Abstract
Hepatitis E virus (HEV) can lead to chronic infection in solid-organ transplant patients. Ribavirin is efficient for treatment of chronically infected patients. Recently, the1634R mutation in the HEV polymerase has been associated with treatment failure. However, it is unclear if this mutation can be used as a prognostic marker of treatment outcome. We studied the prevalence of the 1634R mutation in the HEV polymerase of patients starting ribavirin therapy, the influence of the 1634R variants on the viral response, the frequency of the 1634R mutation in patients whose treatment failed, and its impact on ribavirin retreatment. We analyzed pretreatment samples from 63 solid-organ transplant patients with chronic hepatitis E using deep sequencing; 42 patients had a sustained virologic response (SVR), and 21 were non-SVR patients. We detected the 1634R variant by deep sequencing in 36.5% (23/63) of the patients (proportions, 1.3 to 100%). The 1634R variant was detected in 31.0% (13/42) of baseline plasma samples from patients with SVR and in 47.6% (10/21) in the other patients (P = 0.2). The presence of this mutation did not influence the initial decrease in viral RNA. Lastly, a second prolonged ribavirin treatment led to SVR in 70% of the patients who initially did not have SVR, despite the presence of the 1634R variant. We conclude that the presence of the 1634R variant at ribavirin initiation does not lead to absolute ribavirin resistance. Although its proportion increased in patients whose treatment failed, the presence of the 1634R variant did not compromise the response to a second ribavirin treatment.
INTRODUCTION
Hepatitis E virus (HEV) is one of the most common causes of acute hepatitis worldwide. HEV belongs to the Hepeviridae family. It is a small, unenveloped virus with a positive-sense, single-stranded, 7.2-kb-long RNA genome. It contains 3 open reading frames (ORF).ORF1 encodes a nonstructural protein of about 1,693 amino acids (aa) with at least four putative functional domains: methyltransferase, papain-like cysteine protease (PCP), helicase, and RNA-dependent RNA polymerase (RdRp). It also has domains that are homologous to those of other plant and animal positive-strand RNA viruses: the Y domain, the polyproline region (PPR), and a macro domain. ORF2 encodes the capsid protein and ORF3 a protein involved in virus egress (1). Strains infecting humans are classified into 4 major genotypes, HEV1 to HEV4. These genotypes belong to the genus Orthohepevirus (2). HEV1 and HEV2 infect only humans and are responsible for waterborne outbreaks in developing countries. HEV3 and HEV4 are transmitted zoonotically from animal reservoirs and cause sporadic cases in developed countries (3). Domestic pigs represent the major animal reservoir. Zoonotic transmission is due to consumption of uncooked or undercooked infected pork or game (wild boar or deer) meat (4). Rabbit strains that are close to HEV3 have recently been described in both rabbits and humans (5). Direct transmission after contact with HEV-infected animals also seems possible (3). Finally, transfusion-transmitted HEV infections have been reported (6).
Although most HEV infections are asymptomatic, HEV can cause acute hepatitis with severe forms in patients with preexisting liver disease and in pregnant women in developing countries (7). Extrahepatic manifestations such as hematological manifestations (8–10), acute pancreatitis (11), kidney injuries (12), or neurological manifestations can also occur (13–15).
HEV3 infections can become chronic and progress rapidly to cirrhosis in immunocompromised patients such as patients with human immunodeficiency virus infection (16), patient with hematologic cancers receiving chemotherapy (17), and solid-organ transplant recipients (18, 19). For solid-organ transplant patients with chronic hepatitis E, reducing immunosuppressive drug treatment, especially immunosuppressants that target T cells, is the first-line therapy (20). If this fails, the treatment of choice for most patients is ribavirin monotherapy for a mean duration of 3 months (21, 22). Although the sustained virological response (SVR) rate was 78% in a multicenter study, treatment failure did occur, either as a partial response to ribavirin or as viral recurrence after therapy cessation (23). A G1634R mutation in the C-terminal region of the HEV polymerase was found recently in 2 solid-organ transplant patients who failed to clear HEV after ribavirin treatment (24). In vitro experiments using a subgenomic replicon, infectious virus, and competition assays indicated that the 1634R variant of HEV3 replicates more efficiently than HEV3 lacking this mutation, but it is unclear whether this mutation can be used as a prognostic marker to adjust the dose and duration of ribavirin therapy.
We therefore determined the prevalence of the 1634R mutation in the HEV polymerase of patients starting ribavirin therapy using deep sequencing, the influence of the variants harboring this mutation on the viral response, the frequency of the 1634R mutation in patients whose treatment failed, and its impact on retreatment with ribavirin.
MATERIALS AND METHODS
Ethics statement.
Biological materials and clinical data were obtained for a standard virus diagnosis, following physicians' orders. This noninterventional study involved no additional procedures (no specific sampling, no modification of the sampling protocol, and no questions in addition to the national standardized questionnaire). Data were analyzed using an anonymized database. Such a protocol does not require written informed consent according to French public health law (CSP Art L 1121-1.1).
Patients and clinical samples.
Sixty-three solid-organ transplant patients chronically infected with HEV3 included 40 patients who were included in a previous multicenter study (21) and 23 who were recruited independently. The ribavirin dosage was adjusted according to renal clearance. All patients were given ribavirin for a median of 3 months (range, 3 to 18 months), and 42 of them had a sustained virologic response defined as negative plasma HEV RNA at the end of treatment and 6 months after treatment withdrawal. Patients with a partial response (n = 2) were defined as those who had a viremia decline but were plasma HEV RNA positive at the end of treatment. Patients with relapse (n = 19) was defined as those who achieved an end-of-treatment response (undetectable plasma HEV RNA) but whose HEV RNA reappeared 1 to 3 months later. This study was approved by the institutional review boards at Toulouse University Hospital.
Blood samples were collected and stored at −80°C. The HEV polymerase sequences of all patients were determined before starting treatment (baseline) and at the end of ribavirin therapy for partial responders or at the time of the relapse for the other patients.
Plasma HEV RNA concentrations and genotype determination.
The concentrations of HEV RNA in the plasma samples were measured by real-time PCR targeting ORF3 as previously described (25). HEV genotype was determined by sequencing a 348-nucleotide (nt) fragment within the ORF2 gene and phylogenetic analysis with HEV reference strains (GenBank) (26).
Deep sequencing of the HEV polymerase C-terminal region.
Deep sequencing was performed on a 454 GS Junior instrument. A 451-nucleotide fragment encompassing the C-terminal region of the HEV polymerase was generated by nested reverse transcription-PCR (RT-PCR). RT-PCR was performed in duplicate with sense primer AGTGYGGCATGCCCCAGTGGCTTATCCG and antisense primer AGGGGTTGGTTGGATGAA under the following conditions: 1 h at 55°C, 2 min at 94°C, and 45 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min 30 s at 68°C. The RT-PCR products were pooled, and nested PCR was performed with Phusion (Thermo Scientific Finnzymes, Illkirch Graffenstaden, France) with sense primer TGAAGGTYGAYTAYCGGCCTAT and antisense primer GCCGGTGGCGCGGGCAGCATAGGCA under the following conditions: 30 s at 98°C; 45 cycles of 5 s at 98°C, 10 s at 55°C, and 30 s at 72°C; and a final extension at 72°C for 5 min. The amplified PCR products were purified using Agencourt Ampure PCR purification beads (Beckman Coulter, Brea, CA) and quantified with the Quant-iT Picogreen double-stranded DNA (dsDNA) assay kit (Invitrogen) on a LightCycler 480 (Roche). Nested PCR products were clonally amplified on capture beads in water-in-oil emulsion microreactors. A total of 500,000 enriched-DNA beads were deposited in the wells of a full GS Junior Titanium PicoTiterPlate device and pyrosequenced in both the forward and reverse directions. The 200 nucleotide cycles were performed in a 10-h sequencing run. Phylogenetic analyses excluded any possibility of sample contamination.
The sequence reads of the C-terminal region of the HEV polymerase were quantified using GS amplicon variant analyzer (AVA) software, version 2.5p1 (Roche). The AVA software assigns each read to the proper amplicon and sample using multiplex identifiers. The sequence reads were aligned with the HEV3f TLS09-0 consensus sequence (GenBank accession no. KC166967), and sequence alignments were manually edited to correct for insertions or deletions in homopolymeric regions that would result in a frameshift.
Sensitivity of ultradeep pyrosequencing for detecting 1634R variants.
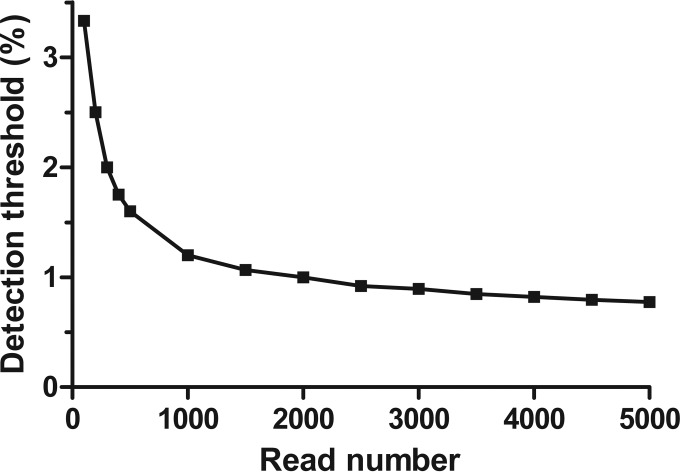
We assessed the frequency of errors resulting from HEV polymerase amplification and GS Junior pyrosequencing by analyzing the data from a panel of 5 plasmid clones of HEV polymerase previously sequenced by the Sanger method. The mean error rate of PCR and pyrosequencing was 0.38% (99% confidence interval, 0.17% to 0.45%). The upper confidence limit of the error rate was used to calculate the sensitivity of pyrosequencing for a given number of reads. The Poisson distribution was used to distinguish authentic variants from artifactual 1634R sequences due to errors arising during PCR amplification and pyrosequencing. P values of <0.001 were considered to be statistically significant. The detection threshold for 1634R variants varied according to the number of reads of HEV polymerase for each sample (Fig. 1). The number of reads ranged from 500 (sensitivity, 1.6%) to 4,000 (sensitivity, 0.8%).
FIG 1.
Sensitivity of pyrosequencing for detecting 1634R variants as a function of the number of reads.
Direct nucleotide sequencing.
Nested PCR products were sequenced on both strands by the dideoxy chain termination method (Prism Ready Reaction AmpliTaq Fs and BigDye terminator; Applied Biosystems, Paris, France) on an ABI 3130XL analyzer (Applied Biosystems, Foster City, CA, USA) using the same sense and antisense primers as used for deep sequencing. Electropherogram data were analyzed using Sequencher 4.8 (Gene Codes Corporation).
Statistical analysis.
Quantitative variables were compared using the Wilcoxon rank sum test. The Wilcoxon signed-rank test was used for matched pairs. Categorical variables were tested using the χ2 test or Fisher's exact test. The Wilcoxon signed-rank test was used for matched pairs. A statistically significant difference was defined as a P value of <0.05.
RESULTS
Detection of the 1634R mutation before ribavirin initiation.
We studied 63 solid-organ transplant recipients with chronic hepatitis E who started their first ribavirin treatment. The 1634R variant was detected at baseline in 36.5% (23/63) of plasma samples in proportions from 1.3% to 100% (median, 8.2%). Patients infected with a 1634R-containing virus had a higher plasma HEV RNA concentration at baseline than did patients in whom the 1634R mutation was not detected (Table 1). Patients infected with HEV3e also tended to be more frequently infected with 1634R viruses.
TABLE 1.
Characteristics of patients before initiation of ribavirin therapy
| Characteristic | Value for group with 1634R variant: |
P value | |
|---|---|---|---|
| Present (n = 23) | Absent (n = 40) | ||
| No. of males/females | 16/7 | 32/8 | 0.37 |
| Median (range) age, yr | 48.5 (20–73) | 48 (7–83) | 0.54 |
| No. with transplantation type: | 0.38 | ||
| Kidney | 17 | 28 | |
| Liver | 3 | 7 | |
| Heart | 3 | 2 | |
| Lung | 0 | 3 | |
| No. with immunosuppressive regimen at start of ribavirin: | |||
| Tacrolimus/cyclosporine | 19/0 | 29/2 | 0.54 |
| Mycophenolic acid | 18 | 30 | 1 |
| mTORa inhibitors | 3 | 6 | 1 |
| Steroids | 19 | 30 | 0.54 |
| Median (range) time between diagnosis and ribavirin therapy, mo | 13 (1–54) | 4 (0.5–47) | 0.29 |
| Median (range) ribavirin dose, mg/kg/day | 9.8 (4.3–13.3) | 8.1 (0.01–16.3) | 0.21 |
| Median (range) ribavirin duration, mo | 3 (3–18) | 3 (3–6) | 0.65 |
| No. (%) with HEV genotype: | 0.07 | ||
| 3c | 3 (8.6) | 14 (35) | |
| 3e | 4 (17.4) | 1 (2.5) | |
| 3f | 13 (56.5) | 21 (52.5) | |
| Undetermined | 3 (8.6) | 4 (10) | |
| Median (range) plasma HEV RNA concn, log copies/ml | 6.2 (4.4–7.5) | 5.8 (3.5–7.4) | 0.05 |
mTOR, mammalian target of rapamycin.
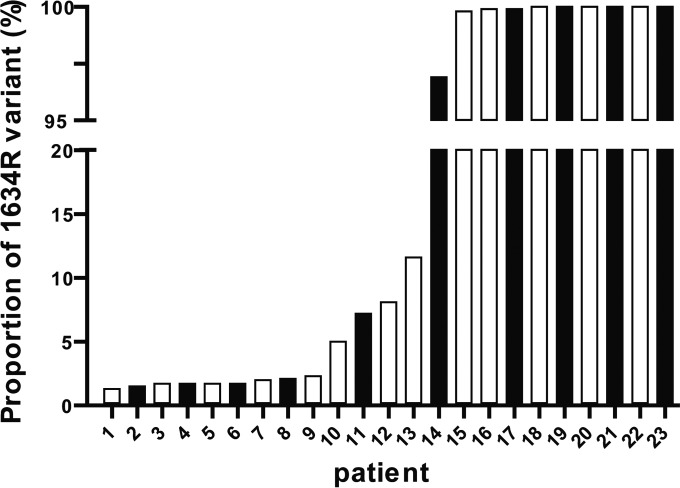
The 63 solid-organ transplant patients included 42 patients (66.7%) who had a sustained virologic response (SVR), two patients who did not achieve a negative viremia at the end of treatment (partial responders), and 19 patients who had a negative viremia at the end of treatment but whose infection recurred 1 to 3 months after therapy (relapsers). We found the 1634R variant in 31.0% (13/42) of baseline plasma samples from patients with SVR and in 47.6% (10/21) of baseline plasma samples from the other patients (P = 0.20). The median proportions of the 1634R variant within the quasispecies were 8.2% (range, 1.3 to 100%) in patients with SVR and 52.1% (range, 1.5 to 100%) in the other patients. The proportions of 1634R variant in the two groups of patients (with or without SVR) were not different (P = 0.85), as the bars were intermixed (Fig. 2).
FIG 2.
Percentages of the 1634R variant in patients with (white bars) or without (black bars) a sustained virologic response.
In addition, the SVR of patients without the 1634R variant was 72%, and it was 61% in those whose proportion of the 1634R variant was below 20% and 50% in those whose proportion of 1634R variant was >20%. However, these differences were not significant (P = 0.36).
Direct population sequencing with the Sanger method showed that 5/42 patients (11.9%) with SVR harbored 1634R variants, while 5/21 patients (23.8%) without SVR harbored 1634R variants (P = 0.28). Direct sequencing did not detect the 1634R variant when the proportion of 1634R variants detected by deep sequencing was below 20%.
Lastly, the duration of ribavirin treatment was analyzed, as it could influence therapy outcome. The median treatment time for the SVR group was 3 months (range, 3 to 18 months), and that for the non-SVR group was 3 months (range, 3 to 6 months) (P = 0.65). We also studied the relationship between the median treatment time and the presence of the 1634R variant. The median treatment time for the 1634R group was 3 months (range, 3 to 18 months), and that for the group of patients without the 1634R variant was 3 months (range, 3 to 11 months) (P = 0.2).
Impact of the 1634R mutation on early virologic response.
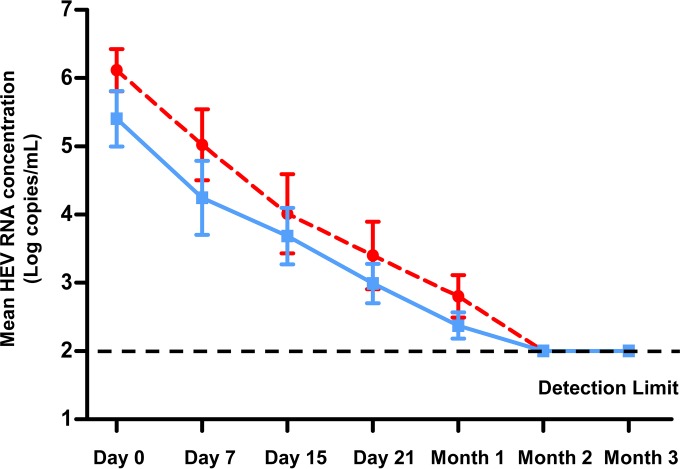
We analyzed the influence of the 1634R variant on the early rate of virus decrease under ribavirin therapy in a subgroup of 16 patients on the same immunosuppressive regime (tacrolimus, mycophenolic acid, and steroids). Half of these patients (8/16) were infected with a 1634R-containing virus. Of each group of 8 patients, 4 had an SVR and 4 had no SVR. The HEV RNA concentrations in the 2 groups decreased similarly (Fig. 3). The drops between days 0 and 7 were −0.7 (range, −2.2 to −0.4) log copies/ml in 1634R variant-infected patients and −0.9 (−1.5 to 0.1) log copies/ml in patients who were not (P = 0.87), those between days 0 and 15 were −2.2 (−3.3 to −0.7) log copies/ml and −1.7 (−3.2 to −0.7) log copies/ml (P = 0.25), those between days 0 and 21 were −2.7 (−3.3 to −0.7) log copies/ml and −2.2 (−4.9 to −1.5) log copies/ml (P = 0.16), and those between days 0 and 30 were −3.2 (−4.4 to −0.2.4) log copies/ml and −2.9 (−5.5 to 1.5) log copies/ml (P = 0.09).
FIG 3.
Decrease in HEV RNA concentration in patients with (red circles) or without (blue squares) 1634R variants. Data are means ± standard errors of the means (SEM).
Detection of the 1634R variant in non-SVR patients after treatment withdrawal.
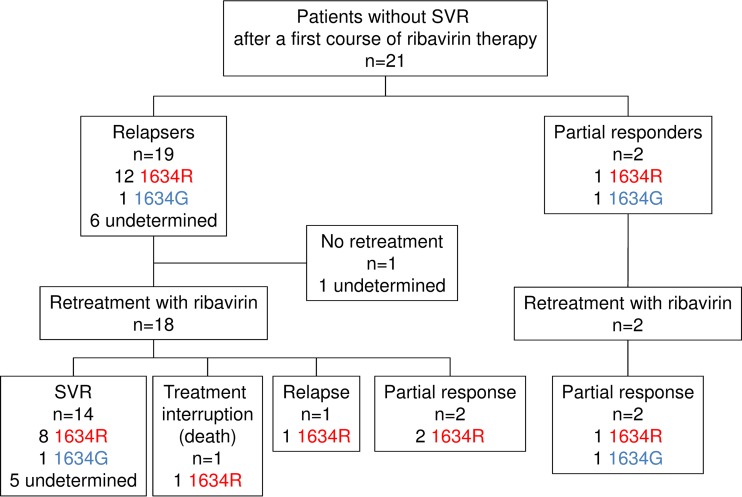
Among the 21 patients who had no SVR, the 1634R variant was detected in one of the two partial responders at the end of treatment and in 92.3% (12/13) of relapsers at the time of relapse. The genomic region encoding the HEV RNA polymerase could not be amplified in 6 relapsers due to the low virus concentration. Many (13/15; 87%) of the patients who were tested for the mutation before retreatment were found to harbor it after their initial unsuccessful treatment. This proportion was higher than that found before the first treatment (26/63; 37%) (P < 0.01).
The 1634R variant emerged in 7 patients for whom the mutation was not detected at baseline (including 1 partial responder and 6 relapsers), with a median proportion of 6.1% (range, 2.3 to 45.6%). In the 6 patients whose 1634R variant was detected at baseline, the median proportion at relapse (73.0%; range, 3.9 to 100%) was higher than the median proportion at baseline (2.1%; range, 1.2 to 100%) (P < 0.01).
Twenty patients (2 partial responders and 18 relapsers) were retreated with ribavirin for a median duration of 6 months (range, 5 to 11 months).The patients were retreated as soon as the relapse was diagnosed. The median proportion of the 1634R variant before first treatment was 0% (range, 0 to 100%), and it was 17.3% (0 to 100%) at the beginning of the second. Thus, the proportion of the 1634R variants in relapsers was higher at the beginning of the second treatment (P < 0.01). The 1634R variant emerged in one partial responder and was still present at the beginning of the second treatment.
The majority (14/20; 70.0%) had a sustained virologic response, and they included 8 patients infected with a 1634R variant, 1 who was not, and 5 in whom it was undetermined (Fig. 4). Neither of the two partial responders had a negative viremia after the second treatment. The 1634R variant was detected in the same patient in whom the 1634R variant emerged after the first treatment (Fig. 4).
FIG 4.
Detection of the 1634R variant by deep sequencing analysis and outcomes for patients without SVR after a first course of ribavirin therapy.
DISCUSSION
In this study, we used deep sequencing analysis to determine the prevalence and the impact of the 1634R mutation in the HEV polymerase gene in 63 solid-organ transplant recipients chronically infected with HEV who initiated ribavirin treatment.
The distributions of the 1634R variant in SVR and non-SVR patients were similar when assayed using deep sequencing and direct sequencing with the Sanger method. Thus, the detection of the 1634R mutation does not indicate absolute ribavirin resistance.
The 1634R variant was found in about one-third of the HEV-infected patients and tends to be more frequently associated with HEV3e infection. The complete genome sequence data available in GenBank indicate that this mutation is present in all HEV3e (7/7), in 6/21 HEV3f, and in 0/6 HEV3c/3i strains. However, there is no evidence that HEV3 pathogenesis is virus dependent (27). Also the HEV RNA concentration in the plasma of patients infected with the 1634R variant virus was higher at the beginning of ribavirin treatment. The difference is slight, but this agrees well with in vitro experiments demonstrating that the replicative capacities of HEV3 and HEV1 with the 1634R mutation are higher than the replicative capacities of the same strains without this mutation (24). Thus, 1634R mutation confers only a limited replicative advantage in vivo. A recent study showed that hepatitis C virus that has increased replicative fitness is also partially resistant to inhibitors that target virus or cell proteins, including alpha interferon (IFN-α), ribavirin, telaprevir, and daclatasvir (28). It was suggested that prolonged replication of HCV in the liver environment during chronic infection could lead to an increase in viral fitness. Consequently, higher drug concentrations or longer treatment duration could be required to achieve antiviral effects comparable to those attained with a low-replicative-fitness virus. In our study, there was no difference in duration of infection before ribavirin therapy between patients infected with a variant harboring or not harboring the 1634R mutation. Thus, the appearance of the 1634R variant does not seem to be associated with the duration of the infection. Other HEV variants with improved replicative capacity in cell culture system were recently isolated from both stool and plasma samples (29–31). These variants were recombinant HEV3 with insertion of part of either a human gene or the HEV genome in the PPR. These recombinant viruses were isolated exclusively in chronically infected solid-organ transplant patients during the chronic phase, suggesting that adaptation of HEV can occur long after the acute phase. Alternatively, such recombinant events could be the consequence of a modification in the host environment. However, the sensitivity of these variants to ribavirin is unknown.
One report suggested that the unique F415Y mutation in the HCV polymerase results in ribavirin resistance, indicating that mutation in the polymerase can confer intrinsic resistance to ribavirin (32). We found that the presence of the 1634R mutation in the HEV polymerase does not influence the early rate of HEV replication, which is a key parameter of SVR for ribavirin (33). It was shown that a decrease of ≥0.5 log copies/ml in the HEV RNA concentration within the first week of ribavirin treatment is an independent predictive factor for SVR (33). As the presence of the 1634R variant does not influence the early kinetics, it seems better to monitor the decrease in HEV RNA concentration to predict treatment outcome. In addition, a prolonged fecal shedding of HEV in patients on ribavirin therapy was also associated with relapse (34).
We detected 1634R variants in most patients at relapse or at the end of the first course of ribavirin treatment. The proportion of this variant increased after the initial unsuccessful therapy. However, a second, more prolonged ribavirin treatment lead to an SVR in 70% of the patients harboring the 1634R variant at relapse, which is quite similar to the percentage of SVR obtained in patients initiating a first-line therapy. Thus, the presence of the 1634R variant does not impair ribavirin treatment success, even after a first exposure. Another recent study showed that prolonged retreatment successfully cleared of HEV a patient whose first course of ribavirin failed to do so. The 1634R mutation was detected in this patient at relapse (35). However, the retreatment was longer than the first treatment, suggesting that the longer treatment period could at least in part explain this favorable outcome. The 1634R variant appeared after ribavirin treatment but was unlikely to be the cause of treatment failure. Perhaps it is one cofactor making it harder to achieve SVR. Quasispecies heterogeneity could also contribute to ribavirin success. In solid-organ transplant patients, it was shown that spontaneous HEV RNA clearance was associated with a lower quasispecies heterogeneity (36, 37). The quasispecies heterogeneity could play a role in patients on ribavirin therapy. A recent study showed that exposure to ribavirin in vitro increases quasispecies diversity in a murine norovirus model by increasing nucleotide substitution (38). This could favor the emergence/increase of the 1634R variant.
To date, the mechanisms by which ribavirin exerts its antiviral activity on HEV are unclear. In vitro experiments have shown that ribavirin inhibits HEV replication, depleting intracellular GTP pools through inhibition of host IMP dehydrogenase (IMPDH). In vitro experiments have shown that mycophenolic acid (MPA) also inhibits HEV replication by depleting the intracellular GTP pools through inhibition of IMPDH (39). However, the use of mycophenolate mofetil (MMF), a prodrug of MPA, in transplant patient with chronic hepatitis E was not associated with a better antiviral response (33). Thus, even though in vitro antiviral activity of MPA and ribavirin relies on the depletion of GTP pool, other mechanisms may also contribute to an antiviral effect in vivo.
Ribavirin could also induce lethal mutagenesis, i.e., increase the number of mutations per genome until it is too large for newly generated variants to be viable (40). Yet another possible mechanism of ribavirin action is inhibition of eukaryote initiation factor 4E (eIF4E). eIF4E is a component of the translation initiation complex, and ribavirin would act as an analogue of the 7-methylguanosine mRNA cap (41). Ribavirin antiviral activity could also be due to immunomodulation. In this schema, ribavirin increases T helper 1 responses, reverses the regulatory T cell (Treg)-mediated suppression of CD4 effector T cells, or improves the interferon signaling cascade, leading to modulation of interferon-stimulated gene (ISG) expression (42, 43). Lastly, the production of IFN-γ by NK cells could be modulated by ribavirin (44). Implication of the immune system in an optimal response to ribavirin treatment is also suggested by the fact that the lymphocyte count at the initiation of ribavirin treatment is an independent predictive factor associated with an SVR (21).
One limitation of our study is the small number of patients; our results need to be confirmed in larger cohorts of patients. At baseline, the 1634R variant was more frequent in patients with ribavirin treatment failure than in patients with SVR. However, this difference failed to reach significance.
We conclude that although in vitro and in vivo data indicate that variants harboring the 1634R mutation have a high replicative capacity, the presence of this mutation before initiation of ribavirin therapy does not lead to absolute ribavirin resistance, as depicted previously in single case reports. In addition, its presence in relapsers does not preclude virus eradication by more prolonged therapy. Further studies are needed to decipher the mechanism involved in HEV resistance to ribavirin.
ACKNOWLEDGMENTS
We thank Vincent Mallet, Stanislas Pol, and Simona Tripon (Cochin, Assistance Publique-Hôpitaux de Paris), Audrey Coilly and Anne-Marie Roque-Afonso (Paris Sud, Assistance Publique-Hôpitaux de Paris), Sophie Radenne and Mary-Anne Trabaud (Croix Rousse, Lyon University Hospital), Jerome Dumortier (Edouard Herriot, Lyon University Hospital), Elisabeth Garnier and Maryvonne Hourmant (Nantes University Hospital), Sophie Hilaire (Foch Hospital, Suresne), and Louis d'Alteroche, Matthias Buchler, and Catherine Gaudy-Graffin (Tours University Hospital) for their help with the follow-up of the patients. We thank Owen Parkes for editing the English text.
We have no conflict of interest to disclose.
Funding Statement
This work was supported by Institut National de la Santé et de la Recherche Médicale. The funders had no role in study design, data collection or interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Holla RP, Ahmad I, Ahmad Z, Jameel S. 2013. Molecular virology of hepatitis E virus. Semin Liver Dis 33:3–14. doi: 10.1055/s-0033-1338110. [DOI] [PubMed] [Google Scholar]
- 2.Smith DB, Simmonds P, International Committee on Taxonomy of Viruses Hepeviridae Study Group, Jameel S, Emerson SU, Harrison TJ, Meng XJ, Okamoto H, Van der Poel WH, Purdy MA. 2014. Consensus proposals for classification of the family Hepeviridae. J Gen Virol 95:2223–2232. doi: 10.1099/vir.0.068429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng XJ. 2013. Zoonotic and foodborne transmission of hepatitis E virus. Semin Liver Dis 33:41–49. doi: 10.1055/s-0033-1338113. [DOI] [PubMed] [Google Scholar]
- 4.Legrand-Abravanel F, Kamar N, Sandres-Saune K, Garrouste C, Dubois M, Mansuy JM, Muscari F, Sallusto F, Rostaing L, Izopet J. 2010. Characteristics of autochthonous hepatitis E virus infection in solid-organ transplant recipients in France. J Infect Dis 202:835–844. doi: 10.1086/655899. [DOI] [PubMed] [Google Scholar]
- 5.Izopet J, Dubois M, Bertagnoli S, Lhomme S, Marchandeau S, Boucher S, Kamar N, Abravanel F, Guerin JL. 2012. Hepatitis E virus strains in rabbits and evidence of a closely related strain in humans, France. Emerg Infect Dis 18:1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colson P, Coze C, Gallian P, Henry M, De Micco P, Tamalet C. 2007. Transfusion-associated hepatitis E, France. Emerg Infect Dis 13:648–649. doi: 10.3201/eid1304.061387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aggarwal R. 2013. Hepatitis E: clinical presentation in disease-endemic areas and diagnosis. Semin Liver Dis 33:30–40. doi: 10.1055/s-0033-1338112. [DOI] [PubMed] [Google Scholar]
- 8.Colson P, Payraudeau E, Leonnet C, De Montigny S, Villeneuve L, Motte A, Tamalet C. 2008. Severe thrombocytopenia associated with acute hepatitis E virus infection. J Clin Microbiol 46:2450–2452. doi: 10.1128/JCM.02295-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fourquet E, Mansuy JM, Bureau C, Recher C, Vinel JP, Izopet J, Peron JM. 2010. Severe thrombocytopenia associated with acute autochthonous hepatitis E. J Clin Virol 48:73–74. doi: 10.1016/j.jcv.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 10.Shah SA, Lal A, Idrees M, Hussain A, Jeet C, Malik FA, Iqbal Z, Rehman H. 2012. Hepatitis E virus-associated aplastic anaemia: the first case of its kind. J Clin Virol 54:96–97. doi: 10.1016/j.jcv.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Deniel C, Coton T, Brardjanian S, Guisset M, Nicand E, Simon F. 2011. Acute pancreatitis: a rare complication of acute hepatitis E. J Clin Virol 51:202–204. doi: 10.1016/j.jcv.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Kamar N, Weclawiak H, Guilbeau-Frugier C, Legrand-Abravanel F, Cointault O, Ribes D, Esposito L, Cardeau-Desangles I, Guitard J, Sallusto F, Muscari F, Peron JM, Alric L, Izopet J, Rostaing L. 2012. Hepatitis E virus and the kidney in solid-organ transplant patients. Transplantation 93:617–623. doi: 10.1097/TP.0b013e318245f14c. [DOI] [PubMed] [Google Scholar]
- 13.Kamar N, Bendall RP, Peron JM, Cintas P, Prudhomme L, Mansuy JM, Rostaing L, Keane F, Ijaz S, Izopet J, Dalton HR. 2011. Hepatitis E virus and neurologic disorders. Emerg Infect Dis 17:173–179. doi: 10.3201/eid1702.100856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Eijk JJ, Madden RG, van der Eijk AA, Hunter JG, Reimerink JH, Bendall RP, Pas SD, Ellis V, van Alfen N, Beynon L, Southwell L, McLean B, Jacobs BC, van Engelen BG, Dalton HR. 2014. Neuralgic amyotrophy and hepatitis E virus infection. Neurology 82:498–503. doi: 10.1212/WNL.0000000000000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van den Berg B, van der Eijk AA, Pas SD, Hunter JG, Madden RG, Tio-Gillen AP, Dalton HR, Jacobs BC. 2014. Guillain-Barre syndrome associated with preceding hepatitis E virus infection. Neurology 82:491–497. doi: 10.1212/WNL.0000000000000111. [DOI] [PubMed] [Google Scholar]
- 16.Dalton HR, Bendall RP, Keane FE, Tedder RS, Ijaz S. 2009. Persistent carriage of hepatitis E virus in patients with HIV infection. N Engl J Med 361:1025–1027. doi: 10.1056/NEJMc0903778. [DOI] [PubMed] [Google Scholar]
- 17.Ollier L, Tieulie N, Sanderson F, Heudier P, Giordanengo V, Fuzibet JG, Nicand E. 2009. Chronic hepatitis after hepatitis E virus infection in a patient with non-Hodgkin lymphoma taking rituximab. Ann Intern Med 150:430–431. doi: 10.7326/0003-4819-150-6-200903170-00026. [DOI] [PubMed] [Google Scholar]
- 18.Kamar N, Dalton HR, Abravanel F, Izopet J. 2014. Hepatitis E virus infection. Clin Microbiol Rev 27:116–138. doi: 10.1128/CMR.00057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamar N, Selves J, Mansuy JM, Ouezzani L, Peron JM, Guitard J, Cointault O, Esposito L, Abravanel F, Danjoux M, Durand D, Vinel JP, Izopet J, Rostaing L. 2008. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med 358:811–817. doi: 10.1056/NEJMoa0706992. [DOI] [PubMed] [Google Scholar]
- 20.Kamar N, Abravanel F, Selves J, Garrouste C, Esposito L, Lavayssiere L, Cointault O, Ribes D, Cardeau I, Nogier MB, Mansuy JM, Muscari F, Peron JM, Izopet J, Rostaing L. 2010. Influence of immunosuppressive therapy on the natural history of genotype 3 hepatitis-E virus infection after organ transplantation. Transplantation 89:353–360. doi: 10.1097/TP.0b013e3181c4096c. [DOI] [PubMed] [Google Scholar]
- 21.Kamar N, Izopet J, Tripon S, Bismuth M, Hillaire S, Dumortier J, Radenne S, Coilly A, Garrigue V, D'Alteroche L, Buchler M, Couzi L, Lebray P, Dharancy S, Minello A, Hourmant M, Roque-Afonso AM, Abravanel F, Pol S, Rostaing L, Mallet V. 2014. Ribavirin for chronic hepatitis E virus infection in transplant recipients. N Engl J Med 370:1111–1120. doi: 10.1056/NEJMoa1215246. [DOI] [PubMed] [Google Scholar]
- 22.Pischke S, Hardtke S, Bode U, Birkner S, Chatzikyrkou C, Kauffmann W, Bara CL, Gottlieb J, Wenzel J, Manns MP, Wedemeyer H. 2013. Ribavirin treatment of acute and chronic hepatitis E: a single-centre experience. Liver Int 33:722–726. doi: 10.1111/liv.12114. [DOI] [PubMed] [Google Scholar]
- 23.Kamar N, Mallet V, Izopet J. 2014. Ribavirin for chronic hepatitis E virus infection. N Engl J Med 370:2447–2448. doi: 10.1056/NEJMoa1215246. [DOI] [PubMed] [Google Scholar]
- 24.Debing Y, Gisa A, Dallmeier K, Pischke S, Bremer B, Manns M, Wedemeyer H, Suneetha PV, Neyts J. 2014. A mutation in the hepatitis E virus RNA polymerase promotes its replication and associates with ribavirin treatment failure in organ transplant recipients. Gastroenterology 147:1008–1011. doi: 10.1053/j.gastro.2014.08.040. [DOI] [PubMed] [Google Scholar]
- 25.Abravanel F, Sandres-Saune K, Lhomme S, Dubois M, Mansuy JM, Izopet J. 2012. Genotype 3 diversity and quantification of hepatitis E virus RNA. J Clin Microbiol 50:897–902. doi: 10.1128/JCM.05942-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legrand-Abravanel F, Mansuy JM, Dubois M, Kamar N, Peron JM, Rostaing L, Izopet J. 2009. Hepatitis E virus genotype 3 diversity, France. Emerg Infect Dis 15:110–114. doi: 10.3201/eid1501.080296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith DB, Ijaz S, Tedder R, Hogema B, Zaaijer H, Izopet J, Bradley-Stewart A, Gunson R, Harvala H, Kokki I, Simmonds P. 2015. Variability and pathogenicity of hepatitis E virus genotype 3 variants. J Gen Virol doi: 10.1099/jgv.0.000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheldon J, Beach NM, Moreno E, Gallego I, Pineiro D, Martinez-Salas E, Gregori J, Quer J, Esteban JI, Rice CM, Domingo E, Perales C. 2014. Increased replicative fitness can lead to decreased drug sensitivity of hepatitis C virus. J Virol 88:12098–12111. doi: 10.1128/JVI.01860-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lhomme S, Abravanel F, Dubois M, Sandres-Saune K, Mansuy JM, Rostaing L, Kamar N, Izopet J. 2014. Characterization of the polyproline region of the hepatitis E virus in immunocompromised patients. J Virol 88:12017–12025. doi: 10.1128/JVI.01625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen HT, Torian U, Faulk K, Mather K, Engle RE, Thompson E, Bonkovsky HL, Emerson SU. 2012. A naturally occurring human/hepatitis E recombinant virus predominates in serum but not in faeces of a chronic hepatitis E patient and has a growth advantage in cell culture. J Gen Virol 93:526–530. doi: 10.1099/vir.0.037259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shukla P, Nguyen HT, Torian U, Engle RE, Faulk K, Dalton HR, Bendall RP, Keane FE, Purcell RH, Emerson SU. 2011. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc Natl Acad Sci U S A 108:2438–2443. doi: 10.1073/pnas.1018878108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young KC, Lindsay KL, Lee KJ, Liu WC, He JW, Milstein SL, Lai MM. 2003. Identification of a ribavirin-resistant NS5B mutation of hepatitis C virus during ribavirin monotherapy. Hepatology 38:869–878. doi: 10.1002/hep.1840380413. [DOI] [PubMed] [Google Scholar]
- 33.Kamar N, Lhomme S, Abravanel F, Cointault O, Esposito L, Cardeau-Desangles I, Del Bello A, Dorr G, Lavayssiere L, Nogier MB, Guitard J, Ribes D, Goin AL, Broue P, Metsu D, Saune K, Rostaing L, Izopet J. 2015. An early viral response predicts the virological response to ribavirin in hepatitis E virus organ transplant patients. Transplantation doi: 10.1097/TP.0000000000000850. [DOI] [PubMed] [Google Scholar]
- 34.Abravanel F, Lhomme S, Rostaing L, Kamar N, Izopet J. 2015. Protracted fecal shedding of HEV during ribavirin therapy predicts treatment relapse. Clin Infect Dis 60:96–99. doi: 10.1093/cid/ciu742. [DOI] [PubMed] [Google Scholar]
- 35.Galante A, Pischke S, Polywka S, Luetgehethmann M, Suneetha PV, Gisa A, Hiller J, Dienes HP, Nashan B, Lohse AW, Sterneck M. 2015. Relevance of chronic hepatitis E in liver transplant recipients: a real-life setting. Transpl Infect Dis 17:617–622. doi: 10.1111/tid.12411. [DOI] [PubMed] [Google Scholar]
- 36.Lhomme S, Garrouste C, Kamar N, Saune K, Abravanel F, Mansuy JM, Dubois M, Rostaing L, Izopet J. 2014. Influence of polyproline region and macro domain genetic heterogeneity on HEV persistence in immunocompromised patients. J Infect Dis 209:300–303. doi: 10.1093/infdis/jit438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lhomme S, Abravanel F, Dubois M, Sandres-Saune K, Rostaing L, Kamar N, Izopet J. 2012. Hepatitis E virus quasispecies and the outcome of acute hepatitis E in solid-organ transplant patients. J Virol 86:10006–10014. doi: 10.1128/JVI.01003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Julian TR, Baugher JD, Rippinger CM, Pinekenstein R, Kolawole AO, Mehoke TS, Wobus CE, Feldman AB, Pineda FJ, Schwab KJ. 2016. Murine norovirus (MNV-1) exposure in vitro to the purine nucleoside analog Ribavirin increases quasispecies diversity. Virus Res 211:165–173. doi: 10.1016/j.virusres.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 39.Debing Y, Emerson SU, Wang Y, Pan Q, Balzarini J, Dallmeier K, Neyts J. 2014. Ribavirin inhibits in vitro hepatitis E virus replication through depletion of cellular GTP pools and is moderately synergistic with alpha interferon. Antimicrob Agents Chemother 58:267–273. doi: 10.1128/AAC.01795-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, Andino R, Cameron CE. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med 6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 41.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. 2004. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A 101:18105–18110. doi: 10.1073/pnas.0406927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hultgren C, Milich DR, Weiland O, Sallberg M. 1998. The antiviral compound ribavirin modulates the T helper (Th) 1/Th2 subset balance in hepatitis B and C virus-specific immune responses. J Gen Virol 79:2381–2391. doi: 10.1099/0022-1317-79-10-2381. [DOI] [PubMed] [Google Scholar]
- 43.Langhans B, Nischalke HD, Arndt S, Braunschweiger I, Nattermann J, Sauerbruch T, Spengler U. 2012. Ribavirin exerts differential effects on functions of Cd4+ Th1, Th2, and regulatory T cell clones in hepatitis C. PLoS One 7:e42094. doi: 10.1371/journal.pone.0042094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Werner JM, Serti E, Chepa-Lotrea X, Stoltzfus J, Ahlenstiel G, Noureddin M, Feld JJ, Liang TJ, Rotman Y, Rehermann B. 2014. Ribavirin improves the IFN-gamma response of natural killer cells to IFN-based therapy of hepatitis C virus infection. Hepatology 60:1160–1169. doi: 10.1002/hep.27092. [DOI] [PMC free article] [PubMed] [Google Scholar]