Abstract
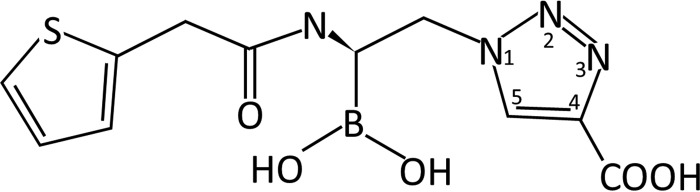
Resistance to expanded-spectrum cephalosporins and carbapenems has rendered certain strains of Klebsiella pneumoniae the most problematic pathogens infecting patients in the hospital and community. This broad-spectrum resistance to β-lactamases emerges in part via the expression of KPC-2 and SHV-1 β-lactamases and variants thereof. KPC-2 carbapenemase is particularly worrisome, as the genetic determinant encoding this β-lactamase is rapidly spread via plasmids. Moreover, KPC-2, a class A enzyme, is difficult to inhibit with mechanism-based inactivators (e.g., clavulanate). In order to develop new β-lactamase inhibitors (BLIs) to add to the limited available armamentarium that can inhibit KPC-2, we have structurally probed the boronic acid transition state analog S02030 for its inhibition of KPC-2 and SHV-1. S02030 contains a boronic acid, a thiophene, and a carboxyl triazole moiety. We present here the 1.54- and 1.87-Å resolution crystal structures of S02030 bound to SHV-1 and KPC-2 β-lactamases, respectively, as well as a comparative analysis of the S02030 binding modes, including a previously determined S02030 class C ADC-7 β-lactamase complex. S02030 is able to inhibit vastly different serine β-lactamases by interacting with the conserved features of these active sites, which includes (i) forming the bond with catalytic serine via the boron atom, (ii) positioning one of the boronic acid oxygens in the oxyanion hole, and (iii) utilizing its amide moiety to make conserved interactions across the width of the active site. In addition, S02030 is able to overcome more distantly located structural differences between the β-lactamases. This unique feature is achieved by repositioning the more polar carboxyl-triazole moiety, generated by click chemistry, to create polar interactions as well as reorient the more hydrophobic thiophene moiety. The former is aided by the unusual polar nature of the triazole ring, allowing it to potentially form a unique C—H…O 2.9-Å hydrogen bond with S130 in KPC-2.
INTRODUCTION
β-Lactamases, ubiquitous resistance determinants, provide bacteria with a nearly impenetrable defense against the lethal action of β-lactam antibiotics. Klebsiella pneumoniae, an aerobic Gram-negative pathogen known to be the agent of serious infections, is capable of expressing numerous β-lactamases, including KPC-2 and SHV-1. The KPC-2 β-lactamase is particularly worrisome due to its ability to confer resistance to carbapenems, a class of “last-resort antibiotics,” and its rapid dissemination via plasmid transfer not only between but also across species. KPC-2-producing organisms, such as K. pneumoniae and Escherichia coli, have caused a number of outbreaks across different continents (1–4). Currently, 24 KPC variants are identified (http://www.lahey.org/Studies/), with KPC-2 being the dominant variant in most countries (5).
To counteract the presence of β-lactamases, β-lactam antibiotics are often coadministered with a β-lactamase inhibitor (BLI). This successful strategy has significantly prolonged the clinical efficacy of ampicillin, amoxicillin, piperacillin, cefoperazone, and ticarcillin (6). Unfortunately, KPC-2 carbapenemase is notoriously difficult to inhibit (7). Currently, there are 4 clinically approved β-lactamase inhibitors, with only the most recently FDA-approved inhibitor, avibactam, having inhibitory potency against KPC-2 (8). Additional inhibitors, such as the boronic acid-containing RPX7009, are in advanced stages of clinical development (as reviewed in reference 9). The crystal structure of avibactam bound to KPC-2 was recently determined (10). However, having only one potent inhibitor currently available for KPC-2, an enzyme that was shown to have the ability to develop avibactam resistance variants (11), suggests that exploring additional inhibitors of KPC-2 is warranted to anticipate future resistance threats.
As stated above, an emerging notion in the discovery of novel BLIs is to utilize boronic acid transition state analogs (BATSIs) to inhibit serine β-lactamases such as KPC-2 and SHV-1 (12–14). Such an approach has led to several potent inhibitor compounds (15, 16). Recently, the BATSI S02030 (Fig. 1) was found to inhibit ADC-7 β-lactamase from Acinetobacter baumannii by forming a transition state boron-mediated bond with the catalytic serine (17). We have extended the structural investigations of S02030 and observed that it readily inhibits SHV-1 and KPC-2 β-lactamases (see also the companion article by Rojas et al. [18]). We present here the 1.54- and 1.87-Å resolution crystal structures of S02030 bound to SHV-1 and KPC-2 β-lactamases, respectively, as well as an in-depth comparative analysis of the S02030 binding modes, including the ADC-7 S02030 complex.
FIG 1.
Chemical structure of S02030.
MATERIALS AND METHODS
The chemical synthesis of S02030 was previously described (17). The structure of S02030 is represented in Fig. 1.
Protein expression, purification, crystallization, and crystal preparation.
The KPC-2 and SHV-1 enzymes were expressed and purified as previously published (10, 13). The KPC-2–S02030 complex was obtained by cocrystallization; the KPC-2 β-lactamase and the S02030 inhibitor were incubated overnight, with a molar ratio of protein and inhibitor of 1:10. Initial cocrystallization screening was carried out using a JCSG+ screen kit (from Molecular Dimension) on a 96-well tray (protein was 15 mg/ml). The ratio of protein mixture to reservoir was 1:1. The cocrystallization condition was 30% polyethylene glycol 8000 (PEG 8000), 0.2 M lithium sulfate, and 0.1 M sodium acetate (pH 4.5). Once KPC-2–S02030 cocrystals grew to their final size, they were mounted and cryoprotected with perfluoropolyether oil (from Hampton Research) prior to being flash-frozen in liquid nitrogen.
In contrast to the case with KPC-2, the SHV-1–S02030 complex was obtained by soaking the ligand in SHV-1 crystals. Apo SHV-1 crystals were first obtained using 20 to 30% PEG 6000, 100 mM Tris (pH 7.5), and 0.56 mM Cymal-6 using the vapor diffusion sitting drop crystallization method (19, 20). SHV-1 crystals were soaked for 30 min with 5 mM S02030-containing mother liquor solution and subsequently cryoprotected in perfluoropolyether oil prior to freezing in liquid nitrogen.
Data collection and structure determination.
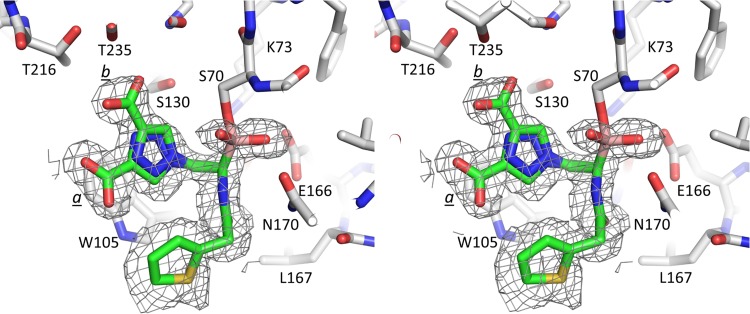
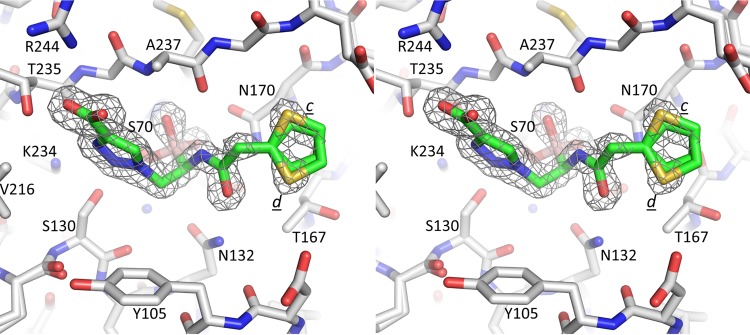
Data for the KPC-2–S02030 complex structure were collected on the in-house Rigaku Micromax-007 HF diffraction system. The SHV-1–S02030 data were collected at Stanford Synchrotron Radiation Lightsource (SSRL) beamline 7-1. Both data sets (Table 1) were processed using HKL2000 (21). The S02030 protein complex structures were refined using CCP4 suite program REFMAC (22), and the program COOT (23) was used for model fitting. The initial search models for KPC-2–S02030 complex and SHV-1–S02030 structures were PDB codes 3RXX and 2H5S, respectively. The PRODRG (24) server was used to generate the parameters and topology files for the S02030 ligand that were observed in the electron density maps (Fo − Fc) in the active site for both β-lactamases (Fig. 2 and 3). The final coordinates for both S02030 SHV-1 and KPC-2 complex structures were validated using PROCHECK (25).
TABLE 1.
X-ray diffraction data and refinement statistics of KPC-2 and SHV-1 S02030 complexes
| Parameter | Value for: |
|
|---|---|---|
| KPC-2–S02030 complex | SHV-1–S02030 complex | |
| Data collection | ||
| Space group | P21 | P212121 |
| Unit cell dimensions (Å) | 51.85, 77.88, 64.77, 90, 108.67, 90 | 49.57, 55.19, 83.50, 90, 90, 90 |
| Wavelength (Å) | 1.5418 | 1.1271 |
| Resolution (Å) | 50–1.85 | 50–1.54 |
| Redundancy | 2.9 (2.3) | 3.5 (3.5) |
| Unique reflections | 39,177 | 33,737 |
| <I>/<σ(I)> | 9.0 (2.2) | 16.5 (2.8) |
| Rmerge (%) | 10.7 (39.3) | 7.0 (42.0) |
| Completeness (%) | 96.7 (90.2) | 97.0 (98.3) |
| Refinement | ||
| Resolution range (Å) | 32.9–1.87 | 33–1.54 |
| R factor (%) | 16.6 | 14.9 |
| Rfree (%) | 20.1 | 17.5 |
| RMSD deviation from ideality | ||
| Bond length (Å) | 0.012 | 0.012 |
| Angle (°) | 1.72 | 1.71 |
| Ramachandran plot statistics (%) | ||
| Core regions | 93.4 | 91.3 |
| Allowed regions | 6.2 | 8.2 |
| Additionally allowed regions | 0.4 | 0.4 |
| Disallowed regions | 0.0 | 0.0 |
FIG 2.
Stereo diagram of difference electron density of the active site of KPC-2 β-lactamase, showing bound S02030. Alternate conformations of the triazole-carboxylic acid moiety of S02030 are indicated (a and b). The difference density (Fo − Fc) map was calculated after 10 rounds of REFMAC refinement, with S02030 removed from the refinement and structure factor calculations. Density was contoured at 3σ.
FIG 3.
Stereo diagram of difference electron density of the active site of SHV-1, showing S02030. Alternate conformations of the thiopene moiety are indicated (c and d). The map was calculated and contoured as described for Fig. 2.
Protein structure accession numbers.
The final coordinates for both S02030 SHV-1 and KPC02 complex structures together with the structure factors were deposited with the Protein Data Bank under PDB codes 5EE8 and 5EEC, respectively.
RESULTS AND DISCUSSION
In this study, we have probed the mode of binding of S02030 to two β-lactamases that are common to K. pneumoniae, KPC-2 and SHV-1. KPC-2 is clinically the most important carbapenemase (5); the crystal structure has been determined, as well as its structure in complex with several different inhibitors (10, 13, 26). SHV-1 is part of a large class of more than 190 SHV β-lactamase variants, some of which are inhibitor resistant (IR) or have an extended-spectrum β-lactamase (ESBL) phenotype. SHV-1 represents a typical class A β-lactamase, and its inclusion in this study allows a comparison with both the carbapenemase KPC-2 and the class C ADC-7 β-lactamase (10, 19, 27–30).
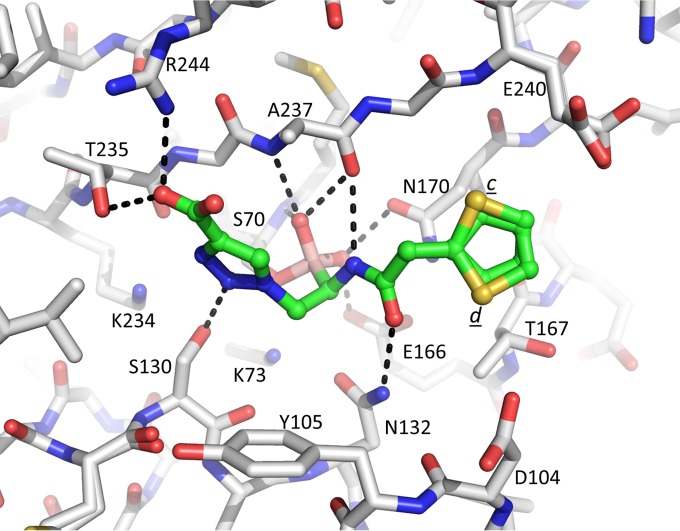
The asymmetric unit of the KPC-2–S02030 structure contained two non-crystallographically related molecules, A and B. The electron density difference (Fo − Fc) map in the KPC-2 β-lactamase active site of molecule A revealed the presence of a fully occupied S02030 molecule refined with two conformations for the carboxyl-triazole moiety (conformations a and b, with occupancies of 0.6 and 0.4, respectively) (Fig. 2 and 4). S02030 bound to KPC-2 molecule B was less well occupied, with an overall occupancy of 0.6; this S02030 molecule adopted a conformation similar to that in molecule A, with conformation b for the carboxyl-triazole moiety (data not shown).
FIG 4.
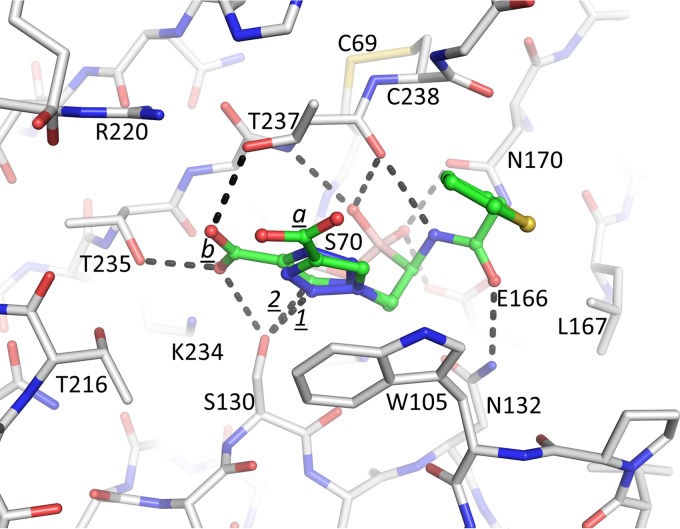
Interactions of S02030 in the active site of KPC-2 β-lactamase. Hydrogen bonds are depicted by dashed lines. The hydrogen bond between the N3 ring nitrogen and S130 in conformation a is labeled “1”; the C—H…O hydrogen bond between the C5 carbon atom and S130 in conformation b is labeled “2.”
The observed S02030 binding modes each have the boron atom attached to the catalytic S70 residue (Fig. 4). One of the oxygens of the boronic acid moiety occupies the oxyanion hole pocket, and the other oxygen is positioned in the pocket usually known to harbor the deacylation water; the latter pocket is comprised of residues E166 and N170. The amide moiety of S02030 interacts with both sides of the active site via interactions with the side chain of N132 and the main-chain carbonyl of T237 (Fig. 4). The carboxyl-triazole moiety of S02030 interacts, in conformation a, with S130 via one of the triazole ring nitrogens (N2) involving a 3.1-Å hydrogen bond (labeled “1” in Fig. 4). In conformation b, the carboxyl group of this moiety makes hydrogen bonds with S130 as well as with T235 and T237. Intriguingly, the C5 ring carbon of the triazole moiety in this conformation b is at a 2.9-Å distance from the Oγ of S130 (labeled “2” in Fig. 4), suggesting a C—H…O hydrogen bonding interaction. Such a C—H-mediated hydrogen bond is unanticipated due to its nonpolar nature, yet in the triazole moiety with its 3 linked nitrogens, the C—H is known to be polar enough to function as a hydrogen bond donor (31, 32). If indeed a C—H-mediated hydrogen bond is present, to our knowledge, this would be the first observed C—H…O hydrogen bond in a triazole-moiety containing ligand with a protein. This could be an important feature to be exploited for future efforts toward discovery of triazole-based drugs in particular, as this moiety is a key product used in click chemistry-based synthesis efforts (32). In addition to the hydrogen bonds, the carboxyl-triazole moiety in both conformations in molecule A makes π stacking interactions with W105 (Fig. 4); in molecule B, this W105 is repositioned in a different orientation and does not make such an interaction likely due to the lower occupancy of the S02030 in that molecule. Note that although negatively charged, the carboxyl-triazole moiety also does not make a salt bridge in either of its two conformations, although there are some electrostatic interactions, as K234 and R220 are at 3.8- and 3.9-Å distances, respectively, from one of the oxygen atoms of the carboxyl moiety in conformation b. At the other end of S02030, the thiophene moiety makes more limited interactions via a 3.6-Å van der Waals interaction with G239.
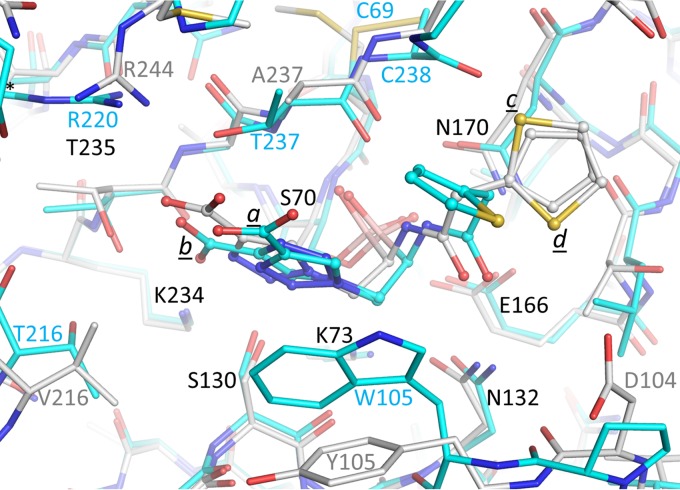
In the SHV-1 complex structure, S02030 adopts a conformation similar to that of KPC-2 (Fig. 3, 5, and 6). The boronic acid moiety and amide moieties of S02030 all make interactions very similar to what was observed with KPC-2. The carboxyl-triazole adopts a single conformation, unlike what was observed when complexed to KPC-2; this conformation is most similar to that of conformation a in molecule A of the KPC-2 complex structure, although overall this moiety is somewhat shifted (Fig. 6). With the SHV-1 complex, the carboxyl-triazole moiety makes a 2.8-Å salt bridge with SHV-1 residue R244 (Fig. 5); this is a difference from the KPC-2 structure, as KPC-2 does not have an arginine at this position. An additional difference compared to KPC-2 is that the π stacking interaction with residue 105 (Y105 in the case of SHV-1) is not present. The thiophene moiety, however, adopts two conformations in SHV-1 (labeled “c” and “d” in Fig. 3 and 5) and is in a conformation different from that observed in KPC-2 (Fig. 6). This thiophene moiety of S02030 makes several van der Waals interactions in the active site of SHV-1, including with carbon atoms of residues N170, T167, and E240. The observation of relatively similar modes of binding ofS02030 to KPC-2 and SHV-1 is in agreement with the 50% inhibitory concentrations (IC50s) that were measured for KPC-2 and SHV-1, which are 0.080 ± 0.002 and 0.130 ± 0.002 μM, respectively (18). The minor difference in IC50 could in part be attributed to S02030's ability to have π stacking interactions with large aromatic residues, e.g., W105, in KPC-2, whereas such an interaction is not observed in SHV-1. It is interesting that a somewhat similar boronic acid inhibitor, RPX7009, has an amide-thiophene moiety yet a different carboxyl-containing moiety (33). The RPX7009 structure complexed to a different class A β-lactamase, CTX-M-15, revealed an orientation of the inhibitor generating similar active interactions by its amide-thiophene moieties and carboxyl group (33).
FIG 5.
Interactions of S02030 in the active site of SHV-1 β-lactamase.
FIG 6.
Superposition of SHV-1- and KPC-2-bound S02030 structures. The protein atoms are shown in stick representation and the ligands are shown in ball-and-stick representation. The KPC-2–S02030 structure is depicted with the carbon atoms in cyan; the SHV-1–S02030 structure is depicted with the carbon atoms in white. The Cα atoms of residues 68 to 84, 121 to 140, 167 to 172, and 233 to 238 of SHV-1 were superimposed on the identical residues of KPC-2, yielding a root mean square deviation (RMSD) of 0.51 Å.
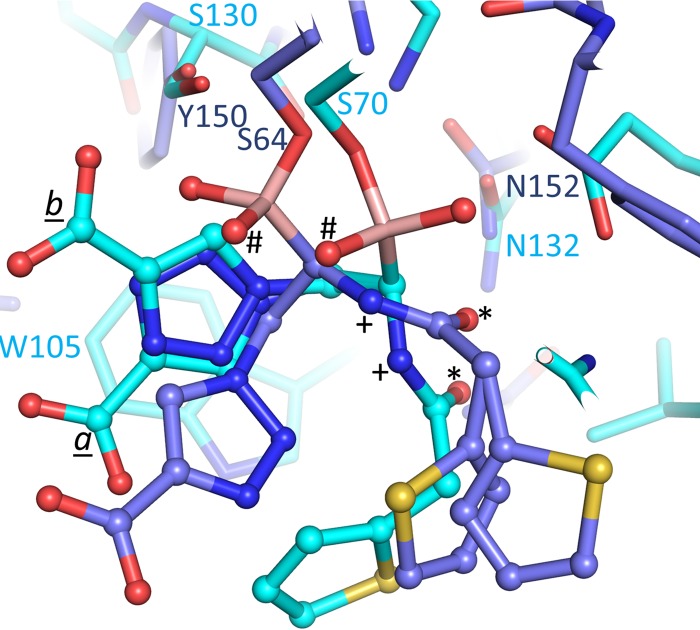
Previously, the structure of S02030 bound to the class C ADC-7 β-lactamase from A. baumannii was determined (17), allowing us now to compare the modes of binding of S02030 to the two class A β-lactamases determined herein. The ADC-7–S02030 complex structure (PDB code 4U0X) was comprised of 4 independently refined ADC-7 molecules in the asymmetric unit, each having an S02030 molecule bound. Analyses of these 4 copies of S02030 bound to ADC-7 showed variability in the positions and orientations of the carboxyl-triazole and thiophene moieties, whereas the boronic acid and amide moieties were bound in a similar fashion (17). Superpositioning of KPC-2 onto one of the ADC-7 S02030 copies revealed interesting differences and similarities (Fig. 7). Key similarities are the presence of the boron bond with the catalytic serine. In addition, the amide moieties provide similar interactions bridging the active site: the amide nitrogen (labeled “+” in Fig. 7) interacts with the carbonyl oxygen of T237 or the equivalent residue in ADC-7; the amide oxygen (labeled “*”) interacts with N132 or the equivalent N152 in ADC-7 (Fig. 7). An important difference between the S02030 binding modes in KPC-2 and ADC-7 is the orientation of the boronic acid moiety, placing the two boron oxygens in different positions. Notably, class C β-lactamases do not have a deacylation pocket comprised of E166/N170, forcing the boron oxygen that occupies this pocket in KPC-2 to now reorient itself such that it occupies the oxyanion hole in ADC-7 (both oxyanion hole-occupying boron oxygens are labeled “#” in Fig. 7). The other boron oxygen of S02030, the one that occupies the oxyanion hole in KPC-2, has swung away to make space to accommodate the other oxygen in ADC-7. The latter conformational difference is achieved by having a different torsion angle around the Cβ—Oγ bond of the catalytic serine (S70 and S64 in KPC-2 and ADC-7, respectively). The carboxyl-triazole moiety is situated in a somewhat similar position in KPC-2 and ADC-7; the position of the S02030 carboxyl moiety in ADC-7 is closest to that of conformation a in KPC-2, although the orientation of triazole ring is flipped (Fig. 7). The thiophene moieties are in the same general vicinity although not in identical positions. Note that the carboxyl-triazole and thiophene moieties in ADC-7 already show variability in orientation and position when the 4 independently refined molecules in the ADC-7 structure are compared (17). Note that since an IC50 was not determined for S02030 inhibition of ADC-7 (a Ki of 44.5 ± 2.2 nM was measured instead [17]), we cannot go into detailed structural comparisons to explain differences in S02030 affinity for ADC-7 compared to KPC-2 and SHV-1.
FIG 7.
Superposition of ADC-7- and KPC-2-bound S02030 structures. The protein is represented in stick and the inhibitors in ball-and-stick representations; KPC-2 is shown with cyan carbon atoms, whereas theADC-7 carbon coordinates are shown in medium blue. The Cα atoms of residues 67 to 84, 132 to 134, 231 to 238, and 243 to 249 of KPC-2 were superimposed onto ADC-7 residues 61 to 78, 152 to 154, 309 to 316, and 319 to 325, respectively, with an RMSD of 0.98 Å.
Overall, upon comparing the binding modes of S02030 in KPC-2, SHV-1, and ADC-7, a common theme emerges, as S02030 is able to recognize and inhibit vastly different serine β-lactamases. First, S02030 has the ability to interact with the conserved features of these active sites, and that includes forming the bond with catalytic serine via the boron atom, positioning one of the boronic acid oxygens in the oxyanion hole, as well as utilizing its amide moiety to make conserved interactions across the width of the active site. Second, in regions that are a bit more distant from this very conserved central active-site region, S02030 is able overcome the structural differences between the β-lactamases to maintain its relative broad specificity. These important interactions are achieved by reorienting the more polar carboxyl-triazole moiety to generate polar interactions as well as reorient the more hydrophobic thiophene moiety depending on what active site it is bound in. The polar carboxyl-triazole moiety seems particularly well suited for this former ability, as by reorienting and changing its torsions angles, different hydrogen bonds and salt bridges can be made in different directions depending on the particular active site it needs to bind to; the presence of the carboxyl moiety is well chosen, as the β-lactamase active sites are all known to attract and bind carboxyl moiety-containing β-lactam substrates. This remarkable structural plasticity of S02030 is likely very important, as it confers the ability to recognize different β-lactamases.
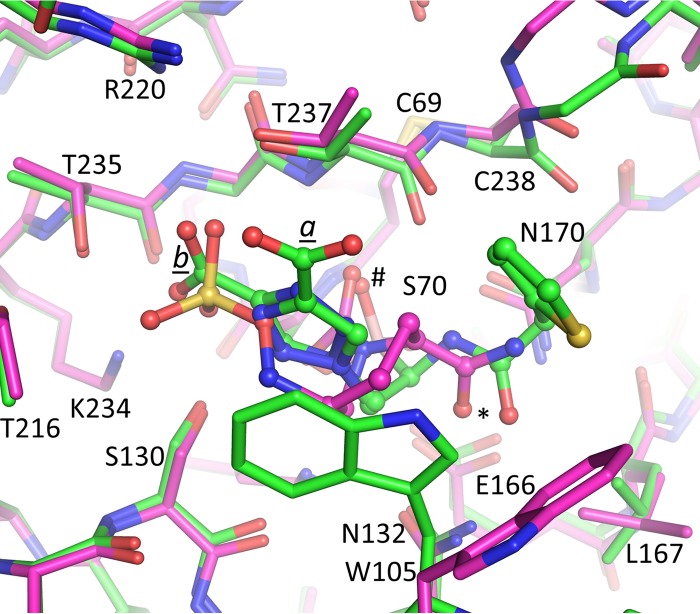
The structure of KPC-2 complexed with avibactam was recently determined (10). Avibactam is a diazabicyclooctane non-β-lactam BLI. Although avibactam is very different than the BATSI S02030, a comparison between the two inhibitors might elucidate some common features of efficient inhibition of KPC-2. Superpositioning of KPC-2-bound S02030 onto KPC-2-bound avibactam reveals that both inhibitors utilize and occupy the oxyanion hole via an oxygen atom (atoms labeled “#” in Fig. 8). In addition, both avibactam and S02030 have an amide oxygen atom that interacts with residue N132, although the nitrogen atom of this amide in avibactam does not make a direct protein interaction. An additional similarity is that both S02030 and avibactam use a negatively charged moiety to interact with the class A carboxyl recognition pocket (34). This negatively charged inhibitor moiety is a carboxylate in S02030 and a sulfate moiety in avibactam (Fig. 8); each were found to interact with residues T235, T237, and S130 (with R220 and R234 situated at around a 4-Å distance to provide some electrostatic attraction).
FIG 8.
Superposition of S02030- and avibactam-bound KPC-2 structures. The protein is in stick representation, whereas the inhibitors are in ball-and-stick representation; the KPC-2–S02030 carbon coordinates are in green, whereas the KPC-2–avibactam structure has its carbon atoms colored magenta.
In summary, the BATSI S02030 possesses structural plasticity and can interact efficiently with conserved and variable regions of related β-lactamases. This is in part due to its triazole moiety, which is very polar as a result of the three electronegative nitrogens on one side of the ring; this polar nature allows the carbon on the opposite site of the ring to have the potential to be a unique C—H hydrogen bond donor to complement the already-present hydrogen bond acceptor abilities of the ring nitrogens. S02030 utilizes features and interactions similar to those used by a completely different BLI, avibactam, to arrive at efficient inhibition of K. pneumoniae β-lactamases KPC-2 and SHV-1.
ACKNOWLEDGMENTS
Research reported in this publication was supported by the Harrington Foundation, National Institute of Allergy and Infectious Diseases of the National Institutes of Health, under award numbers R01AI100560 and R01AI063517 to R.A.B. This study was also supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs to R.A.B., by Veterans Affairs Merit Review Program Award 1I01BX001974 to R.A.B., and by Geriatric Research Education and Clinical Center VISN 10 to R.A.B.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
For a companion article on this topic, see doi:10.1128/AAC.02641-15.
REFERENCES
- 1.Silva KE, Cayo R, Carvalhaes CG, Patussi Correia Sacchi F, Rodrigues-Costa F, Ramos da Silva AC, Croda J, Gales AC, Simionatto S. 2015. Coproduction of KPC-2 and IMP-10 in carbapenem-resistant Serratia marcescens isolates from an outbreak in a Brazilian teaching hospital. J Clin Microbiol 53:2324–2328. doi: 10.1128/JCM.00727-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu WL, Lee MF, Tang HJ, Chang MC, Walther-Rasmussen J, Chuang YC. 2015. Emergence of KPC new variants (KPC-16 and KPC-17) and ongoing outbreak in southern Taiwan. Clin Microbiol Infect 21:347.e5–347.e8. [DOI] [PubMed] [Google Scholar]
- 3.Bratu S, Landman D, Haag R, Recco R, Eramo A, Alam M, Quale J. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch Intern Med 165:1430–1435. doi: 10.1001/archinte.165.12.1430. [DOI] [PubMed] [Google Scholar]
- 4.Perez F, Endimiani A, Ray AJ, Decker BK, Wallace CJ, Hujer KM, Ecker DJ, Adams MD, Toltzis P, Dul MJ, Windau A, Bajaksouzian S, Jacobs MR, Salata RA, Bonomo RA. 2010. Carbapenem-resistant Acinetobacter baumannii and Klebsiella pneumoniae across a hospital system: impact of post-acute care facilities on dissemination. J Antimicrob Chemother 65:1807–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nordmann P, Poirel L. 2014. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 6.Drawz SM, Bonomo RA. 2010. Three decades of β-lactamase inhibitors. Clin Microbiol Rev 23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watkins RR, Papp-Wallace KM, Drawz SM, Bonomo RA. 2013. Novel beta-lactamase inhibitors: a therapeutic hope against the scourge of multidrug resistance. Front Microbiol 4:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Durand-Reville TF, Lahiri S, Thresher J, Livchak S, Gao N, Palmer T, Walkup GK, Fisher SL. 2013. Kinetics of avibactam inhibition against class A, C, and D beta-lactamases. J Biol Chem 288:27960–27971. doi: 10.1074/jbc.M113.485979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush K. 2015. A resurgence of beta-lactamase inhibitor combinations effective against multidrug-resistant Gram-negative pathogens. Int J Antimicrob Agents 46:483–493. doi: 10.1016/j.ijantimicag.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Krishnan NP, Nguyen NQ, Papp-Wallace KM, Bonomo RA, van den Akker F. 2015. Inhibition of Klebsiella beta-lactamases (SHV-1 and KPC-2) by avibactam: a structural study. PLoS One 10:e0136813. doi: 10.1371/journal.pone.0136813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papp-Wallace KM, Winkler ML, Taracila MA, Bonomo RA. 9 February 2015. Variants of the KPC-2 beta-lactamase which are resistant to inhibition by avibactam. Antimicrob Agents Chemother doi: 10.1128/AAC.04406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winkler ML, Rodkey EA, Taracila M, Drawz SM, Bethel C, Papp-Wallace K, Smith K, Xu Y, Dwulit-Smith J, Romagnoli C, Caselli E, Prati F, van den Akker F, Bonomo RA. 2013. Design and exploration of novel boronic acid inhibitors reveals important interactions with a clavulanic acid-resistant sulfhydryl-variable (SHV) β-lactamase. J Med Chem 56:1084–1097. doi: 10.1021/jm301490d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ke W, Bethel CR, Papp-Wallace KM, Pagadala SR, Nottingham M, Fernandez D, Buynak JD, Bonomo RA, van den Akker F. 2012. Crystal structures of KPC-2 β-lactamase in complex with 3-nitrophenyl boronic acid and the penam sulfone PSR-3-226. Antimicrob Agents Chemother 56:2713–2718. doi: 10.1128/AAC.06099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ke W, Sampson JM, Ori C, Prati F, Drawz SM, Bethel CR, Bonomo RA, van den Akker F. 2011. Novel insights into the mode of inhibition of class A SHV-1 β-lactamases revealed by boronic acid transition state inhibitors. Antimicrob Agents Chemother 55:174–183. doi: 10.1128/AAC.00930-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eidam O, Romagnoli C, Dalmasso G, Barelier S, Caselli E, Bonnet R, Shoichet BK, Prati F. 2012. Fragment-guided design of subnanomolar beta-lactamase inhibitors active in vivo. Proc Natl Acad Sci U S A 109:17448–17453. doi: 10.1073/pnas.1208337109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eidam O, Romagnoli C, Caselli E, Babaoglu K, Pohlhaus DT, Karpiak J, Bonnet R, Shoichet BK, Prati F. 2010. Design, synthesis, crystal structures, and antimicrobial activity of sulfonamide boronic acids as β-lactamase inhibitors. J Med Chem 53:7852–7863. doi: 10.1021/jm101015z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powers RA, Swanson HC, Taracila MA, Florek NW, Romagnoli C, Caselli E, Prati F, Bonomo RA, Wallar BJ. 2014. Biochemical and structural analysis of inhibitors targeting the ADC-7 cephalosporinase of Acinetobacter baumannii. Biochemistry 53:7670–7679. doi: 10.1021/bi500887n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rojas LJ, Taracila MA, Papp-Wallace KM, Bethel CR, Caselli E, Romagnoli C, Winkler ML, Spellberg B, Prati F, Bonomo RA. 2016. Boronic acid transition state inhibitors active against KPC and other class A beta-lactamases: structure-activity relationships as a guide to inhibitor design. Antimicrob Agents Chemother 60:1751–1759. doi: 10.1128/AAC.02641-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padayatti PS, Helfand MS, Totir MA, Carey MP, Hujer HM, Carey PR, Bonomo RA, van den Akker F. 2004. Tazobactam forms a stoichiometric trans-enamine intermediate in the E166A variant of SHV-1 β-lactamase: 1.63 Å crystal structure. Biochemistry 43:843–848. doi: 10.1021/bi035985m. [DOI] [PubMed] [Google Scholar]
- 20.Kuzin AP, Nukaga M, Nukaga Y, Hujer AM, Bonomo RA, Knox JR. 1999. Structure of the SHV-1 β-lactamase. Biochemistry 38:5720–5727. doi: 10.1021/bi990136d. [DOI] [PubMed] [Google Scholar]
- 21.Minor W, Tomchick D, Otwinowski Z. 2000. Strategies for macromolecular synchrotron crystallography. Structure 8:R105–R110. doi: 10.1016/S0969-2126(00)00139-8. [DOI] [PubMed] [Google Scholar]
- 22.Winn MD, Murshudov GN, Papiz MZ. 2003. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol 374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Schüttelkopf AW, van Aalten DM. 2004. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 2001. PROCHECK—a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291. [Google Scholar]
- 26.Ke W, Bethel CR, Thomson JM, Bonomo RA, van den Akker F. 2007. Crystal structure of KPC-2: insights into carbapenemase activity in class A β-lactamases. Biochemistry 46:5732–5740. doi: 10.1021/bi700300u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ke W, Rodkey EA, Sampson JM, Skalweit MJ, Sheri A, Pagadala SR, Nottingham MD, Buynak JD, Bonomo RA, van den AF. 2012. The importance of the trans-enamine intermediate as a β-lactamase inhibition strategy probed in inhibitor-resistant SHV β-lactamase variants. Chem Med Chem 7:1002–1008. doi: 10.1002/cmdc.201200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sampson JM, Ke W, Bethel CR, Pagadala SR, Nottingham MD, Bonomo RA, Buynak JD, van den Akker F. 2011. Ligand-dependent disorder of W-loop observed in extended-spectrum SHV-type β-lactamase. Antimicrob Agents Chemother 55:2303–2309. doi: 10.1128/AAC.01360-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Totir MA, Padayatti PS, Helfand MS, Carey MP, Bonomo RA, Carey PR, van den Akker F. 2006. Effect of the inhibitor-resistant M69V substitution on the structures and populations of trans-enamine beta-lactamase intermediates. Biochemistry 45:11895–11904. doi: 10.1021/bi060990m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padayatti PS, Sheri A, Totir MA, Helfand MS, Carey MP, Anderson VE, Carey PR, Bethel CR, Bonomo RA, Buynak JD, van den Akker F. 2006. Rational design of a β-lactamase inhibitor achieved via stabilization of the trans-enamine intermediate: 1.28Å crystal structure of wt SHV-1 complex with a penam sulfone. J Am Chem Soc 128:13235–13242. doi: 10.1021/ja063715w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar A, Pandey PS. 2008. Anion recognition by 1,2,3-triazolium receptors: application of click chemistry in anion recognition. Org Lett 10:165–168. doi: 10.1021/ol702457w. [DOI] [PubMed] [Google Scholar]
- 32.Hua Y, Flood AH. 2010. Click chemistry generates privileged CH hydrogen-bonding triazoles: the latest addition to anion supramolecular chemistry. Chem Soc Rev 39:1262–1271. doi: 10.1039/b818033b. [DOI] [PubMed] [Google Scholar]
- 33.Hecker SJ, Reddy KR, Totrov M, Hirst GC, Lomovskaya O, Griffith DC, King P, Tsivkovski R, Sun D, Sabet M, Tarazi Z, Clifton MC, Atkins K, Raymond A, Potts KT, Abendroth J, Boyer SH, Loutit JS, Morgan EE, Durso S, Dudley MN. 2015. discovery of a cyclic boronic acid beta-lactamase inhibitor (RPX7009) with utility vs class A serine carbapenemases. J Med Chem 58:3682–3692. doi: 10.1021/acs.jmedchem.5b00127. [DOI] [PubMed] [Google Scholar]
- 34.Rodkey EA, Drawz SM, Sampson JM, Bethel CR, Bonomo RA, van den Akker F. 2012. Crystal structure of a pre-acylation complex of the β-lactamase inhibitor sulbactam bound to a sulfenamide bond-containing thiol-beta-lactamase. J Am Chem Soc 134:16798–16804. doi: 10.1021/ja3073676. [DOI] [PMC free article] [PubMed] [Google Scholar]