Abstract
The prevalence of intrinsic and acquired resistance among colonizing Candida isolates from patients after candidemia was investigated systematically in a 1-year nationwide study. Patients were treated at the discretion of the treating physician. Oral swabs were obtained after treatment. Species distributions and MIC data were investigated for blood and posttreatment oral isolates from patients exposed to either azoles or echinocandins for <7 or ≥7 days. Species identification was confirmed using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) and internal transcribed spacer (ITS) sequencing, susceptibility was examined by EUCAST EDef 7.2 methodology, echinocandin resistance was examined by FKS sequencing, and genetic relatedness was examined by multilocus sequence typing (MLST). One hundred ninety-three episodes provided 205 blood and 220 oral isolates. MLST analysis demonstrated a genetic relationship for 90% of all paired blood and oral isolates. Patients exposed to azoles for ≥7 days (n = 93) had a significantly larger proportion of species intrinsically less susceptible to azoles (particularly Candida glabrata) among oral isolates than among initial blood isolates (36.6% versus 12.9%; P < 0.001). A similar shift toward species less susceptible to echinocandins among 85 patients exposed to echinocandins for ≥7 days was not observed (4.8% of oral isolates versus 3.2% of blood isolates; P > 0.5). Acquired resistance in Candida albicans was rare (<5%). However, acquired resistance to fluconazole (29.4%; P < 0.05) and anidulafungin (21.6%; P < 0.05) was common in C. glabrata isolates from patients exposed to either azoles or echinocandins. Our findings suggest that the colonizing mucosal microbiota may be an unrecognized reservoir of resistant Candida species, especially C. glabrata, following treatment for candidemia. The resistance rates were high, raising concern in general for patients exposed to antifungal drugs.
INTRODUCTION
Candidemia due to Candida species which are intrinsically less susceptible to fluconazole is becoming proportionally more common at the expense of Candida albicans (1–5). Prophylactic antifungal use has been demonstrated to significantly drive the species distributions in candidemia patients toward less susceptible species, such as Candida glabrata in patients exposed to fluconazole and Candida parapsilosis in patients exposed to caspofungin (6–8). Indeed, intrinsic resistance and the changing epidemiology are the main concerns. On the other hand, acquired resistance is rare in epidemiologic candidemia studies, perhaps in part because such studies include only the initial blood isolate from each candidemia patient. Thus, subsequent and potentially resistant isolates may be overlooked. Resistance selection may be facilitated by a longer duration of treatment (9) and subtherapeutic drug levels (10, 11). Hence, abdominal candidiasis and Candida species at mucosal surfaces may represent a nest and reservoir for resistant isolates (10, 12). Typing studies have illustrated that the concomitant and predominant colonizing Candida isolate in candidemia patients is also the invasive Candida species (90 to 95% concordance), and most patients are still colonized despite the infection being cleared (7, 9, 13–17). Thus, the persistent colonizing microbiota is a relevant reservoir for resistant Candida strains and may impose a risk to this group of patients concerning reinfection, but also to individuals in other patient settings, where infection may follow prior antifungal drug exposure. The present study was a systematic nationwide multicenter study investigating the colonizing microbiota obtained in candidemia patients after antifungal drug exposure. The purpose was to elucidate the prevalence of Candida species less susceptible to antifungals and the extent of acquired resistance in the colonizing mucosal microbiota of patients treated for candidemia, since the increasing number of reports addressing these topics includes mainly case reports or single-center studies (18).
MATERIALS AND METHODS
Patients, isolates, and treatment.
All candidemia patients in Denmark diagnosed within the 1-year study period (March 2013 to February 2014) were eligible for inclusion in this study. Blood isolates were referred to the Mycology Reference Laboratory at Statens Serum Institut as part of the ongoing national surveillance of fungemia from all departments of clinical microbiology in Denmark. Corresponding candidemia patients were located through the Danish Microbiology Database (MiBa), and one oral swab was requested after discontinuation of antifungal treatment. Separate candidemia episodes were included only if they occurred >21 days apart. Information on antifungal treatment for invasive candidiasis (IC) but not empirical treatment was sought. Patients were excluded if an oral swab or IC treatment information was not obtained.
Prevalence of intrinsic resistance.
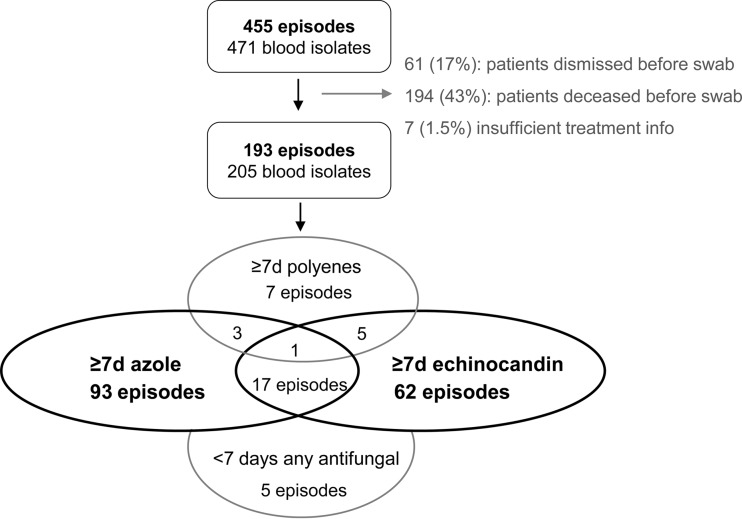
For patients (n = 93) treated with azoles for ≥7 days but with a drug from another drug class for <7 days (Fig. 1), species distributions in blood isolates (before IC treatment) and oral isolates (after IC treatment) were determined, and the proportions of species intrinsically less susceptible to azoles were compared. Likewise, for patients (n = 62) treated with echinocandins for ≥7 days but with a drug from another drug class for <7 days (Fig. 1), the proportions of species intrinsically less susceptible to echinocandins among blood and oral isolates were compared. The 7-day cutoff for significant exposure was selected based on previous findings identifying this duration as a critical driver of altered species distribution (9). Because coadministration of different drug classes (combination or sequential) for ≥7 days might bias the evaluation of each drug class's impact on posttreatment species distribution, patients undergoing such coadministration (n = 26) were excluded from this evaluation.
FIG 1.
Flowchart illustrating the numbers of eligible patients, included patients, and episodes, divided into those for patients receiving ≥7 days of exposure to azoles (93), echinocandins (62), polyenes (7), or a combination thereof (26). The total numbers of episodes for patients exposed to azoles, echinocandins, and polyenes for ≥7 days (irrespective of sequential or concomitant additional treatment) were 114, 85, and 16, respectively.
Species-specific prevalence of acquired resistance.
Rates of acquired resistance among initial blood and posttreatment oral isolates were evaluated for C. albicans and C. glabrata isolates. Susceptibility data for oral isolates from patients exposed to either azoles (n = 114; 109/114 patients exposed to fluconazole and 5/114 exposed to voriconazole) or echinocandins (n = 85) for ≥7 days were evaluated for both species. Susceptibility data for blood isolates as well as oral isolates exposed to either azoles (n = 79) or echinocandins (n = 108), respectively, for <7 days were included for both species for comparison. Coadministration of unrelated drug classes was not regarded as a driver of acquired resistance. Therefore, the evaluation of acquired resistance to a given drug class included all patients, with and without exposure to alternative drug classes.
Ethical considerations.
This study was approved by the Research Ethics Committees of the Capital Region of Denmark (protocol no. H-4-2012-FSP 104), the Danish Data Protection Agency (protocol no. 2013-41-1606), and the Danish Health and Medicines Authority (protocol no. 3-3013-302/1/).
Microbiology.
Blood isolates and oral swabs were inoculated onto CHROMagar (CHROMagar Co., Paris, France) for 2 to 4 days at 37°C. Species identifications were done by conventional methods and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis (Bruker Daltonics, Germany), supplemented with carbon assimilation with ATB strips (ID32C; bioMérieux, Marcy l'Etoile, France) or internal transcribed spacer (ITS) DNA sequencing when necessary (19). Polyfungal samples were identified by colony color differentiation (also within the same species) and separated by subculturing. Isolates were considered to be unique if a different species was detected or if the same species had different genotypes or susceptibility profiles (2-fold dilution MIC difference of ≥3 for any antifungal tested). MiBa was further used to assess microbiological data.
Susceptibility testing was done according to EUCAST EDef 7.2 (azoles and anidulafungin) (20) and by Etest (amphotericin B). Susceptibility interpretation was performed by applying the defined EUCAST breakpoints (21, 22; http://www.eucast.org/clinical_breakpoints/).
Statistical analyses.
Species distributions were compared by unpaired proportion analyses (chi-square tests). Candida krusei, C. glabrata, Candida guilliermondii, Candida pelliculosa, Saccharomyces cerevisiae, and Cryptococcus neoformans were classified as species intrinsically less susceptible to azoles. Similarly, C. parapsilosis, C. guilliermondii, and S. cerevisiae were classified as less susceptible to the echinocandins. S. cerevisiae was regarded as less susceptible to both drug classes because the modal MIC is comparable to that for C. glabrata for azoles (EUCAST data) but at least one step higher for echinocandins (unpublished data). When multiple species were found in blood or swabs, the isolate of the species with the highest epidemiological cutoff value (ECOFF) was counted. Numbers of negative oral cultures or polyfungal oral cultures were compared as a subanalysis between patients exposed to either azoles or echinocandins to determine the degree of fungal colonization after antifungal exposure.
Fisher's exact test of unpaired data was applied to compare proportions of acquired resistance among C. albicans and C. glabrata isolates from blood and oral swabs from patients exposed to an antifungal for <7 days or ≥7 days. The difference in mean MICs for each drug class was also determined for C. glabrata and C. albicans and analyzed by the Wilcoxon rank sum test, assuming a nonnormal distribution of MIC data. All polyfungal isolates were included in these analyses.
All P values of <0.05 were considered significant. R software was used for all statistical calculations.
MLST.
Multilocus sequence typing (MLST) was performed as previously described for the six major species C. albicans (23), C. glabrata (24), C. krusei (25), Candida tropicalis (26), Candida dubliniensis (27), and S. cerevisiae (28), and microsatellite typing was used for C. parapsilosis (29). MLST profiles were assigned according to corresponding pubMLST databases, when available, or assigned as novel for S. cerevisiae, starting from “1” for each allele. CLC Main Workbench (CLC-Bio, Qiagen) bioinformatics software with the MLST plug-in was applied to analyze sequence data. One allelic difference were considered microevolution between paired isolates, based on previous findings, and thus isolates were regarded as genetically unrelated if they differed at two or more alleles for all MLST schemes (30, 31).
Resistance mechanisms.
Underlying molecular resistance mechanisms were characterized for echinocandin resistance by sequencing of the gene encoding β-1,3-glucan synthase (FKS1).
RESULTS
Demographics.
During the 1-year study period, 471 unique blood isolates from 455 candidemia cases were referred to the reference laboratory. Oral swabs were obtained from 200 of these patients from all 12 major hospitals in Denmark; 63 (31.5%) were from intensive care units (ICUs), 55 (27.5%) from surgical units, 73 (36.5%) from medical units, and 9 (4.5%) from other units. In total, 194 of 455 patients (42.6%) deceased before the swab was obtained, and thus 76.6% (200 of 261 patients) of all eligible candidemia patients were included (Fig. 1). The remaining 61 patients were discharged before a mouth swab was obtained. Neither median age (67 versus 68 years) nor the proportion of men (59.6% versus 57.7%) varied significantly between enrolled patients and the total population of candidemia patients. Due to insufficient treatment information, seven patients were excluded. In total, 190 patients (including one neonate) and 193 episodes provided 205 blood isolates and 220 swab isolates included for statistical analyses. The total IC treatment median duration (range) was 17 days (5 to 61 days). The duration of the individual drug class for patients treated with azoles or echinocandins separately for ≥7 days was 14 days (7 to 40 days) for azoles and 15 days (7 to 56 days) for echinocandins.
Species distributions.
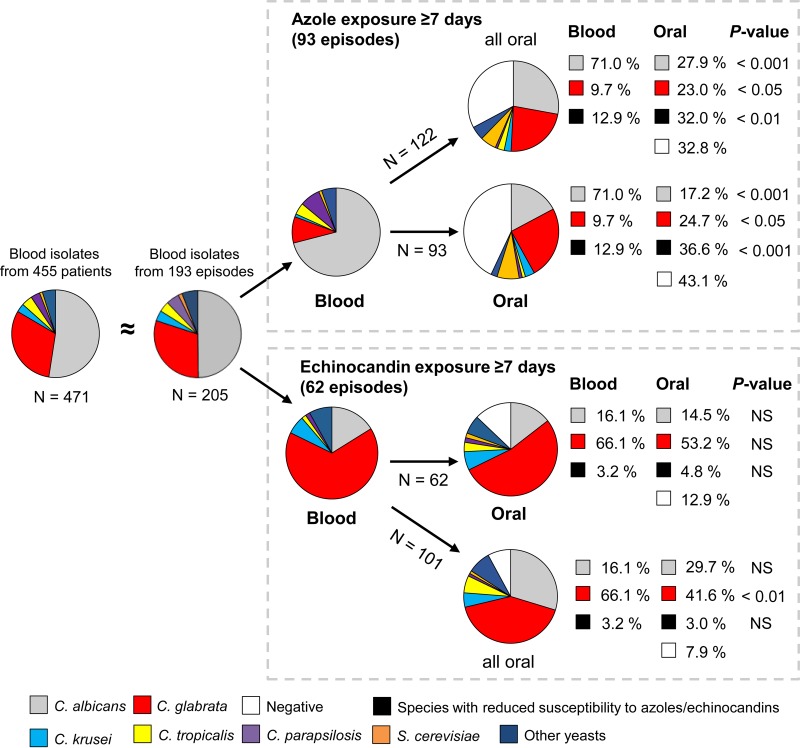
The species distribution among blood isolates from included patients was similar to the distribution for all candidemia cases (Fig. 2). Among 93 patients treated with azoles for ≥7 days but with another drug class for <7 days, 66 (71.0%) had C. albicans in the original blood culture, while among the 62 patients treated with echinocandins for ≥7 days and with another drug class for <7 days, 41 (66.1%) had C. glabrata in the blood (Fig. 2). In azole-exposed patients, the species distributions of blood and oral isolates were significantly different, with 71.0% of blood isolates being C. albicans and fewer than 20% of oral isolates being C. albicans (Fig. 2). On the other hand, C. glabrata and S. cerevisiae constituted about 10% and 1%, respectively, of blood isolates but about 25% and 7.5%, respectively, of oral isolates. The overall number of isolates less susceptible to fluconazole was significantly larger (P < 0.001) for the oral isolates (34/93 isolates [36.6%]) than for the blood isolates (12/93 isolates [19.9%]). For the echinocandin group, minor differences were observed in comparing species distributions among blood and oral isolates from exposed patients, with smaller proportions of C. albicans and C. glabrata observed in the oral isolates. Only 2 (3.2%) isolates categorized as intrinsically less susceptible to echinocandins were found among the blood isolates (1 C. parapsilosis and 1 Geotrichum candidum isolate), in contrast to three (4.8%) (1 C. parapsilosis, 1 C. guilliermondii, and 1 S. cerevisiae isolate) among the oral isolates.
FIG 2.
Pie charts displaying species distributions in the indicated groups. N, number of isolates. Blood isolates represented baseline colonization in patients subsequently exposed to either azoles or echinocandins. Oral isolates (≥7 days of exposure) represented end-of-treatment colonization. NS, not significant. On the patient level (pie charts on the same horizontal level), only the species with the highest ECOFF value was counted in case of polyfungal samples; however, the distribution of all oral isolates is also shown (above for azoles and below for echinocandins). Proportion analysis was performed by chi-square and Fisher's exact tests.
Among patients treated with echinocandins, 87% (54/62 patients) had culture-positive oral swabs and 63% (39/62 patients) had polyclonal colonization, compared to 57% (53/93 patients) and 31% (29/93 patients), respectively, of patients treated with azoles. Both proportions of culture-positive oral swabs and polyclonal colonization were significantly higher in the echinocandin group (P < 0.001 and P < 0.001, respectively).
MLST analyses.
Among 193 patients, 104 (53.9%) patients harbored a total of 112 paired isolates, with the same species found before and after antifungal exposure, 32 patients (16.5%) had a different colonizing species posttreatment, 57 patients (29.5%) were swab culture negative, and 72 patients (37.3%) had two or more isolates in the oral samples. Only 43/100 (43%) patients with C. albicans in the blood were colonized with C. albicans after treatment, two patients with C. albicans in the blood had two different C. albicans isolates in the oral sample, and one patient with two separate C. glabrata candidemia episodes had C. albicans in both oral swabs taken 10 weeks apart (see Table S1 in the supplemental material). Among the total of 45 paired C. albicans isolates, MLST analysis showed that 38 (84.4%) had similar genotypes. Similarly, 52 of 62 (83.9%) patients with C. glabrata were colonized with C. glabrata after treatment, four C. glabrata patients had two different C. glabrata isolates in the oral sample, and among a total of 56 paired C. glabrata isolates, 55 (98%) had similar genotypes. Overall, 101/112 (90%) paired Candida isolates had similar genotypes (Table 1; see Table S1).
TABLE 1.
Species distribution for isolates from blood and oral swab samples and numbers of paired isolates sharing similar genotypes
| Species | No. (%) of isolates |
No. of isolates of same species in paired blood and swab samples | No. (%) of isolates with similar genotypesa | |
|---|---|---|---|---|
| Blood | Swabs | |||
| C. albicans | 102 (49.8) | 84 (38.2) | 45b | 38 (84.4) |
| C. dubliniensis | 3 (1.5) | 8 (3.6) | 1 | 1 (100) |
| C. glabrata | 62 (30.2) | 82 (37.3) | 56 | 55 (98.2) |
| C. krusei | 8 (3.9) | 11 (5.0) | 4 | 4 (100.0) |
| C. tropicalis | 8 (3.9) | 12 (5.5) | 3 | 1 (33.3) |
| C. parapsilosis | 10 (4.9) | 3 (1.4) | 1 | 1 (100) |
| S. cerevisiae | 3 (1.5) | 12 (5.5) | 2 | 1 (50.0) |
| Other Candida/yeast species | 9c (4.4) | 8d (3.6) | (1e) | NAe |
| Total | 205 (100) | 220 (100) | 112 | 101 (90.2) |
Isolates with 5/6 or 6/7 identical MLST alleles or with the same fragment length for the microsatellite-based typing of C. parapsilosis.
One pair of C. albicans isolates came from one patient's swab samples from two separate C. glabrata candidemia episodes.
C. lusitaniae (4), C. guilliermondii (1), C. inconspicua (1), C. neoformans (1), C. utilis (1), and C. pelliculosa (1).
C. guilliermondii (3), C. kefyr (1), C. lusitaniae (2), C. pulcherrima (1), and Geotrichum candidum (1).
C. lusitaniae (1); the isolate was not genotyped and thus not counted.
Susceptibility and resistance.
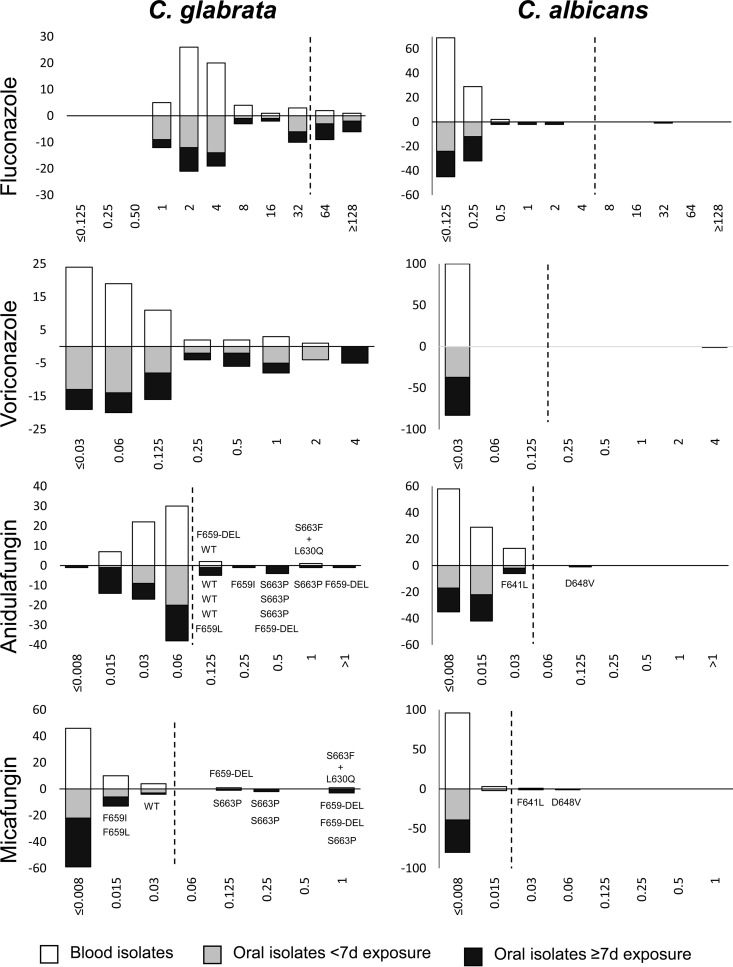
Fluconazole MIC distributions for the 62 blood and 82 oral C. glabrata isolates were bimodal, with peaks at 2 to 4 mg/liter and 32 to 64 mg/liter (Fig. 3). The proportion of resistant isolates (MICs of >32 mg/liter) was significantly higher among oral isolates from azole-exposed patients (29.4%) than among initial blood isolates (4.8%) (P < 0.01) as well as oral isolates from less exposed patients (10.4%) (P < 0.05) (Table 2). Accordingly, the geometric mean MIC of fluconazole was significantly higher for isolates from azole-exposed patients (Table 2). A similar trend was observed for voriconazole MIC distributions, although no clinical breakpoint has yet been established for C. glabrata (Fig. 3). For anidulafungin, the modal MIC was 0.06 mg/liter, and again, the proportion of isolates with MICs above the breakpoint was significantly higher among oral isolates from patients exposed to echinocandins (21.6%) than among blood and oral isolates from patients exposed to echinocandins for <7 days (4.8% and 3.2%, respectively) (Table 2). No statistical differences were observed in comparing geometric means (Table 2). Among the 11 C. glabrata isolates (21.6%) with anidulafungin MICs above the breakpoint (Table 2), three isolates carried wild-type FKS (with an anidulafungin MIC of 0.125 mg/liter), while eight isolates harbored well-known mutations responsible for the observed resistance (MICs of 0.125 to >1 mg/liter) (Fig. 3). Micafungin MICs were above the breakpoint for six oral isolates from patients exposed to echinocandins, all six of which harbored FKS mutations. For two FKS mutants (F659L and F659I), the micafungin MIC was below the breakpoint (0.015 mg/liter for both). Two C. glabrata isolates from blood harbored FKS hot spot mutations (conferring amino acid substitutions F659-DEL and S663F+L630Q).
FIG 3.
MIC distributions for C. glabrata (left) and C. albicans (right). MICs are given in milligrams per liter. White bars show data for blood isolates, gray bars are for swab isolates obtained after <7 days of exposure to either azoles or echinocandins, and black bars are for swab isolates obtained after ≥7 days of exposure to either drug class. The rows display MIC distributions for (top to bottom) fluconazole, voriconazole, anidulafungin, and micafungin. The y axis indicates the number of isolates either from blood (positive) or from swabs (negative).
TABLE 2.
Acquired resistance to fluconazole and anidulafungin in C. glabrata isolates from blood and swab samples
| Isolate group and parameter | Value for isolatesa |
||
|---|---|---|---|
| Oral (≥7 days)b | Blood (no exposure) | Oral (<7 days) | |
| Isolates with exposure to azoles | |||
| No. of isolates with fluconazole MIC above BP/total no. of isolates (%) | 10/34 (29.4) | 3/62 (4.8)<0.01 | 5/48 (10.4)<0.05 |
| Fluconazole geometric mean MIC (mg/liter) | 10.01 | 3.66<0.05 | 4.83<0.05 |
| Isolates with exposure to echinocandins | |||
| No. of isolates with anidulafungin MIC above BP/total no. of isolates (%) | 11/51 (21.6) | 3/62 (4.8)<0.01 | 1/31 (3.2)<0.05 |
| Anidulafungin geometric mean MIC (mg/liter) | 0.053 | 0.043NS | 0.048NS |
Superscript numbers indicate significant P values. NS, not significant. BP, EUCAST clinical breakpoint for resistance.
Reference column for statistical comparisons. Isolates were exposed to either azole or echinocandins for ≥7 days before a swab was obtained.
Only 1 of 47 (2.1%) C. albicans oral isolates, from a patient exposed to 16 days of fluconazole treatment, was azole resistant, while 0 of 37 oral isolates from unexposed patients and 0 of 100 blood isolates were resistant. Two of 43 (4.7%) C. albicans isolates from patients exposed to echinocandins harbored an FKS mutation (causing an F641L or D648V amino acid substitution) and were micafungin resistant, and one of them had cross-resistance to anidulafungin (Fig. 3). Both FKS mutants were paired with a previous isogenic wild-type isolate, and they were derived from patients who had received 28 and 15 days of caspofungin before the resistant isolate was obtained (see Table S1 in the supplemental material). MIC distributions of genetically linked blood and oral isolates for both C. albicans and C. glabrata displayed MIC histograms equivalent to those for the overall susceptibility data, which also included isolates solely found in either blood or oral swabs (see Fig. S1).
MIC data for other Candida species showed that two patients had persistent C. krusei isolates displaying anidulafungin resistance (MIC of 0.125 mg/liter) in blood and oral swabs. Both isolates harbored the Fks1 L701M substitution (Table 3). No other Candida species displayed acquired resistance to echinocandins. Two oral isolates were fluconazole resistant (1 of 8 C. dubliniensis isolates and 1 of 3 C. parapsilosis isolates), while 1 of 8 C. dubliniensis isolates was categorized as intermediate; all of these were from patients exposed to fluconazole for >14 days. In addition, 1 of 8 C. tropicalis isolates from blood and 2 of 12 C. tropicalis oral isolates displayed trailing phenotypes (partial growth inhibition), and these 3 isolates were acquired from patients with no information on previous fluconazole exposure (Table 3).
TABLE 3.
MIC distributions of Candida species other than C. albicans and C. glabrataa
| Species | Sample type | n | FLC |
VRC |
ANI |
MICA |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC range (mg/liter) | MIC50 (mg/liter) | No. (%) of resistant isolatesb | MIC range (mg/liter) | MIC50 (mg/liter) | MIC range (mg/liter) | MIC50 (mg/liter) | No. (%) of resistant isolatesb | Range (mg/liter) | MIC50 (mg/liter) | |||
| C. dubliniensis | Blood | 3 | ≤0.125–1 | NA | 0 | ≤0.03 | NA | 0.015–0.03 | NA | NA | ≤0.008–0.015 | NA |
| Swab | 8 | ≤0.125–≥128 | NA | 1c (12.5) | ≤0.03–>4 | NA | 0.015–0.06 | NA | NA | ≤0.008–0.06 | NA | |
| C. tropicalis | Blood | 8 | ≤0.125–16 | NA | 1d (12.5) | ≤0.03–4 | NA | ≤0.008–0.06 | NA | 0 | ≤0.008–0.03 | NA |
| Swab | 12 | 0.25–≥128 | 0.5 | 2e (16.7) | ≤0.03–>4 | ≤0.03 | ≤0.008–0.06 | 0.03 | 0 | ≤0.008–0.03 | 0.015 | |
| C. krusei | Blood | 8 | 16–≥128 | NA | Intrinsic | 0.125–2 | NA | 0.015–0.125 | NA | 2 (25)g | 0.06–0.125 | NA |
| Swab | 11 | 16–≥128 | 32 | Intrinsic | 0.125–2 | 0.25 | 0.03–0.125 | 0.06 | 2 (18.2)g | 0.06–0.25 | 0.125 | |
| S. cerevisiae | Blood | 2 | 16 | NA | NA | 0.25–0.5 | NA | 0.06–0.25 | NA | NA | 0.125 | NA |
| Swab | 12 | 2–16 | 4 | NA | 0.06–0.25 | 0.125 | 0.03–0.5 | 0.125 | NA | 0.06–0.25 | 0.125 | |
| C. parapsilosis | Blood | 10 | 0.25–1 | 0.5 | 0 | ≤0.03 | ≤0.03 | 0.5–>1 | 1 | NA | 0.5–>1 | 1 |
| Swab | 3 | 0.5–8 | NA | 1f (33.3) | ≤0.03–0.125 | NA | 1–>1 | NA | NA | 0.5–>1 | NA | |
FLC, fluconazole; VRC, voriconazole; ANI, anidulafungin; MICA, micafungin; NA, not applicable (too few isolates).
Only applicable if a breakpoint exists (EUCAST breakpoints were used).
One additional isolate was FLC intermediate, with a MIC of 4 mg/liter. Both patients with these isolates were exposed to fluconazole for >14 days.
Trailing phenotype (50% growth inhibition in entire FLC MIC range) and no information on prophylactic treatment for this patient.
Trailing phenotype. Both patients received caspofungin monotherapy (>14 days). One patient had a genetically similar and susceptible C. tropicalis isolate in blood.
A C. parapsilosis isolate with an FLC MIC of 8 mg/liter was found in a patient exposed to fluconazole for >14 days.
C. krusei isolates harboring the Fks1 L701M substitution were obtained from 2 persistently colonized patients (same species and genotype in the blood and swab), one treated with caspofungin for >14 days and the other exposed to the following three drugs for >7 days: fluconazole, voriconazole, and amphotericin B.
DISCUSSION
This study investigated the extent of intrinsic and acquired antifungal resistance of the oral fungal community after antifungal treatment. Overall, this study presented a large proportion of species less susceptible to azoles in the colonizing microbiota (primarily C. glabrata) for patients treated with azoles, as well as high frequencies of acquired resistance to both azoles and echinocandins among colonizing C. glabrata isolates.
Our findings corroborate previous reports demonstrating increased proportions of intrinsically less azole-susceptible species (primarily C. glabrata) among invasive (6) as well as colonizing Candida isolates (7) following azole prophylaxis. In contrast, only discrete and nonsignificant differences in species distribution were noted for patients treated with echinocandins. Echinocandin prophylaxis was previously demonstrated to significantly drive the invasive Candida species toward intrinsically less susceptible species (mainly C. parapsilosis), but that was shown in regions where the background prevalence of C. parapsilosis was higher than that in the current settings (1, 6, 8, 32, 33).
Interestingly, azole treatment resulted in more culture-negative and fewer polyfungal posttreatment oral cultures than those from echinocandin-exposed patients, despite the same median treatment duration of 14 days. Because pretreatment oral swabs could not be obtained for logistical reasons, we cannot rule out that more patients treated with fluconazole theoretically might have been culture negative in the oral cavity before treatment and thus that subsequent differences were not related to the choice of antifungal treatment. In our view, however, it is more likely that azoles clear the colonizing mucosal Candida microbiota more effectively than the echinocandins do. This could be explained partly by the high protein binding affinity of echinocandins and the associated lower drug concentrations at the mucosal surfaces. Another hypothesis for a lower positive rate for echinocandin-treated patients is that C. glabrata is potentially a more persistent colonizer (34). Indeed, the dominating species was C. glabrata in swabs from both azole- and echinocandin-treated patients, despite the different initial species distributions in blood. This would not explain the prevalence of polyfungal oral swabs, however, and further studies are thus warranted to evaluate these hypotheses (10, 30, 35, 36).
One approximation for this study was to interpret the blood isolates as surrogate markers for baseline colonization and to compare this to subsequent oral colonization. Genotyping revealed a 90% genetic correlation for species acquired from both sites, supporting the general concept that the invasive species was also a colonizing agent, as previously shown (15, 37–40). In addition, the MIC data for paired blood and oral isolates mirrored those for the overall isolate collection for both C. albicans and C. glabrata. The stronger genetic association between C. glabrata isolates from blood and oral isolates provided more support for the assumption of C. glabrata as a highly persistent colonizer, but this could potentially be ascribed to a less discriminatory MLST scheme (34, 41). In contrast to the findings for C. albicans, where most isolates had unique genotypes, identical genotypes were found for C. glabrata isolates, despite no geographical or historical link, but again, this can probably be ascribed to the low discriminatory power rather than to clonal expansion (41, 42).
The Fks substitutions found in C. glabrata were all well-known and previously associated with echinocandin resistance (18). The observed differential susceptibility to echinocandins found here for C. glabrata has been described previously, with some FKS mutations conferring resistance to anidulafungin but not to micafungin (43, 44). The observed resistance figures were higher than those reported from surveillance studies, where <5% acquired resistance has generally been observed (1, 5, 45–47). However, acquired echinocandin resistance rates of 10 to 30% have been reported for C. glabrata upon echinocandin exposure at certain centers, and depending on the site of infection (10, 48–50). Likewise, fluconazole resistance rates ranged from 13.7% in cancer patients (49) to 33.3% in candidemia patients (6) and >40% in the colonizing microbiota (7) of patients with previous exposure to azoles. Our study suggests that even in a nationwide study of unselected candidemic patients from district as well as university hospitals, acquired azole and echinocandin resistance in C. glabrata is common and therefore adds to the concerns that this organism is becoming an important “multidrug-resistant” yeast challenge of our time (51–53).
In contrast, the resistance rates among colonizing C. albicans and other Candida species remained low in exposed patients and equivalent to resistance rates from surveillance studies (1). Still, as supported by genotyping, the observed acquisition of azole or echinocandin resistance is probably an example of in vivo resistance selection upon antifungal exposure, as demonstrated previously (12, 54).
Our study has several limitations. Ideally, initial oral swabs from all patients should have been included as a baseline comparator, yet logistically this would require that every patient at risk of candidemia be sampled whenever blood is cultured. This is a much more comprehensive, if not impossible, task. Moreover, ideally, any prior exposure to antifungals unrelated to the treatment of the index candidemia case should have been recorded. However, despite these limitations, the current systematic study provides a new understanding of the level of resistance following treatment which constitutes a reservoir for future resistant Candida infections (15, 16). Together with previous reports of emerging resistance, our data highlight that the risk of generating antifungal resistance should not be neglected and underscore the importance of antifungal stewardship and of susceptibility testing not only for epidemiological purposes but also for daily clinical management.
Supplementary Material
ACKNOWLEDGMENTS
The highly collaborative staff at all involved hospital departments, especially the clinical microbiological laboratories as well as secretaries, nurses, and doctors, are acknowledged for the collection of swab samples. Birgit Brandt is acknowledged for her invaluable assistance with culturing, identification, and susceptibility testing, with support from the entire Mycology Laboratory at Statens Serum Institut. The DNA laboratory is acknowledged for their assistance with genotyping analyses.
H.K.J. was supported by the Novo Nordisk Foundation via a clinical research stipend.
This work was supported by Gilead Sciences (research grant IX-DK-131-0286) and by a research grant (2013) from the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) to R.H.J. M.C.A. has received research grants and travel grants from and has been paid for talks on behalf of Astellas, Basilea, Gilead, Merck Sharp & Dohme, and Pfizer. She has been on the advisory board for Merck and Gilead. K.M.T.A. has received travel grants from Gilead and Pfizer. L.E.L. has received travel grants from Astellas, Merck Sharp & Dohme, and Pfizer to attend conferences. H.K.J., L.M.S., F.S.R., L.N., B.O., L.K., and E.D. have no conflicts of interest.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01763-15.
REFERENCES
- 1.Arendrup MC, Dzajic E, Jensen RH, Johansen HK, Kjaeldgaard P, Knudsen JD, Kristensen L, Leitz C, Lemming LE, Nielsen L, Olesen B, Rosenvinge FS, Røder BL, Schønheyder HC. 2013. Epidemiological changes with potential implication for antifungal prescription recommendations for fungaemia: data from a nationwide fungaemia surveillance programme. Clin Microbiol Infect 19:E343–E353. doi: 10.1111/1469-0691.12212. [DOI] [PubMed] [Google Scholar]
- 2.Diekema D, Arbefeville S, Boyken L, Kroeger J, Pfaller M. 2012. The changing epidemiology of healthcare-associated candidemia over three decades. Diagn Microbiol Infect Dis 73:45–48. doi: 10.1016/j.diagmicrobio.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Tortorano AM, Prigitano A, Lazzarini C, Passera M, Deiana ML, Cavinato S, De Luca C, Grancini A, Lo Cascio G, Ossi C, Sala E, Montagna MT. 2013. A 1-year prospective survey of candidemia in Italy and changing epidemiology over one decade. Infection 41:655–662. doi: 10.1007/s15010-013-0455-6. [DOI] [PubMed] [Google Scholar]
- 4.Trick WE, Fridkin SK, Edwards JR, Hajjeh RA, Gaynes RP. 2002. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989–1999. Clin Infect Dis 35:627–630. doi: 10.1086/342300. [DOI] [PubMed] [Google Scholar]
- 5.Lockhart SR, Iqbal N, Cleveland AA, Farley MM, Harrison LH, Bolden CB, Baughman W, Stein B, Hollick R, Park BJ, Chiller T. 2012. Species identification and antifungal susceptibility testing of Candida bloodstream isolates from population-based surveillance studies in two U.S. cities from 2008 to 2011. J Clin Microbiol 50:3435–3442. doi: 10.1128/JCM.01283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lortholary O, Desnos-Ollivier M, Sitbon K, Fontanet A, Bretagne S, Dromer F. 2011. Recent exposure to caspofungin or fluconazole influences the epidemiology of candidemia: a prospective multicenter study involving 2,441 patients. Antimicrob Agents Chemother 55:532–538. doi: 10.1128/AAC.01128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mann PA, McNicholas PM, Chau AS, Patel R, Mendrick C, Ullmann AJ, Cornely OA, Patino H, Black TA. 2009. Impact of antifungal prophylaxis on colonization and azole susceptibility of Candida species. Antimicrob Agents Chemother 53:5026–5034. doi: 10.1128/AAC.01031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchard E, Lortholary O, Boukris-Sitbon K, Desnos-Ollivier M, Dromer F, Guillemot D. 2011. Prior caspofungin exposure in patients with hematological malignancies is a risk factor for subsequent fungemia due to decreased susceptibility in Candida spp.: a case-control study in Paris, France. Antimicrob Agents Chemother 55:5358–5361. doi: 10.1128/AAC.00690-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arendrup MC, Sulim S, Holm A, Nielsen L, Nielsen SD, Knudsen JD, Drenck NE, Christensen JJ, Johansen HK. 2011. Diagnostic issues, clinical characteristics, and outcomes for patients with fungemia. J Clin Microbiol 49:3300–3308. doi: 10.1128/JCM.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shields RK, Nguyen MH, Press EG, Clancy CJ. 2014. Abdominal candidiasis is a hidden reservoir of echinocandin resistance. Antimicrob Agents Chemother 58:7601–7605. doi: 10.1128/AAC.04134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah DN, Yau R, Lasco TM, Weston J, Salazar M, Palmer HR, Garey KW. 2012. Impact of prior inappropriate fluconazole dosing on isolation of fluconazole-nonsusceptible Candida species in hospitalized patients with candidemia. Antimicrob Agents Chemother 56:3239–3243. doi: 10.1128/AAC.00019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen RH, Astvad KMT, Silva LV, Sanglard D, Jørgensen R, Nielsen KF, Mathiasen EG, Doroudian G, Perlin DS, Arendrup MC. 2015. Stepwise emergence of azole, echinocandin and amphotericin B multidrug resistance in vivo in Candida albicans orchestrated by multiple genetic alterations. J Antimicrob Chemother 70:2551–2555. doi: 10.1093/jac/dkv140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lockhart SR, Fritch JJ, Meier AS, Schroppel K, Srikantha T, Galask R, Soll DR. 1995. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J Clin Microbiol 33:1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gammelsrud KW, Lindstad BL, Gaustad P, Ingebretsen A, Høiby EA, Brandtzaeg P, Sandven P. 2012. Multilocus sequence typing of serial Candida albicans isolates from children with cancer, children with cystic fibrosis and healthy controls. Med Mycol 50:619–626. doi: 10.3109/13693786.2012.675088. [DOI] [PubMed] [Google Scholar]
- 15.Brillowska-Dabrowska A, Bergmann O, Jensen IM, Jarløv JO, Arendrup MC. 2010. Typing of Candida isolates from patients with invasive infection and concomitant colonization. Scand J Infect Dis 42:109–113. doi: 10.3109/00365540903348336. [DOI] [PubMed] [Google Scholar]
- 16.Lau AF, Kabir M, Chen SC-A, Playford EG, Marriott DJ, Jones M, Lipman J, McBryde E, Gottlieb T, Cheung W, Seppelt I, Iredell J, Sorrell TC. 2015. Candida colonization as a risk marker for invasive candidiasis in mixed medical-surgical intensive care units: development and evaluation of a simple, standard protocol. J Clin Microbiol 53:1324–1330. doi: 10.1128/JCM.03239-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odds FC, Jacobsen MD. 2008. Multilocus sequence typing of pathogenic Candida species. Eukaryot Cell 7:1075–1084. doi: 10.1128/EC.00062-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arendrup MC, Perlin DS. 2014. Echinocandin resistance: an emerging clinical problem? Curr Opin Infect Dis 27:484–492. doi: 10.1097/QCO.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Bolchacova E, Voigt K, Crous PW, Miller AN, Wingfield MJ, Aime MC, An K-D, Bai F-Y, Barreto RW, Begerow D, Bergeron M-J, Blackwell M, Boekhout T, Bogale M, Boonyuen N, Burgaz AR, Buyck B, Cai L, Cai Q, Cardinali G, Chaverri P, Coppins BJ, Crespo A, Cubas P, Cummings C, Damm U, de Beer ZW, de Hoog GS, Del-Prado R, Dentinger B, Dieguez-Uribeondo J, Divakar PK, Douglas B, Duenas M, Duong TA, Eberhardt U, Edwards JE, Elshahed MS, Fliegerova K, Furtado M, Garcia MA, Ge Z-W, Griffith GW, Griffiths K, Groenewald JZ, Groenewald M, Grube M, Gryzenhout M, Guo L-D, Hagen F, Hambleton S, Hamelin RC, Hansen K, Harrold P, Heller G, Herrera C, Hirayama K, Hirooka Y, Ho H-M, Hoffmann K, Hofstetter V, Hognabba F, Hollingsworth PM, Hong S-B, Hosaka K, Houbraken J, Hughes K, Huhtinen S, Hyde KD, James T, Johnson EM, Johnson JE, Johnston PR, Jones EBG, Kelly LJ, Kirk PM, Knapp DG, Koljalg U, Kovacs GM, Kurtzman CP, Landvik S, Leavitt SD, Liggenstoffer AS, Liimatainen K, Lombard L, Luangsa-Ard JJ, Lumbsch HT, Maganti H, Maharachchikumbura SSN, Martin MP, May TW, McTaggart AR, Methven AS, Meyer W, Moncalvo J-M, Mongkolsamrit S, Nagy LG, Nilsson RH, Niskanen T, Nyilasi I, Okada G, Okane I, Olariaga I, Otte J, Papp T, Park D, Petkovits T, Pino-Bodas R, Quaedvlieg W, Raja HA, Redecker D, Rintoul TL, Ruibal C, Sarmiento-Ramirez JM, Schmitt I, Schussler A, Shearer C, Sotome K, Stefani FOP, Stenroos S, Stielow B, Stockinger H, Suetrong S, Suh S-O, Sung G-H, Suzuki M, Tanaka K, Tedersoo L, Telleria MT, Tretter E, Untereiner WA, Urbina H, Vagvolgyi C, Vialle A, Vu TD, Walther G, Wang Q-M, Wang Y, Weir BS, Weiss M, White MM, Xu J, Yahr R, Yang ZL, Yurkov A, Zamora J-C, Zhang N, Zhuang W-Y, Schindel D. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arendrup MC, Cuenca-Estrella M, Lass-Flörl C, Hope W, EUCAST-AFST . 2012. EUCAST technical note on the EUCAST definitive document EDef 7.2: method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts EDef 7.2 (EUCAST-AFST). Clin Microbiol Infect 18:E246–E247. doi: 10.1111/j.1469-0691.2012.03880.x. [DOI] [PubMed] [Google Scholar]
- 21.Arendrup MC, Cuenca-Estrella M, Lass-Flörl C, Hope WW. 2014. EUCAST technical note on Candida and micafungin, anidulafungin and fluconazole. Mycoses 57:377–379. doi: 10.1111/myc.12170. [DOI] [PubMed] [Google Scholar]
- 22.Arendrup MC, Cuenca-Estrella M, Lass-Flörl C, Hope WW. 2013. Breakpoints for antifungal agents: an update from EUCAST focussing on echinocandins against Candida spp. and triazoles against Aspergillus spp. Drug Resist Updat 16:81–95. doi: 10.1016/j.drup.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Bougnoux M-E, Morand S, Enfert C, d'Enfert C. 2002. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J Clin Microbiol 40:1290–1297. doi: 10.1128/JCM.40.4.1290-1297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dodgson AR, Pujol C, Denning DW, Soll DR, Fox AJ. 2003. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J Clin Microbiol 41:5709–5717. doi: 10.1128/JCM.41.12.5709-5717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobsen MD, Gow NAR, Maiden MCJ, Shaw DJ, Odds FC. 2007. Strain typing and determination of population structure of Candida krusei by multilocus sequence typing. J Clin Microbiol 45:317–323. doi: 10.1128/JCM.01549-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tavanti A, Davidson AD, Johnson EM, Maiden MCJ, Shaw DJ, Gow NAR, Odds FC. 2005. Multilocus sequence typing for differentiation of strains of Candida tropicalis. J Clin Microbiol 43:5593–5600. doi: 10.1128/JCM.43.11.5593-5600.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McManus BA, Coleman DC, Moran G, Pinjon E, Diogo D, Bougnoux M-E, Borecká-Melkusova S, Bujdákova H, Murphy P, D'Enfert C, Sullivan DJ. 2008. Multilocus sequence typing reveals that the population structure of Candida dubliniensis is significantly less divergent than that of Candida albicans. J Clin Microbiol 46:652–664. doi: 10.1128/JCM.01574-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muñoz R, Gómez A, Robles V, Rodríguez P, Cebollero E, Tabera L, Carrascosa AV, Gonzalez R. 2009. Multilocus sequence typing of oenological Saccharomyces cerevisiae strains. Food Microbiol 26:841–846. doi: 10.1016/j.fm.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Diab-Elschahawi M, Forstner C, Hagen F, Meis JF, Lassnig AM, Presterl E, Klaassen CHW. 2012. Microsatellite genotyping clarified conspicuous accumulation of Candida parapsilosis at a cardio-thoracic surgery intensive care unit. J Clin Microbiol 50:3422–3426. doi: 10.1128/JCM.01179-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enache-Angoulvant A, Bourget M, Brisse S, Stockman-Pannier C, Diancourt L, François N, Rimek D, Fairhead C, Poulain D, Hennequin C. 2010. Multilocus microsatellite markers for molecular typing of Candida glabrata: application to analysis of genetic relationships between bloodstream and digestive system isolates. J Clin Microbiol 48:4028–4034. doi: 10.1128/JCM.02140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen K-W, Chen Y-C, Lo H-J, Odds FC, Wang T-H, Lin C-Y, Li S-Y. 2006. Multilocus sequence typing for analyses of clonality of Candida albicans strains in Taiwan. J Clin Microbiol 44:2172–2178. doi: 10.1128/JCM.00320-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sipsas NV, Lewis RE, Tarrand J, Hachem R, Rolston KV, Raad II, Kontoyiannis DP. 2009. Candidemia in patients with hematologic malignancies in the era of new antifungal agents (2001–2007): stable incidence but changing epidemiology of a still frequently lethal infection. Cancer 115:4745–4752. doi: 10.1002/cncr.24507. [DOI] [PubMed] [Google Scholar]
- 33.Forrest GN, Weekes E, Johnson JK. 2008. Increasing incidence of Candida parapsilosis candidemia with caspofungin usage. J Infect 56:126–129. doi: 10.1016/j.jinf.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 34.Spampinato C, Leonardi D. 2013. Molecular fingerprints to identify Candida species. Biomed Res Int 2013:923742. doi: 10.1155/2013/923742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perlin DS, Shor E, Zhao Y. 2015. Update on antifungal drug resistance. Curr Clin Microbiol Rep 2:84–95. doi: 10.1007/s40588-015-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lott TJ, Frade JP, Lockhart SR. 2010. Multilocus sequence type analysis reveals both clonality and recombination in populations of Candida glabrata bloodstream isolates from U.S. surveillance studies. Eukaryot Cell 9:619–625. doi: 10.1128/EC.00002-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hattori H, Iwata T, Nakagawa Y, Kawamoto F, Tomita Y, Kikuchi A, Kanbe T. 2006. Genotype analysis of Candida albicans isolates obtained from different body locations of patients with superficial candidiasis using PCRs targeting 25S rDNA and ALT repeat sequences of the RPS. J Dermatol Sci 42:31–46. doi: 10.1016/j.jdermsci.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Gammelsrud KW, Sandven P, Høiby EA, Sandvik L, Brandtzaeg P, Gaustad P. 2011. Colonization by Candida in children with cancer, children with cystic fibrosis, and healthy controls. Clin Microbiol Infect 17:1875–1881. doi: 10.1111/j.1469-0691.2011.03528.x. [DOI] [PubMed] [Google Scholar]
- 39.Karaman M, Firinci F, Karaman O, Uzuner N, Hakki Bahar I. 2013. Long-term oropharyngeal colonization by C. albicans in children with cystic fibrosis. Yeast 30:429–436. doi: 10.1002/yea.2977. [DOI] [PubMed] [Google Scholar]
- 40.Soll DR. 2000. The ins and outs of DNA fingerprinting the infectious fungi. Clin Microbiol Rev 13:332–370. doi: 10.1128/CMR.13.2.332-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Odds FC, Hanson MF, Davidson AD, Jacobsen MD, Wright P, Whyte JA, Gow NAR, Jones BL. 2007. One year prospective survey of Candida bloodstream infections in Scotland. J Med Microbiol 56:1066–1075. doi: 10.1099/jmm.0.47239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lott TJ, Frade JP, Lyon GM, Iqbal N, Lockhart SR. 2012. Bloodstream and non-invasive isolates of Candida glabrata have similar population structures and fluconazole susceptibilities. Med Mycol 50:136–142. doi: 10.3109/13693786.2011.592153. [DOI] [PubMed] [Google Scholar]
- 43.Shields RK, Nguyen MH, Press EG, Kwa AL, Cheng S, Du C, Clancy CJ. 2012. The presence of an FKS mutation rather than MIC is an independent risk factor for failure of echinocandin therapy among patients with invasive candidiasis due to Candida glabrata. Antimicrob Agents Chemother 56:4862–4869. doi: 10.1128/AAC.00027-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arendrup MC, Perlin DS, Jensen RH, Howard SJ, Goodwin J, Hope W. 2012. Differential in vivo activities of anidulafungin, caspofungin, and micafungin against Candida glabrata isolates with and without FKS resistance mutations. Antimicrob Agents Chemother 56:2435–2442. doi: 10.1128/AAC.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hesstvedt L, Gaustad P, Andersen CT, Haarr E, Hannula R, Haukland HH, Hermansen N-O, Larssen KW, Mylvaganam H, Ranheim TE, Sandven P, Nordøy I. 2015. Twenty-two years of candidaemia surveillance—results from a Norwegian national study. Clin Microbiol Infect 21:938–945. doi: 10.1016/j.cmi.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 46.Pfaller MA, Moet GJ, Messer SA, Jones RN, Castanheira M. 2011. Candida bloodstream infections: comparison of species distributions and antifungal resistance patterns in community-onset and nosocomial isolates in the SENTRY Antimicrobial Surveillance Program, 2008–2009. Antimicrob Agents Chemother 55:561–566. doi: 10.1128/AAC.01079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, Zhou S, Pan A, Li J, Liu B. 2015. Surveillance of antifungal susceptibilities in clinical isolates of Candida species at 36 hospitals in China from 2009 to 2013. Int J Infect Dis 33:1–4. doi: 10.1016/j.ijid.2014.12.033. [DOI] [PubMed] [Google Scholar]
- 48.Alexander BD, Johnson MD, Pfeiffer CD, Jiménez-Ortigosa C, Catania J, Booker R, Castanheira M, Messer SA, Perlin DS, Pfaller MA. 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis 56:1724–1732. doi: 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farmakiotis D, Tarrand JJ, Kontoyiannis DP. 2014. Drug-resistant Candida glabrata infection in cancer patients. Emerg Infect Dis 20:1833–1840. doi: 10.3201/eid2011.140685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shields RK, Nguyen MH, Press EG, Updike CL, Clancy CJ. 2013. Anidulafungin and micafungin MIC breakpoints are superior to that of caspofungin for identifying FKS mutant Candida glabrata strains and echinocandin resistance. Antimicrob Agents Chemother 57:6361–6365. doi: 10.1128/AAC.01451-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panackal AA, Gribskov JL, Staab JF, Kirby KA, Rinaldi M, Marr KA. 2006. Clinical significance of azole antifungal drug cross-resistance in Candida glabrata. J Clin Microbiol 44:1740–1743. doi: 10.1128/JCM.44.5.1740-1743.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. 2012. Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev 36:288–305. doi: 10.1111/j.1574-6976.2011.00278.x. [DOI] [PubMed] [Google Scholar]
- 53.Rodrigues CF, Silva S, Henriques M. 2014. Candida glabrata: a review of its features and resistance. Eur J Clin Microbiol Infect Dis 33:673–688. doi: 10.1007/s10096-013-2009-3. [DOI] [PubMed] [Google Scholar]
- 54.Jensen RH, Johansen HK, Arendrup MC. 2013. Stepwise development of homozygous S80P substitution in Fks1p, conferring echinocandin resistance in Candida tropicalis. Antimicrob Agents Chemother 57:614–617. doi: 10.1128/AAC.01193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.