Abstract
In two pairs of clinical colistin-susceptible/colistin-resistant (Csts/Cstr) Acinetobacter baumannii strains, the Cstr strains showed significantly decreased biofilm formation in static and dynamic assays (P < 0.001) and lower relative fitness (P < 0.05) compared with those of the Csts counterparts. The whole-genome sequencing comparison of strain pairs identified a mutation converting a stop codon to lysine (*241K) in LpsB (involved in lipopolysaccharide [LPS] synthesis) in one Cstr strain and a frameshift mutation in CarO and the loss of a 47,969-bp element containing multiple genes associated with biofilm production in the other.
TEXT
Colistin-resistant (Cstr) Acinetobacter baumannii clinical isolates are increasingly recovered worldwide (1), which is causing major clinical concerns. Thus far, two primary colistin resistance mechanisms have been described in A. baumannii, (i) modification of the lipid A moiety of lipopolysaccharide (LPS) mediated by mutations and/or overexpression in the two-component regulatory system pmrAB and (ii) loss of LPS caused by either mutations or insertional inactivation of lipid A biosynthesis genes (2).
The development of colistin resistance in clinical and laboratory-derived Cstr A. baumannii due to changes in the PmrAB system has been correlated with impaired fitness and virulence (3, 4) and lower infectivity (5, 6). Also, reduced biofilm formation has been observed in laboratory-generated Cstr A. baumannii (7, 8).
We previously reported the characteristics of two pairs of colistin-susceptible (Csts)/Cstr A. baumannii clinical strains (Ab248/Ab249 and Ab299/Ab347; colistin MICs of 0.5/128 and 0.5/32 μg/ml, respectively) sequentially obtained from two patients after prolonged colistin exposure (6). Briefly, compared to Csts strains, Cstr strains harbored a single pmrB mutation (P233S for Ab249 and P170L for Ab347) and had significantly slower growth. Strains within each pair had identical pulsed-field gel electrophoresis (PFGE) profiles, and all were assigned to the international clone 2. Important differences in the antibiotic resistance phenotypes other than colistin were not observed (6). One Cstr strain underexpressed the CsuA/B and CsuC proteins, which are involved in biofilm formation (6). In the present follow-up study, we compared these strain pairs in terms of fitness, biofilm-forming ability, and whole-genome sequencing (WGS) focusing specifically on genes involved in virulence and biofilm formation.
In vitro competition assays were performed in triplicate at 5 h and 20 h (4), and relative fitness was calculated as previously described (9). To study adhesion (6 h) and biofilm formation (24 h) under static conditions, the crystal violet method was used (10) with some modifications. Briefly, inoculum was prepared by adjusting exponential cultures grown in Luria-Bertani (LB) broth to a 0.5 McFarland standard, and this was followed by a 1:10 dilution in LB broth. Then, 200-μl aliquots (approximately 2 × 106 CFU) were loaded into a 96-well polystyrene microplate (8 replicates/strain) and were incubated for 6 h or 24 h at 37°C. The plates were washed 3 times with sterile phosphate-buffered saline (PBS), fixated with methanol, and stained with 0.2% crystal violet. The biomass was quantified by eluting the dye in 33% acetic acid, and then the optical density at 620 nm (OD620) was measured using a Multiskan FC photometer (Thermo Fisher Scientific, Bremen, Germany). Assays were performed in triplicate and at three independent time points. A. baumanni ATCC 19606 was utilized as a positive control and uninoculated wells as negative controls.
Initial adhesion or early biofilm formation under dynamic conditions was also determined using the BioFlux system (Fluxion Biosciences Inc., South San Francisco, CA) (10) with some modifications. Briefly, inocula containing approximately 106 CFU of each strain were loaded in a BioFlux plate (5 replicates/strain) and were allowed to attach for 30 min. The plate was incubated for 6 h at 37°C with a flow speed of 0.5 dyne/cm2, and the biomass was stained by Live/Dead BacLight stain (Invitrogen, Life Technologies) and was visualized by fluorescence microscopy (Observer Z1; Carl Zeiss Inc., Oberkochen, Germany). Three independent experiments were performed. Biofilms were quantified using the ZEN 2012 (Zeiss) and ImageJ (http://imagej.nih.gov/ij/) software, and fluorescence levels were recorded as integrated density (IntDen) on the total area of the channel.
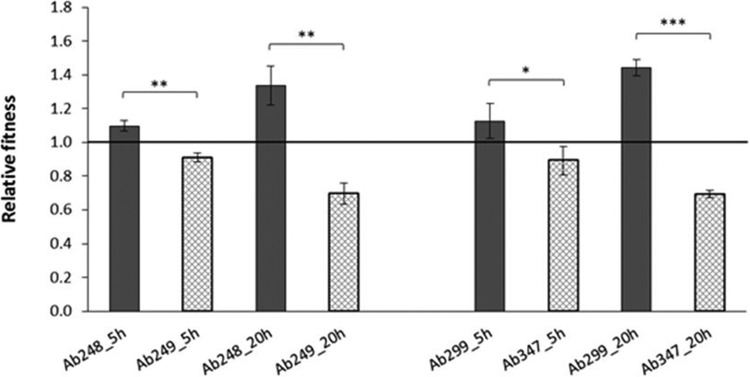
In vitro competition experiments within the two pairs showed a significant fitness burden in the Cstr strains (Fig. 1). The Cstr strain Ab249 showed an average fitness reduction of 17.0% at 5 h (relative fitness, 0.91; standard deviation [SD], 0.03; P = 0.001) and of 47.9% at 20 h (relative fitness, 0.70; SD, 0.06; P = 0.001) compared to the Csts competitor Ab248. Similarly, the Cstr strain Ab347 exhibited an average fitness decrease of 20.7% at 5 h (relative fitness, 0.89; SD, 0.08; P = 0.038) and of 51.9% at 20 h (relative fitness, 0.69; SD, 0.02; P < 0.001) relative to the Csts strain Ab299.
FIG 1.
Relative fitness of Csts/Cstr A. baumannii strains. Values represent the means ± SD from three independent experiments performed at 5 and 20 h. Fitness values below 1 denote fitness cost, while those above 1 denote a fitness benefit. Asterisks indicate statistically significant differences between strains (*, P < 0.05; **, P = 0.001; ***, P < 0.001).
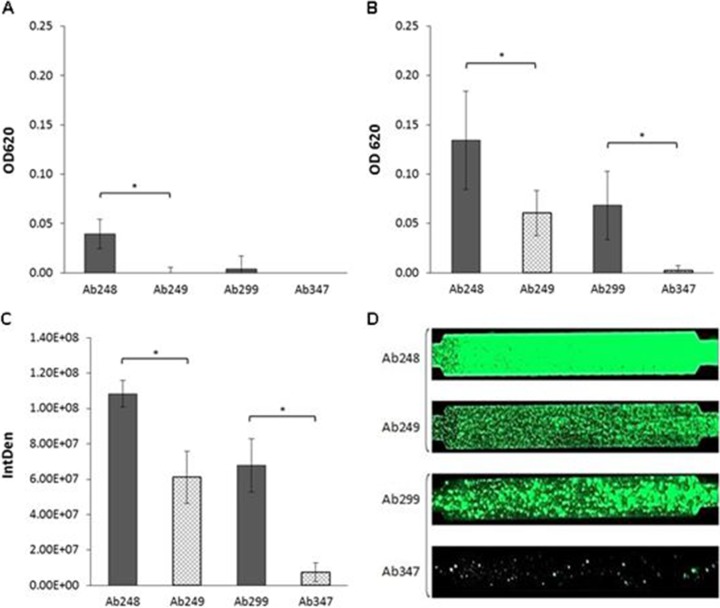
The two Cstr A. baumannii strains showed significantly lower biofilm production in the 24-h static assay than Csts strains. In particular, the Cstr strain Ab249 produced 55.1% less biomass than its Csts counterpart Ab248 (Ab249, OD620 = 0.060 and SD = 0.023; Ab248, OD620 = 0.134 and SD = 0.05; P < 0.001) (Fig. 2B). The Cstr strain Ab347 showed a 96.4% reduction relative to Csts strain Ab299 (Ab347, OD620 = 0.002 and SD = 0.005; Ab299, OD620 = 0.068 and SD = 0.035; P < 0.001) (Fig. 2B). Our findings for these clinical strains support previous studies, which showed that laboratory-generated Cstr A. baumannii strains were weaker biofilm producers than their Csts counterparts (7, 8). We also utilized a 6-h static assay protocol to test for differences in the initial adhesion capacity of the strain pairs; Ab248 did show higher biomass than that of Ab249 (P < 0.001), while the Ab299/Ab347 pair showed a visible growth delay compared to the growth of the control strain (ATCC 19606); the two strains essentially did not form biofilms (Fig. 2A). In the more sensitive 6-h dynamic assay, the Cstr strain Ab249 had 43.6% lower surface coverage than its Csts counterpart Ab248 (Ab249, IntDen = 6.11E + 07 and SD = 1.48E + 07; Ab248, IntDen = 1.08E + 08 and SD = 7.51E + 06; P < 0.001). The Cstr strain Ab347 demonstrated 89.2% less surface coverage (P < 0.001) than Ab299 (Ab347, IntDen = 7.36E + 06 and SD = 5.35E + 06; Ab299, IntDen = 6.78E + 07 and SD = 1.50E + 07; P < 0.001) (Fig. 2C and D).
FIG 2.
Adhesion and biofilm formation by Csts/Cstr A. baumannii strains at 6 h (A) and 24 h (B) in a static model. Results represent the mean values ± SD from three independent experiments (OD, optical density). Asterisks indicate statistically significant differences between strains (*, P < 0.001). (C) Biofilm formation in a dynamic model; results represent the mean values ± SD from three independent experiments (IntDen, integrated density). Asterisks indicate statistically significant differences between strains (*, P < 0.001). (D) Fluorescence microscopy images of BioFlux channels.
WGS was performed using 2 × 150 b paired-end sequencing (Nextera XT sample preparation kit and MiSeq; Illumina). Sequences were independently assembled using SPAdes v3.1.0 (11), and scaffolds were aligned against the reference genome of the ACICU strain that is available under the GenBank accession number CP000863. The obtained pseudochromosomes were compared using Mauve v2.3.1 for genome rearrangements. Next, the two Cstr strains were compared to their Csts counterparts as reference templates, and reference mapping and single nucleotide polymorphism (SNP) extractions were performed using the CLC Genomics Workbench v7.5.1 (CLC bio, Denmark) with default parameters. All changes identified by WGS were confirmed by PCR and Sanger sequencing.
Intrapair WGS comparisons identified the previously detected single amino acid change in the PmrB protein in each of the two Cstr strains (6). Also, a mutation converting a stop codon to lysine (*241K) in LpsB, a highly conserved glycosyltransferase that is involved in the biosynthesis of the LPS core (12) and is potentially important for A. baumannii virulence and colistin resistance (13), was observed in the Cstr strain Ab249. Interestingly, in the second Cstr strain Ab347, we observed the loss of a 47,969-bp genomic region containing, among others, the genes mrkC, mrkD, modA, modB, modC, modD, and ppk, which have been previously associated with biofilm production in Enterobacteriaceae and in Pseudomonas strains (14–16). Of note is the removal of the genes mrkC (pilin) and mrkD (assembly chaperone), which are part of chaperone-usher (CU) system assembling pili. It was previously shown that the disruption of such pili CU systems, like Csu or Fim, induce a severe decrease in biofilm formation in A. baumannii (17, 18). The loss of these genes and of the genomic element was confirmed by Sanger sequencing and by whole-genome mapping (data not shown) (19). Also, in the strain Ab347, a frameshift mutation (A19fs) was observed in the outer membrane protein CarO, which was previously implicated in biofilm formation (20). In concordance, proteomic data generated for strain Ab347 also showed significant (−28.34-fold change; P = 5.00 × 10−15) underexpression of CarO (6). The above genetic modifications identified in Ab347 may partly explain the almost complete absence of biofilm production observed in this strain.
Our study demonstrated that acquisition of colistin resistance by these two clinical A. baumannii strains was associated with a significant loss of biofilm forming capacity. To the best of our knowledge, there are no studies comparing the impact of colistin resistance on biofilm formation among clinical Csts/Cstr A. baumannii strains, and the only available surveys investigated laboratory-generated Cstr mutants, which also produced less biofilm than their Csts counterparts (7, 8). Also, in vitro competition experiments showed that Cstr A. baumannii strains demonstrated considerably lower relative fitness than their Csts ancestors. Genome-wide analysis identified, in the two pairs, unique changes. As the changes observed in the Cstr strains were primarily localized in genes affecting the surface properties, changes in initial adhesion and therefore in biofilm formation capabilities may be expected. In addition, the decreased fitness of these strains measured in competitive growth experiments also raises the possibility of a reduced growth rate of the Cstr strains being the cause of the reduced biomass of these stains in the biofilm experiments. From a clinical perspective, reduced biofilm formation as well as fitness may affect the infectivity and actually facilitate treatment of infections caused by Cstr A. baumannii.
Overall, the findings of the current study support previous findings (6) regarding the reduced clinical invasiveness of Cstr strains. However, further research with a larger set of isolates is needed to fully elucidate the relationship between colistin resistance and biofilm formation and the underlying mechanisms responsible for these developments in clinical strains.
(Part of this work was presented at the 25th European Congress of Clinical Microbiology and Infectious Diseases, Copenhagen, Denmark, 25 to 28 April 2015, and at the 10th International Symposium on the Biology of Acinetobacter, Athens, Greece, 3 to 5 June 2015.)
ACKNOWLEDGMENTS
K.D. was supported by a grant from the Federation of European Microbiological Societies (FEMS Research Fellowship). B.B.X. is supported by University of Antwerp Research Funds (BOF-DOCPRO 2012-27450).
We declare no conflict of interest.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors and was supported by internal funding.
REFERENCES
- 1.Cai Y, Chai D, Wang R, Liang B, Bai N. 2012. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother 67:1607–1615. doi: 10.1093/jac/dks084. [DOI] [PubMed] [Google Scholar]
- 2.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.López-Rojas R, McConnell MJ, Jiménez-Mejías ME, Domínguez-Herrera J, Fernández-Cuenca F, Pachón J. 2013. Colistin resistance in a clinical Acinetobacter baumannii strain appearing after colistin treatment: effect on virulence and bacterial fitness. Antimicrob Agents Chemother 57:4587–4589. doi: 10.1128/AAC.00543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beceiro A, Moreno A, Fernández N, Vallejo JA, Aranda J, Adler B, Harper M, Boyce JD, Bou G. 2014. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob Agents Chemother 58:518–526. doi: 10.1128/AAC.01597-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rolain JM, Roch A, Castanier M, Papazian L, Raoult D. 2011. Acinetobacter baumannii resistant to colistin with impaired virulence: a case report from France. J Infect Dis 204:1146–1147. doi: 10.1093/infdis/jir475. [DOI] [PubMed] [Google Scholar]
- 6.Pournaras S, Poulou A, Dafopoulou K, Chabane YN, Kristo I, Makris D, Hardouin J, Cosette P, Tsakris A, Dé E. 2014. Growth retardation, reduced invasiveness, and impaired colistin-mediated cell death associated with colistin resistance development in Acinetobacter baumannii. Antimicrob Agents Chemother 58:828–832. doi: 10.1128/AAC.01439-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernández-Reyes M, Rodríguez-Falcón M, Chiva C, Pachón J, Andreu D, Rivas L. 2009. The cost of resistance to colistin in Acinetobacter baumannii: a proteomic perspective. Proteomics 9:1632–1645. doi: 10.1002/pmic.200800434. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Nation RL, Owen RJ, Wong S, Spelman D, Franklin C. 2007. Antibiograms of multidrug-resistant clinical Acinetobacter baumannii: promising therapeutic options for treatment of infection with colistin-resistant strains. Clin Infect Dis 45:594–598. doi: 10.1086/520658. [DOI] [PubMed] [Google Scholar]
- 9.Gillespie SH. (ed). 2001. Antibiotic resistance: methods and protocols. Humana Press, Totowa, NJ. [Google Scholar]
- 10.Vanhommerig E, Moons P, Pirici D, Lammens C, Hernalsteens JP, De Greve H, Kumar-Singh S, Goossens H, Malhotra-Kumar S. 2014. Comparison of biofilm formation between major clonal lineages of methicillin resistant Staphylococcus aureus. PLoS One 9(8):e104561. doi: 10.1371/journal.pone.0104561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luke NR, Sauberan SL, Russo TA, Beanan JM, Olson R, Loehfelm TW, Cox AD, St Michael F, Vinogradov EV, Campagnari AA. 2010. Identification and characterization of a glycosyltransferase involved in Acinetobacter baumannii lipopolysaccharide core biosynthesis. Infect Immun 78:2017–2023. doi: 10.1128/IAI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hood MI, Becker KW, Roux CM, Dunman PM, Skaar EP. 2013. Genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect Immun 81:542–551. doi: 10.1128/IAI.00704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burmølle M, Bahl MI, Jensen LB, Sørensen SJ, Hansen LH. 2008. Type 3 fimbriae, encoded by the conjugative plasmid pOLA52, enhance biofilm formation and transfer frequencies in Enterobacteriaceae strains. Microbiology 154:187–195. doi: 10.1099/mic.0.2007/010454-0. [DOI] [PubMed] [Google Scholar]
- 15.Pederick VG, Eijkelkamp BA, Ween MP, Begg SL, Paton JC, McDevitt CA. 2014. Acquisition and role of molybdate in Pseudomonas aeruginosa. Appl Environ Microbiol 80:6843–6852. doi: 10.1128/AEM.02465-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rashid MH, Rumbaugh K, Passador L, Davies DG, Hamood AN, Iglewski BH, Kornberg A. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97:9636–9641. doi: 10.1073/pnas.170283397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 18.Rumbo-Feal S, Gómez MJ, Gayoso C, Álvarez-Fraga L, Cabral MP, Aransay AM, Rodríguez-Ezpeleta N, Fullaondo A, Valle J, Tomás M, Bou G, Poza M. 2013. Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells. PLoS One 8(8):e72968. doi: 10.1371/journal.pone.0072968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xavier BB, Sabirova J, Pieter M, Hernalsteens JP, de Greve H, Goossens H, Malhotra-Kumar S. 2014. Employing whole genome mapping for optimal de novo assembly of bacterial genomes. BMC Res Notes 7:484. doi: 10.1186/1756-0500-7-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabral MP, Soares NC, Aranda J, Parreira JR, Rumbo C, Poza M, Valle J, Calamia V, Lasa I, Bou G. 2011. Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J Proteome Res 10:3399–3417. doi: 10.1021/pr101299j. [DOI] [PubMed] [Google Scholar]