Abstract
This work describes the diversity and evolution of Tn5801 among enterococci, staphylococci, and streptococci based on analysis of the 5,073 genomes of these bacterial groups available in gene databases. We also examined 610 isolates of Enterococcus (from 10 countries, 1987 to 2010) for the presence of this and other known CTn-tet(M) elements due to the scarcity of data about Tn5801 among enterococci. Genome location (by ICeu-I–pulsed-field gel electrophoresis [PFGE] hybridization/integration site identification), conjugation and fitness (by standard methods), Tn5801 characterization (by long-PCR mapping/sequencing), and clonality (by PFGE/multilocus sequence typing [MLST]) were studied. Twenty-three Tn5801 variants (17 unpublished) clustered in two groups, designated “A” (25 kb; n = 14; predominant in Staphylococcus aureus) and “B” (20 kb; n = 9; predominant in Streptococcus agalactiae). The percent GC content of the common backbone suggests a streptococcal origin of Tn5801 group B, with further acquisition of a 5-kb fragment that resulted in group A. Deep sequence analysis allowed identification of variants associated with clonal lineages of S. aureus (clonal complex 8 [CC8], sequence type 239 [ST239]), S. agalactiae (CC17), Enterococcus faecium (ST17/ST18), or Enterococcus faecalis (ST8), local variants, or variants located in different species and geographical areas. All Tn5801 elements were chromosomally located upstream of the guaA gene, which serves as an integration hot spot. Transferability was demonstrated only for Tn5801 type B among E. faecalis clonal backgrounds, which eventually harbored another Tn5801 copy. The study documents early acquisition of Tn5801 by Enterococcus, Staphylococcus, and Streptococcus. Clonal waves of these pathogens seem to have contributed to the geographical spread and local evolution of the transposon. Horizontal transfer, also demonstrated, could explain the variability observed, with the isolates often containing sequences of different origins.
INTRODUCTION
The extended use of tetracycline since its introduction into the therapeutic arsenal in 1948 seems to have resulted in an evolutionary bottleneck in the population structure of some genera of Firmicutes that are of interest in human health. Recent studies have associated the acquisition and further fixation of tetracycline resistance (Tetr) by pathogenic clones of group B Streptococcus (GBS) with the global increase of high-mortality GBS neonatal infections in the last decades (1, 2). Similarly, some Staphylococcus aureus lineages are enriched in Tetr elements (3–5).
Tetracycline resistance in major Gram-positive human opportunistic pathogens is caused mainly by the acquisition of integrative and conjugative elements (ICE) of the Tn916 family carrying the tet(M) gene (5–7). These elements display a common synteny but differ in the integrase (int) and the excisionase (xis) sequences, insertion site specificity, and host range (8, 9). Variants of some Tn916-tet(M) members, namely Tn916, Tn5397, Tn6000, or Tn5801, are widely spread among several genera of Firmicutes, apparently suggesting a successful multihost dissemination of these elements (7).
Tn5801 has a site-specific tyrosine recombinase that recognizes the 3′ end of the GMP synthase gene (guaA) (10, 11), an insertion hot spot of genomic islands coding for pathogenicity or antibiotic resistance in different Firmicutes (12–14). Tn5801 was originally detected in the vancomycin-resistant Staphylococcus aureus (VRSA) clinical strain Mu50 recovered in Japan in 1997 (15), but several studies have documented the presence of platforms highly similar to Tn5801 in early isolates of Streptococcus agalactiae (France, 1953) (1), S. aureus (Denmark, 1963; designated Tn6014) (3), and Clostridium perfringens (United States, 1977; a truncated element designated CW459tet(M)] (16). The diversity of Tn5801-like backbones in a few isolates of different species (3, 17–20) collected throughout more than 50 years suggests the evolutionary interplay of selective events, horizontal dissemination, and the genetic plasticity of this transposon, allowing its spread in different clonal backgrounds and environments. However, little is known about Tn5801, at either the molecular or the epidemiological level.
In this study, we created a data set comprising all the genomes of Firmicutes with the presence of a Tn5801-like ICE. Due to the relevance of enterococci in the spread of Tetr and the low number of sequenced genomes in public databases at the time this study was started, we also analyzed the presence and genetic background of Tn5801 in a large collection of enterococcal strains from different geographical areas as well as its transferability. This comprehensive phylogenetic and genomic analysis allowed documentation of the genetic variability of Tn5801-like elements among genomes of enterococci, staphylococci, streptococci, and other bacterial species through years and across continents. The results highlight the relevance of comprehensive analysis at local and global levels to accurately establish the transmission and evolvability of elements at different scales. It also proved the transferability of the element in different genetic contexts.
MATERIALS AND METHODS
Screening of Tn5801 in GenBank databases and creation of data sets.
The DNA sequence from S. aureus Mu50 (GenBank accession number BA000017) was used as a reference to screen for the presence of Tn5801 among 5,073 draft and complete genomes from Firmicutes (4,130 S. aureus, 345 Enterococcus faecalis, 301 S. agalactiae, 254 Enterococcus faecium, 23 Streptococcus mitis, 14 Lactococcus garvieae, 4 Staphylococcus pseudintermedius, 2 Enterococcus villorum, and 1 Streptococcus thoraltensis) obtained from the NCBI archive (last updated January 2015). BLAST searches identified different sequences showing homology with Tn5801-like elements. They were further stored and analyzed by using the MUSCLE (multiple-sequence comparison by log expectation) algorithm (http://www.ebi.ac.uk/Tools/msa/muscle/) and Artemis comparison tool (21) (http://www.sanger.ac.uk/resources/software/act/). ICE maps were created with Vector NTI (ThermoFisher Scientific) and Inkscape (http://inkscape.org/) software.
Phylogenetic analyses were accomplished using MEGA version 6 (22). A common region of 13,895 bp was identified among all the sequences with homology with Tn5801 (here called “Tn5801 variant sequences”). The region comprises 16 open reading frames (ORFs) (intTn5801-orf24; orf22, orf18, orf16, orf15, orf12-10, and orf8-4), which were trimmed and concatenated to be further analyzed. A maximum-likelihood phylogenetic tree was constructed with 1,000 bootstrap replications in order to infer the relationship among all Tn5801 variants. Due to the identification of modules with different GC contents and with recombination landmarks, the Tn5801 variants were further split into five core fragments (intTn5801, orf26-19, orf18-14, orf12-8, and orf7-4), with each fragment being separately analyzed as mentioned before. The orf26-19 fragment was excluded from this analysis because of the variability observed among variants due to frequent indels and recombinatorial events. An arbitrary number was attributed to different sequences of each core fragment, and Tn5801 sequences with unique number profiles were used to calculate a consensus tree. The data set created included epidemiological information for isolates (host/source, year of isolation, country, and sequence type [ST]). To identify the STs of the isolates, we used the MLST 1.8 software of the Center of Genomic Epidemiology server (23).
Epidemiological background of bacterial strains and identification of tet(M) transposons among enterococci.
A collection of 610 enterococcal isolates from humans (n = 320; hospitalized patients [HP], n = 195; healthy human volunteers [HV], n = 125); animals (n = 236; poultry [P], n = 164; swine/piggeries [SP], n = 72), and hospital/urban sewage (SW) (n = 54) collected in different countries over a 23-year period (1987 to 2010) was included (9, 24). All isolates were tested for susceptibility to tetracycline by the disk diffusion and/or agar dilution method following CLSI guidelines (25). The presence of genes coding for tetracycline resistance [tet(M), tet(L), tet(S), tet(K), and tet(O)] was assayed by PCR as previously described (26). The presence of the main transposons of the Tn916 family (Tn5801, Tn916, Tn5397, Tn5386, and Tn6000) was investigated with a multiplex PCR assay designed here for detecting integrases, excisionases, and other specific sequences of known transposons. The susceptibility and specificity of the assay were validated using appropriate controls. Primers and conditions are listed in Table S1 in the supplemental material. The clonal relationship among E. faecium and E. faecalis isolates harboring the integrase of Tn5801 (intTn5801) was established by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) (http://pubmlst.org/) (27, 28). Population genetic analysis was performed using BAPS software with E. faecium isolates as previously described (29, 30).
PCR characterization of Tn5801 backbones in Enterococcus.
The Tn5801 backbone was fully characterized by a PCR mapping assay based on the Tn5801 sequence of S. aureus Mu50 (GenBank accession number BA000017). Amplified fragments obtained from positive intTn5801 strains were further sequenced (Table 1; see Fig. S1 in the supplemental material). Genomic location of Tn5801 was assessed by hybridization of I-CeuI (TaKaRa Biotechnology, Dalian, China) digested genomic DNA using specific intTn5801 and 23S rRNA probes (31). Amplification of the intTn5801-guaA region was performed in order to characterize the integration site of Tn5801 (Table 1; see Fig. S1 in the supplemental material) (14).
TABLE 1.
Characterization of Tn5801: PCR strategy and conditions
| Primer | Sequence (5′→3′) | Position in Tn5801 (bp) | PCR | Sequence amplified | Amplicon size (bp) | Amplification conditions |
|---|---|---|---|---|---|---|
| gmp1 | CCTGGTCTTGGTATTCGTGT | 25763–25783a | gmp1-gmp2 | guaA | 170 | 1 cycle of 95°C for 10 min; 25 cycles of 94°C for 30 s, 54°C for 30 s, 72°C for 30 s; 1 cycle of 72°C for 10 min |
| gmp2 | GTAGTCTCCCATAACACC | 25609–25627a | ||||
| Int1 | CTGTTTCCGATATTGAGC | 24490–24473a | Int1-gmp1 | intTn5801-guaA | 1,311 | 35 cycles of 96°C for 30 s, 45–59°C (according to primer pair) for 1 min, 72°C for 7 min; 1 cycle of 72°C for 10 min |
| P1 | GTTTCGCAAGTAGTCTACAG | 25310–25329a | P1-P2 | orf25-intTn5801 | 2,085 | |
| P2 | GTAAAGGCGACAGATGG | 23244–23260a | ||||
| P3 | CTTAGAGATGAGTTTCGTTTC | 23391–23411a | P3-P4 | orf22-orf25 | 1,455 | |
| P4 | GGATAGTTCTTTGTCTGTAAAG | 21956–21977a | ||||
| P5 | CGATTTTAGAGCCGTTGGTTTAG | 22210–22232a | P5-P6 | tet(M)-orf22 | 2,448 | |
| P6 | GAACGTCAGAGAGGAATTAC | 19784–19803a | ||||
| P7 | GGGAATCCCCATTTTCCTAA | 19986–20005a | P7-P8 | orf15-tet(M) | 6,498 | |
| P8 | ACCCACTCGCTGTTTAATCG | 13507–13526a | ||||
| P9 | CTTGCAAGGCTAGGTTGGAG | 14162–14181a | P9-P10 | orf7-orf15 | 5,993 | |
| P10 | ATGTCTGAAATGGGCTTTGG | 8188–8207a | ||||
| P11 | CTTTCACTTCGTGCGGTACA | 8382–8401a | P11-P12 | orf3-orf7 | 4,631 | |
| P12 | GGGTGGGGACAATACATCAG | 3770–3789a | ||||
| P13 | CAGCCATGTAGCGTCTTTGA | 4315–4334a | P13-P14 | orf1-orf3 | 4,228 | |
| P14 | ACGGAGTTAACGGCTTTCCT | 106–125a | ||||
| P15 | CCGTATGTTCTTTCAACCACT | 1030–1051b | P15-P11 | orf5-orf7 | 2,675 | |
| P16 | GCTGACAGTTCCAGTATCC | 18096–18115c | P16-P5 | orf23′-orf22 | 1,209 | |
| P17 | CAGAACCAGCCATTACC | 18561–18578c | P17-P18 | orf23′-orf19′ | 3,938 | |
| P18 | GAAGATACGAGAAACCAATAG | 14602–14623c | ||||
| P19 | GGTGGCAATTCAAGTGTTCC | 14859–14878c | P19-P20 | orf19′-orf17 | 1,957 | |
| P20 | GGCGTATGACAAAGCTGG | 12921–12939c |
Reference sequence, Staphylococcus aureus Mu50 (GenBank accession number BA000017).
Reference sequence, Streptococcus agalactiae COH1 (GenBank accession number AAJR01000021.1).
Reference sequence, Enterococcus faecalis JH2-2 (GenBank accession number AXOI01000011.1).
Transferability of Tn5801.
Transferability of Tn5801 was screened by filter mating using different recipient strains of E. faecalis (JH2-2, OG1RF, and OG1SSp) and E. faecium (64/3, BM4105RF, and BM4105SS), all being resistant to rifampin and fusidic acid (designated RF) or streptomycin and spectinomycin (designated SS) and negative for the presence of the tet(M) gene (32). The mating assays were performed at 37°C for 36 h using aliquots of broth cultures of donors and recipients grown up to the exponential phase (McFarland 0.5) and mixed at a ratio of 1:1 (32). Transconjugants were selected on brain heart infusion (BHI) agar (Pronadisa, Laboratorios Conda, SA, Madrid, Spain) plates supplemented with tetracycline (10 mg/liter) plus rifampin (30 mg/liter) and fusidic acid (25 mg/liter) or streptomycin (250 mg/liter) and spectinomycin (250 mg/liter) (Sigma-Aldrich, Inc., St. Louis, MO). Unexpectedly, we identified a Tn5801 Δtet(M) variant in the laboratory recipient E. faecalis strain JH2-2, which is an emblematic receptor strain (see below). Secondary filter mating assays were performed using E. faecalis JH2-2 transconjugants as donors and the E. faecalis OG1SS and E. faecium BM4105SS strains as recipients. Transconjugants were characterized according to the susceptibility to tetracycline (Etest; bioMérieux SA, France), the presence of Tn5801 [intTn5801 and tet(M)], the transposon backbone (PCR overlapping), and comparison of SmaI-digested DNA PFGE profiles of transconjugants with those of recipients and wild-type strains. The location of Tn5801 in the recipient genomes was assessed by hybridization of SmaI-digested DNA with specific guaA, intTn5801, and tet(M) probes. PCR of the guaA-int5801 fragment was performed in all transconjugants.
Fitness cost assays.
Transconjugants and the corresponding recipient strain were cultured at 37°C in BHI broth overnight. Grown cultures were diluted 1:1,000 into BHI broth to obtain an inoculum of approximately 105 CFU/ml. An aliquot (300 μl) of this dilution was transferred into a multiwell plate and incubated at 37°C, and the growth rate was estimated from the interval of optical density at 600 nm estimated to be exponential (0.01 to 0.1) by using the GrowthRates 2.1 program (33). The measurement was performed every 15 min for 22 h in a Microbiology Workstation Bioscreen C (ThermoLabSystems, Helsinki, Finland). To guarantee culture optical homogeneity, the plates were shaken for 10 s before all measurements. Three biological and technical replicates were assayed for each strain. Relative values for each parameter were calculated by dividing the average corresponding growth rate of a given transconjugant by the average corresponding parameter of the donor in the same experiment (34).
Nucleotide sequence accession number.
The A4 variant of Tn5801 detected in this study is available in the GenBank database with accession number KP001176.
RESULTS
Genetic diversity of Tn5801 in the genomes of four major Gram-positive genera.
Twenty-three Tn5801 variants were identified among the 5,073 genomes screened, 17 of which had not been previously published. The 23 distinct Tn5801 backbones corresponded to 225 strains (139 S. aureus, 43 Streptococcus agalactiae, 24 E. faecium, 15 E. faecalis, 1 Streptococcus mitis, 1 Enterococcus villorum, 1 Streptococcus thoraltensis, and 1 Lactococcus garvieae). Table 2 and Table S2 in the supplemental material show the epidemiological data and the data set of the analyzed genomes.
TABLE 2.
Presence of Tn5801 types A and B in Firmicutes sequences deposited in the GenBank genome database as of January 2015
| Species | No. of genomes studied | No. with Tn5801 type: |
|
|---|---|---|---|
| A | B | ||
| Streptococcus agalactiae | 301 | 1 | 42 |
| Streptococcus mitis | 23 | 1 | |
| Streptococcus thoraltensis | 1 | 1 | |
| Enterococcus faecalis | 344 | 1 | 13 |
| Enterococcus faecium | 254 | 16 | 7 |
| Enterococcus villorum | 2 | 2 | |
| Staphylococcus aureus | 4,130 | 139 | |
| Staphylococcus pseudintermedius | 4 | 1 | |
| Lactococcus garvieae | 14 | 1 | |
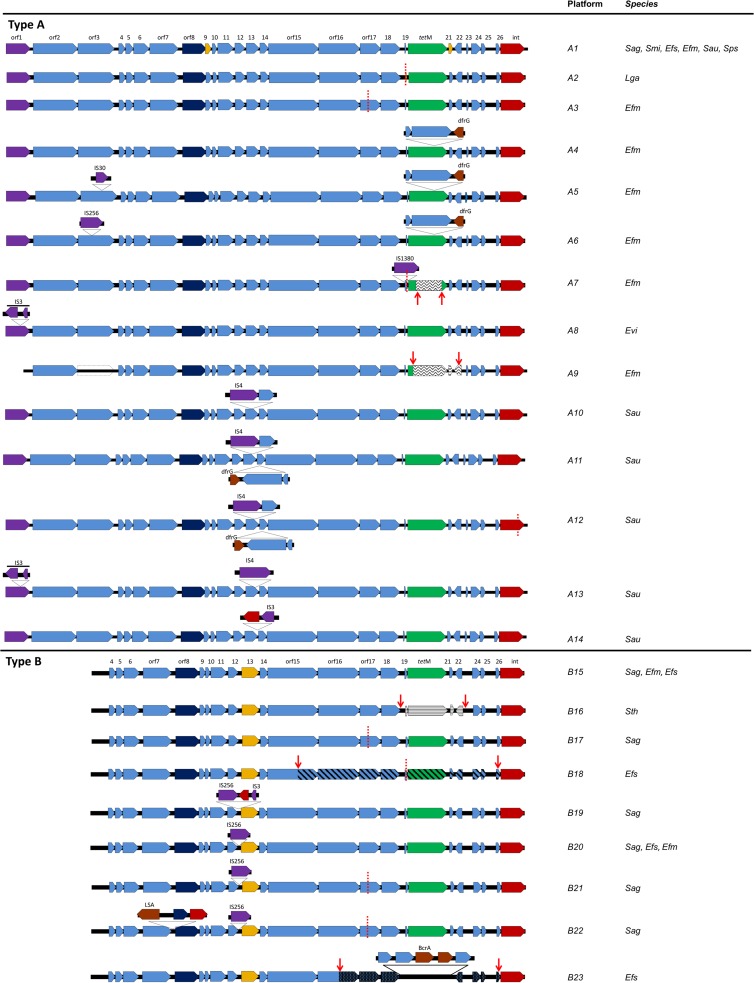
The Tn5801 backbones were classified in two main groups arbitrarily designated by capital letters as “group A” (14 types, A1 to A14) and “group B” (9 types, B15 to B23), which are shown in Fig. 1. These two groups differ in length (20 to 25 kb, due to the presence of an extra 5-kb fragment at the left arm of the group A transposons), nucleotide sequence, and the presence of indels and rearrangements in different regions. Variants within each group were designated by the corresponding capital letter followed by a number. The detailed analysis of the core sequences facilitated detection of differences and common features within a given type (Fig. 2; see Table S2 in the supplemental material). The GC percentage of the common Tn5801 backbone (36%) contrasted with that of the 5-kb fragment found in type A sequences (32%) (see Fig. S2 in the supplemental material). An alignment of selected variants is shown in Fig. 3.
FIG 1.
Diversity of Tn5801-like elements, types A and B, based on strains used in this study and sequences deposited in the GenBank genome database. In silico comparative analysis was made using BLAST and MUSCLE software available at the BLAST (http://www.ncbi.nlm.nih.gov/blast//) and MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/) websites. Dashed red lines, deletion sequences; red ORFs, ORFs encoding integrase-like proteins; dark blue ORFs, ORFs encoding replication proteins; light blue ORFs, ORFs encoding hypothetical proteins; green ORFs, tetracycline resistance gene tet(M); yellow ORFs, orf9 and orf21 are absent in Staphylococcus aureus Mu50 (reference genome) and orf13 in type B differs from orf13 in type A; brown ORFs, other resistance genes; purple ORFs, insertion sequences (IS). Wavy lines, recombination with Tn5397; straight lines, recombination with Tn916; diagonal lines, recombination with type A variant. ORFs with spots, B23 sequence differing from type B variants. dfrG, dihydrofolate reductase; LSA, lincosamides and streptogramins A; BcrA, bacitracin transport; Sag, Streptococcus agalactiae; Smi, Streptococcus mitis; Efs, Enterococcus faecalis; Efm, Enterococcus faecium; Sau, Staphylococcus aureus; Sps, Staphylococcus pseudintermedius. All IS correspond to IS families: IS1062 (IS30 family, A5), IS1542 (IS256 family, A6), IS1678 (IS1380 family, A7), ISEfa8 (IS3 family, A8), IS1542 (IS256 family; B19, B20, B21, and B22), ISLgar5/ISEfm2 (IS256 family, orf1).
FIG 2.
Phylogenetic analysis of Tn5801 variants in Firmicutes. The 4 numbers separated by lines indicate the four core fragments (intTn5801, orf18-14, orf12-8, and orf7-4) representing the common region in all Tn5801 variants described in the GenBank database. These sequences were aligned, concatenated, trimmed, and further analyzed. A maximum-likelihood phylogenetic tree is represented, with 1,000 bootstrap replications. The orf26-19 fragment was excluded from this analysis because of its variability (resulting from both mutation and recombination events). The distribution of Tn5801 variants by species and geographical regions is also represented. Countries within each continent are represented by different tones of gray, black and white. Sau, Staphylococcus aureus; Sps, Staphylococcus pseudintermedius; Efm, Enterococcus faecium; Efs, Enterococcus faecalis; Evi, Enterococcus villorum; Sag, Streptococcus agalactiae, Smi, Streptococcus mitis, Sth, Streptococcus thoraltensis; Lga, Lactococcus garvieae; ARG, Argentina; AUS, Australia; BEL, Belgium; BRA, Brazil; CHE, Switzerland; CHN, China; DEU, Germany; DNK, Denmark; FRA, France; GBR, Great Britain; HKG, Hong Kong; IRL, Ireland; JPN, Japan; MYS, Malaysia; NLD, The Netherlands; PRT, Portugal; ESP, Spain; TWN, Taiwan; USA, United States; unk, unknown.
FIG 3.
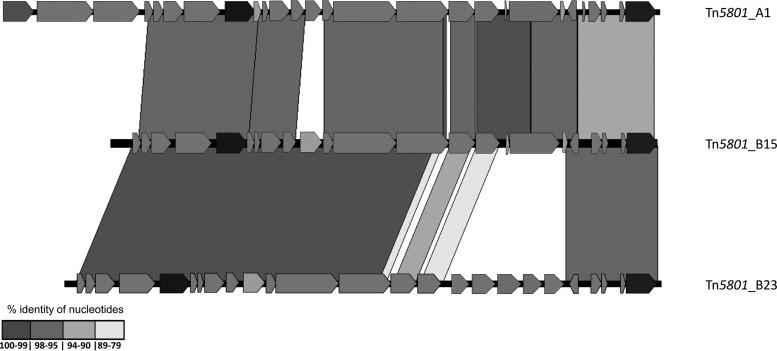
Alignment of selected transposons (Tn5801_A1, Tn5801_B15, and Tn5801_B23) by using the Artemis comparison tool (ACT). Only regions of >50 bp are represented. Gray areas mark matching regions of the same orientation among ICE from different species, indicating an identity between 79 and 100%. Only the areas with the highest percentages of similarity are shown.
Below we analyze in detail the features of variants of these clusters.
(i) Tn5801 group A.
Fourteen variants (A1 to A14) were identified among the available sequences (Fig. 1 and Table 3; see Table S2 in the supplemental material). Phylogenetic analysis allowed us to group them in different subgroups of highly related variants (Fig. 2).
TABLE 3.
Epidemiological features of the enterococcal isolates carrying Tn5801-like ICEa
| Species | Tn5801 platform (n)b | MLST (BAPS) | No. of isolates | Yr | Source(s) (n) | Country (n) | Tetr phenotype(s) (n) | Tetr gene(s) (n) | Other Tn(s) (n) |
|---|---|---|---|---|---|---|---|---|---|
| E. faecalis | B15 (2) | ST9/CC9 | 3 | 2001 | HP | ESP, ARG | + | tet(M) (3), tet(L) (3) | |
| A1 | ST30/CC30 | 1 | 1997 | HP | AUS | + | tet(M) | ||
| B15 (1) | ST318/CC9 | 2 | ND | HP | BRA | + | tet(M) | ||
| B15 | ST55/CC55 | 1 | 2001 | HV | PRT | + | tet(M) | Tn916, Tn6000 | |
| A1 | ST445 | 1 | 2001 | SP | PRT | + | tet(M), tet(L), tet(S) | ||
| B15 (3) | ND/ND | 5 | 2001/2002 | HP (2), HV (2), SW (1) | BRA (2), PRT (3) | + | tet(M) | Tn916 (3) | |
| E. faecium | + | ST17/CC17 (3.32) | 7 | 1998–2005 | HP (5), SW (2) | ESP (3), AUS (1), POL (1), PRT (2) | + (6), − (1) | tet(M) (5), tet(L) (3) | |
| + | ST16/CC17 (3.32) | 2 | 1999 | HP | AUS (2) | + | tet(M) | ||
| A1 (1), A4 (2) | ST18/CC17 (3.31) | 9 | 1996–2005 | HP (7), SW (2) | PRT (8), SER (1) | + | tet(M) (7), tet(L) (7) | Tn916 (2), Tn5397 (2) | |
| + | ST64/CC17 (3.31) | 4 | 2003–2004 | HP | CHI | + | tet(M) (3) | ||
| + | ST173/CC17 (3.31) | 1 | 2000 | HP | AUS | + | tet(M), tet(L) | ||
| + | ST80/CC17 (2.1a) | 1 | 2004 | HP | HUN | + | |||
| + | ST132/CC17 (3.31) | 1 | 2001 | HP | PRT | + | tet(M), tet(L) | Tn5397 | |
| A1 | ST182/CC17 (7) | 1 | 1992 | HP | USA | + | tet(M) | ||
| + | ST202/CC17 (3.32) | 1 | 2005 | HP | POL | + | tet(M), tet(L) | ||
| A1 | ST50/CC9 (2.1a) | 1 | NA | HP | BRA | + | tet(M) | ||
| + | ST366 (5) | 1 | 2000 | HP | PRT | tet(M) | Tn916 | ||
| + | ST368 (3.31) | 1 | 2001 | SW | PRT | ||||
| + | ND/ND | 11 | 2001–2002 | HP (4), HV (1), SW (3), P (2), SP (1) | PRT (9), ESP (1), TUR (1) | + (8), − (3) | tet(M) (8), tet(L) (5) | Tn916 (2), Tn5397 (1) | |
| Enterococcus sp. | + | ND/ND | 7 | 2001–2006 | HV (4), SW (2), SP (1) | PRT | + (3), − (4) | tet(M) (4), tet(L) (1) | Tn916 (1) |
Abbreviations: ST, sequence type; BAPS, Bayesian analysis of population structure; HV, healthy volunteer; HP, hospital patient; SW, sewage; P, poultry; SP, swine/piggeries; ARG, Argentina; AUS, Australia; BRA, Brazil; CHI, China; HUN, Hungary; POL, Poland; PRT, Portugal; ESP, Spain; SRB, Serbia; TUR, Turkey; ND, not determined; Tetr, tetracycline resistance; Tn(s), transposon(s).
+, Tn5801-like platform which has not been characterized in the corresponding type.
Tn5801 type A1 was considered the paradigm of the group. It was detected mostly in Staphylococcus (96%; n = 107 S. aureus/S. pseudintermedius isolates out of 112 isolates with Tn5801 type A1) and was also the predominant Tn5801 variant within this genus (76%; n = 107/140 Staphylococcus isolates carrying Tn5801). Variant A1 was also present in clonally unrelated E. faecium and E. faecalis isolates and species of Streptococcus (S. agalactiae and S. mitis), as reported previously (19). It is of note that most Tn5801 type A1 isolates showing particular mutations were associated with specific geographical locations and/or from specific clonal backgrounds (Fig. 2; see Tables S2 and S3 in the supplemental material) (see below). For example, most S. aureus isolates (83%; n = 88/106) carrying Tn5801 A1 belonged to CC8 (ST8, ST507, and ST609; many of them were recovered in the United States), and only a few isolates were associated with other clonal lineages (ST5 and ST664).
Variant A4 contained a 3,285-bp insertion within the tet(M) gene that was flanked by direct and inverted repeats that comprised dfrG (encoding a dihydrofolate reductase) and two hypothetical protein sequences. Such an element confers resistance to trimethoprim, and it was also detected within orf14 of Tn5801 types A11 and A12 (see below), in the Tn916-like transposon (Tn6198) of Listeria monocytogenes, in the plasmid pMG1 from E. faecium, and also in Streptococcus pyogenes isolates from India (35–37). The backbone of A4 was identical to that of Tn5801 types A5 and A6, which also showed additional insertion sequences (IS) interrupting the uvrD (orf3) gene (IS30 or IS256, respectively). All A4, A5, and A6 variants were found only among E. faecium ST18 isolates. Interestingly, the core sequences of A4, A5, and A6 variants clustered with type A1 sequences from human isolates of E. faecium and S. aureus from Japan and different European countries and shared specific mutations (G2336A_orf17, A3918T_orf16, A6574T_orf15, and T13303C_orf6), suggesting a common origin for them (Fig. 2; see Table S3 in the supplemental material).
Types A2, A7, A8, A13, and A14 and some A1 subvariants also clustered together (Fig. 2). Types A7 and A8 from enterococci and A2 from L. garvieae were highly similar according to the backbone consensus sequence (only 1 nucleotide [nt] of difference, G2644A_orf17). Types A2 and A7 (and also B18, an “A+B” Tn5801 hybrid [see below]) shared a 125-bp deletion in orf19 (orf12Tn916-like) that also appeared in the Tn5801-like element originally called CW459tet(M) from C. perfringens (16). In A7, such a deletion is upstream of tet(M) and next to an IS1380/IS1678 insertion. A7 also showed a mosaic tet(M) gene with Tn5397 tet(M) (GenBank accession number NG_034213.1). Variant A8 identified in a genome of E. villorum had the orf1 (identified as ISLgar5/ISEfm2 belonging to the IS256 family) truncated by a fragment comprising two ORFs identified as ISEfa8 (IS3 family). Variants A13 (ST5) and A14 (ST247) from S. aureus isolates exhibited insertions of partial and complete copies of the IS4 family and ISEfa8 (IS3 family) sequences in orf14 and orf1 (variant A13) or an insertion with an integrase core domain within orf14 (variant A14). A2, A7, A8, A13, A14, and A1 shared 5 mutations, namely, G1457A and T1804C in orf18, G3043C_orf17, G70008T_orf15, and C13758T_orf4. Some isolates collected in common geographical areas also had specific additional amino acid changes (see Table S3 in the supplemental material).
Variants A10, A11, and A12 clustered together (Fig. 2) and contained insertions within orf13 and orf14 at different positions, which include an ATP binding protein and a hypothetical protein encoded within orf13 (A10, A11, and A12) or the cassette comprising dfrG mentioned above in orf14 (A11 and A12). A12 also had a 107-bp deletion in the integrase. They were mostly observed in S. aureus belonging to ST239 (84%; n = 26/31) and shared four nucleotide change mutations (T7199C_orf15, G7884A_orf14, T9310A_orf11, and C12423T_orf7) (see Table S3 in the supplemental material). Specific mutations were detected among isolates from specific geographical areas (variant A10 from China had an A4479G_orf16 mutation which was absent in A10 from the United States or Australia).
Variant A3, detected in a few isolates of E. faecium (ST50) collected in France during the 1990s, clustered separately and showed an 81-bp deletion in orf17 (Fig. 2). Variant A9, previously described as Tn6086 (GenBank accession no. HM636636.1), was a chimera of Tn5397 [region tet(M)-orf21-orf22] and Tn5801 and also had an A/T mutation at nt 985 of the uvrD (orf3) gene that resulted in a stop codon and lack of orf1. It is of note that the A9 platform detected in E. faecium TC6 clustered with other A1 variants from S. mitis and E. faecalis isolated in Germany and Denmark, respectively, all sharing three distinct mutations (T3046A, T3586A, and C4861T). In fact, E. faecium TC6 is an E. faecium D344SRF (ST25) transconjugant from strain C68 that contains the same A9 variant (38), thus reflecting transferability of the element. Independent recombination events between Tn5801 and Tn5397 seem to have occurred in A9 and A7.
(ii) Tn5801 group B.
Group B comprises nine Tn5801 elements identified among available sequences of streptococci and enterococci (B15 to B23) (Fig. 1 and Table 3; see Table S2 in the supplemental material). They split into three main clusters represented by platforms B15 to B22 (mainly in streptococci) and B18 and B23 (both in enterococci).
The B15 variant (also designated Tn5801.Sag) (20) was considered the paradigm of the Tn5801 type and was detected among E. faecalis (this study), E. faecium, and S. agalactiae isolates. Some type B Tn5801 elements shared a deletion of 81 bp in orf17 (B17, B21, and B22 from S. agalactiae; also detected in A3 from E. faecium) or an IS256-like insertion upstream of orf13 (B20, B21, and B22). The B19 sequence had this IS256 insertion followed by an integrase core domain and an IS3 family element. The B22 variant had an insertion of a 5,227 bp between orf7 and orf8, which comprises a phage-specific recombinase gene, a replication protein gene, and a lsa(C) gene encoding resistance to lincosamide, streptogramin A, and pleuromutilins (39). Such an insertion is identical to another found in S. agalactiae UCN70 (GenBank accession number HM990671.1) and similar to that of a Tn916-like element from S. mitis, although it lacks the lsa(C) gene (18, 40).
Type B23 has a region of five ORFs (encoding a transcriptional regulator, an histidine kinase, a bacitracin ABC transporter, a bacitracin transport permease, and an ABC transporter permease-like protein) instead of orf19-tet(M)-orf21. The sequence between orf26 and orf16 shared only a 92% similarity with the other type B sequences. This B23 variant was restricted to clonal complex 8 (CC8) (ST8, ST64, and ST90) E. faecalis isolates, one of the most ancient E. faecalis lineages (41). It included the oldest Tetr strain described to date (an ST90 isolate from 1954, part of the historical collection of the Laboratory of Streptococcal Diseases at the National Institute of Allergy and Infectious Diseases in the United States) and ST8-related isolates of an emblematic strain widely used as a recipient (strains FA2 and JH2-2) which predate 1973 (41). The B18 variant is a mosaic of type A (region between orf26 and orf15) and type B (integrase, orf4 to part of orf15) platforms and exhibits the 125-bp deletion in orf19 also observed in type A variants (A2 and A7). The integrases of B18 and B23 share three unique nucleotide changes. Finally, variant B16, detected in an S. thoraltensis isolate (GenBank accession no. ARCI01000001.1), revealed a recombination event that resulted in the replacement of the orf19-orf22 region of Tn5801 by that of Tn916.
Epidemiological background of Tn5801 among enterococci.
intTn5801 was detected in 10% of the enterococcal isolates analyzed (n = 61/610; 41 E. faecium, 13 E. faecalis, and 7 Enterococcus spp.) (Table 3). A large proportion of the intTn5801-positive isolates (84%; n = 51/61) were resistant to tetracycline, and most of them carried tet(M), tet(L), and/or tet(S) (77% [n = 47/61], 38% [n = 23/61], and 2% [n = 1/61], respectively). Specific transposon sequences such as int/xisTn916 (16%; n = 10/61), resolvaseTn5397 (7%, n = 4/61), or int/xisTn6000 (2%; n = 1/61) were also observed. The Tn5801 platforms identified in isolates of this collection correspond to A1, A4, and B15 variants. Epidemiological features of the intTn5801-positive isolates of E. faecium and E. faecalis are shown in Table 3.
Location of Tn5801.
All Tn5801 platforms identified in enterococci in this study were chromosomally located at the 3′ end of guaA, as previously observed for three emblematic Tn5801 elements of staphylococci, Clostridium, and S. agalactiae (13, 28). In the available sequences of the Tn5801 variants was the 11-bp sequence described previously as direct repeats (DRs) situated within the 3′ end of guaA (13). All guaA sequences within the same species had high similarity (99 to 100%), which was somewhat lower (70 to 80%) when comparing isolates from different species.
Tn5801 transfer in Enterococcus.
The ability of Tn5801 to be transferred was studied in mating assays using enterococcal isolates carrying the Tn5801 variants A1, A4, and B15 as donors. Only Tn5801 variant B15 was successfully transferred by conjugation to E. faecalis strains JH2-2 and OG1RF. The conjugation frequency was similar for the two receptors despite the presence of the B23 variant in the genome of the JH2-2 recipient (Table 4). Hybridization of SmaI-digested DNA from transconjugants with specific probes for guaA, intTn5801, and tet(M) allowed us to infer the location and the size of the transferred region (see Fig. S3 in the supplemental material). A single band of 150 kb that hybridized with the three probes tested was observed for all E. faecalis OG1RF transconjugants obtained in primary mating experiments. For E. faecalis JH2-2 transconjugants, the sizes of the bands that hybridized with the probes were 200 and 250 kb (the estimated size of the transferred region was ca. 20 kb for OG1RF and ca. 50 or 100 kb for the two JH2-2 transconjugants). Mating assays using these E. faecalis JH2-2::Tn5801_1 (50 kb) and E. faecalis JH2-2::Tn5801_2 (100 kb) transconjugants as donors and E. faecalis OG1SS as the recipient yielded E. faecalis OG1SS::Tn5801 transconjugants with one or two copies of a ca. 20-kb transposable element. All transconjugants that contain two Tn5801 copies showed positive hybridization of two bands with the intTn5801 and tet(M) genes, but only one band hybridized with the guaA probe (see Fig. S3 in the supplemental material). Moreover, a PCR mapping assay demonstrated the presence of a B15 Tn5801-like element in all primary and secondary transconjugants, while amplification of specific B23 fragments was observed only for one primary and one secondary transconjugant, E. faecalis JH2-2::Tn5801_2 and E. faecalis OG1SS::Tn5801_2.3, respectively (Table 4; see Fig. S3 in the supplemental material).
TABLE 4.
Results of filter mating assays with Enterococcus faecalis strainsa
| Transconjugant | Donorb | Recipient | No. of Tn5801 copies detected | Hybridized SmaI-digested DNA band size (added fragment size), kb | Hybridized probe |
Transfer rate per donor | Growth rate, mean/SD | Relative growth rate, % (transconjugant/recipientc) | Tet MIC, μg/ml | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| int | tet | gua | |||||||||
| JH2-2::Tn5801_1 | E. faecalis Ef1 | JH2-2 | 1 | 200 (50) | + | + | + | 1.6 × 10−8 | 0.15437/0.01096 | 3.66385 | 48 |
| JH2-2::Tn5801_2 | E. faecalis Ef1 | JH2-2 | 2 | 250 (100) | + | + | + | 1.6 × 10−8 | 0.15489/0.01568 | 3.34281 | 96 |
| OG1SS::Tn5801_1.1 | JH2-2::Tn5801_1 | OG1SS | 1 | 150 (20) | + | + | + | 1.81 × 10−7 | 0.16787/0.00576 | 3.16674 | 32 |
| OG1SS::Tn5801_1.2 | JH2-2::Tn5801_1 | OG1SS | 2 | 340 (20) | + | + | − | 1.01 × 10−8 | 0.16773/0.00321 | 3.24749 | 96 |
| 150 (20) | + | + | + | ||||||||
| OG1RF::Tn5801_1 | E. faecalisEf1 | OG1RF | 1 | 150 (20) | + | + | + | 3.3 × 10−8 | 0.16087/0.00769 | 0.16068 | 24 |
| OG1RF::Tn5801_2 | E. faecalis Ef1 | OG1RF | 1 | 150 (20) | + | + | + | 3.3 × 10−8 | NDd | ND | 48 |
| OG1RF::Tn5801_3 | E. faecalis Ef1 | OG1RF | 1 | 150 (20) | + | + | + | 3.3 × 10−8 | ND | ND | 64 |
| OG1SS::Tn5801_2.1 | JH2-2::Tn5801_2 | OG1SS | 1 | 150 (20) | + | + | + | 2 × 10−7 | 0.17217/0.00709 | 0.68706 | 96 |
| OG1SS::Tn5801_2.2 | JH2-2::Tn5801_2 | OG1SS | 2 | 150 (20) | + | + | + | 2 × 10−7 | 0.17717/0.00661 | −2.19576 | 96 |
| 120 (20) | + | + | − | ||||||||
| OG1SS::Tn5801_2.3 | JH2-2::Tn5801_2 | OG1SS | 2 | 200 (50) | + | + | − | 1.6 × 10−5 | 0.17283/0.00776 | 0.30828 | 96 |
| 170 (50) | + | + | + | ||||||||
| OG1SS::Tn5801_2.4 | JH2-2::Tn5801_2 | OG1SS | 1 | 150 (20) | + | + | + | 1.6 × 10−5 | 0.16903/0.00645 | 2.50019 | 32 |
Following the order presented in Fig. S3 in the supplemental material.
Enterococcus faecalis Ef1 tetracycline MIC, 48 μg/ml.
Recipient growth rates (means/standard deviations): 0.16024/0.01179 (E. faecalis JH2-2), 0.17336/0.00362 (E. faecalis OG1SS), and 0.16112/0.00771 (E. faecalis OG1RF).
ND, not determined.
Fitness cost of the transconjugants.
The growth rates of E. faecium transconjugants containing B15 variants ranged from a 2.2% increase to a 3.7% reduction in the growth rate of the recipient strain (growth rates and standard deviations are indicated in Table 4). Detailed analysis of the data revealed that the secondary transconjugants (using JH2-2 as donor and OG1SS as recipient) that apparently acquired two entire Tn5801 elements yielded different values of fitness increase or fitness reduction in comparison with the donor strain. They correspond to transconjugants that showed slightly different PFGE types, showed hybridization patterns that indicate insertion of the two acquired Tn5801 at different genomic locations, and had a high MIC of tetracycline (96 μg/ml) (Table 4).
DISCUSSION
This study represents the first description of Tn5801-like elements in enterococci and one of the first studies addressing the evolutionary changes of antibiotic resistance transposons throughout decades. To date, only scarce Tn5801-like backbones of a few S. aureus and Streptococcus strains have been identified in separate studies (3, 15, 18–20), which did not analyze the clonal context of the isolates, thus limiting the understanding of the evolvability of this element.
Sequence analysis of type A and B variants revealed a common Tn5801 backbone compatible with an S. agalactiae origin on the basis of the GC content (36%) and suggested the further acquisition of a 5-kb extra fragment by staphylococci (GC content = 32%). Such acquisition might have occurred through an IS256 family insertion event (orf1 of the group A platforms) and would have resulted in the emergence of type A variants.
The apparent confinement of Tn5801 type A or B in particular clonal backgrounds is remarkable. A recent study suggests that the introduction and massive use of tetracycline since the 1950s could have contributed to a positive selection of neonatal hypervirulent clonal lineages (CC17, CC23, or CC10) of S. agalactiae carrying tet(M)-CTn, as Tn5801, which we here designated Tn5801 B type platforms (1). We were also able to detect Tn5801 in predominant antibiotic-resistant clonal lineages associated with human infections, such as S. aureus (CC8, CC5, and ST239) and E. faecium (ST17 and ST18) from the 1990s, or early lineages of E. faecalis (CC8). However, our data and available knowledge necessitate a carefully interpretation of the relationship between the emergence and persistence of antibiotic resistance and selective processes of particular clones due to antimicrobial treatments (42). In this respect, we should take into account therapeutic strategies and relevant bacterial features that could have facilitated the selective acquisition of certain elements by particular clones. For example, anti-restriction-modification (anti-RM) occurring in Tn916-like elements often influences selective horizontal gene transfer between staphylococcal populations and other species of Firmicutes (43, 44). Putative anti-RM detected in Tn5801 (orf12) acts against staphylococcal type I RM systems which are present in S. aureus CC5 and CC8 (the backgrounds in which Tn5801 seems to have initially been detected in the late 1950s) (3, 15) and ST239 (an emergent hybrid lineage of ST8 and ST30) (10, 17, 45).
Deep sequence analysis is a powerful tool to accurately establish transmission of pathogens (46). In this study, the comprehensive analysis of Tn5801 variants identified some types associated with sympatric populations of major clonal lineages (as reflected by the presence of specific nucleotide changes among isolates from particular geographical areas). This seems clear for S. aureus ST239 (which carries highly related A10, A11, and A12 variants) and ST8/ST609 human isolates from the United States recovered from 1994 to date (carrying A1), for which differences in the genome sequences of regional endemic clones have also been reported (17, 44). Besides clonal spread of strains carrying Tn5801, the presence of some variants in isolates of different species recovered in different countries and, eventually, different hosts indicates that horizontal gene transfer has also contributed to the current occurrence of Tn5801. This study demonstrates the transference of Tn5801 between different E. faecalis backgrounds, even if the strain already harbored a Tn5801 element. Although the guaA-associated islands theoretically integrate site specifically, tyrosine recombinases can be functional at secondary integration sites (11, 12). This would explain the insertion of elements at different genomic locations (transconjugants E. faecalis OG1SS::Tn5801_1.2, OG1SS::Tn5801_2.2, and OG1SS::Tn5801_2.3). Moreover, integration at the same site cannot be discarded if a consensus hot spot remains available for new integration events after the first Tn5801 acquisition. For example, two E. faecalis genomes (E. faecalis Com1 and E. faecalis Com2; GenBank accession no. AJES01000002.1 and AJBL01000002.1, respectively) harbored a composite structure that comprises Tn5801 B15 and a putative transposable element (GC content of 27.3%, compatible with a clostridial origin [47]), both located upstream of the guaA gene. The clostridial element seems to have been inserted at the 11-bp hot spot of a guaA-intTn5801 region. Finally, recombination cannot be discarded for some cases, as described for other elements in E. faecalis (48). Such events increase the number (if they include different genes) or the level of resistance (if the gene dosage is increased as we observed here), with a variable fitness cost to the cell, influencing the probability of being selected and stably maintained.
Although the classical JH2-2 lab strain was serendipitously useful for this work, as it allowed detection of the acquisition of more than one Tn5801 element by a single strain, the presence of conjugative genetic elements that can interfere with the transferability of other elements under study opens discussion about the suitability of certain classic laboratory strains for use in transfer experiments.
CTn-tet(M) elements with the same genetic backbone may carry different determinants associated with resistance (e.g., to bacitracin, lincosamides/streptogramins, and trimethoprim in our study) and/or ISs (IS3, IS4, IS30, IS256, and IS1380 families) which are frequently designated with different numbers in other studies (7, 20, 49). However, all these variants reflect the dynamics of a basic particular element circulating among bacterial communities. In this study, ISs with intact inverted repeats (IR) were observed within different genes (orf1, orf3, orf13, and orf19) of all Tn5801 type A isolates of S. aureus and E. faecium, suggesting their recent acquisition. Among type B variants, only the mutator family IS256 was consistently detected upstream of orf13 (types B20, B18, and B19), and this stability persisted even when some particular variants (e.g., B20) were disseminated across genera. Some ISs from the IS256 family have been implicated as modulators of gene expression, suggesting that future studies might reveal the role of this IS in the function of type B Tn5801-like elements (50–53).
In summary, this study shows that Tn5801-like variants have been present among Firmicutes since at least the 1950s, being predominant in microorganisms obtained from human hosts (54). Different waves of expansion in human populations of certain clones of Staphylococcus, Enterococcus, and Streptococcus seem to play an important role in their occurrence and evolvability, although transfer events between different clonal backgrounds would also contribute to their spread, as demonstrated experimentally here for E. faecalis. The detection of genetic rearrangements in the four functional regions highlights once more the plasticity of CTns, potentially influencing microevolutionary events eliciting adaptive functions in microbial populations and communities in common or heterogeneous hosts and environments (55–58).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Instituto de Salud Carlos III (PI12-01581) for the support of R.L.S. We also acknowledge the European Development Regional Fund “A way to achieve Europe” (ERDF) for cofounding the Plan Nacional de I+D+ I 2012-2015 (PI12-01581) and CIBER actions (CIBER in Epidemiology and Public Health, CIBERESP; CB06/02/0053).
Funding Statement
European Commission, Seven Framework Program (EVOTARFP7-HEALTH-282004), provided support to Fernando Baquero and Teresa M. Coque. The Ministry of Economy and Competitiveness of Spain, Plan Nacional de I+D+ I 2012-2015, Instituto de Salud Carlos III (PI12-01581), provided support to Teresa M. Coque. The Ministry of Economy and Competitiveness of Spain, CIBERESP (CB06/02/0053), provided support to Fernando Baquero. The European Development Regional Fund “A way to achieve Europe” (ERDF) cofunds ISCIII and CIBER actions. The Regional Government of Madrid (PROMPT-S2010/BMD2414) provided support to Fernando Baquero. The Fundação para a Ciência e a Tecnologia, FCT UID/MULTI/04378/2013, provided support to Luisa Peixe and Carla Novais.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01864-15.
REFERENCES
- 1.Da Cunha V, Davies MR, Douarre P-E, Rosinski-Chupin I, Margarit I, Spinali S, Perkins T, Lechat P, Dmytruk N, Sauvage E, Ma L, Romi B, Tichit M, Lopez-Sanchez M-J, Descorps-Declere S, Souche E, Buchrieser C, Trieu-Cuot P, Moszer I, Clermont D, Maione D, Bouchier C, McMillan DJ, Parkhill J, Telford JL, Dougan G, Walker MJ, Holden MTG, Poyart C, Glaser P. 2014. Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat Commun 5:4544. doi: 10.1038/ncomms5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edmond KM, Kortsalioudaki C, Scott S, Schrag SJ, Zaidi AKM, Cousens S, Heath PT. 2012. Group B streptococcal disease in infants aged younger than 3 months: systematic review and meta-analysis. Lancet 379:547–556. doi: 10.1016/S0140-6736(11)61651-6. [DOI] [PubMed] [Google Scholar]
- 3.de Vries LE, Christensen H, Skov RL, Aarestrup FM, Agersø Y. 2009. Diversity of the tetracycline resistance gene tet(M) and identification of Tn916- and Tn5801-like (Tn6014) transposons in Staphylococcus aureus from humans and animals. J Antimicrob Chemother 64:490–500. doi: 10.1093/jac/dkp214. [DOI] [PubMed] [Google Scholar]
- 4.Soge OO, Beck NK, White TM, No DB, Roberts MC. 2008. A novel transposon, Tn6009, composed of a Tn916 element linked with a Staphylococcus aureus mer operon. J Antimicrob Chemother 62:674–680. doi: 10.1093/jac/dkn255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts AP, Mullany P. 2011. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol Rev 35:856–871. doi: 10.1111/j.1574-6976.2011.00283.x. [DOI] [PubMed] [Google Scholar]
- 6.Rice LB. 1998. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob Agents Chemother 42:1871–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clewell DB, Weaver KE, Dunny GM, Coque TM, Francia MV, Hayes F. 2014. Extrachromosomal and mobile elements in enterococci: transmission, maintenance, and epidemiology. In Gilmore M, Clewell D, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 8.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. 2010. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect 16:541–554. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 9.Novais C, Freitas AR, Silveira E, Baquero F, Peixe L, Roberts AP, Coque TM. 2012. Different genetic supports for the tet(S) gene in enterococci. Antimicrob Agents Chemother 56:6014–6018. doi: 10.1128/AAC.00758-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smyth DS, McDougal LK, Gran FW, Manoharan A, Enright MC, Song J-H, de Lencastre H, Robinson DA. 2010. Population structure of a hybrid clonal group of methicillin-resistant Staphylococcus aureus, ST239-MRSA-III. PLoS One 5:e8582. doi: 10.1371/journal.pone.0008582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smyth DS, Robinson DA. 2009. Integrative and sequence characteristics of a novel genetic element, ICE6013, in Staphylococcus aureus. J Bacteriol 191:5964–5975. doi: 10.1128/JB.00352-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song L, Pan Y, Chen S, Zhang X. 2012. Structural characteristics of genomic islands associated with GMP synthases as integration hotspot among sequenced microbial genomes. Comput Biol Chem 36:62–70. doi: 10.1016/j.compbiolchem.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Wipf JRK, Schwendener S, Nielsen JB, Westh H, Perreten V. 2015. The new macrolide-lincosamide-streptogramin B resistance gene erm(45) is located within a genomic island in Staphylococcus fleurettii. Antimicrob Agents Chemother 59:3578–3581. doi: 10.1128/AAC.00369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd DA, Cabral T, Van Caeseele P, Wylie J, Mulvey MR. 2002. Molecular characterization of the vanE gene cluster in vancomycin-resistant Enterococcus faecalis N00-410 isolated in Canada. Antimicrob Agents Chemother 46:1977–1979. doi: 10.1128/AAC.46.6.1977-1979.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. 2001. Whole-genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240. doi: 10.1016/S0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 16.Roberts AP, Johanesen PA, Lyras D, Mullany P, Rood JI. 2001. Comparison of Tn5397 from Clostridium difficile, Tn916 from Enterococcus faecalis and the CW459tet(M) element from Clostridium perfringens shows that they have similar conjugation regions but different insertion and excision modules. Microbiology 147:1243–1251. doi: 10.1099/00221287-147-5-1243. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto T, Takano T, Higuchi W, Iwao Y, Singur O, Reva I, Otsuka Y, Nakayashiki T, Mori H, Reva G, Kuznetsov V, Potapov V. 2012. Comparative genomics and drug resistance of a geographic variant of ST239 methicillin-resistant Staphylococcus aureus emerged in Russia. PLoS One 7:e29187. doi: 10.1371/journal.pone.0029187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brenciani A, Tiberi E, Tili E, Mingoia M, Palmieri C, Varaldo PE, Giovanetti E. 2014. Genetic determinants and elements associated with antibiotic resistance in viridans group streptococci. J Antimicrob Chemother 69:1197–1204. doi: 10.1093/jac/dkt495. [DOI] [PubMed] [Google Scholar]
- 19.Denapaite D, Brückner R, Nuhn M, Reichmann P, Henrich B, Maurer P, Schähle Y, Selbmann P, Zimmermann W, Wambutt R, Hakenbeck R. 2010. The genome of Streptococcus mitis B6—what is a commensal? PLoS One 5:e9426. doi: 10.1371/journal.pone.0009426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mingoia M, Morici E, Tili E, Giovanetti E, Montanari MP, Varaldo PE. 2013. Characterization of Tn5801.Sag, a variant of Staphylococcus aureus Tn916 family transposon Tn5801 that is widespread in clinical isolates of Streptococcus agalactiae. Antimicrob Agents Chemother 57:4570–4574. doi: 10.1128/AAC.00521-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silveira E, Freitas AR, Antunes P, Barros M, Campos J, Coque TM, Peixe L, Novais C. 2014. Co-transfer of resistance to high concentrations of copper and first-line antibiotics among Enterococcus from different origins (humans, animals, the environment and foods) and clonal lineages. J Antimicrob Chemother 69:899–906. doi: 10.1093/jac/dkt479. [DOI] [PubMed] [Google Scholar]
- 25.CLSI. 2014. Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement. CLSI, Wayne, PA. [Google Scholar]
- 26.Aarestrup FM, Agerso Y, Gerner-Smidt P, Madsen M, Jensen LB. 2000. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn Microbiol Infect Dis 37:127–137. doi: 10.1016/S0732-8893(00)00130-9. [DOI] [PubMed] [Google Scholar]
- 27.Coque TM, Willems RJL, Fortún J, Top J, Diz S, Loza E, Cantón R, Baquero F. 2005. Population structure of Enterococcus faecium causing bacteremia in a Spanish university hospital: setting the scene for a future increase in vancomycin resistance? Antimicrob Agents Chemother 49:2693–2700. doi: 10.1128/AAC.49.7.2693-2700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willems RJL, Top J, van Schaik W, Leavis H, Bonten M, Sirén J, Hanage WP, Corander J. 2012. Restricted gene flow among hospital subpopulations of Enterococcus faecium. mBio 3:e00151-12. doi: 10.1128/mBio.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tedim AP, Ruiz-Garbajosa P, Corander J, Rodríguez CM, Cantón R, Willems RJ, Baquero F, Coque TM. 2015. Population biology of intestinal Enterococcus isolates from hospitalized and nonhospitalized individuals in different age groups. Appl Environ Microbiol 81:1820–1831. doi: 10.1128/AEM.03661-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freitas AR, Tedim AP, Novais C, Ruiz-Garbajosa P, Werner G, Laverde-Gomez JA, Cantón R, Peixe L, Baquero F, Coque TM. 2010. Global spread of the hylEfm colonization-virulence gene in megaplasmids of the Enterococcus faecium CC17 polyclonal subcluster. Antimicrob Agents Chemother 54:2660–2665. doi: 10.1128/AAC.00134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Werner G, Freitas AR, Coque TM, Sollid JE, Lester C, Hammerum AM, Garcia-Migura L, Jensen LB, Francia MV, Witte W, Willems RJ, Sundsfjord A. 2011. Host range of enterococcal vanA plasmids among Gram-positive intestinal bacteria. J Antimicrob Chemother 66:273–282. doi: 10.1093/jac/dkq455. [DOI] [PubMed] [Google Scholar]
- 33.Hall BG, Acar H, Nandipati A, Barlow M. 2014. Growth rates made easy. Mol Biol Evol 31:232–238. doi: 10.1093/molbev/mst187. [DOI] [PubMed] [Google Scholar]
- 34.Foucault M-L, Depardieu F, Courvalin P, Grillot-Courvalin C. 2010. Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc Natl Acad Sci U S A 107:16964–16969. doi: 10.1073/pnas.1006855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergmann R, Sagar V, Nitsche-Schmitz DP, Chhatwal GS. 2012. First detection of trimethoprim resistance determinant dfrG in Streptococcus pyogenes clinical isolates in India. Antimicrob Agents Chemother 56:5424–5425. doi: 10.1128/AAC.01284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekiguchi J, Tharavichitkul P, Miyoshi-Akiyama T, Chupia V, Fujino T, Araake M, Irie A, Morita K, Kuratsuji T, Kirikae T. 2005. Cloning and characterization of a novel trimethoprim-resistant dihydrofolate reductase from a nosocomial isolate of Staphylococcus aureus CM.S2 (IMCJ1454). Antimicrob Agents Chemother 49:3948–3951. doi: 10.1128/AAC.49.9.3948-3951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergmann R, van der Linden M, Chhatwal GS, Nitsche-Schmitz DP. 2014. Factors that cause trimethoprim resistance in Streptococcus pyogenes. Antimicrob Agents Chemother 58:2281–2288. doi: 10.1128/AAC.02282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice LB, Lakticová V, Carias LL, Rudin S, Hutton R, Marshall SH. 2009. Transferable capacity for gastrointestinal colonization in Enterococcus faecium in a mouse model. J Infect Dis 199:342–349. doi: 10.1086/595986. [DOI] [PubMed] [Google Scholar]
- 39.Douarre P-E, Sauvage E, Poyart C, Glaser P. 2015. Host specificity in the diversity and transfer of lsa resistance genes in group B Streptococcus. J Antimicrob Chemother 70:3205–3213. doi: 10.1093/jac/dkv277. [DOI] [PubMed] [Google Scholar]
- 40.Malbruny B, Werno AM, Murdoch DR, Leclercq R, Cattoir V. 2011. Cross-resistance to lincosamides, streptogramins A, and pleuromutilins due to the lsa(C) gene in Streptococcus agalactiae UCN70. Antimicrob Agents Chemother 55:1470–1474. doi: 10.1128/AAC.01068-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McBride SM, Fischetti VA, Leblanc DJ, Moellering RC, Gilmore MS. 2007. Genetic diversity among Enterococcus faecalis. PLoS One 2:e582. doi: 10.1371/journal.pone.0000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Day T, Huijben S, Read AF. 2015. Is selection relevant in the evolutionary emergence of drug resistance? Trends Microbiol 23:126–133. doi: 10.1016/j.tim.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts GA, Chen K, Bower EKM, Madrzak J, Woods A, Barker AM, Cooper LP, White JH, Blakely GW, Manfield I, Dryden DTF. 2013. Mutations of the domain forming the dimeric interface of the ArdA protein affect dimerization and antimodification activity but not antirestriction activity. FEBS J 280:4903–4914. doi: 10.1111/febs.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts GA, Houston PJ, White JH, Chen K, Stephanou AS, Cooper LP, Dryden DTF, Lindsay JA. 2013. Impact of target site distribution for type I restriction enzymes on the evolution of methicillin-resistant Staphylococcus aureus (MRSA) populations. Nucleic Acids Res 41:7472–7484. doi: 10.1093/nar/gkt535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sadykov MR. 2014. Restriction-modification systems as a barrier for genetic manipulation of Staphylococcus aureus, p 9–23. In Bose JL. (ed), The genetic manipulation of staphylococci. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 46.Worby CJ, Lipsitch M, Hanage WP. 2015. Shared genomic variants: identification of transmission routes using pathogen deep sequence data. bioRxiv doi: 10.1101/032458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lassalle F, Périan S, Bataillon T, Nesme X, Duret L, Daubin V. 2015. GC-content evolution in bacterial genomes: the biased gene conversion hypothesis expands. PLoS Genet 11:e1004941. doi: 10.1371/journal.pgen.1004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice LB, Carias LL, Marshall SH, Hutton-Thomas R, Rudin S. 2007. Characterization of Tn5386, a Tn916-related mobile element. Plasmid 58:61–67. doi: 10.1016/j.plasmid.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Palmieri C, Mingoia M, Massidda O, Giovanetti E, Varaldo PE. 2012. Streptococcus pneumoniae transposon Tn1545/Tn6003 changes to Tn6002 due to spontaneous excision in circular form of the erm(B)- and aphA3-containing macrolide-aminoglycoside-streptothricin (MAS) element. Antimicrob Agents Chemother 56:5994–5997. doi: 10.1128/AAC.01487-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maki H, Murakami K. 1997. Formation of potent hybrid promoters of the mutant llm gene by IS256 transposition in methicillin-resistant Staphylococcus aureus. J Bacteriol 179:6944–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benson MA, Ohneck EA, Ryan C, Alonzo F, Smith H, Narechania A, Kolokotronis S-O, Satola SW, Uhlemann A-C, Sebra R, Deikus G, Shopsin B, Planet PJ, Torres VJ. 2014. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol Microbiol 93:664–681. doi: 10.1111/mmi.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McEvoy CRE, Tsuji B, Gao W, Seemann T, Porter JL, Doig K, Ngo D, Howden BP, Stinear TP. 2013. Decreased vancomycin susceptibility in Staphylococcus aureus caused by IS256 tempering of WalKR expression. Antimicrob Agents Chemother 57:3240–3249. doi: 10.1128/AAC.00279-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fléchard M, Gilot P. 2014. Physiological impact of transposable elements encoding DDE transposases in the environmental adaptation of Streptococcus agalactiae. Microbiology 160:1298–1315. doi: 10.1099/mic.0.077628-0. [DOI] [PubMed] [Google Scholar]
- 54.Agersø Y, Pedersen AG, Aarestrup FM. 2006. Identification of Tn5397-like and Tn916-like transposons and diversity of the tetracycline resistance gene tet(M) in enterococci from humans, pigs and poultry. J Antimicrob Chemother 57:832–839. doi: 10.1093/jac/dkl069. [DOI] [PubMed] [Google Scholar]
- 55.Bellanger X, Payot S, Leblond-Bourget N, Guédon G. 2014. Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev 38:720–760. doi: 10.1111/1574-6976.12058. [DOI] [PubMed] [Google Scholar]
- 56.Bellanger X, Roberts AP, Morel C, Choulet F, Pavlovic G, Mullany P, Decaris B, Guédon G. 2009. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J Bacteriol 191:2764–2775. doi: 10.1128/JB.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burrus V, Pavlovic G, Decaris B, Guédon G. 2002. Conjugative transposons: the tip of the iceberg. Mol Microbiol 46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 58.Roberts AP, Mullany P. 2009. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol 17:251–258. doi: 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.