Abstract
A high fosfomycin resistance rate was observed in Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae (KPC-KP) in our previous study, but little is known about its mechanisms. In this study, we explored the prevalence of plasmid-mediated fosfomycin resistance determinants among fosfomycin-resistant KPC-KP strains from a Chinese university hospital and determined the complete sequence of a novel fosA3-carrying plasmid isolated from an epidemic K. pneumoniae sequence type (ST) 11 strain. A total of 97 KPC-KP strains were studied, of which 57 (58.8%) were resistant to fosfomycin, including 44 (45.4%) harboring fosA3 and 1 harboring fosA. All fosA3-positive strains belonged to the dominant ST11-pulse type (PT) A clone according to multilocus sequence typing and pulsed-field gel electrophoresis, suggesting clonal dissemination. The fosA-positive isolate belonged to ST11-PTE. The fosA3-carrying plasmid pKP1034 is 136,848 bp in length and is not self-transmissible. It is a multireplicon plasmid belonging to IncR-F33:A−: B−. Besides fosA3, a variety of other resistance determinants, including blaKPC-2, rmtB, blaCTX-M-65, and blaSHV-12, are identified in pKP1034, which would allow for coselection of fosA3 by most β-lactams and/or aminoglycosides and facilitate its dissemination despite limited use of fosfomycin in China. Detailed comparisons with related plasmids revealed that pKP1034 is highly mosaic and might have evolved from alarming recombination of the blaKPC-2-carrying plasmid pKPC-LK30 from Taiwan and the epidemic fosA3-carrying plasmid pHN7A8 from mainland China.
INTRODUCTION
Increasing prevalence of Klebsiella pneumoniae carbapenemase–producing K. pneumoniae (KPC-KP) has presented an alarming clinical threat. Fosfomycin seems to retain in vitro activity against many carbapenem-resistant Enterobacteriaceae and has been recently reintroduced into the fight against these superbugs, including KPC-KP strains (1–4). A previous study showed that only 43.4% of KPC-KP strains retained susceptibility to fosfomycin in a Chinese university hospital (5). Recently, Jiang et al. reported a comparable fosfomycin susceptibility rate (39.2%) in KPC-KP collected from 12 hospitals in China, and fosfomycin resistance could be attributed to the plasmid-mediated fosA3 gene in 94 (55.6% of fosfomycin-resistant KPC-KP) strains (6).
FosA3 inactivates fosfomycin by glutathione S-transferase activity and was characterized first in CTX-M-producing Escherichia coli in Japan (7). Thereafter, it has been detected in food-producing animals, pets, patients, and healthy individuals in Asian countries (8–11). In most cases, the fosA3 gene is associated with blaCTX-M genes and sometimes with rmtB as well, and it resides on IS26-composite transposons, facilitating its dissemination. More recently, fosA3 has been characterized in two atypical blaKPC-carrying plasmids (6, 12). Here we explored the prevalence of plasmid-mediated fosfomycin resistance determinants among KPC-KP strains from a Chinese university hospital and determined the complete sequence of a novel fosA3-carrying plasmid pKP1034 from an epidemic KPC-KP sequence type (ST) 11 strain. To the best of our knowledge, this is the first fully sequenced plasmid from the dominant KPC-KP ST11 strain cocarrying fosA3, blaKPC-2, rmtB, blaCTX-M-65, and blaSHV-12.
MATERIALS AND METHODS
A total of 97 nonduplicate KPC-KP strains were collected from the first affiliated hospital, Zhejiang University School of Medicine, between January 2010 and February 2013. MICs of fosfomycin were determined by the agar dilution method on Mueller-Hinton agar supplemented with glucose-6-phosphate (G-6-P) (25 mg/liter) and interpreted according to the Clinical and Laboratory Standards Institute guidelines (13). Only fosfomycin-resistant strains were included for further investigation. PCR was conducted to confirm the presence of blaKPC and plasmid-mediated fosfomycin determinants (fosA, fosA3, fosB, fosC2), as described previously (9, 14). Since rmtB was prevalent in KPC-KP in our hospital, it was screened with PCR in all fosfomycin-resistant strains as well (5). Pulsed-field gel electrophoresis (PFGE) was performed using the XbaΙ restriction enzyme and interpreted by the criteria proposed by Tenover et al. (15). Multilocus sequence typing (MLST) was carried out according to protocols on the MLST website for K. pneumoniae (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html), and wzi sequencing was conducted to predict the capsular (K) types of these K. pneumoniae isolates (16).
Conjugation experiments were carried out by broth mating for a representative fosA3-positive strain KP1034 (ST11-PTA1) with E. coli J53Azi as the recipient. Transconjugants were selected on lysogenic agar plates supplemented with sodium azide (150 mg/liter), fosfomycin (100 mg/liter), and G-6-P (25 mg/liter). The transformation experiment was conducted using total plasmids extracted from KP1034 and E. coli DH5α competent cells (TaKaRa Bio, Japan) as the recipient, according to the user manual. Transformants were then selected on LB agar containing fosfomycin (100 mg/liter) and G-6-P (25 mg/liter) after overnight incubation. Antimicrobial susceptibility (to imipenem, meropenem, ertapenem, piperacillin-tazobactam, cefotaxime, ceftazidime, cefepime, aztreonam, gentamicin, amikacin, ciprofloxacin, colistin, tigecycline, and fosfomycin) was determined for KP1034 and its E. coli DH5α transformant KP1034-TF by Etest, except for fosfomycin, for which the MICs were determined as described above. The aforementioned resistance determinants were detected in KP1034-TF by PCR as well. Nuclease S1-PFGE was performed for both KP1034 and KP1034-TF to make sure that only one plasmid was transferred to KP1034-TF and to determine the approximate size of the plasmid.
Finally, fosA3-carrying plasmid pKP1034 was extracted from KP1034-TF using the Qiagen plasmid maxi kit (Qiagen, Valencia, CA). The plasmid was sequenced by a whole-genome shotgun approach, as previously described (17). In brief, pKP1034 DNA was randomly sheared using a Hydroshear DNA shearing device (Gene Machines, USA), and 2.0- to 4.0-kb fragments were cloned into a pMD19-T vector (TaKaRa Bio, Japan) and then transformed into E. coli DH5α. End sequencing was conducted using an ABI 3730xl sequencer (Applied Biosystems, USA). The average coverage was approximately 7× across the plasmid. Sequence data were assembled to 25 contigs, including 3 bigger ones, with the Phred/Phrap/Consed software suite (University of Washington, Seattle, WA, USA). Six pairs of primers were designed according to ends of the 3 bigger contigs, and gaps were filled with PCR and Sanger sequencing. The plasmid sequence was initially annotated with RAST (http://rast.nmpdr.org) and then curated manually using the BLASTn and BLASTp algorithms (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Easyfig 2.0 was used to map whole plasmids and to conduct limited comparisons between closely related plasmids. The multiresistance region (MRR) on pKP1034 was mapped with reference to the MRRs of pHN7A8, pKPC-LK30, pKP048, and pHS102707 (GenBank accession no. JN232517, KC405622, FJ628167, and KF701335, respectively).
Nucleotide sequence accession number.
The annotated sequence of pKP1034 has been deposited in GenBank under accession number KP893385.
RESULTS AND DISCUSSION
Of the 97 KPC-KP strains, 57 (58.8%) were resistant to fosfomycin, including 44 (45.4%) harboring fosA3 and 1 (1.0%) harboring fosA. The fosfomycin resistance rate in KPC-KP was comparable to that (60.8%) recently reported by Jiang et al., although a higher breakpoint for fosfomycin resistance (256 mg/liter) was adopted in our study (6). Consistent with the previous study, the high-level incidence of fosfomycin resistance in KPC-KP in our study was mainly due to acquisition of fosA3 (77.2%). Molecular typing identified two STs (ST11 and ST494) based on MLST and five pulse types (PTA through PTE) based on PFGE (Table 1; see also Fig. S1 in the supplemental material). However, PTA comprised five subtypes, PTA1 to PTA5, and PTA1 was the major subtype. Note that all minor subtypes but PTA2 were found only in strains collected in 2013 (data not shown), suggesting increasing diversity of the genetic backgrounds of these fosfomycin-resistant KPC-KP strains over time. All fosA3-positive strains belonged to the dominant clone ST11-PTA, indicating clonal dissemination. Eleven of the 12 fosA- and fosA3-negative strains also fell into the ST11 clone. Six of them were grouped into PTA, 4 into PTB, and 1 into PTC, with the remaining one belonging to ST494-PTD. The fosA-positive strain belonged to ST11-PTE. The results of wzi sequencing showed high-level consistency with that of PFGE (Table 1). A total of 4 wzi alleles (wzi209, wzi64, wzi108, and wzi141) were found, and the main allele wzi209 was first reported and showed relatively low-level identity to alleles reported by Brisse et al. (16). Thus, the K type of the dominant clone could not been predicated according to the reference database, and classical K typing should be performed in the future (16). However, wzi sequencing did serve as a reliable typing method and seemed to demonstrate a discriminatory power lower than that of PFGE but higher than that of MLST. According to wzi sequencing, all fosA3-harboring isolates clustered in the novel wzi209 allele except one, which had a distinct wzi sequence (wzi64). The fosA-harboring isolate could not be typed by wzi sequencing (Table 1).
TABLE 1.
Characterization of 57 fosfomycin-resistant KPC-KP strains
| Strain | PTa | STb | wzi allele | No. | Resistance determinant |
|---|---|---|---|---|---|
| fos negativec (n = 12) | A1 | 11 | wzi209 | 1 | rmtB-blaKPC |
| wzi209 | 1 | blaKPC | |||
| wzi209 | 3 | rmtB-blaKPC | |||
| A2 | 11 | wzi209 | 1 | rmtB-blaKPC | |
| B | 11 | wzi108 | 4 | rmtB-blaKPC | |
| C | 11 | wzi108 | 1 | blaKPC | |
| D | 494 | wzi141 | 1 | blaKPC | |
| fos positive (n = 45) | A1 | 11 | wzi209 | 34 | fosA3-rmtB-blaKPC |
| A2 | 11 | wzi209 | 2 | fosA3-rmtB-blaKPC | |
| A3 | 11 | wzi64 | 1 | fosA3-rmtB-blaKPC | |
| A4 | 11 | wzi209 | 3 | fosA3-rmtB-blaKPC | |
| 3 | fosA3-blaKPC | ||||
| A5 | 11 | wzi209 | 1 | fosA3-rmtB-blaKPC | |
| E | 11 | UTd | 1 | fosA-blaKPC |
PT, pulse type.
ST, sequence type.
Strains negative for all plasmid-mediated fosfomycin resistance determinants screened in this study.
UT, untypeable.
Transfer of the fosA3-carrying plasmid by conjugation was unsuccessful for KP1034, despite repeated attempts. Two plasmids were extracted from KP1034, of which the bigger one (∼135 kb), carrying blaKPC, fosA3, and rmtB, was transformed into the E. coli DH5α transformant (see Fig. S1 in the supplemental material), conferring resistance to β-lactams, fosfomycin, and aminoglycosides, respectively (Table 2).
TABLE 2.
MICs for K. pneumoniae KP1034 and its E. coli DH5α transformant coharboring fosA3, blaKPC-2, and rmtB
| Strain | MICa (mg/liter) of: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPM | MEM | ETP | TZP | CTX | CAZ | FEP | ATM | GEN | AMK | CIP | FOS | TGC | CST | |
| K. pneumoniae KP1034 | 32 | 64 | 64 | >256 | >32 | >32 | 16 | >256 | >256 | >256 | >32 | >512 | 1 | 0.38 |
| KP1034-TFb | 1.5 | 1 | 2 | 128 | >32 | >32 | 3 | >256 | >256 | >256 | 0.012 | >512 | 0.5 | 0.13 |
| E. coli DH5α | 0.125 | 0.016 | 0.125 | 2 | 0.125 | 0.25 | 0.02 | 0.02 | 0.38 | 0.75 | 0.012 | 1 | 0.25 | 0.13 |
All MICs were determined by Etest except for fosfomycin, for which the MIC was determined by agar dilution method. AMK, amikacin; ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; CST, colistin; CTX, cefotaxime; ETP, ertapenem; FEP, cefepime; GEN, gentamicin; FOS, fosfomycin; IPM, imipenem; MEM, meropenem; TGC, tigecycline; TZP, piperacillin-tazobactam.
KP1034-TF, E. coli DΗ5α transformant harboring pKP1034.
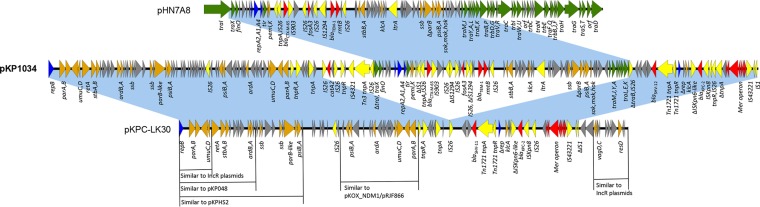
The 136,848-bp plasmid pKP1034 has an average GC content of 54.5% and harbors 191 predicted open reading frames. It is a multireplicon plasmid and belongs to IncR-F33:A−: B− according to PCR-based replicon typing and replicon sequence typing schemes (18–20). The overall structure of the plasmid is highly mosaic and can be divided into three genetically and physically distinct modules (Fig. 1): an ∼10.0-kb composite transposon carrying resistance determinant catA2 (chloramphenicol) flanked by two inversely oriented IS26 elements at each end, an ∼49.1-kb module essentially homologous to the epidemic fosA3-carrying plasmid pHN7A8 reported in mainland China (17), and an ∼77.7-kb region closely related to the blaKPC-carrying plasmid pKPC-LK30 from Taiwan (21).
FIG 1.
Major structural features of plasmid pKP1034, indicated with bold type, compared with fosA3-positive plasmid pHN7A8 (JN232517.1) and blaKPC-2-positive plasmid pKPC-LK30 (KC405622.1). Light-blue shading indicates shared regions with a high degree of homology. Open reading frames (ORFs) are portrayed by arrows and colored according to their putative functions. Dark-blue arrows indicate replication-associated genes. Genes associated with plasmid conjugal transfer are indicated by green arrows, and genes involved in plasmid stability are indicated by brown arrows. Red and yellow arrows indicate antimicrobial resistance genes and mobile element genes, respectively. Gray arrows indicate genes for hypothetical proteins as well as proteins of unknown function.
The ∼49.1-kb pHN7A8-derived module contains an incomplete IncFII replicon and a multidrug resistance region similar to the MRR of pHN7A8, with the former encompassing the following: (i) a replication region composed of repA1, repA2, and repA4 genes, (ii) genes responsible for plasmid maintenance and stability, including parB (plasmid partition), stbAB (plasmid stabilization), sok-mok-hok (postsegregation killing), psiAB (plasmid SOS inhibition protein), and pemIK (toxin-antitoxin system), and (iii) an incomplete tra region comprising traX, traM, traJ, traY, traA, traL, traE, and traK genes and remnants of traI and traB. The incomplete tra region may explain why pKP1034 could not transfer conjugatively to recipients like pHN7A8. All of these scaffold genes are 100% identical to their counterparts on pHN7A8 except for the traA gene and traB remnant, where a 1-nucleotide difference is observed for each. pHN7A8 is an F33:A−: B− type epidemic plasmid coharboring blaCTX-M-65, blaTEM-1, rmtB, and fosA3 (17). It was originally isolated from a dog in Guangzhou, China. Recently, pHN7A8-like plasmids have disseminated widely in food-producing animals and pets in China, playing an important role in the spread of resistance genes blaCTX-M-65, rmtB, and fosA3 (9, 17, 22, 23). To date, pHN7A8- and pHN7A8-like plasmids have not been detected in human strains yet. However, the fosA3-carrying plasmid pFOS-HK151325, related (88.0% query coverage, 98.0% identity by BLASTn analysis at NCBI) to pHN7A8, has been detected in a human E. coli strain in Hong Kong recently, and it is virtually identical (>99.9% query coverage, >99.9% identity by BLASTn analysis at NCBI) to the plasmid pHK23a that originated from a slaughter pig in the same region, indicating potential zoonotic transmission (8, 10). In our study, the pHN7A8-derived module on pKP1034 also suggests the transfer of plasmid pHN7A8 from an animal source to a human, and the plasmid appears to have been subjected to extensive rearrangements afterward.
The ∼77.7-kb pKPC-LK30-like module is a chimera encompassing an abundance of scaffold genes and another multidrug resistance region. The IncR region containing genes responsible for replication (repB), partition (parAB), and stability (umuCD) is identified on this module, which is conserved among most sequenced IncR plasmids, such as pEFER, pKP1780, pKPN101-IT, pK245, pKPS30, and pKPS77 (GenBank accession no. CU928144, JX424614, JX283456, DQ449578, KF793937, and KF954150, respectively). However, vagCD (toxin-antitoxin system) and resD (multimer resolution) genes commonly located upstream of repB on IncR plasmids are missing in pKP1034, probably due to IS1-mediated deletion. The downstream region contains additional genes for plasmid stability, including stbAB, ardAB (antirestriction protein), parB-like gene, and psiAB, which together with the IncR region exhibits the highest identity (>99%) to that of plasmid pKPHS2 (GenBank accession no. CP003224), a blaKPC-carrying plasmid identified in K. pneumoniae from Shanghai. Located next to the pKPHS2-like region was a region closely related to those of two blaNDM-1-carrying plasmids pKOX-NDM1 and pRJF866 (GenBank accession no. JQ314407 and KF732966, respectively), found in Klebsiella oxytoca and K. pneumoniae, respectively. This region consists of psiAB, ardA, umuCD (involved in SOS mutagenesis), and parAB. Although lack of a functional transfer system has likely prevented extensive spread of pKP1034 across various K. pneumoniae lineages, the abundance of genes for plasmid maintenance and stability has likely ensured its stable inheritance in strains harboring it, leading to clonal dissemination of fosA3-carrying KPC-KP in our hospital.
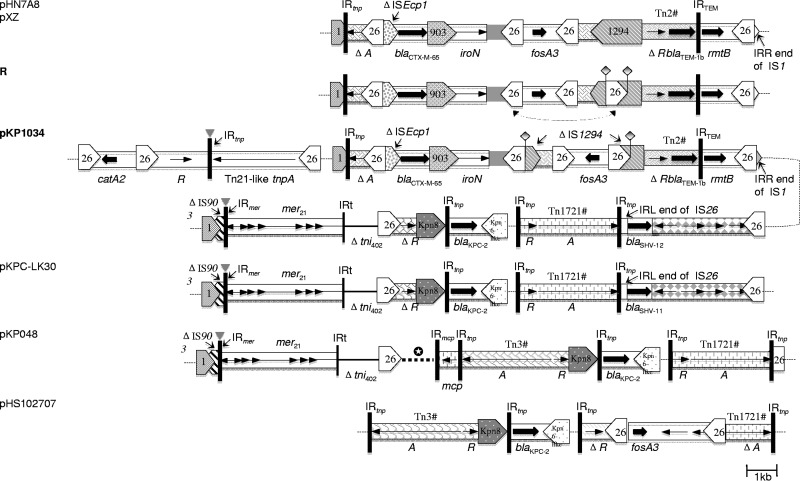
Besides fosA3, a variety of other resistance determinants, including blaKPC-2, rmtB, blaCTX-M-65, blaSHV-12, blaTEM-1, and catA2, are present in pKP1034, which allows coselection of fosA3 by most β-lactams and/or aminoglycosides frequently used in clinic and, therefore, has further facilitated stable inheritance and wide dissemination of fosA3 in our hospital despite limited use of fosfomycin. The mosaic, modular MRR in pKP1034 could be divided into three segments of distinct phylogenetic origins (Fig. 2). The first one is located upstream of the pHN7A8-derived module and carries catA2 flanked by two inversely oriented IS26 elements at each end. In detail, this segment contains a small composite transposon carrying catA2 bracketed by directly oriented copies of IS26, which has been commonly found on several fully sequenced plasmids, such as pENT-8a4, pEC-IMPQ, pEC-IMP, pVC1447, and pK245 (GenBank accession no. CP008899.1, EU855788.1, EU855787.1, KM083064.1, and DQ449578.1, respectively). However, compared with its counterparts in the above plasmids, 11 nucleotides immediately downstream of the first IS26 are missing in pKP1034, indicating that the small IS26-flanked composite transposon in pKP1034 might have originated from an independent capture event. The downstream structure encodes a resolvase, a Tn21-like transposase, and an inversely oriented IS26, which constitutes a bigger composite transposon together with the small one upstream. catA2 could be horizontally transferred via either the small composite transposon or the bigger one. The second segment is almost identical to the MRR in pHN7A8 and contains fosA3, blaCTX-M-65, blaTEM-1, and rmtB. However, the IS1294 adjacent to fosA3 in pKP1034 was interrupted by IS26, and homologous recombination (HR) occurred afterward between the newly inserted IS26 and the one located upstream of fosA3, inverting the intervening structure. The third segment carries blaKPC-2, blaSHV-12, and a mercury resistance operon, which is highly related to the MRR in pKPC-LK30. However, blaSHV-12 instead of blaSHV-11 is identified in pKP1034. blaSHV-12 is carried by an IS26-flanked composite transposon, as previously reported (24, 25). Nonetheless, the IS26 adjacent to Tn1721 was truncated, probably by Tn1721, at the same location as its counterpart in pKPC-LK30. The immediate genetic surroundings of blaKPC-2 originated from the typical Tn3-Tn4401 integration structure reported in China (6), and, like pKPC-LK30 and pKPHS2, Tn3 was truncated at the Tn3 tnpR as well, probably due to IS26-mediated deletion. So far, only two plasmids, pFOS18 and pHS102707, coharboring blaKPC-2 and fosA3 have been fully sequenced, and they were identified from K. pneumoniae and E. coli, respectively (6, 12). In both cases, fosA3 was acquired by two atypical plasmids carrying only blaKPC-2 via IS26-composite transposons embedding it, which is different from pKP1034 in our study.
FIG 2.
The MRR of plasmid pKP1034, indicated with bold type, compared with related MRRs of fosA3-positive plasmids pHN7A8 (JN232517.1) and pXZ (JF927996.1), blaKPC-2-positive plasmids pKPC-LK30 (KC405622.1) and pKP048 (FJ628167.2), and plasmid pHS102707 (KF701335.1) coharboring fosA3 and blaKPC-2. Various transposons and other modules have different shading and are generally labeled once for each plasmid. A pound sign (#) indicates that a transposon is incomplete. IS elements are pointed boxes labeled with the number/name. Tall bars represent the 38-bp inverted repeat (IR) of transposons, as indicated. Positions/orientations of selected resistance and other genes are indicated by arrows, generally labeled once for each plasmid. A, tnpA; R, tnpR. Gray triangles indicate the positions of IS4321 elements that have been removed for ease of comparison. Dashed lines represent plasmid backbone. The star inside a black circle indicates more MRR components of the plasmid pKP048 between the surrounding elements, including resistance genes mph2, armA, sul1, blaDHA-1, and qnrB4. The structure named R above pKP1034 is hypothetical and is intended to facilitate comparison between related MRRs. Paired squares represent flanking sequences that are reverse complementary to one another. Dashed arrowed lines indicate where homologous recombination (HR) happens to invert the region between two IS26 elements in opposite orientations.
A total of 11 copies of IS26 are dispersed through pKP1034, mainly in the MRR. Nevertheless, no target duplication repeats flanking IS26 are observed. This indicates that HR might play a key role in the generation of pKP1034. Detailed comparisons suggest that pKP1034 may have evolved from several recombination events of the two closely related plasmids pHN7A8 and pKPC-LK30. First, in plasmid pHN7A8, the composite transposon carrying catA2 and an extra copy of IS26 in the same orientation as the IS26 at the boundary of catA2 were inserted and truncated genes traI and traB, respectively. Second, HR between the two directly oriented IS26 elements occurred, deleting a circular molecule containing the intervenient structure plus one copy of IS26. Afterward, the excised circular molecule was inserted next to the IS26 downstream of blaSHV-11 in plasmid pKPC-LK30 via another HR event between IS26 elements. Last, insertion of an extra IS1 element between the repB gene and the resD gene in pKPC-LK30 followed by HR between the directly oriented IS1 elements led to generation of a pKP1034-like plasmid. The third and fourth recombination events could have occurred in either order.
In conclusion, we report a high fosfomycin resistance rate in KPC-KP strains in a Chinese university hospital, mainly driven by clonal spread of fosA3-carrying KPC-KP ST11 strains. pKP1034, a novel fosA3-carrying plasmid from an representative KPC-KP strain, is a multireplicon plasmid belonging to IncR-F33:A−: B−. Besides fosA3, it carries a variety of other resistance determinants, including blaKPC-2, rmtB, blaCTX-M-65, blaSHV-12, blaTEM-1, and catA2. It is highly mosaic and could evolve from an alarming recombination of the blaKPC-carrying plasmid pKPC-LK30 from Taiwan and the epidemic fosA3-bearing plasmid pHN7A8 reported in mainland China.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Scientific and Technological Major Progects of China (2013ZX10004-904 and 2014ZX10004-008).
We thank the team of curators of the Institut Pasteur MLST and genome databases for curating the data and making them publicly available at http://bigsdb.web.pasteur.fr.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01488-15.
REFERENCES
- 1.Pontikis K, Karaiskos I, Bastani S, Dimopoulos G, Kalogirou M, Katsiari M, Oikonomou A, Poulakou G, Roilides E, Giamarellou H. 2014. Outcomes of critically ill intensive care unit patients treated with fosfomycin for infections due to pandrug-resistant and extensively drug-resistant carbapenemase-producing Gram-negative bacteria. Int J Antimicrob Agents 43:52–59. doi: 10.1016/j.ijantimicag.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Sbrana F, Malacarne P, Viaggi B, Costanzo S, Leonetti P, Leonildi A, Casini B, Tascini C, Menichetti F. 2013. Carbapenem-sparing antibiotic regimens for infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae in intensive care unit. Clin Infect Dis 56:697–700. doi: 10.1093/cid/cis969. [DOI] [PubMed] [Google Scholar]
- 3.Kaase M, Szabados F, Anders A, Gatermann SG. 2014. Fosfomycin susceptibility in carbapenem-resistant Enterobacteriaceae from Germany. J Clin Microbiol 52:1893–1897. doi: 10.1128/JCM.03484-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porres-Osante N, Azcona-Gutierrez JM, Rojo-Bezares B, Undabeitia E, Torres C, Saenz Y. 2014. Emergence of a multiresistant KPC-3 and VIM-1 carbapenemase-producing Escherichia coli strain in Spain. J Antimicrob Chemother 69:1792–1795. doi: 10.1093/jac/dku055. [DOI] [PubMed] [Google Scholar]
- 5.Li JJ, Sheng ZK, Deng M, Bi S, Hu FS, Miao HF, Ji ZK, Sheng JF, Li LJ. 2012. Epidemic of Klebsiella pneumoniae ST11 clone coproducing KPC-2 and 16S rRNA methylase RmtB in a Chinese university hospital. BMC Infect Dis 12:373. doi: 10.1186/1471-2334-12-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang Y, Shen P, Wei Z, Liu L, He F, Shi K, Wang Y, Wang H, Yu Y. 2015. Dissemination of a clone carrying a fosA3-harboring plasmid mediates high fosfomycin resistance rate of KPC-producing Klebsiella pneumoniae in China. Int J Antimicrob Agents 45:66–70. doi: 10.1016/j.ijantimicag.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Wachino J, Yamane K, Suzuki S, Kimura K, Arakawa Y. 2010. Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob Agents Chemother 54:3061–3064. doi: 10.1128/AAC.01834-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho PL, Chan J, Lo WU, Law PY, Chow KH. 2013. Plasmid-mediated fosfomycin resistance in Escherichia coli isolated from pig. Vet Microbiol 162:964–967. doi: 10.1016/j.vetmic.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 9.Hou J, Huang X, Deng Y, He L, Yang T, Zeng Z, Chen Z, Liu JH. 2012. Dissemination of the fosfomycin resistance gene fosA3 with CTX-M beta-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob Agents Chemother 56:2135–2138. doi: 10.1128/AAC.05104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho PL, Chan J, Lo WU, Lai EL, Cheung YY, Lau TC, Chow KH. 2013. Prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes among blood and urinary Escherichia coli isolates. J Med Microbiol 62:1707–1713. doi: 10.1099/jmm.0.062653-0. [DOI] [PubMed] [Google Scholar]
- 11.Sato N, Kawamura K, Nakane K, Wachino J, Arakawa Y. 2013. First detection of fosfomycin resistance gene fosA3 in CTX-M-producing Escherichia coli isolates from healthy individuals in Japan. Microb Drug Resist 19:477–482. doi: 10.1089/mdr.2013.0061. [DOI] [PubMed] [Google Scholar]
- 12.Li G, Zhang Y, Bi D, Shen P, Ai F, Liu H, Tian Y, Ma Y, Wang B, Rajakumar K, Ou HY, Jiang X. 2015. First report of a clinical, multidrug resistant Enterobacteriaceae isolate coharboring fosfomycin resistance gene fosA3 and cabarpenemase gene blaKPC-2 on the same transposon, Tn1721. Antimicrob Agents Chemother 59:338–343. doi: 10.1128/AAC.03061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. 2011. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard— 8th ed CLSI M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 14.Arca P, Reguera G, Hardisson C. 1997. Plasmid-encoded fosfomycin resistance in bacteria isolated from the urinary tract in a multicenter survey. J Antimicrob Chemother 40:393–399. doi: 10.1093/jac/40.3.393. [DOI] [PubMed] [Google Scholar]
- 15.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, Decré D. 2013. wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol 51:4073–4078. doi: 10.1128/JCM.01924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He L, Partridge SR, Yang X, Hou J, Deng Y, Yao Q, Zeng Z, Chen Z, Liu JH. 2013. Complete nucleotide sequence of pHN7A8, an F33:A−: B− type epidemic plasmid carrying blaCTX-M-65, fosA3, and rmtB from China. J Antimicrob Chemother 68:46–50. doi: 10.1093/jac/dks369. [DOI] [PubMed] [Google Scholar]
- 18.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Fernández A, Fortini D, Veldman K, Mevius D, Carattoli A. 2009. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother 63:274–281. [DOI] [PubMed] [Google Scholar]
- 21.Chen YT, Lin JC, Fung CP, Lu PL, Chuang YC, Wu TL, Siu LK. 2014. KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother 69:628–631. doi: 10.1093/jac/dkt409. [DOI] [PubMed] [Google Scholar]
- 22.Chan J, Lo WU, Chow KH, Lai EL, Law PY, Ho PL. 2014. Clonal diversity of Escherichia coli isolates carrying plasmid-mediated fosfomycin resistance gene fosA3 from livestock and other animals. Antimicrob Agents Chemother 58:5638–5639. doi: 10.1128/AAC.02700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Liu W, Liu Y, Wang J, Lv L, Chen X, He D, Yang T, Hou J, Tan Y, Xing L, Zeng Z, Liu JH. 2014. F33: A−: B−, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and blaCTX-M-55/-14/-65 in Escherichia coli from chickens in China. Front Microbiol 5:688. doi: 10.3389/fmicb.2014.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Compain F, Frangeul L, Drieux L, Verdet C, Brisse S, Arlet G, Decre D. 2014. Complete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniae. Antimicrob Agents Chemother 58:4207–4210. doi: 10.1128/AAC.02773-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen YT, Liao TL, Liu YM, Lauderdale TL, Yan JJ, Tsai SF. 2009. Mobilization of qnrB2 and ISCR1 in plasmids. Antimicrob Agents Chemother 53:1235–1237. doi: 10.1128/AAC.00970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.