125 flanked by two 4-bp repeats (CTAA) (Fig. 2). This suggests that pOM26-1 evolved from the pKpN01-NDM7-like plasmid as a result of an ∼1-kb ΔISAba125-bearing element deletion.
The first blaNDM-harboring IncX3 plasmid (pNDM-HN380, blaNDM-1) was reported in 2012 from a K. pneumoniae isolate discovered in China (36). Since then, IncX3 plasmids containing different blaNDM alleles, including blaNDM-1, blaNDM-4 (e.g., pJEG027), blaNDM-5 (e.g., pNDM_MGR194), and blaNDM-7 (e.g., pKpN01-NDM7 [current study] and pOM26-1) have been reported from different geographical regions (e.g., China, India, Australia, Germany, Canada, and Oman) and in different species (34–36, 44), dramatizing the significant role played by plasmids in the rapid worldwide dissemination of NDM-type carbapenemases. Our study showed that NDM-7 is present in several Enterobacteriaceae genera isolated in the Calgary region due to a single identical blaNDM-7-harboring IncX3 plasmid, pKpN01-NDM7. The close sequence identities among these blaNDM-harboring IncX3 plasmids also show their genetic evolution. Espedido et al. (35) hypothesized that pJEG027 (with blaNDM-4) may have arisen from a pNDM-HN380-like plasmid (with blaNDM-1) ancestor as a result of a different IS5 insertion, an IS26-mediated flanking deletion of cutA1-groL, and acquisition of the A460C mutation in blaNDM-1 (Fig. 2). pNDM_MGR194 (blaNDM-5) may have also evolved from pJEG027 by accumulation of an additional mutation (G262T). Our study suggested that pKpN01-NDM7 had arisen from a pJEG027-like plasmid (carrying blaNDM-4) through acquisition of a different mutation (G388A) and that pOM26-1 is descended from pKpN01-NDM7 through an ∼1-kb deletion.
In summary, this study describes the characteristics of Enterobacteriaceae with blaNDM-7 isolated from 4 Canadian patients from Calgary, Alberta without apparent epidemiological linkages. The 46,161-bp IncX3 plasmid with blaNDM-7 is highly similar to other blaNDM-harboring IncX3 plasmids. The NDM-7-containing plasmids from this study, interestingly, showed identical structures within the different Enterobacteriaceae isolates (i.e., K. pneumoniae, E. coli, Serratia marcescens, and E. hormaechei). This suggests that very effective horizontal transfer events had occurred previously between these patients or that there were connections with patients from settings where these organisms are endemic that we were unable to establish. To the best of our knowledge, this is the first report of S. marcescens and E. hormaechei with blaNDM-7.
ACKNOWLEDGMENTS
We thank Joseph Kim for providing some of the clinical information on patient 2.
J.D.P. had previously received research funds from Merck and Astra Zeneca.
REFERENCES
- 1.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson AP, Woodford N. 2013. Global spread of antibiotic resistance: the example of New Delhi metallo-beta-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol 62:499–513. doi: 10.1099/jmm.0.052555-0. [DOI] [PubMed] [Google Scholar]
- 3.Netikul T, Sidjabat HE, Paterson DL, Kamolvit W, Tantisiriwat W, Steen JA, Kiratisin P. 2014. Characterization of an IncN2-type blaNDM-1-carrying plasmid in Escherichia coli ST131 and Klebsiella pneumoniae ST11 and ST15 isolates in Thailand. J Antimicrob Chemother 69:3161–3163. doi: 10.1093/jac/dku275. [DOI] [PubMed] [Google Scholar]
- 4.Rimrang B, Chanawong A, Lulitanond A, Wilailuckana C, Charoensri N, Sribenjalux P, Phumsrikaew W, Wonglakorn L, Kerdsin A, Chetchotisakd P. 2012. Emergence of NDM-1- and IMP-14a-producing Enterobacteriaceae in Thailand. J Antimicrob Chemother 67:2626–2630. doi: 10.1093/jac/dks267. [DOI] [PubMed] [Google Scholar]
- 5.Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol 19:588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 6.van der Bij AK, Pitout JD. 2012. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J Antimicrob Chemother 67:2090–2100. doi: 10.1093/jac/dks214. [DOI] [PubMed] [Google Scholar]
- 7.Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother 55:5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 9.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordmann P, Poirel L. 2014. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 11.Andrade LN, Novais A, Branquinho R, Peixe L. 2015. Species reassignment of Enterobacter cloacae complex isolates by in silico phylogenetic analysis, abstr 2692. FEMS 2015, Maastricht, The Netherlands, 7 to 11 June 2015. [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement CLSI M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13.Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JD. 2012. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol 50:3877–3880. doi: 10.1128/JCM.02117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd DA, Tyler S, Christianson S, McGeer A, Muller MP, Willey BM, Bryce E, Gardam M, Nordmann P, Mulvey MR. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob Agents Chemother 48:3758–3764. doi: 10.1128/AAC.48.10.3758-3764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 16.Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 17.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyoshi-Akiyama T, Hayakawa K, Ohmagari N, Shimojima M, Kirikae T. 2013. Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS One 8:e66358. doi: 10.1371/journal.pone.0066358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L, Hu H, Chavda KD, Zhao S, Liu R, Liang H, Zhang W, Wang X, Jacobs MR, Bonomo RA, Kreiswirth BN. 2014. Complete sequence of a KPC-producing IncN multidrug-resistant plasmid from an epidemic Escherichia coli sequence type 131 strain in China. Antimicrob Agents Chemother 58:2422–2425. doi: 10.1128/AAC.02587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJ, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuzon G, Bonnin RA, Nordmann P. 2013. First identification of novel NDM carbapenemase, NDM-7, in Escherichia coli in France. PLoS One 8:e61322. doi: 10.1371/journal.pone.0061322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottig S, Hamprecht AG, Christ S, Kempf VA, Wichelhaus TA. 2013. Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-beta-lactamase with increased carbapenemase activity. J Antimicrob Chemother 68:1737–1740. doi: 10.1093/jac/dkt088. [DOI] [PubMed] [Google Scholar]
- 28.Rahman M, Shukla SK, Prasad KN, Ovejero CM, Pati BK, Tripathi A, Singh A, Srivastava AK, Gonzalez-Zorn B. 2014. Prevalence and molecular characterisation of New Delhi metallo-beta-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int J Antimicrob Agents 44:30–37. doi: 10.1016/j.ijantimicag.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Lee CS, Vasoo S, Hu F, Patel R, Doi Y. 2014. Klebsiella pneumoniae ST147 coproducing NDM-7 carbapenemase and RmtF 16S rRNA methyltransferase in Minnesota. J Clin Microbiol 52:4109–4110. doi: 10.1128/JCM.01404-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizuno Y, Yamaguchi T, Matsumoto T. 2014. A first case of New Delhi metallo-beta-lactamase-7 in an Escherichia coli ST648 isolate in Japan. J Infect Chemother 20:814–816. doi: 10.1016/j.jiac.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Seara N, Oteo J, Carrillo R, Perez-Blanco V, Mingorance J, Gomez-Gil R, Herruzo R, Perez-Vazquez M, Astray J, Garcia-Rodriguez J, Ruiz-Velasco LM, Campos J, de Burgos C, Ruiz-Carrascoso G. 2015. Interhospital spread of NDM-7-producing Klebsiella pneumoniae belonging to ST437 in Spain. Int J Antimicrob Agents 46:169–173. doi: 10.1016/j.ijantimicag.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Hammerum AM, Littauer P, Hansen F. 2015. Detection of Klebsiella pneumoniae co-producing NDM-7 and OXA-181, Escherichia coli producing NDM-5 and Acinetobacter baumannii producing OXA-23 in a single patient. Int J Antimicrob Agents 46:597–598. doi: 10.1016/j.ijantimicag.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Yang Q, Fang L, Fu Y, Du X, Shen Y, Yu Y. 2015. Dissemination of NDM-1-producing Enterobacteriaceae mediated by the IncX3-type plasmid. PLoS One 10:e0129454. doi: 10.1371/journal.pone.0129454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishnaraju M, Kamatchi C, Jha AK, Devasena N, Vennila R, Sumathi G, Vaidyanathan R. 2015. Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J Med Microbiol 33:30–38. doi: 10.4103/0255-0857.148373. [DOI] [PubMed] [Google Scholar]
- 35.Espedido BA, Dimitrijovski B, van Hal SJ, Jensen SO. 2015. The use of whole-genome sequencing for molecular epidemiology and antimicrobial surveillance: identifying the role of IncX3 plasmids and the spread of blaNDM-4-like genes in the Enterobacteriaceae. J Clin Pathol 68:835–838. doi: 10.1136/jclinpath-2015-203044. [DOI] [PubMed] [Google Scholar]
- 36.Ho PL, Li Z, Lo WU, Cheung YY, Lin CH, Sham PC, Cheng VC, Ng TK, Que TL, Chow KH. 2012. Identification and characterization of a novel incompatibility group X3 plasmid carrying blaNDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg Microbes Infect 1:e39. doi: 10.1038/emi.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wailan AM, Paterson DL, Kennedy K, Ingram PR, Bursle E, Sidjabat HE. 19 October 2015. Genomic characteristics of NDM-producing Enterobacteriaceae in Australia and their blaNDM genetic contexts. Antimicrob Agents Chemother doi: 10.1128/AAC.01243-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho PL, Cheung YY, Lo WU, Li Z, Chow KH, Lin CH, Chan JF, Cheng VC. 2013. Molecular characterization of an atypical IncX3 plasmid pKPC-NY79 carrying blaKPC-2 in a Klebsiella pneumoniae. Curr Microbiol 67:493–498. doi: 10.1007/s00284-013-0398-2. [DOI] [PubMed] [Google Scholar]
- 39.Zurfluh K, Poirel L, Nordmann P, Klumpp J, Stephan R. 2015. First detection of Klebsiella variicola producing OXA-181 carbapenemase in fresh vegetable imported from Asia to Switzerland. Antimicrob Resist Infect Control 4:38. doi: 10.1186/s13756-015-0080-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Feng Y, Wu W, Xie Y, Wang X, Zhang X, Chen X, Zong Z. 2015. First report of OXA-181-producing Escherichia coli in China and characterization of the isolate using whole-genome sequencing. Antimicrob Agents Chemother 59:5022–5025. doi: 10.1128/AAC.00442-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norman A, Hansen LH, She Q, Sorensen SJ. 2008. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60:59–74. doi: 10.1016/j.plasmid.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Avila P, Nunez B, de la Cruz F. 1996. Plasmid R6K contains two functional oriTs which can assemble simultaneously in relaxosomes in vivo. J Mol Biol 261:135–143. doi: 10.1006/jmbi.1996.0447. [DOI] [PubMed] [Google Scholar]
- 43.Nunez B, Avila P, de la Cruz F. 1997. Genes involved in conjugative DNA processing of plasmid R6K. Mol Microbiol 24:1157–1168. doi: 10.1046/j.1365-2958.1997.4111778.x. [DOI] [PubMed] [Google Scholar]
- 44.Feng J, Qiu Y, Yin Z, Chen W, Yang H, Yang W, Wang J, Gao Y, Zhou D. 2015. Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of Citrobacter freundii. J Antimicrob Chemother 70:2987–2991. doi: 10.1093/jac/dkv232. [DOI] [PubMed] [Google Scholar]