Abstract
Strategies to prevent the development of antibiotic resistance in bacteria are needed to reduce the threat of infectious diseases to human health. The de novo acquisition of resistance due to mutations and/or phenotypic adaptation occurs rapidly as a result of interactions of gene expression and mutations (N. Handel, J. M. Schuurmans, Y. Feng, S. Brul, and B. H. Ter Kuile, Antimicrob Agents Chemother 58:4371–4379, 2014, http://dx.doi.org/10.1128/AAC.02892-14). In this study, the contribution of several individual genes to the de novo acquisition of antibiotic resistance in Escherichia coli was investigated using mutants with deletions of genes known to be involved in antibiotic resistance. The results indicate that recA, vital for the SOS response, plays a crucial role in the development of antibiotic resistance. Likewise, deletion of global transcriptional regulators, such as gadE or soxS, involved in pH homeostasis and superoxide removal, respectively, can slow the acquisition of resistance to a degree depending on the antibiotic. Deletion of the transcriptional regulator soxS, involved in superoxide removal, slowed the acquisition of resistance to enrofloxacin. Acquisition of resistance occurred at a lower rate in the presence of a second stress factor, such as a lowered pH or increased salt concentration, than in the presence of optimal growth conditions. The overall outcome suggests that a central cellular mechanism is crucial for the development of resistance and that genes involved in the regulation of transcription play an essential role. The actual cellular response, however, depends on the class of antibiotic in combination with environmental conditions.
INTRODUCTION
Antibiotic-resistant bacteria pose a serious threat to human health, as the costs of therapy of infections caused by such bacteria increase and the treatment outcome is negatively affected. Bacteria can become resistant de novo by genetic or phenotypic changes and also through the acquisition of resistance-conferring mobile genetic elements. Resistance to antibiotics is rapidly induced as a result of exposure to stepwise increasing sublethal drug concentrations (1). In less than 100 generations, bacterial cells developed genetic mutations and permanent transcriptional changes (2). On the one hand, these cellular modifications allow the population to grow in the presence of high antibiotic concentrations, but on the other hand, they may decrease fitness or cause a metabolic burden (1–3). This metabolic cost does not necessarily come in the form of an increased energy requirement. For example, the adaptation of Escherichia coli to amoxicillin was accompanied by a reduced ecological range, because resistant cells were less able to grow well under adverse external conditions (4).
In order to devise measures to prevent or at least slow the development of antibiotic resistance, it is essential to understand the reaction of bacteria to drug exposure at the molecular level. Genes that were permanently differentially regulated in E. coli cells made resistant to amoxicillin, enrofloxacin, or tetracycline compared to their regulation in their sensitive ancestor (2) are likely to play a role in the development of resistance. For example, gadABC and hdeA, which confer resistance to acidic conditions, were differentially expressed in cells made permanently resistant to amoxicillin, enrofloxacin, or tetracycline by exposure to stepwise increasing antibiotic levels. The change in the expression of gadABC and hdeA was the only overlap between all cells made resistant to these three antibiotics. gadE is a common regulator of gadABC and hdeA and may therefore influence the de novo acquisition of antibiotic resistance (5). Other genes that have been documented to affect antibiotic resistance are ompF, which codes for an outer membrane porin (6, 7); the transcriptional regulator soxS (8), which is involved in superoxide removal; and recA, which codes for a nucleoprotein that is involved in the SOS response (2, 7, 9). The first aim of this study was to establish whether these genes, in addition to influencing resistance itself, also affect the acquisition of resistance. Therefore, the rates of adaptation of ΔgadE, ΔsoxS, ΔompF, and ΔrecA mutants were compared to those of the wild type.
The acquisition of antibiotic resistance under optimized culture conditions and the underlying molecular mechanisms causing the cellular response are well documented (2, 10–12). The second aim of this study was to investigate whether additional stress factors, such as low pH or increased NaCl concentrations, influence the ability of the cell to acquire antibiotic resistance de novo. In general, environmental stress is known to induce bacterial stress responses that can in turn increase mutagenesis rates and therefore may be beneficial for bacteria exposed to antibiotics (13). Nonetheless, E. coli cells made resistant to amoxicillin were more affected by sublethal antibiotic concentrations in the presence of a second stress factor, indicating that the cell could manage exposure to certain levels of antibiotics or adverse environmental conditions but not both simultaneously (4). To elucidate whether the development of resistance in E. coli is hampered by the application of additional environmental stress, the effects of a lowered pH or an increased salt concentration on adaptation to amoxicillin and enrofloxacin were documented. These antibiotics were chosen for their different modes of antimicrobial action, whereby the β-lactam amoxicillin interferes with cell wall synthesis and the fluoroquinolone enrofloxacin inhibits DNA replication by binding to gyrA. The combined results of this study provide insight into the cellular processes involved in the acquisition of resistance.
MATERIALS AND METHODS
Bacterial strains, growth media, antibiotics, and MIC measurement.
The drug-sensitive wild-type strains E. coli MG1655 (F− λ− rph-1) and E. coli BW25113 were used as controls throughout this study. Both BW25113 and MG1655 are derived from the W1485 strain background and are therefore closely related. Single-knockout (ΔrecA ΔgadE ΔsoxS ΔompF) strains were obtained from a BW25113 [Δ(araD-araB)567 Δ(rhaD-rhaB)568 ΔlacZ4787 (::rrnB-3) hsdR514 rph-1] deletion library (14, 15). Cells were grown in 100-ml flasks at 37°C in a phosphate-buffered (100 mM Na2H2PO4) defined minimal medium containing 55 mM glucose and shaken at 200 rpm (16). If not stated otherwise, the pH was set to 6.9 with 4 N NaOH. In experiments performed under various environmental conditions, the pH was set to 6.0 or an additional 2% NaCl was added to the medium. In experiments to study the development of resistance under nonoptimal conditions, wild-type cells were adapted for 1 week by the daily transfer of cells at an optical density at 600 nm (OD600) of 0.1 into fresh medium, resulting in approximately 70 generations for each condition tested: (i) pH 6.9 and 0% additional NaCl, (ii) pH 6.0 and 0% additional NaCl, and (iii) pH 6.9 and 2% additional NaCl. As ΔrecA cells were not able to grow in defined mineral medium, these cells were grown in LB medium, as were the wild-type strains, when they were used as a control for the ΔrecA mutant. LB medium consisted of 5 g/liter NaCl (Sigma-Aldrich, Steinheim, Germany), 2.5 g yeast extract (Scharlau-Microbiology, Barcelona, Spain), and 5 g Bacto tryptone (Brunschwig Chemie, Amsterdam, The Netherlands).
Amoxicillin and enrofloxacin stock solutions (10 mg/ml) were filter sterilized through a 0.2-μm-pore-size filter and stored at 4°C. The wild-type strains and the deletion mutants were grown with 1.25 μg/ml amoxicillin or 0.125 μg/ml enrofloxacin to induce the buildup of resistance, corresponding to 0.25× MIC. The starting enrofloxacin concentration for the ΔrecA deletion mutant was reduced to 0.03125 μg/ml, as cells showed no growth with higher concentrations of this antibiotic. Resistance to antibiotics was induced by stepwise increasing the drug concentration with every transfer cycle when almost normal growth occurred (1). With every increase in antibiotic concentration, 0.5 ml of culture that had been grown overnight was mixed with 0.5 ml of 60% sterile glycerol and stored at −80°C for further analysis. The MIC values were measured by following the growth in 96-well plates as described previously (17), using duplicate serial dilutions of a factor of 2 ranging from 0 to 1,024 μg/ml of each antibiotic. The MIC was defined as the lowest concentration of antibiotic that reduced the growth to an OD595 of 0.2 or less after 23 h.
Amplification and sequencing of resistance-conferring gene loci.
After plating of samples from the −80°C glycerol stock on LB agar plates, 2 colonies were randomly chosen for sequencing of the resistance-conferring gene loci. Amplification was performed in 50-μl working volumes with Taq DNA polymerase (Thermo Scientific), using the following parameters: denaturation at 95°C for 5 min, followed by 35 cycles of 35 s at 95°C, 55 s at 49°C, and 90 s at 72°C and, finally, a 90-s extension at 72°C. An MSB Spin PCRapace kit (Invitek) was used to purify the amplified PCR products. The oligonucleotide primers used for amplification of the resistance-conferring regions were identical to those used in a previous study (2). The PCR products were sequenced by Macrogen Europe.
RNA isolation and qRT-PCR.
E. coli precultures were grown overnight without antibiotics in defined minimal medium under different conditions. Cells were inoculated to an OD600 of 0.2 in fresh medium with or without 0.125 μg/ml enrofloxacin. When the cultures reached an OD600 of 1.0, the cells were harvested and directly placed into RNAlater RNA stabilization reagent (Qiagen). After centrifugation for 3 min at 4,000 rpm and 4°C, the pellets were flash-frozen in liquid nitrogen and stored at −80°C. The total RNA was extracted by adding 500 μl of RNeasy lysis buffer containing 1% mercaptoethanol and incubated at room temperature for 5 min. The lysed cells were extracted twice with acid phenol, followed by two chloroform extractions. Total RNA was precipitated with isopropanol, incubated overnight at −80°C, and centrifuged for 30 min at 4°C. The pellet was washed with ice-cold 75% ethanol and redissolved in 100 μl RNase-free water. The RNA samples were purified with an RNeasy kit (Qiagen), and RNA quality was verified on a 1% agarose gel to ensure the absence of RNA degradation. The amount of RNA was measured on a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). DNA residues were removed with the help of Ambion Turbo DNA-free DNase treatment and removal reagents according to the manufacturer's instructions. First-strand cDNA was synthesized from RNA (1 μg) using a RevertAid first-strand cDNA synthesis kits (Fermentas). The final concentrations of cDNA and primers in a total volume of 50 μl were 20 ng and 50 nM, respectively. The optimal primer concentration was verified beforehand for each primer pair. The primers used for quantitative real-time reverse transcription-PCR (qRT-PCR) used in this study are shown in Table 1. qRT-PCR was performed with a 7300 real-time PCR system (Applied Biosystems) and universal cycling conditions (2 min at 50°C, 10 min at 95°C, 40 cycles of 15 s at 95°C and 1 min at 60°C) using the Power SYBR green PCR master mix (Life Technologies). Cycle threshold (CT) values were determined by automated threshold analysis with ABI Prism (version 1.0) software. The relative abundance was calculated and normalized by the ΔΔCT method, using idnT as the reference gene, as described by Zhou et al. (18). Stable expression of the reference gene was verified with expression data obtained in a microarray study comparing the levels of expression by antibiotic-exposed sensitive and antibiotic-resistant E. coli cells to the levels of expression by wild-type E. coli cells (2). The performance of the qRT-PCR under the above-mentioned conditions was validated according to the manufacturer's instructions using serial dilutions of template cDNA.
TABLE 1.
Primers used for qRT-PCR in this study
| Gene | Orientation | Oligonucleotide sequence |
|---|---|---|
| idnT | Forward | 5′-CGCCACTACGCTGATTGCT-3′ |
| Reverse | 5′-TCACTAGCGCCCATTGCA-3′ | |
| rpoS | Forward | 5′-TCGCCGCCGGATGA-3′ |
| Reverse | 5′-GGCGGGCAATTTTTACCA-3′ | |
| soxS | Forward | 5′-GGCCGCCGTTGAGTTG-3′ |
| Reverse | 5′-GGTCCATTGCGATATCAAAAATC-3′ | |
| umuD | Forward | 5′-TGATATTGTCATCGCTGCTGTTG-3′ |
| Reverse | 5′-CGGGCGTAGTTGCAATTTTT-3′ |
RESULTS
Effect of gadE, soxS, and ompF deletions on acquisition of antibiotic resistance.
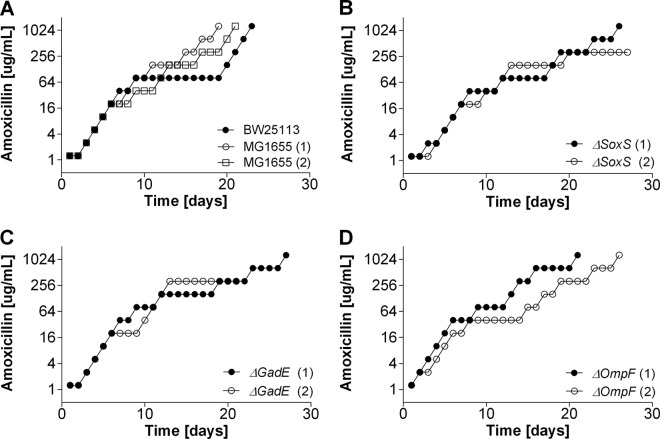
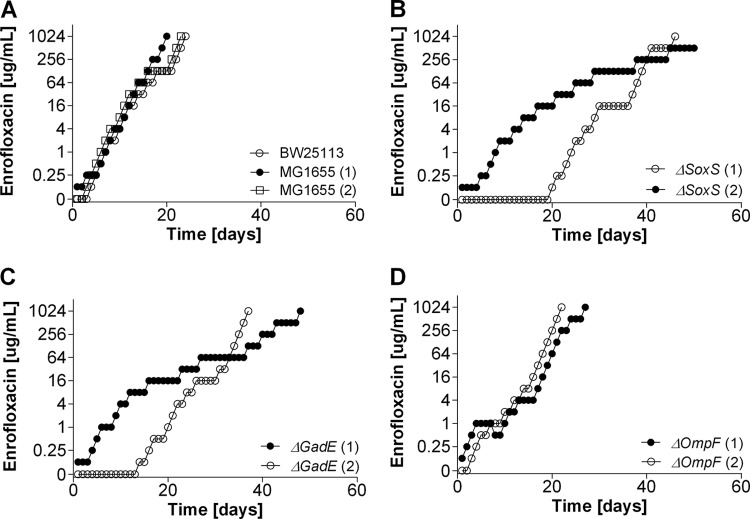
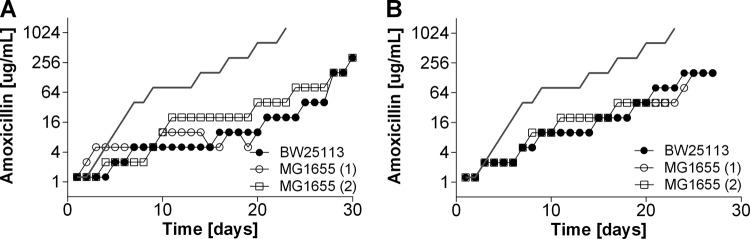
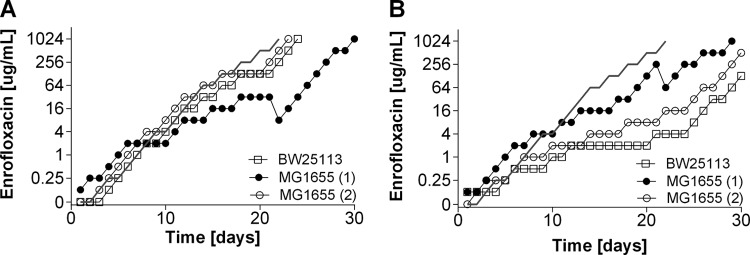
The single deletion of gadE (which is involved in internal pH regulation), soxS (which plays a role in superoxide removal), and ompF (which is an outer membrane porin) had only a marginal effect on sensitivity to amoxicillin compared to that of the wild-type strains (Table 2). The MICs of enrofloxacin for the ΔsoxS and ΔgadE strains were reduced 8- and 4-fold, respectively. The induction of resistance to amoxicillin and enrofloxacin was affected in different ways by these deletions. During the acquisition of resistance to amoxicillin, adaptation rates were similar for all strains (Fig. 1), but the ΔgadE and ΔsoxS mutants adapted to enrofloxacin slower than the wild-type strains (Fig. 2). Adaptation to the bacteriostatic agent tetracycline was very similar in the wild-type strains and the ΔsoxS mutants (data not shown). Cells of the ΔompF strain containing a deletion of outer membrane protein F had rates of adaptation to increasing enrofloxacin levels similar to those of the wild-type strains. Sequencing of resistance-conferring regions revealed that the mutations in the wild-type strains and mutants were the same as those usually reported to accompany acquired resistance to a member of the penicillin group or a fluoroquinolone (Tables 3 and 4).
TABLE 2.
MICs of amoxicillin and enrofloxacin for the E. coli MG1655 and BW25113 wild-type strains and deletion mutants grown in defined mineral or LB medium
| Medium and strain | MIC (μg/ml) |
|
|---|---|---|
| Amoxicillin | Enrofloxacin | |
| Defined mineral medium | ||
| Wild type | 4 | 0.5 |
| ΔsoxS mutant | 8 | 0.0625 |
| ΔgadE mutant | 4 | 0.125 |
| ΔompF mutant | 4 | 1 |
| LB medium | ||
| Wild type | 16 | 0.25 |
| ΔrecA mutant | 8 | 0.03125 |
FIG 1.
Acquisition of resistance to amoxicillin in the E. coli MG1655 and BW25113 wild-type strains and ΔsoxS, ΔompF, and ΔgadE knockout strains in mineral medium. Cells were adapted by stepwise increasing the drug concentration by a factor of 2 when growth was comparable to that of the wild-type cells.
FIG 2.
Acquisition of resistance to enrofloxacin in E. coli MG1655 and BW25113 wild-type cells and ΔsoxS, ΔompF, and ΔgadE cells in mineral medium.
TABLE 3.
Mutations found in ampC promoter region of cells of wild-type strains BW25113 and MG1655 and deletion mutants during acquisition of resistance to amoxicillin in mineral medium or LB mediuma
| Amoxicillin concn (μg/ml) | Mutation found in the indicated strain in the following medium: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EV |
LB medium |
|||||||||||||
| BW25113 | MG1655 |
ΔsoxS mutant |
ΔgadE mutant |
ΔompF mutant in EV |
BW25113 | MG1655 |
ΔrecA mutant |
|||||||
| 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | |||
| 0 | ||||||||||||||
| 2.5 | Interbox* | −35 box | ||||||||||||
| 10 | Interbox | −10 box, −35 box | −35 box | −10 box* | −10 box, attenuator | −35 box | −10 box | −35 box | −10 box | Interbox* | −10 box | −10 box | −10 box | −10 box, interbox |
| 20 | Interbox | −10 box, −35 box | −35 box | Attenuator | −10 box, attenuator | −35 box | −10 box | −35 box | −10 box | Interbox | Interbox | −10 box, interbox | −10 box, interbox | Interbox |
| 320 | −10 box, interbox | −10 box, −35 box | Attenuator | Attenuator, interbox | −35 box, attenuator | −35 box | −10 box | −35 box* | −35 box, attenuator | Interbox | −10 box, attenuator | −10 box, interbox | −10 box, interbox | |
| 1,280 | −10 box, interbox | −10 box, −35 box, interbox | Attenuator | Attenuator, interbox | −35 box, attenuator | −35 box | −10 box | −35 box | Attenuator | Interbox | −10 box, attenuator | −10 box, interbox | −10 box | |
The ampC promoter region of E. coli consists of regulatory sequences located −10 and −35 bp upstream of the transcription start site (−10 and −35 box). The nucleotide sequence between the −10 and −35 box is referred to as the interbox. Expression of ampC in E. coli wild-type cells is kept at a low level by a growth rate-dependent attenuator mechanism, whereby mutations in this region can weaken RNA polymerase attenuation and, hence, increase ampC expression. Results for two independent biological replicates (1 and 2) are given for all strains except the BW25113 strain, for which the results are for only one assay and which served as an additional control for the MG1655 wild type. Results were observed in PCR products from two colonies unless indicated otherwise with an asterisk, in which case it was found in one out of two colonies. EV, Evans defined medium.
TABLE 4.
Mutations found in resistance-conferring regions of gyrA and parC in the wild-type strains and deletion mutants during acquisition of resistance to enrofloxacin in mineral mediuma
| Enrofloxacin concn (μg/ml) | Mutations(s) found in the following region of the indicated strain: |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
gyrA |
parC |
|||||||||||||||||
| BW25113 | MG1655 |
ΔsoxS mutant |
ΔgadE mutant |
ΔompF mutant |
BW25113 | MG1655 |
ΔsoxS mutant |
ΔgadE mutant |
ΔompF mutant |
|||||||||
| 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | |||
| 0 | ||||||||||||||||||
| 0.125 | D87G | S83L | S83L | S83L | D87G | D87Y | D87G | S83L | ||||||||||
| 0.25 | S83L | S83L | S83L, D87N | D87Y | S83L | G81C, R38R* | S83L, D87Y* | D87G | S83L | |||||||||
| 0.5 | S83L, D87N | S83L | S83L, D87N | G81D | S83L, D87G | G81C | S83L | D87G, R38R* | S83L, D87G | |||||||||
| 16 | S83L, D87N | S83L, D87G | S83L, D87N | G81D | S83L, D87G | G81D | S83L | S83L, D87G | S83L, D87G | S80I, E84V | S80I | S80I, E84V | S80I | S80R | S80I | S80I | ||
| 1,024 | S83L, D87N, H91T | S83L, D87G | S83L, D87N | G81D | S83L, D87G | G81D | S83L, D87N | S83L, D87G | S83L, D87G | S80I, E84V | S80I | S80I, E84V | S80I, D79A | S80I | S80R | S80I | E84G | G78D |
Results are for two independent replicates (1 and 2) except for the BW25113 wild type, for which the results are for only one assay. *, only one out of two colonies carried the mutation.
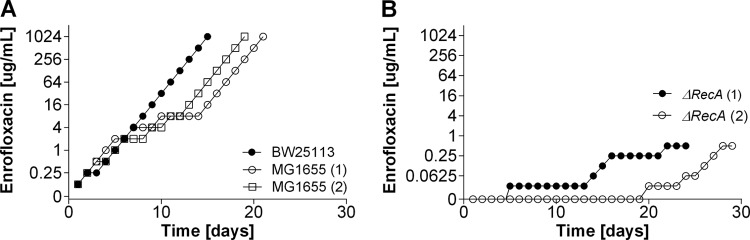
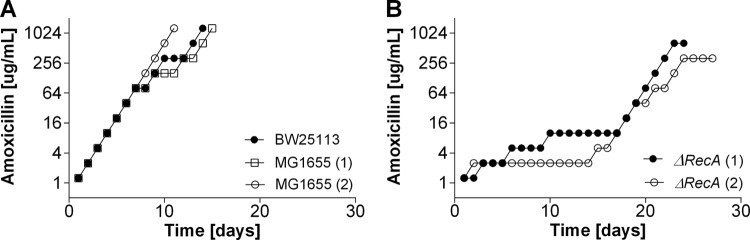
Acquisition of antibiotic resistance in the ΔrecA mutant with an ineffective SOS response.
To study the development of antibiotic resistance in cells with an ineffective SOS response, complex LB medium had to be used, as the ΔrecA strain was not able to grow in mineral medium. The time that wild-type cells needed to adapt in mineral medium and complex medium was similar to that needed by the ΔrecA strain (compare Fig. 1 to 4). If anything, adaptation of wild-type cells to amoxicillin seemed to benefit from complex medium, as growth in the presence of 1,280 μg/ml had already occurred after 11 days (Fig. 3). Adaptation to amoxicillin concentrations higher than 2.5 μg/ml in mineral medium always resulted in at least one mutation in the ampC promoter region, but in LB medium with amoxicillin at 320 and 1,280 μg/ml, one of the wild-type replicates did not have any of the usual mutations (Table 3). The MIC of amoxicillin for ΔrecA cells was only marginally affected (Table 2). The buildup of resistance to amoxicillin in ΔrecA cells was temporarily halted at 2 and 8 μg/ml (Fig. 3), corresponding to the range of concentrations in which resistance-conferring mutations occur in the ampC promoter region (2). After some time, ΔrecA cells were able to adapt to the same amoxicillin concentrations as the wild-type strains. This adaptation was accompanied by two of the regularly observed mutations (Table 3).
FIG 4.
Acquisition of resistance to enrofloxacin in E. coli MG1655 and BW25113 wild-type cells and ΔrecA cells in complex LB medium.
FIG 3.
Acquisition of resistance to amoxicillin in E. coli MG1655 and BW25113 wild-type cells and ΔrecA cells in complex LB medium.
During the development of enrofloxacin resistance, the wild-type strains had very similar rates of increase in their MICs and basically the same mutations in gyrA or ampC in LB medium and in mineral medium (Fig. 2 and 4 and Tables 4 and 5). The MIC value of enrofloxacin for the ΔrecA strain was already reduced 8-fold compared to that for the wild-type strains (Table 2). As a consequence, the starting sub-MIC had to be lowered from 0.125 μg/ml for wild-type cells to 0.03125 μg/ml for ΔrecA cells. After 6 days of growth in LB medium and, subsequently, 5 days of growth in fresh medium containing 0.03125 μg/ml enrofloxacin, sufficient growth was obtained to continue the process of adaptation (Fig. 4, replicate 1). After 20 days in one replicate or 30 days in the other replicate, ΔrecA cells were adapted to a concentration of only 0.5 μg/ml, twice the MIC for the wild-type strains in LB medium. In contrast to the wild-type strains, which showed a first resistance-conferring mutation in gyrA with 0.125 μg/ml enrofloxacin, the first mutation in gyrA of ΔrecA cells was identified at a concentration of only 0.25 μg/ml (Table 5).
TABLE 5.
Mutations found in resistance-conferring regions of gyrA and parC in the wild-type strains and ΔrecA mutant during the acquisition of resistance to enrofloxacin in LB mediuma
| Enrofloxacin concn (μg/ml) | Mutations(s) found in the following region of the indicated strain: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
gyrA |
parC |
|||||||||
| BW25113 | MG1655 |
ΔrecA mutant |
BW25113 | >MG1655 |
ΔrecA mutant |
|||||
| 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | |||
| 0 | ||||||||||
| 0.125 | S83L | G81D | S83L | |||||||
| 0.25 | S83L | S83L | S83L | D87Y | S83L | |||||
| 0.5 | S83L | S83L | S83L | D87Y | D87Y | |||||
| 16 | S83L, D87N | S83W | S83L | ND | ND | S80I, E84V | G78D | G78D | ND | ND |
| 1,024 | S83L, D87N | S83L | S83L, D87N | ND | ND | S80I, E84V | S80R | S80R | ND | ND |
Results are for two replicates (1 and 2) except for the BW25113 wild type, for which the results are for only one assay. ND, not determined, as the ΔrecA mutant could not grow at these concentrations.
Acquisition of antibiotic resistance in suboptimal environments.
The development of resistance under nonoptimal conditions, such as in the presence of increased salt concentrations or reduced pH, differed from the adaptation achieved in optimized mineral medium (pH 6.9, 0% additional NaCl) (Fig. 5 and 6). The maximum specific growth rate of wild-type cells in defined mineral medium decreased from 0.7 h−1 to 0.54 h−1 upon addition of 2% NaCl. A change of the pH from 6.9 to 6.0 lowered the maximum specific growth rate to 0.39 h−1. The counts of a culture grown overnight (in numbers of CFU per milliliter) did not differ significantly between pH 6.9 and 6.0 (a P value of ≥0.05 by Student's t test was considered significant; for pH 6.9 and 0% additional NaCl, P = 3.8 × 109; for pH 6.0 and 0% additional NaCl, P = 2 × 109; for pH 6.9 and 2% additional NaCl, P = 3.7 × 109). In all incubations, the final OD600 was between 4 and 7, indicating that a similar number of divisions had occurred. Therefore, the increase in the MIC per generation could be compared between the various incubations. Adaptation to amoxicillin (Fig. 5) was far more affected by these slightly adverse conditions than adaptation to enrofloxacin (Fig. 6). The mutations that accompanied the acquisition of resistance to amoxicillin were the same as those found in the wild-type strains under standard conditions (data not shown). In contrast, sequencing of resistance-conferring regions in gyrA revealed different patterns in the accumulation of genetic mutations leading to enrofloxacin resistance (compare Table 6 to Table 4). In the presence of a reduced pH or increased salt concentrations, no mutations in gyrA were found when cells were grown in the presence of 0.125 μg/ml enrofloxacin; mutations in gyrA were found only when cells were grown in the presence of enrofloxacin at a concentration of 0.25 μg/ml or higher. The mutations observed in the presence of additional stress were almost never the usual ones. In contrast, wild-type cells cultured in LB or mineral medium under optimized conditions already showed the usual mutations at the lowest enrofloxacin concentration applied (Table 4). Once cells were able to grow in the presence of 16 μg/ml enrofloxacin under optimized conditions, the usual G78D mutation in parC was observed. Under nonoptimal conditions, mutations in parC were found only at higher enrofloxacin concentrations. Cells made resistant to 1,024 μg/ml enrofloxacin in medium with a pH of 6 or an increased salt concentration showed either no mutation in parC or one of four unique mutations that were not found under optimal conditions.
FIG 5.
Acquisition of resistance to amoxicillin in the presence of pH 6 (A) or 2% NaCl (B) in the E. coli MG1655 and BW25113 wild-type strains. The lines without points represent the averages for the three controls (MG1655 twice and BW25113 once) at pH 6.9 without additional NaCl. Cells were adapted to pH 6 or 2% NaCl by daily passaging to fresh medium without any antibiotic for 7 days.
FIG 6.
Acquisition of resistance to enrofloxacin in the presence of pH 6 (A) or 2% NaCl (B) in the E. coli MG1655 and BW25113 wild-type strains. The lines without points represent the average for the three controls at pH 6.9 without additional NaCl. Cells were adapted to pH 6 or 2% NaCl by daily passaging to fresh medium without any antibiotic for 7 days.
TABLE 6.
Mutations found in resistance-conferring regions of gyrA and parC in wild-type cells adapted to enrofloxacin in mineral medium under different conditionsa
| Enrofloxacin concn (μg/ml) | Mutations(s) found in the following region of the indicated strain under the indicated condition: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
gyrA |
parC |
||||||||||
| BW25113, pH 6 | MG1655, pH 6 |
BW25113, 2% salt gyrA | MG1655, 2% salt gyrA1 |
BW25113, pH 6 | MG1655, pH 6 parC | BW25113, 2% salt parC | MG1655 2% salt, parC1 |
||||
| 1 | 2 | 1 | 2 | 1 | 2 | ||||||
| 0 | |||||||||||
| 0.125 | S83 deletionb | ||||||||||
| 0.25 | G81D | D87G | D87N | D87T | |||||||
| 0.5 | G81D | D87N | D87N | ||||||||
| 16 | D87N | S83W | D87N | D87G | S83L | D87G | S80I | S80R | |||
| 1,024 | G81D, D87N | G81D | D87N | D87N | S83L, D87G | S83L, D87G | E84G | S80R | G78D | E84G | S80I |
Results are for two independent biological replicates (1 and 2) for all strains except the BW25113 strain, for which the results are for only one assay.
The S83 deletion was observed in only one out of two colonies.
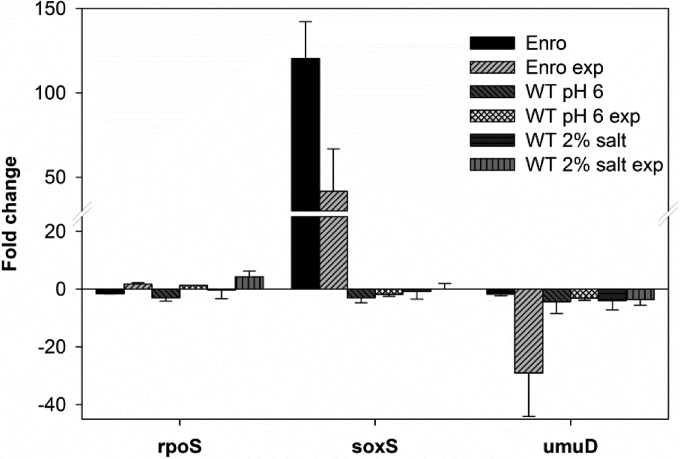
Resistance to enrofloxacin strongly depends on mutations in gyrA and parC. Bacteria induce transient mutagenesis mechanisms in response to stress (19). To test if changes in the cellular stress response account for the different mutational patterns observed during the acquisition of resistance under various environmental conditions, the levels of rpoS (general stress), soxS (oxidative stress), and umuD (SOS response) expression were investigated using qRT-PCR (Fig. 7). In agreement with previous results obtained in a microarray study (4), expression of soxS was upregulated more than 100 times in enrofloxacin-adapted cells. Under nonoptimized conditions, whether or not they were used in combination with exposure to 0.125 μg/ml enrofloxacin, expression of soxS did not change in the wild type. There was little variation in the expression of the general master stress regulator rpoS. The largest effect was a 4-fold induction in cells grown in the presence of additional salt (2% NaCl) and exposed to 0.125 μg/ml enrofloxacin. The SOS response, measured as the expression of umuD, was strongly reduced when enrofloxacin-resistant cells were exposed to that drug. In addition, umuD was downregulated at least 3-fold under suboptimal growth conditions with and without enrofloxacin.
FIG 7.
Change in expression levels of selected genes in enrofloxacin-resistant (Enro) and E. coli MG1655 wild-type (WT) cells cultured in mineral medium at pH 6 or with additional salt (2% NaCl) compared to that in the wild-type strain (pH 6.9, 0% additional NaCl). Expression levels were compared to those by the wild type in the absence or presence (exp) of 0.125 μg/ml enrofloxacin. Data are the means ± SDs for two biological replicates.
DISCUSSION
The first conclusion to be drawn from the results presented above is that E. coli acquires de novo resistance to amoxicillin in a very different manner than to enrofloxacin. While the development of resistance to enrofloxacin was affected in all mutants examined except for the ΔompF mutant, the acquisition of resistance to amoxicillin was delayed only in ΔrecA cells, which have an incomplete SOS response. On the one hand, this outcome seems to concur with the different modes of action of both antibiotics. On the other hand, both are biocidal drugs and as such may have a common mechanism for disrupting the cell involving reactive oxygen species (ROS) (10, 20). E. coli cells adapted to amoxicillin faster in rich medium than in mineral medium, but in the case of enrofloxacin, there was no difference in the time of adaptation in the two media. This observation is in line with the analysis of the difference in gene expression in E. coli cells growing on mineral medium and E. coli cells growing on rich medium (21), because adaptation to amoxicillin involves more drastic changes in gene expression than adaptation to enrofloxacin. The cause for the more rapid adaptation is that the E. coli cells produced more β-lactamase in rich medium than mineral medium. Thus, the nutrient-rich environment may promote the development of antibiotic resistance if the resistance mechanism depends on the overproduction of predominantly antibiotic-degrading enzymes, such as β-lactamases. In the case of enrofloxacin, resistance increases once mutations in gyrA and/or parC that change the confirmation of the antibiotic target can be identified. Thus, depending on the resistance mechanism, the rate of acquisition of resistance can be influenced by external conditions, such as nutrient availability. In addition, cells experience less osmotic stress in a rich medium than in a mineral medium, so defects in the cell wall caused by β-lactam antibiotics are less harmful.
The delay in amoxicillin resistance buildup caused by the incomplete SOS response of ΔrecA mutants suggests that a well-functioning SOS system increases the chance that mutations upregulating β-lactamase expression will occur (2). The lack of an effect of the other mutations tested indicates that pH regulation, superoxide removal, or the OmpF outer membrane porin is not involved in the de novo acquisition of β-lactam resistance under our test conditions. The effect of internal pH regulation, however, may not be black and white. The strongly reduced buildup of resistance in the presence of a lower pH or increased salt levels indicates that E. coli cannot simultaneously handle well the challenges of suboptimal environmental conditions and antibiotics. This is in line with the suggestion that the costs of resistance can consist of a reduced ecological range (4). An alternative explanation could be that β-lactamase, which is an ectoenzyme, itself has reduced specific activity under these conditions. Sensitive, intermediately resistant, and highly resistant cells can coexist within a growing clonal population (22, 23). In biofilm matrices, for example, the overproduction of chromosomal β-lactamases degraded β-lactam antibiotics before they reached sensitive cells located in the biofilm center (24, 25). Thus, in a complex medium even within a clonal population, a small fraction of active β-lactamase producers could protect the susceptible cells by altruism.
The SOS response is clearly essential for the development of resistance to enrofloxacin, as the ΔrecA mutants could barely adapt. Both reduced superoxide removal, which is done by soxS, and regulation of the internal pH, which is done by gadE, influenced the ability to acquire resistance. In general, acidic conditions are associated with the upregulation of oxidative damage-related genes (26), so these effects may not be independent. Fluoroquinolones can enter cells via multiple porins (OmpA, OmpC, and OmpF) as well as via nonporin pathways, directly passing through the lipid bilayer (27). The single ompF deletion mutant has been associated with increased fluoroquinolone resistance several times (6, 27, 28). In this study, the absence of this porin in the outer membrane played no role, possibly because alternative entries were available. Since resistance to fluoroquinolones is initially determined by point mutations in gyrA and resistance at higher levels is determined by point mutations in parC (28, 29), this is not unexpected. The porin is less likely than superoxide or internal pH to influence the rates of mutation. The slowdown of the acquisition of effective mutations in the mutants is, to a certain extent, counterintuitive, as it would be expected that mutation rates would increase in the absence of protection against stressors. Either this is not the case or the accumulated mutations, which are different from those found in the wild type, are less effective. The additional stress factors of reduced pH and increased salt caused other mutations to be acquired, and certainly in the case of additional salt, these were less effective than the usual ones.
The delay in the appearance of mutations observed in cultures adapting to enrofloxacin under nonoptimized conditions is in agreement with results obtained with the recA deletion mutant and provides supporting evidence for the importance of the SOS response in the acquisition of antibiotic resistance. Adaptation to acidic conditions reduced the level of soxS expression and lowered the level of expression of umuD, which depends on RecA activation, creating a genetic background similar to that in the recA deletion mutant and thereby possibly causing the delayed genetic change in gyrA and parC. Inhibition of the SOS response could prolong the efficacy of fluoroquinolone antibiotics because of its crucial role in modulating adaptation, mutation (30), and horizontal transfer (31) and provides a means to combat the evolution of bacterial resistance to antibiotics.
Overall, the process of adaptation to antibiotics is complex and consists of an initial response of regulatory networks changing their gene expression and subsequent mutations. Induction of resistance to enrofloxacin was slowed in ΔsoxS mutants, indicating that the cells were hampered by the increased production of superoxide due to exposure to bactericidal antibiotics. Even though we did not observe any slowdown in the development of amoxicillin resistance in ΔsoxS cells, ROS production may still contribute to bactericidal cell killing, as shown previously (10). However, ROS production can be encountered by multiple pathways using different sets of enzymes, for example, catalase or superoxide dismutase. The preference for certain ROS-scavenging enzymes could be accounted for by regulatory overlaps between the genetic responses caused by oxidative or antibiotic stress. SoxR regulates the expression of soxS and the multidrug efflux pump acrAB (32). Enrofloxacin-resistant cells may profit more by activating this cellular pathway to encounter elevated ROS production. Taken together, our data strongly suggest that the development of resistance to bactericidal antibiotics is caused by the activation of common underlying mechanisms but that the actual cellular response, as well as the contribution of single genes and environmental manipulations, varies for each class of antibiotic depending on the mode of action.
ACKNOWLEDGMENTS
The students Suzanne van Wouw, Kavel Ozturk, Lachmie Toewar, and Marc Russchenberg performed experiments as part of their degree requirements.
We have no competing financial or professional interests.
Funding Statement
This work was financed by The Netherlands Food and Consumer Product Safety Authority.
REFERENCES
- 1.van der Horst MA, Schuurmans JM, Smid MC, Koenders BB, Ter Kuile BH. 2011. De novo acquisition of resistance to three antibiotics by Escherichia coli. Microb Drug Resist 17:141–147. doi: 10.1089/mdr.2010.0101. [DOI] [PubMed] [Google Scholar]
- 2.Handel N, Schuurmans JM, Feng Y, Brul S, Ter Kuile BH. 2014. Interaction between mutations and regulation of gene expression during development of de novo antibiotic resistance. Antimicrob Agents Chemother 58:4371–4379. doi: 10.1128/AAC.02892-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez JL, Rojo F. 2011. Metabolic regulation of antibiotic resistance. FEMS Microbiol Rev 35:768–789. doi: 10.1111/j.1574-6976.2011.00282.x. [DOI] [PubMed] [Google Scholar]
- 4.Handel N, Schuurmans JM, Brul S, Ter Kuile BH. 2013. Compensation of the metabolic costs of antibiotic resistance by physiological adaptation in Escherichia coli. Antimicrob Agents Chemother 57:3752–3762. doi: 10.1128/AAC.02096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castanie-Cornet MP, Cam K, Bastiat B, Cros A, Bordes P, Gutierrez C. 2010. Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res 38:3546–3554. doi: 10.1093/nar/gkq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duval V, Nicoloff H, Levy SB. 2009. Combined inactivation of lon and ycgE decreases multidrug susceptibility by reducing the amount of OmpF porin in Escherichia coli. Antimicrob Agents Chemother 53:4944–4948. doi: 10.1128/AAC.00787-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harder KJ, Nikaido H, Matsuhashi M. 1981. Mutants of Escherichia coli that are resistant to certain beta-lactam compounds lack the ompF porin. Antimicrob Agents Chemother 20:549–552. doi: 10.1128/AAC.20.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosel M, Li L, Drlica K, Zhao X. 2013. Superoxide-mediated protection of Escherichia coli from antimicrobials. Antimicrob Agents Chemother 57:5755–5759. doi: 10.1128/AAC.00754-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webber MA, Piddock LJ. 2001. Absence of mutations in marRAB or soxRS in acrB-overexpressing fluoroquinolone-resistant clinical and veterinary isolates of Escherichia coli. Antimicrob Agents Chemother 45:1550–1552. doi: 10.1128/AAC.45.5.1550-1552.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 11.Lazar V, Nagy I, Spohn R, Csorgo B, Gyorkei A, Nyerges A, Horvath B, Voros A, Busa-Fekete R, Hrtyan M, Bogos B, Mehi O, Fekete G, Szappanos B, Kegl B, Papp B, Pal C. 2014. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat Commun 5:4352. doi: 10.1038/ncomms5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oz T, Guvenek A, Yildiz S, Karaboga E, Tamer YT, Mumcuyan N, Ozan VB, Senturk GH, Cokol M, Yeh P, Toprak E. 2014. Strength of selection pressure is an important parameter contributing to the complexity of antibiotic resistance evolution. Mol Biol Evol 31:2387–2401. doi: 10.1093/molbev/msu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poole K. 2012. Bacterial stress responses as determinants of antimicrobial resistance. J Antimicrob Chemother 67:2069–2089. doi: 10.1093/jac/dks196. [DOI] [PubMed] [Google Scholar]
- 14.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grenier F, Matteau D, Baby V, Rodrigue S. 2014. Complete genome sequence of Escherichia coli BW25113. Genome Announc 2(5):e01038-14. doi: 10.1128/genomeA.01038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans CGT, Herbert D, Tempest DW. 1970. The continuous culture of microorganisms. Construction of a chemostat, vol 2 Academic Press, London, United Kingdom. [Google Scholar]
- 17.Schuurmans JM, Nuri Hayali AS, Koenders BB, ter Kuile BH. 2009. Variations in MIC value caused by differences in experimental protocol. J Microbiol Methods 79:44–47. doi: 10.1016/j.mimet.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 18.Zhou K, Zhou L, Lim Q, Zou R, Stephanopoulos G, Too HP. 2011. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol Biol 12:18. doi: 10.1186/1471-2199-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hersh MN, Ponder RG, Hastings PJ, Rosenberg SM. 2004. Adaptive mutation and amplification in Escherichia coli: two pathways of genome adaptation under stress. Res Microbiol 155:352–359. doi: 10.1016/j.resmic.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao H, Bausch C, Richmond C, Blattner FR, Conway T. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol 181:6425–6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider C, Weindel M, Brade V. 1996. Frequency, clonal heterogeneity and antibiotic resistance of methicillin-resistant Staphylococcus aureus (MRSA) isolated in 1992-1994. Zentralbl Bakteriol 283:529–542. doi: 10.1016/S0934-8840(96)80131-9. [DOI] [PubMed] [Google Scholar]
- 23.Lee HH, Molla MN, Cantor CR, Collins JJ. 2010. Bacterial charity work leads to population-wide resistance. Nature 467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciofu O, Beveridge TJ, Kadurugamuwa J, Walther-Rasmussen J, Hoiby N. 2000. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J Antimicrob Chemother 45:9–13. doi: 10.1093/jac/45.1.9. [DOI] [PubMed] [Google Scholar]
- 25.Dibdin GH, Assinder SJ, Nichols WW, Lambert PA. 1996. Mathematical model of beta-lactam penetration into a biofilm of Pseudomonas aeruginosa while undergoing simultaneous inactivation by released beta-lactamases. J Antimicrob Chemother 38:757–769. doi: 10.1093/jac/38.5.757. [DOI] [PubMed] [Google Scholar]
- 26.Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol 187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chapman JS, Georgopapadakou NH. 1988. Routes of quinolone permeation in Escherichia coli. Antimicrob Agents Chemother 32:438–442. doi: 10.1128/AAC.32.4.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tavio MM, Vila J, Ruiz J, Martin-Sanchez AM, Jimenez de Anta MT. 1999. Mechanisms involved in the development of resistance to fluoroquinolones in Escherichia coli isolates. J Antimicrob Chemother 44:735–742. doi: 10.1093/jac/44.6.735. [DOI] [PubMed] [Google Scholar]
- 29.Redgrave LS, Sutton SB, Webber MA, Piddock LJ. 2014. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol 22:438–445. doi: 10.1016/j.tim.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. 2000. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J 19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beaber JW, Hochhut B, Waldor MK. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 32.Lee JO, Cho KS, Kim OB. 2014. Overproduction of AcrR increases organic solvent tolerance mediated by modulation of SoxS regulon in Escherichia coli. Appl Microbiol Biotechnol 98:8763–8773. doi: 10.1007/s00253-014-6024-9. [DOI] [PubMed] [Google Scholar]