Abstract
Burkholderia pseudomallei is the etiologic agent of melioidosis, a difficult-to-treat disease with diverse clinical manifestations. β-Lactam antibiotics such as ceftazidime are crucial to the success of melioidosis therapy. Ceftazidime-resistant clinical isolates have been described, and the most common mechanism is point mutations affecting expression or critical amino acid residues of the chromosomally encoded class A PenA β-lactamase. We previously showed that PenA was exported via the twin arginine translocase system and associated with the spheroplast fraction. We now show that PenA is a membrane-bound lipoprotein. The protein and accompanying β-lactamase activity are found in the membrane fraction and can be extracted with Triton X-114. Treatment with globomycin of B. pseudomallei cells expressing PenA results in accumulation of the prolipoprotein. Mass spectrometric analysis of extracted membrane proteins reveals a protein peak whose mass is consistent with a triacylated PenA protein. Mutation of a crucial lipobox cysteine at position 23 to a serine residue results in loss of β-lactamase activity and absence of detectable PenAC23S protein. A concomitant isoleucine-to-alanine change at position 20 in the signal peptide processing site in the PenAC23S mutant results in a nonlipidated protein (PenAI20A C23S) that is processed by signal peptidase I and exhibits β-lactamase activity. The resistance profile of a B. pseudomallei strain expressing this protein is indistinguishable from the profile of the isogenic strain expressing wild-type PenA. The data show that PenA membrane association is not required for resistance and must serve another purpose.
INTRODUCTION
Burkholderia pseudomallei is a Gram-negative bacterium found in tropical and subtropical regions of the world and the etiologic agent of melioidosis (1). Melioidosis is an often fatal disease that is refractory to antibiotic therapy because of intrinsic resistance to many antimicrobials, including older β-lactam antibiotics such as ampicillin and carbenicillin (2–4). The introduction of ceftazidime in melioidosis therapy in the late 1980s halved mortality rates in patients with severe melioidosis (5). This cephalosporin remains the main antibiotic used for acute-phase therapy (4, 6). Although still rare, ceftazidime resistance as a result of therapy has been observed (7). Ceftazidime-resistant mutants are the consequence of mutations affecting the expression or amino acid sequence of chromosomally encoded class A PenA β-lactamase (8–12) and deletion of the ceftazidime target penicillin binding protein 3 (PBP3) BPSS1219 (13). However, PenA is the dominant player in ceftazidime resistance. Detailed structural and biochemical analyses of the soluble form of PenA (called PenI by R. A. Bonomo's group) provide key insights into the hydrolytic mechanisms of this β-lactamase and determinants of its substrate spectrum (14). The soluble form of PenA used in these studies is a truncated version expressed by a penA gene lacking the first 90 nucleotides, which encode the signal peptide (14). While ceftazidime is a poor substrate for wild-type PenA, point mutations causing amino acid substitutions in several of the Ambler domains result in an enzyme bestowing significant ceftazidime resistance. Previously documented amino acid changes implicated in ceftazidime resistance in clinical isolates include C69Y and P167S (8, 9, 11, 12), but other mutations continue to be identified. We previously demonstrated that PenA is secreted by the twin arginine translocation (TAT) system and that the enzyme was located in the spheroplastic fraction rather than the periplasmic fraction (10). This observation was consistent with an early B. pseudomallei β-lactamase study, which reported a membrane-associated chromosomal cephalosporinase, presumably PenA (15). In this study, we verified the location of PenA in the membrane fraction and examined the molecular factors governing PenA membrane association.
MATERIALS AND METHODS
Bacteria and culture conditions.
B. pseudomallei strains used in this study were Bp82 (16), Bp82.11 (Bp82 ΔpenA) (10), Bp82.22 (Bp82.11::mini-Tn7T-KM-Ptac-penA), Bp82.143 (Bp82 penAC23S [cysteine-to-serine change at position 23 encoded by penA]) and Bp82.188 (Bp82.143 penAI20A C23S). Bp82 is excluded from select-agent regulations (www.selectagents.gov/SelectAgentsandToxinsExclusions.html). All experiments with strain Bp82 and its derivatives were conducted at biosafety level 2 (BSL-2) with Institutional Biosafety Committee approval. Bacteria were routinely cultured in Lennox LB (MO BIO Laboratories, Carlsbad, CA) at 37°C with aeration. Bp82-derived strains were grown in media supplemented with 80 μg/ml adenine. Antibiotics were used at the following concentrations: 100 μg/ml ampicillin (Ap) and 35 μg/ml kanamycin (Km) for Escherichia coli and 1,000 μg/ml Km for B. pseudomallei. YT medium with 15% sucrose and 50 μg/ml 5-bromo-4-chloro-3-indoxyl-β-d-glucuronide (X-Gluc) (Gold Biotechnology, St. Louis, MO) was used for resolution of merodiploids during allelic exchange (17). Expression from the tac promoter was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Gold Biotechnology, St. Louis, MO).
MIC determination.
B. pseudomallei Bp82 and its derivatives were grown in cation-adjusted Mueller-Hinton II broth (MHB) (Becton Dickinson and Company, Sparks, MD) supplemented with 40 μg/ml adenine. The MICs of antibiotics were determined by the standard microdilution method, following CLSI guidelines (18).
Allele replacement.
Allelic replacement was performed using pEXKm5-based mutagenic plasmids, employing Km for merodiploid selection and sucrose counterselection for merodiploid resolution as previously described (17). For construction of strain Bp82.143 expressing PenAC23S, pPS3092 containing penA with a nucleotide 67 T-to-A change was created with mutagenic primer P2594 (5′-TGATCGGCGCCAGCGCGCCGCTG) and the QuikChange II kit (Agilent Technologies, Santa Clara, CA). Similarly, strain Bp82.188 expressing PenAI20A C23S was constructed using pPS3112, containing penA with nucleotide changes at positions 58 (A to G) and 59 (T to C) that collectively lead to the I20A substitution and the T-to-A change at position 67, which causes the C23S substitution. These nucleotide changes were obtained using mutagenic primers P2609 (5′-CCATTTCCACCCCATTGGCCGGCGCCAGCG) and P2610 (5′-CGCTGGCGCCGGCCAATGGGGTGGAAATGG). The mutagenic plasmids were introduced into strain Bp82.11 by electroporation, and successful allele replacement was monitored by PCR amplification of the penA gene and nucleotide sequencing using primers P1687 (5′-GGATCCGACGAGAGCTGATACGCTAG) and P1712 (5′-AAGCTTATACCGGCATCGTTTCGCTG).
Genomic mini-Tn7 insertion.
Mini-Tn7 constructs, e.g., the mini-Tn7-Ptac-penA expression vector, were inserted into the B. pseudomallei genome, and insertions were verified by PCR and sequencing using previously described methods (19). Strains with mini-Tn7 insertions at the glmS2-associated attTn7 site were chosen for consistency.
Globomycin treatment.
A crucial step in bacterial lipoprotein synthesis requires processing of the diacylated prolipoprotein by lipoprotein signal peptidase II (SPase II) (20). Globomycin is a specific inhibitor of SPase II (21), and inhibition of this enzyme results in accumulation of the uncleaved prolipoprotein (22). To assess possible effects of globomycin on posttranslational PenA modification, its expression in bacteria grown in LB medium supplemented with 80 μg/ml adenine was induced by adding IPTG to two 1-ml LB cultures of strain Bp82.22 at an optical density at 600 nm (OD600) of 0.6. One culture was treated with 250 μg globomycin (10 mg/ml in dimethyl sulfoxide [DMSO] [Sigma-Aldrich, St, Louis, MO]), and the other culture was treated with an equal volume of DMSO only for a control. After incubation for 1 h at 37°C, cells from 0.5 ml of the cultures were pelleted by centrifugation in a microcentrifuge and resuspended in Laemmli sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (Bio-Rad Laboratories, Hercules, CA).
Extraction of periplasmic proteins.
Although we attempted several procedures for extraction of periplasmic proteins, the chloroform method (23) was in our hands most efficient and reliable (see additional experimental details and Fig. S1 in the supplemental material). Cells were collected from 2-ml samples from overnight cultures grown in LB medium by centrifugation for 30 s at 14,000 × g at room temperature. Pellets were resuspended in approximately 20 μl of residual medium, 20 μl of chloroform was added, and the cell suspensions were vortexed periodically while incubating at room temperature for 15 min. Two hundred microliters of 10 mM Tris (pH 8) was added, followed by centrifugation at 6,000 × g for 20 min at room temperature. The top, aqueous layers were removed as periplasmic fractions and stored at −20°C.
Membrane fractionation.
For preparation of total membrane fractions, bacteria were grown overnight in 10 ml LB (strain Bp82.11) or LB in the presence of 1 mM IPTG to induce penA expression (strain Bp82.22). Cells were collected by centrifugation at 2,500 × g for 10 min at 4°C and resuspended in 5 ml distilled H2O (diH2O). The cells were then lysed by sonication on ice by using a Sonics Vibracell VC750 sonicator (Newtown, CT) at 30% amplitude. The cells were lysed by four cycles of sonication, with one cycle consisting of 1-s pulses for 1 min followed by a 30-s pause. Cell debris was pelleted by centrifugation at 2,500 × g for 10 min at 4°C. Supernatants were transferred to new tubes, and centrifugation was repeated. Membranes were pelleted by centrifugation for 1 h at 154,000 × g at 4°C in a Sorvall WX Ultra 100 ultracentrifuge (Thermo Fisher Scientific, Waltham, MA), using a TH-641 swinging bucket rotor. Supernatants containing the soluble fractions (cytoplasm and periplasm) were moved to new tubes, and the membrane pellets were resuspended in 1 ml of diH2O and frozen at −20°C.
Phase separation of membrane proteins using Triton X-114.
The procedure used for phase separation of membrane proteins was adapted from the method of Radolf et al. (24). Briefly, total membranes were prepared as described above, but instead of water, membrane pellets were resuspended in 150 μl of 4% Triton X-114 (TX-114) in phosphate-buffered saline (PBS), transferred to microcentrifuge tubes, rocked overnight at 4°C, and centrifuged for 1 h at 25,000 × g and 4°C. Supernatants were moved to new tubes and stored at 4°C, while pellets were resuspended in 150 μl of 4% TX-114 in PBS, rocked for 1 h at 4°C, and centrifuged as described above. Supernatants were combined with those obtained from the overnight extraction, incubated at 37°C for 1 h with occasional rocking, and centrifuged for 1 h at 25,000 × g and 37°C. The upper (aqueous) phases were removed, and an equal volume of ice-cold PBS was added to return the TX-114 concentration to 4%. The 37°C incubation and centrifugation steps were repeated twice. Nine volumes of ice-cold acetone was added to the final detergent phases, and the mixtures were incubated at −20°C overnight. Acetone precipitates were centrifuged for 1 h at 25,000 × g at 4°C, supernatants were discarded, pellets were resuspended in 150 μl ice cold acetone, and centrifugation was repeated. Pellets were air dried completely, resuspended in 200 μl distilled H2O, and frozen at −20°C.
MALDI-TOF mass spectrometry.
For matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry, 1-ml membrane samples in distilled H2O were washed once with 8 volumes of ice-cold acetone and incubated at −20°C overnight, and debris was pelleted by centrifugation (27,000 × g for 1 h at 4°C). Pellets were air dried and resuspended in 50 μl of 50% acetonitrile and 0.1% trifluoroacetic acid. One microliter of each sample was mixed with 1 μl of 10-mg/ml sinapinic acid matrix on the MALDI-TOF mass spectrometry target plate and allowed to air dry overnight. The samples were then analyzed using a Bruker Ultraflex II MALDI-TOF/TOF mass spectrometer and Flex Analysis software (Bruker Daltonics Inc., Billerica, MA).
Protein quantification.
The protein content of samples was quantified using the Pierce bicinchoninic acid (BCA) protein assay reagent kit (Thermo Fisher Scientific, Waltham, MA) and microtiter plates, using bovine serum albumin (BSA) standards and following the manufacturer's instructions. The A570 of each well was read using a Multiskan Spectrum plate reader (Thermo Fisher Scientific, Waltham, MA). Protein concentrations were calculated using Revelation data processing software (Dynex Technologies, Chantilly, VA).
Western blot analysis.
Cellular fractions were diluted 1:1 in 2× Laemmli sample buffer (Bio-Rad Laboratories, Hercules, CA), boiled for 10 min, cooled, and separated on 12% polyacrylamide Tris-glycine SDS-polyacrylamide gels (Bio-Rad Laboratories) for approximately 1.5 h at 200 V. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes using a Bio-Rad Trans-Blot SD semidry cell (Bio-Rad Laboratories, Hercules, CA). Membranes were blocked with 4% BSA in Tris-buffered saline with Tween 20 (TBS-T) (20 mM Tris, 500 mM NaCl, 0.05% Tween 20 [pH 7.5]) rocking for 1 h at room temperature or overnight at 4°C. Blots were washed twice for 5 min each time with water and stained with primary antibody (e.g., rabbit polyclonal anti-PenA [10]) diluted in blocking solution for 1 h with rocking at room temperature. Blots were washed three times for 5 min each time with TBS-T and then incubated with secondary antibody (e.g., goat polyclonal anti-rabbit IgG conjugated to horseradish peroxidase [HRP] [Promega, Madison, WI]) for 30 min with rocking at room temperature. Blots were washed three times with TBS-T and twice with water (5 min per wash). One milliliter of Novex ECL enhanced chemiluminescent substrate (Life Technologies, Carlsbad, CA) was added, and the mixture was rocked for 1 min. Blots were imaged on a ChemiDoc XRS system (Bio-Rad Laboratories, Hercules, CA).
β-Lactamase activity assay.
The presence of β-lactamase activity in samples was detected using the colorimetric substrate nitrocefin. In microtiter plates, 200 μl of 50-μg/ml nitrocefin (TOKU-E, Bellingham, WA) in 100 mM sodium phosphate buffer (pH 7) was added to 20 μl of cellular fractions, mixed by pipette, and incubated for 30 min at 37°C. A486 was read with a Multiskan Spectrum plate reader (Thermo Fisher Scientific, Waltham, MA), and β-lactamase activity from duplicate sample wells was calculated as follows: (average sample A486 − average blank A486)/microgram of protein. Each assay was repeated in three separate technical replicates.
RESULTS AND DISCUSSION
PenA is localized in the membrane fraction.
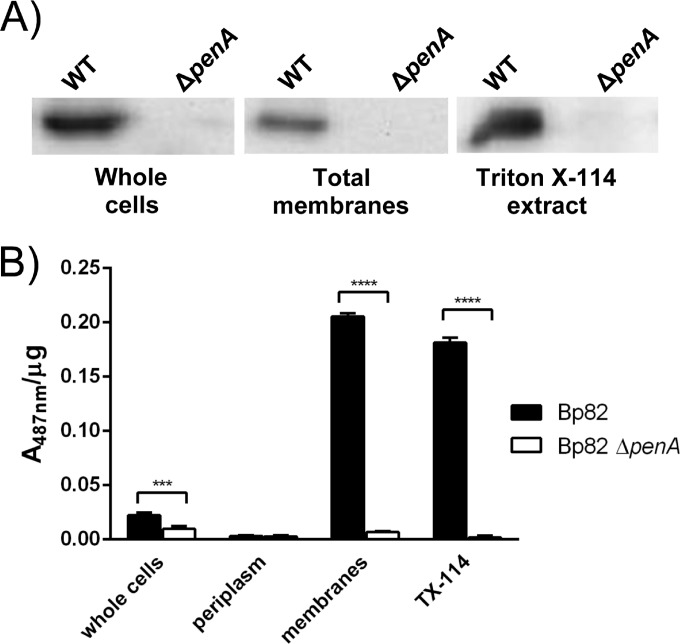
Total membranes and chloroform-extracted periplasmic fractions were isolated and analyzed for the presence of PenA protein and β-lactamase activity. Western blot analysis of whole-cell and total membrane fractions with anti-PenA antibodies revealed the presence of PenA in both fractions from strain Bp82, but not its ΔpenA derivative (Fig. 1A). Only very small amounts of PenA protein could be detected in the periplasmic fraction (see Fig. S2 in the supplemental material). PenA β-lactamase activity paralleled PenA protein localization with the highest specific activity found in the total membrane fraction. Lower amounts of specific β-lactamase activity were found in whole cells, only spurious amounts were found in the periplasmic fraction, and no activity in any of the fractions derived from the ΔpenA mutant (Fig. 1B). Both PenA protein and its β-lactamase activity could be readily extracted from the membrane fraction by Triton X-114, a nonionic detergent with a low cloud point (25). Triton X-114 has been used to enrich for lipoproteins from a variety of bacterial species (24, 26–29). Following a temperature-based TX-114 phase separation, lipoproteins, as well as other lipophilic proteins, are found in the detergent phase, while hydrophilic proteins partition to the aqueous phase. While separation of a protein to the TX-114 detergent phase cannot definitively identify it as a lipoprotein, this hydrophobic behavior supports the model of PenA as a membrane-bound lipoprotein. Attempts were therefore made to assess whether PenA membrane association is a result of posttranslational lipid modification.
FIG 1.
PenA is a membrane protein. (A) Western blots of whole cells, total membranes and Triton X-114 extracts using anti-PenA antibodies. The bacteria analyzed were the wild-type (WT) Bp82 strain and its isogenic ΔpenA derivative. (B) PenA β-lactamase activity in cellular fractions. Nitrocefin assays were performed with the indicated cellular fractions. Specific activity is calculated as A487 per microgram of protein. Values are shown as means plus standard errors of the means (error bars). Values that are statistically significantly different are indicated by bars and asterisks as follows: ***, P ≤ 0.001; ****, P ≤ 0.0001.
PenA is a lipoprotein. (i) Bioinformatic evidence.
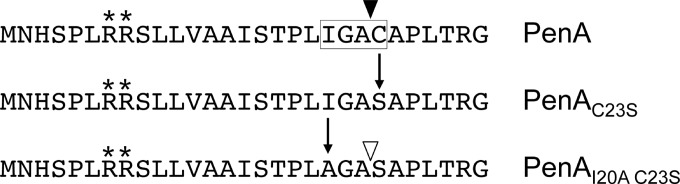
Bacterial lipoproteins contain lipobox motifs that mediate lipid modification and signal peptide cleavage, allowing for anchoring to the inner or outer membrane (20). The online LipoP software predicts whether prokaryotic protein sequences contain lipoprotein signal peptides and identifies any lipoprotein signal peptidase II (SPase II) cleavage sites (30). According to LipoP, PenA is likely a lipoprotein, with SPase II cleaving between amino acids 22 (alanine) and 23 (cysteine) (Fig. 2). The alanine and the essential cysteine are part of a noncanonical lipobox, which is modified by covalent attachment of a diacylglycerol moiety to the thiol group on the cysteine side chain in thioether linkage, resulting in a prolipoprotein (20). After lipidation, SPase II cleaves the signal peptide, leaving the modified cysteine as the new amino-terminal residue, which in Gram-negative bacteria is then further modified by the addition of a fatty acid in amide linkage (20). The bioinformatic analysis suggests that it is highly probable that PenA is a lipoprotein and that the posttranslational lipid modification occurs at cysteine 23 (C23).
FIG 2.
Amino-terminal PenA sequences. The 29 amino acids of the wild-type PenA and its mutant derivatives PenAC23S and PenAI20A C23S are shown. Key features include the twin arginine residues (marked with asterisks), of which arginine 8 is essential for secretion of PenA by the twin arginine transport system (10). The amino acids that constitute the lipobox, including the essential cysteine, are boxed. The vertical arrows mark amino acid changes introduced by successive site-directed mutagenesis steps to arrive at the mutant proteins PenAC23S and PenAI20A C23S, respectively. The open and closed triangles mark signal peptidase I and II cleavage sites, respectively.
(ii) Biochemical evidence.
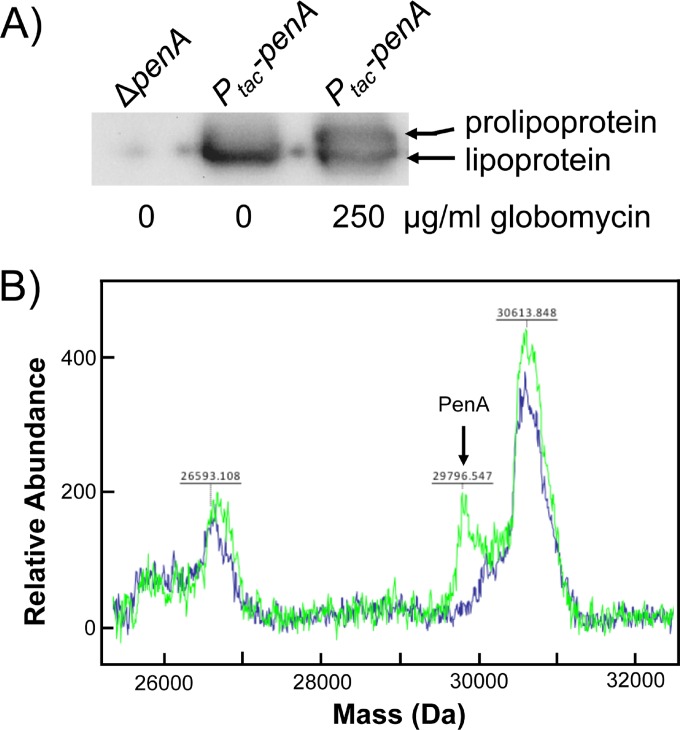
We first used biochemical approaches to obtain evidence for PenA being a lipoprotein. Globomycin is an inhibitor of SPase II (21) and has been used extensively to study bacterial lipoprotein maturation because SPase II inhibition results in accumulation of the uncleaved prolipoprotein. The size difference between this full-length prolipoprotein and the mature lipoprotein lacking the signal sequence can be visualized by SDS-PAGE. Analyses of total cell extracts from untreated cells expressing PenA revealed a single band (Fig. 3A, middle lane). In contrast, globomycin treatment resulted in accumulation of a second higher-molecular-weight band in addition to the one seen in untreated cells, consistent with the presence of a prolipoprotein (Fig. 3A, rightmost lane). The presence of the mature lipoprotein band in addition to the prolipoprotein is frequently observed (29, 31), presumably because either inhibition of SPase II by globomycin is incomplete (22) or mature lipoprotein is still present from before treatment. We next used MALDI-TOF mass spectrometry to obtain evidence for mature PenA being a triacylated lipoprotein. Total membrane fractions from B. pseudomallei Bp82 ΔpenA and Bp82 ΔpenA expressing wild-type penA from the IPTG-inducible tac promoter were isolated, concentrated, and analyzed by MALDI-TOF mass spectrometry (Fig. 3B). Two separate runs revealed a lone difference between the two spectra, a peak with a molecular mass of between 29,790 and 29,800 Da. Assuming a calculated molecular mass of ∼29,800 Da for a triacylated PenA (29,016 Da for processed PenA amino acids [C23 to carboxy terminus], ∼550 Da for diacylglycerol, and ∼250 Da for the amide-linked fatty acid), the experimentally determined value provides solid evidence for mature PenA being a lipoprotein. To our knowledge, there are only two other documented examples of Gram-negative lipoprotein β-lactamase enzymes: BRO-1 (32) and NDM-1 (33, 34).
FIG 3.
PenA is a lipoprotein. (A) Inhibition of signal peptidase II with globomycin results in accumulation of PenA prolipoprotein. Whole cells of strain Bp82 ΔpenA::mini-Tn7-Km-Ptac-penA (Ptac-penA) induced with IPTG, with or without treatment with 250 μg/ml globomycin, were analyzed by Western blotting using anti-PenA antibodies. Untreated whole cells of strain Bp82 ΔpenA were included as a control. The positions of bands corresponding to prolipoprotein and lipoprotein are marked. (B) The molecular mass of PenA is consistent with it being a lipoprotein. Mass spectrometric analysis of membrane proteins extracted from strains Bp82 ΔpenA (blue trace) and Bp82 ΔpenA::mini-Tn7-Km-Ptac-penA induced with IPTG (green trace) reveals a peak whose mass is consistent with a triacylated PenA protein.
(iii) Genetic evidence.
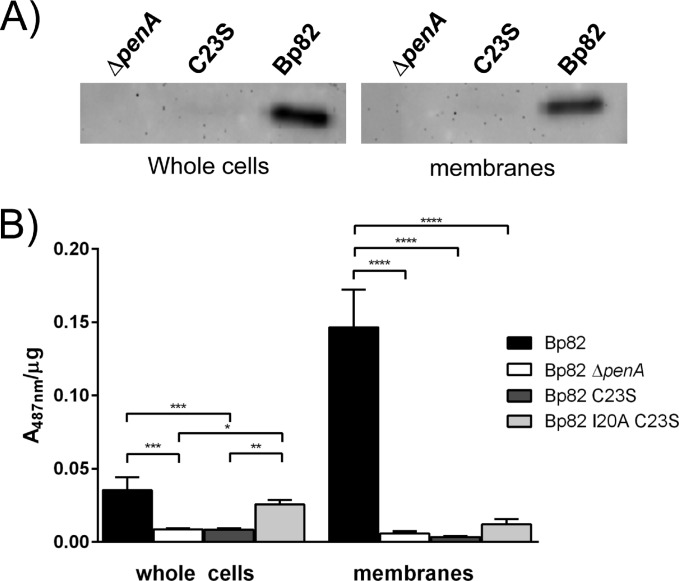
To obtain further evidence for PenA being a lipoprotein, we changed the essential cysteine in the lipobox to a serine, which resulted in the PenAC23S variant (Fig. 2). To our surprise, PenA protein in the Bp82 penAC23S strain was not detected in whole cells or in total membranes (Fig. 4A), although penA transcript levels in the strain expressing either wild-type PenA or PenAC23S were indistinguishable (see Fig. S3 in the supplemental material) and the desired penAC23S mutant allele was present. The absence of PenAC23S was accompanied by the absence of PenA β-lactamase activity in whole cells and total membranes as assessed by enzyme assay (Fig. 4B) and β-lactam susceptibility profile (Table 1). This apparent degradation of the PenAC23S mutant protein indicates that PenA requires lipidation for stability and functionality. It has previously been shown that a NDM-1C26A nonlipidated variant is more prone to proteolysis in the periplasm (34), and it is conceivable that the same is true for nonlipidated PenA.
FIG 4.
Mutations of lipobox residues alter PenA stability and activity. (A) The absence of lipid modification results in the loss of detected PenA. Whole-cell extracts and total membranes of strain Bp82 (wild-type penA), Bp82 ΔpenA, and Bp82 penAC23S were analyzed by Western blotting using anti-PenA antibodies. PenA can be readily detected in strain Bp82, but not in mutants lacking PenA or expressing PenAC23S. (B) The absence of lipid modification results in the loss of PenA β-lactamase activity which can be restored by introduction of a signal peptidase I cleavage site. Nitrocefin assays were performed using whole cells and total membrane fractions of Bp82 (wild-type penA), Bp82 ΔpenA, Bp82 penAC23S, and Bp82 penAI20AC23S. Specific activity is calculated as A487 per microgram of protein. Values are shown as means plus standard errors of the means (error bars). Values that are statistically significantly different are indicated by bars and asterisks as follows: *, P ≤ 0.05, **; P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
TABLE 1.
β-Lactam susceptibility of strains expressing PenA and its mutant derivatives
| Strain | MIC (μg/ml)a |
|||
|---|---|---|---|---|
| Ampicillin | Carbenicillin | Ceftazidime | Imipenem | |
| Bp82 | 128 | 1,024 | 4 | 1 |
| Bp82 ΔpenA | 8 | 16 | 1 | 0.25 |
| Bp82 penAC23S | 16 | 128 | 1b | 0.25 |
| Bp82 penAI20A C23S | 256 | 1,024 | 4 | 1 |
MICs obtained were determined by the broth microdilution method unless indicated otherwise.
This value was obtained by the agar dilution method.
Membrane association of PenA is not required for activity and substrate profile.
To test whether membrane association is required for PenA β-lactamase activity and perhaps substrate specificity, we had hoped that PenAC23S would still be secreted via the TAT system and nonlipidated but processed by SPase I, rather than the lipoprotein processing system and SPase II. Although the C23S change resulted in an ASA that corresponds to the AXA consensus sequence preceding SPase I cleavage sites (AXA↓) (35, 36), the absence of any detectable protein or β-lactamase activity in whole cells indicated that PenAC23S was not processed into a stable form by SPase I. We therefore decided to engineer a new SPase I cleavage site immediately upstream of the S23 by changing the isoleucine at position 20 to an alanine, resulting in an AGA motif (Fig. 2). The resulting PenAI20A C23S exhibited β-lactamase activity (Fig. 4B) and restored β-lactam resistance to levels observed with the wild-type Bp82 PenA strain (Table 1). While PenAI20A C23S β-lactamase activity in whole cells was comparable to that of wild-type PenA, no significant PenAI20A C23S activity was observed in the membrane fraction. Like the wild-type PenA, PenAI20A C23S was primarily active against penicillins and displayed very little activity against ceftazidime and imipenem. These data show that membrane localization is not required for β-lactamase activity and not a determinant of substrate specificity. They are consistent with previous findings that showed hydrolysis of β-lactams by a purified soluble form of PenA lacking the first 90 nucleotides, which encode its TAT secretion signal and the lipobox (14).
Conclusions.
Our results confirm and provide a molecular explanation for the findings of an early study of B. pseudomallei β-lactamase, which reported a membrane-associated chromosomal cephalosporinase, most likely PenA (15). At the onset of our studies, Moraxella (Branhamella) catarrhalis BRO-1 was the only known Gram-negative lipoprotein β-lactamase (32). Recently, it was shown that NDM-1 carbapenemase is also a lipoprotein, which when expressed in E. coli is localized in the outer membrane (33, 34). PenA has an alanine rather than aspartic acid at position +2 from the lipidation signal cleavage site and is thus expected to localize to the outer membrane rather than the inner membrane (37, 38). The functional role of the membrane localization of these enzymes remains largely uncharacterized. Our data show that membrane association is not essential for enzyme activity and does not influence substrate specificity. It must therefore serve another purpose. A recent conference abstract and poster provided evidence that NDM-1 was shed in outer membrane vesicles (OMVs) that displayed carbapenemase activity (34). Incubation of these OMVs with NDM-1 naive E. coli conferred increased imipenem resistance to these cells (34). Secretion of NDM-1 thus constitutes a novel resistance dissemination mechanism, which confers phenotypic rather than genetic resistance on susceptible recipient bacteria (34). We hypothesize that membrane anchoring of B. pseudomallei PenA may serve a similar purpose. For instance, one could envision development of phenotypic resistance in a localized cell population by transforming neighboring B. pseudomallei cells expressing wild-type PenA (inactive on the antibiotic ceftazidime) by fusion with OMVs containing mutated PenA conferring ceftazidime resistance. The PenA lipidation signal is conserved in most sequenced Burkholderia species, e.g., the animal or plant pathogens B. ambifaria, B. cenocepacia, B. dolosa, B. gladioli, B. glumae, B. mallei, B. multivorans, B. thailandensis, and B. vietnamensis, and absent from only a few Burkholderia species, including the plant symbionts B. phymatum and B. phytofirmans (39). The conservation of a lipidation signal and outer membrane localization in clinically significant β-lactamases such as NDM-1 and PenA opens avenues of research, contributing new knowledge in terms of antibiotic resistance and therapeutic development.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH NIAID grant AI065357 to the Rocky Mountain Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research at Colorado State University.
We thank Sheila Nathan from the National University of Malaysia for providing anti-Omp85 antibodies.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02444-15.
REFERENCES
- 1.Wiersinga WJ, Currie BJ, Peacock SJ. 2012. Melioidosis. N Engl J Med 367:1035–1044. doi: 10.1056/NEJMra1204699. [DOI] [PubMed] [Google Scholar]
- 2.Wuthiekanun V, Peacock SJ. 2006. Management of melioidosis. Expert Rev Anti Infect Ther 4:445–455. doi: 10.1586/14787210.4.3.445. [DOI] [PubMed] [Google Scholar]
- 3.Schweizer HP. 2012. Mechanisms of antibiotic resistance in Burkholderia pseudomallei: implications for treatment of melioidosis. Future Microbiol 7:1389–1399. doi: 10.2217/fmb.12.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dance D. 2014. Treatment and prophylaxis of melioidosis. Int J Antimicrob Agents 43:310–318. doi: 10.1016/j.ijantimicag.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White NJ, Dance DA, Chaowagul W, Wattanagoon Y, Wuthiekanun V, Pitakwatchara N. 1989. Halving of mortality of severe melioidosis by ceftazidime. Lancet ii:697–701. [DOI] [PubMed] [Google Scholar]
- 6.Lipsitz R, Garges S, Aurigemma R, Baccam P, Blaney DD, Cheng AC, Currie BJ, Dance D, Gee JE, Larsen J, Limmathurotsakul D, Morrow MG, Norton R, O'Mara E, Peacock SJ, Pesik N, Rogers LP, Schweizer HP, Steinmetz I, Tan G, Tan P, Wiersinga WJ, Wuthiekanun V, Smith TL. 2012. Workshop on treatment of and postexposure prophylaxis for Burkholderia pseudomallei and B. mallei infection, 2010. Emerg Infect Dis 18:e2. doi: 10.3201/eid1812.120638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wuthiekanun V, Amornchai P, Saiprom N, Chantratita N, Chierakul W, Koh GC, Chaowagul W, Day NP, Limmathurotsakul D, Peacock SJ. 2011. Survey of antimicrobial resistance in clinical Burkholderia pseudomallei isolates over two decades in Northeast Thailand. Antimicrob Agents Chemother 55:5388–5391. doi: 10.1128/AAC.05517-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tribuddharat C, Moore RA, Baker P, Woods DE. 2003. Burkholderia pseudomallei class A beta-lactamase mutations that confer selective resistance against ceftazidime or clavulanic acid inhibition. Antimicrob Agents Chemother 47:2082–2087. doi: 10.1128/AAC.47.7.2082-2087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sam IC, See KH, Puthucheary SD. 2009. Variations in ceftazidime and amoxicillin-clavulanate susceptibilities within a clonal infection of Burkholderia pseudomallei. J Clin Microbiol 47:1556–1558. doi: 10.1128/JCM.01657-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rholl DA, Papp-Wallace KM, Tomaras AP, Vasil ML, Bonomo RA, Schweizer HP. 2011. Molecular investigations of PenA-mediated beta-lactam resistance in Burkholderia pseudomallei. Front Microbiol 2:139. doi: 10.3389/fmicb.2011.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarovich DS, Price EP, Von Schulze AT, Cook JM, Mayo M, Watson LM, Richardson L, Seymour ML, Tuanyok A, Engelthaler DM, Pearson T, Peacock SJ, Currie BJ, Keim P, Wagner DM. 2012. Characterization of ceftazidime resistance mechanisms in clinical isolates of Burkholderia pseudomallei from Australia. PLoS One 7:e30789. doi: 10.1371/journal.pone.0030789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarovich DS, Price EP, Limmathurotsakul D, Cook JM, Von Schulze AT, Wolken SR, Keim P, Peacock SJ, Pearson T. 2012. Development of ceftazidime resistance in an acute Burkholderia pseudomallei infection. Infect Drug Resist 5:129–132. doi: 10.2147/IDR.S35529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chantratita N, Rholl DA, Sim B, Wuthiekanun V, Limmathurotsakul D, Amornchai P, Thanwisai A, Chua HH, Ooi WF, Holden MTG, Day NP, Tan P, Schweizer HP, Peacock SJ. 2011. Antimicrobial resistance to ceftazidime involving loss of penicillin-binding protein 3 in Burkholderia pseudomallei. Proc Natl Acad Sci U S A 108:17165–17170. doi: 10.1073/pnas.1111020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papp-Wallace KM, Taracila MA, Gatta JA, Ohuchi N, Bonomo RA, Nukaga M. 2013. Insights into beta-lactamases from Burkholderia species, two phylogenetically related yet distinct resistance determinants. J Biol Chem 288:19090–19102. doi: 10.1074/jbc.M113.458315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livermore DM, Chau PY, Wong AI, Leung YK. 1987. Beta-lactamase of Pseudomonas pseudomallei and its contribution to antibiotic resistance. J Antimicrob Chemother 20:313–321. doi: 10.1093/jac/20.3.313. [DOI] [PubMed] [Google Scholar]
- 16.Propst KL, Mima T, Choi KH, Dow SW, Schweizer HP. 2010. A Burkholderia pseudomallei ΔpurM mutant is avirulent in immunocompetent and immunodeficient animals: candidate strain for exclusion from select-agent lists. Infect Immun 78:3136–3143. doi: 10.1128/IAI.01313-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez CM, Rholl DA, Trunck LA, Schweizer HP. 2009. Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Appl Environ Microbiol 75:6496–6503. doi: 10.1128/AEM.01669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing: twenty-fifth informal supplement M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.Choi K-H, Mima T, Casart Y, Rholl D, Kumar A, Beacham IR, Schweizer HP. 2008. Genetic tools for select agent compliant manipulation of Burkholderia pseudomallei. Appl Environ Microbiol 74:1064–1075. doi: 10.1128/AEM.02430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovacs-Simon A, Titball RW, Michell SL. 2011. Lipoproteins of bacterial pathogens. Infect Immun 79:548–561. doi: 10.1128/IAI.00682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dev IK, Harvey RJ, Ray PH. 1985. Inhibition of prolipoprotein signal peptidase by globomycin. J Biol Chem 260:5891–5894. [PubMed] [Google Scholar]
- 22.Hussain M, Ichihara S, Mizushima S. 1980. Accumulation of glyceride-containing precursor of the outer membrane lipoprotein in the cytoplasmic membrane of Escherichia coli treated with globomycin. J Biol Chem 255:3707–3712. [PubMed] [Google Scholar]
- 23.Ames GF-L, Prody C, Kustu S. 1984. Simple, rapid, and quantitative release of periplasmic proteins by chloroform. J Bacteriol 160:1181–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radolf JD, Chamberlain NR, Clausell A, Norgard MV. 1988. Identification and localization of integral membrane proteins of virulent Treponema pallidum subsp. pallidum by phase partitioning with the nonionic detergent Triton X-114. Infect Immun 56:490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bordier C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem 256:1604–1607. [PubMed] [Google Scholar]
- 26.Sjostedt A, Tarnvik A, Sandstrom G. 1991. The T-cell-stimulating 17-kilodalton protein of Francisella tularensis LVS is a lipoprotein. Infect Immun 59:3163–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janson H, Heden LO, Forsgren A. 1992. Protein D, the immunoglobulin D-binding protein of Haemophilus influenzae, is a lipoprotein. Infect Immun 60:1336–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tibor A, Decelle B, Letesson JJ. 1999. Outer membrane proteins Omp10, Omp16, and Omp19 of Brucella spp. are lipoproteins. Infect Immun 67:4960–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maeda Y, Makino M, Crick DC, Mahapatra S, Srisungnam S, Takii T, Kashiwabara Y, Brennan PJ. 2002. Novel 33-kilodalton lipoprotein from Mycobacterium leprae. Infect Immun 70:4106–4111. doi: 10.1128/IAI.70.8.4106-4111.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valente FM, Pereira PM, Venceslau SS, Regalla M, Coelho AV, Pereira IA. 2007. The [NiFeSe] hydrogenase from Desulfovibrio vulgaris Hildenborough is a bacterial lipoprotein lacking a typical lipoprotein signal peptide. FEBS Lett 581:3341–3344. doi: 10.1016/j.febslet.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 32.Bootsma HJ, Aerts PC, Posthuma G, Harmsen T, Verhoef J, van Dijk H, Mooi FR. 1999. Moraxella (Branhamella) catarrhalis BRO β-lactamase: a lipoprotein of Gram-positive origin? J Bacteriol 181:5090–5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King D, Strynadka N. 2011. Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance. Protein Sci 20:1484–1491. doi: 10.1002/pro.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez L, Bahr G, Bonomo RA, Vila A. 2014. Membrane anchoring of carbapenemase Ndm-1 favors protein stability and resistance transfer, abstr C162c. Abstr 54th Intersci Conf Antimicrob Agents Chemother. American Society for Microbiology, Washington, DC. [Google Scholar]
- 35.Paetzel M, Karla A, Strynadka NC, Dalbey RE. 2002. Signal peptidases. Chem Rev 102:4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 36.Tuteja R. 2005. Type I signal peptidase: an overview. Arch Biochem Biophys 441:107–111. doi: 10.1016/j.abb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 37.Ferrandez Y, Condemine G. 2008. Novel mechanism of outer membrane targeting of proteins in Gram-negative bacteria. Mol Microbiol 69:1349–1357. doi: 10.1111/j.1365-2958.2008.06366.x. [DOI] [PubMed] [Google Scholar]
- 38.Viarre V, Cascales E, Ball G, Michel GP, Filloux A, Voulhoux R. 2009. HxcQ liposecretin is self-piloted to the outer membrane by its N-terminal lipid anchor. J Biol Chem 284:33815–33823. doi: 10.1074/jbc.M109.065938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FSL. 2008. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics 24:2803–2804. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.