Abstract
Plasmid-mediated qnr genes provide only a modest decrease in quinolone susceptibility but facilitate the selection of higher-level resistance. In Escherichia coli strain J53 without qnr, ciprofloxacin resistance often involves mutations in the GyrA subunit of DNA gyrase. Mutations in gyrA were absent, however, when 43 mutants with decreased ciprofloxacin susceptibility were selected from J53(pMG252) with qnrA1. Instead, in 13 mutants, individual and whole-genome sequencing identified mutations in marR and soxR associated with increased expression of marA and soxS and, through them, increased expression of the AcrAB pump, which effluxes quinolones. Nine mutants had increased expression of the MdtE efflux pump, and six demonstrated increased expression of the ydhE pump gene. Many efflux mutants also had increased resistance to novobiocin, another pump substrate, but other mutants were novobiocin hypersusceptible. Mutations in rfaD and rfaE in the pathway for inner core lipopolysaccharide (LPS) biosynthesis were identified in five such strains. Many of the pump and LPS mutants had decreased expression of OmpF, the major porin channel for ciprofloxacin entry. Three mutants had increased expression of qnrA that persisted when pMG252 from these strains was outcrossed. gyrA mutations were also rare when mutants with decreased ciprofloxacin susceptibility were selected from E. coli J53 with aac(6′)-Ib-cr or qepA. We suggest that multiple genes conferring low-level resistance contribute to enhanced ciprofloxacin resistance selected from an E. coli strain carrying qnrA1, aac(6′)-Ib-cr, or qepA because these determinants decrease the effective ciprofloxacin concentration and allow more common but lower-resistance mutations than those in gyrA to predominate.
INTRODUCTION
The plasmid-mediated quinolone resistance (PMQR) gene qnrA1 causes only a modest decrease in ciprofloxacin susceptibility but facilitates the selection of mutants with higher levels of ciprofloxacin resistance (1). In Escherichia coli, ciprofloxacin resistance results from mutations in the quinolone resistance-determining region (QRDR) of the GyrA subunit of DNA gyrase and mutations altering the regulation of efflux pumps, especially AcrAB (2, 3). Gyrase mutations are frequent in ciprofloxacin-resistant mutants of E. coli J53 without qnr but are surprisingly rare in its presence. Cesaro et al. found that only 2 of 200 mutants selected from J53 with qnrA had gyrA mutations, while 27% of those selected from qnr-free E. coli J53 or KL16 had gyrA mutations (4). The aim of this study was to explore the reason for the lack of gyrA mutations and to characterize the causes of non-gyrA-mediated enhanced ciprofloxacin resistance. A variety of mutations affecting susceptibility to ciprofloxacin and other antimicrobials was discovered.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli J53 (met pro azide resistant) (5) and plasmids pMG252 IncA-C, conferring resistance to β-lactams (due to blaFOX-5 and blaPSE-1), chloramphenicol (cmlA1 and catB2), gentamicin, kanamycin, quinolones (qnrA1), streptomycin (aadA2), sulfonamide (sul1), tobramycin, trimethoprim (dhfr), and mercuric chloride (mer) (1); pMG253 (qnrA1 from pMG252 cloned into pBC SK [cat]) (Agilent, Santa Clara, CA) (6); pMG320 [aac(6′)-Ib-cr cloned into pBC SK] (7); pAT851 with cloned qepA (8); and R692 (Ap Su Tc IncA-C) (9) were used. E. coli C600 (B1 leu thr) was used as the recipient for outcrossing from J53(pMG252) Cipr mutants. Spontaneous mutants of J53, J53(pMG252), J53(pMG253), J53(pMG320), and J53(pAT851) were selected with an inoculum of up to ∼1010 organisms on Mueller-Hinton agar plates containing ciprofloxacin at concentrations between the MIC and the mutant prevention concentration (MPC) of each strain after incubation at 37°C for 72 h.
Displacement of pMG252 by incompatibility.
E. coli C600(R692) was mated with J53(pMG252) Cipr mutants with selection on media with growth requirements of the recipient but not the donor and tetracycline at 25 μg/ml, to which the donor but not the recipient was resistant. Purified colonies were shown to lack resistances specific for pMG252 and tested for ciprofloxacin MICs.
Susceptibility testing.
MICs of ciprofloxacin, nalidixic acid, and novobiocin (Sigma-Aldrich) were determined by agar dilution on Mueller-Hinton agar at 37°C with an inoculum of ∼104 CFU according to CLSI guidelines (10). E. coli J53 and ATCC 25922 were used for quality control. Minimal bacterial concentrations (MBCs) for ciprofloxacin were determined by subculturing 200 μl from a microdilution well without visible growth after washing with Mueller-Hinton broth onto antibiotic-free Mueller-Hinton agar (10).
PCR and DNA sequencing.
We amplified by PCR and sequenced the QRDRs of the gyrA, gyrB, and parC genes (and, in some mutants, the entire genes); regulator genes such as marR, robA, acrR, soxR, soxS, and hns and the promoter of gadE; the lon protease gene; the efflux pump genes acrAB, acrEF, acrD, acrS, and mdtF and its promoter; ydhE, also with its promoter; the lipopolysaccharide (LPS) biosynthesis genes rfaE, rfaH, lpcA, and gmhB and the rfaDFCL and rfaQGPSBIJYZU operons; and other genes, including yoaA, surA, topB, and rbsA. DNA templates were prepared by boiling, and the primers used are listed in Table S1 in the supplemental material. We used Maxima Hot Start PCR master mix (Thermo Scientific, Waltham, MA) in a final volume of 50 μl. PCR products were purified by using a PCR purification kit (Qiagen, Valencia, CA) and sent for sequencing by the Tufts University Core Facility, Boston, MA.
Whole-genome sequencing.
Whole-genome sequencing was performed for 50 mutants by different facilities. Forty mutants (and the parental E. coli J53 strain) were sequenced by the Tufts University Genomics Core Facility. Genomic DNA was processed by using the Nextera XT DNA library preparation kit (Illumina). Libraries were size selected with fragment sizes of ≥500 bases recovered. The molar concentration of each library was determined with a fragment analyzer (Advanced Analytical), and libraries were mixed into a single pool at equal molar concentrations. The molar concentration of the pooled libraries was adjusted to 8.5 pM for sequencing on an Illumina MiSeq platform using a MiSeq V2 500 cycle kit, yielding 26.34 million pass-filter-paired reads. Five mutants were analyzed by BaseClear BV (Leiden, the Netherlands) on the Illumina HiSeq2500 platform after library preparation with the Nextera XT kit, and five others [including the J53(pMG252) parental strain] were analyzed by AstraZeneca (Waltham, MA), where they also constructed the libraries by using the Nextera XT kit. Libraries were then quantitated against a quantitative PCR (qPCR)-generated standard curve by Kapa BioSystems (Woburn, MA) to perform sequencing on an Illumina MiSeq platform with the V2 2 × 150 paired-end system. From each source, the raw data in FASTQ format were analyzed with CLC Genomics Workbench software (version 7.5) (CLC Bio, Aarhus, Denmark). After trimming of the sequences, the data were mapped to an E. coli J53 reference genome (GenBank accession number AICK01000001.1) to perform both basic and fixed ploidy variant detection. Variants were filtered for nucleotide changes producing amino acid alterations that occurred in >80% of the countable fragments. If no mutations were found, the frequency was lowered. All detected mutations were confirmed by conventional PCR and DNA sequencing using primers listed in Table S1 in the supplemental material.
Relative expression levels of genes encoding efflux pumps and regulators.
Reverse transcription followed by real-time qPCR (RT-qPCR) was used to determine the expression levels of selected efflux pump genes, including acrA, mdfA, ydhE, acrE, tolC, mdtE, mdtF; regulators such as marA, soxS, fis, dsrA, and evgA; and the porin protein-encoding genes ompC and ompF. Comparison was made to the expression level of the housekeeping gene mdh. Primers used are shown in Table S1 in the supplemental material. The same primers were also employed to generate PCR DNA products to perform qPCR standard plots for each gene tested.
Cultures were inoculated from −80°C stocks into LB broth, grown at 37°C overnight, diluted 1:100 into fresh LB broth, and grown for 2 to 3 h to an optical density at 600 nm (OD600) of ∼0.3 (Genesys 20 visible spectrophotometer). Total RNA in the cultures was stabilized by using RNAprotect bacterial reagent (Qiagen), bacteria were digested with lysozyme-proteinase K, and total RNA was isolated by using an RNeasy minikit (Qiagen) and On-Column DNase I digestion (Qiagen) according to the manufacturer's specifications. RNA purity and concentration were determined by using a NanoDrop 1000 spectrophotometer (Thermo Scientific). Reverse transcription was performed by using a Verso cDNA synthesis kit (Thermo Scientific). The cDNAs obtained were measured after 40 cycles by using a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA) with 20-μl reaction mixtures containing primers (500 nM each), cDNA (3 μl), and SsoFast EvaGreen supermix from Bio-Rad (10 μl). To calculate the relative expression levels, the threshold cycle (CT) value of the target gene was normalized to the CT value of the housekeeping gene, after the ΔCT value of the test sample was normalized to the ΔCT value of the parental strain, and finally, the expression ratio was calculated with the formula 2−ΔΔCT. At least three different assays with three independent cultures and RNA extractions were performed for each gene tested.
Assay for promoter efficiency.
The wild type and putative promoter mutations were cloned into plasmid pRS415 (11) upstream from its promoter-free lac gene, and comparative β-galactosidase activity in ΔlacZ E. coli strain BW25113 was assayed (12) as a measure of promoter efficiency.
RESULTS
When E. coli J53 or J53(pMG252) is plated onto increasing concentrations of ciprofloxacin above its MIC, the number of survivors progressively decreases (Fig. 1). The lowest concentration at which no mutants are observed is the mutant prevention concentration (MPC) (13). It represents the MIC of the least susceptible single-step mutant. For quinolone-susceptible strain J53, the ciprofloxacin MIC was 0.016 μg/ml, and the MPC was 16-fold higher at 0.25 μg/ml. In contrast, J53 containing plasmid pMG252 encoding QnrA1 had a ciprofloxacin MIC of 0.25 to 0.5 μg/ml and an MPC of between 1 and 2 μg/ml. The shape of the survival curves (Fig. 1) suggests that a variety of mutations provide lower-level resistance and that progressively fewer mutations confer higher-level resistance.
FIG 1.
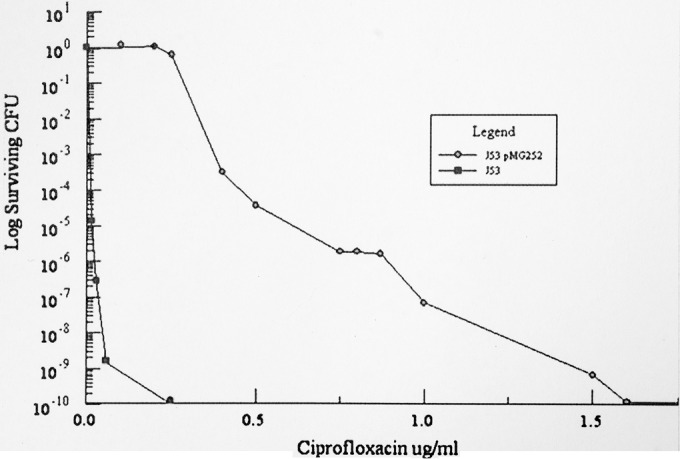
Survival of E. coli J53 and J53(pMG252) with increasing ciprofloxacin concentrations. A large inoculum (1010 CFU) and appropriate dilutions were applied to Mueller-Hinton agar plates containing the indicated concentrations of ciprofloxacin, and surviving colonies were counted after incubation for 72 h at 37°C. (Reproduced from reference 56.)
At 3 times the MIC, most (14 of 15 analyzed) of the ciprofloxacin-resistant mutants selected from J53 had alterations in the QRDR of gyrA. In contrast, none of the >50 spontaneous ciprofloxacin-resistant mutants selected at 3 times the MIC or other multiples from J53(pMG252) had QRDR mutations in gyrA or other topoisomerase genes, nor did any of the 20 ciprofloxacin-resistant mutants selected at up to three times the MIC from J53(pMG253) containing the cloned qnrA1 gene. This lack of gyrA mutations was not qnr specific but occurred with other PMQR genes. Mutations in gyrA were found in only 2 of 9 mutants selected from J53 with pMG320 containing aac(6′)Ib-cr at concentrations of ciprofloxacin up to 20 times the MIC and in only 1 of 10 mutants selected at 3 times the ciprofloxacin MIC from J53(pAT851) carrying qepA. In every case, mutants from the cultures with the highest concentration of ciprofloxacin with surviving colonies were analyzed.
Forty-three mutants derived from J53(pMG252) were studied in detail. Ten mutants were selected at 0.5 μg/ml, 23 were selected at 0.87 μg/ml, and 10 were selected at 1 μg/ml ciprofloxacin. There were no surviving colonies at higher ciprofloxacin concentrations.
Susceptibility.
Table 1 shows that the increases in MICs for the J53(pMG252) ciprofloxacin-resistant mutants ranged from 2- to 8-fold. The ciprofloxacin MBC for J53(pMG252) was 2 times the MIC, and this MBC/MIC ratio was unchanged in several ciprofloxacin-resistant mutants (data not shown). The nalidixic acid MIC was unchanged or increased up to 4-fold except in seven mutants, in which it decreased 4-fold.
TABLE 1.
Ciprofloxacin, nalidixic acid, and novobiocin MICs of E. coli J53(pMG252) mutants
| Strain | Ciprofloxacin concn for selection (μg/ml) | MIC (μg/ml) |
||
|---|---|---|---|---|
| Ciprofloxacin | Nalidixic acid | Novobiocin | ||
| J53 | 0.016 | 8 | 160 | |
| J53(pMG252) | 0.25 | 32 | 160 | |
| J53(pMG252) Cipr mutantsa | ||||
| 2 | 1 | 1.5 | 128 | 1,280 |
| 3 | 1 | 2 | 128 | 1,280 |
| 4 | 1 | 2 | 64 | 1,280 |
| 5 | 1 | 2 | 128 | 1,280 |
| 6 | 1 | 2 | 8 | 40 |
| 7 | 1 | 0.87 | 128 | 320 |
| 8 | 1 | 0.5 | 64 | 320 |
| 9 | 1 | 1.5 | 64 | 640 |
| 10 | 1 | 0.75 | 64 | 1,280 |
| 11 | 1 | 1 | 128 | 160 |
| 12 | 0.87 | 2 | 128 | 1,280 |
| 13 | 0.87 | 2 | 128 | 1,280 |
| 14 | 0.87 | 2 | 128 | 1,280 |
| 15 | 0.87 | 0.87 | 8 | 10 |
| 16 | 0.87 | 0.87 | 8 | 20 |
| 17 | 0.87 | 0.87 | 128 | 1,280 |
| 18 | 0.87 | 0.75 | 8 | 20 |
| 19 | 0.87 | 1 | 8 | 20 |
| 20 | 0.87 | 0.87 | 8 | 10 |
| 21 | 0.5 | 0.87 | 32 | 80 |
| 22 | 0.5 | 1 | 32 | 160 |
| 23 | 0.5 | 0.87 | 32 | 5 |
| 24 | 0.5 | 1.5 | 64 | 1,280 |
| 25 | 0.5 | 1.5 | 64 | 1,280 |
| 26 | 0.5 | 2 | 32 | 160 |
| 27 | 0.5 | 1 | 64 | 320 |
| 28 | 0.5 | 2 | 128 | 1,280 |
| 29 | 0.5 | 0.87 | 8 | 20 |
| 30 | 0.5 | 0.87 | 32 | 1,280 |
| 2-1 | 0.87 | 1 | 32 | 640 |
| 2-2 | 0.87 | 2 | 64 | 640 |
| 3-1 | 0.87 | 1 | 64 | 20 |
| 3-2 | 0.87 | 2 | 64 | 40 |
| 4-1 | 0.87 | 1 | 64 | 640 |
| 5-1 | 0.87 | 1 | 128 | 640 |
| 5-2 | 0.87 | 1 | 64 | 1,280 |
| 6-1 | 0.87 | 1 | 64 | 640 |
| 8-1 | 0.87 | 2 | 32 | 640 |
| 8-2 | 0.87 | 2 | 64 | 320 |
| 9-1 | 0.87 | 0.5 | 64 | 320 |
| 9-2 | 0.87 | 2 | 64 | 5 |
| 10-1 | 0.87 | 1 | 64 | 1,280 |
| 10-2 | 0.87 | 2 | 64 | 1,280 |
Mutants 2 to 30 and 2-1 to 10-2 were selected in separate experiments. Pairs like 2-1 and 2-2 came from the same selection plate.
In attempting to cure pMG252 from the mutants with novobiocin (14), we discovered that 12 of the mutants were hypersusceptible to novobiocin (including all those with increased nalidixic acid susceptibility), while 23 were ≥4-fold more resistant than the parent (Table 1). Both ciprofloxacin and novobiocin target DNA gyrase but act on different subunits, and cross-resistance does not occur from target mutations. Consequently, decreased susceptibility to both ciprofloxacin and novobiocin suggested the activation of an efflux pump exporting both compounds.
Expression of efflux pumps.
We sequenced the pump regulator genes marR, robA, acrR, soxR, and soxS; the lon protease gene, which degrades the products of some of these pump regulators; and hns, encoding a histone-like protein that regulates the expression of the acrEF and mdtEF drug exporter genes. We found that 11 mutants had alterations in MarR (Table 2), including one mutant with a 2-bp change (TA to AC) 22 bp upstream from the marR start codon at the binding site where MarR regulates its own expression (15). The marR mutations were mainly insertions and deletions causing frameshifts in the protein. In ciprofloxacin-resistant mutants 2 and 4, the same 47-bp duplication after bp 196 was found, creating a frameshift at amino acid 81. Mutant 6 had an A residue inserted at bp 126, creating a frameshift at amino acid 42. Four different deletions were observed: an A residue deleted at two different positions (bp 126 and bp 181) for mutants 12 and 20, a sizeable deletion of 144 bp in mutant 25, and a G deletion at bp 186 in mutant 27. Three strains had single point mutations, 2 of which had G→A transitions at bp 141, creating a TGA stop codon at amino acid 47. Mutations in robA, acrR, lon, hns, or soxS were not detected, but two mutants had G121D alterations in SoxR.
TABLE 2.
Mutations in MarA and SoxS found in 13 J53(pMG252) Cipr strains
| J53(pMG252) Cipr strain | Mutated protein (mutation detected)a | Mean fold change in expression relative to J53(pMG252) (SEM) |
||
|---|---|---|---|---|
| marA | soxS | acrA | ||
| 2 | MarR (G81fs) | 44.04 (11.49) | 0.23 (0.07) | 2.40 (0.91) |
| 3 | MarR (TA-AC)b | 95.46 (19.40) | 0.10 (0.04) | 4.87 (1.54) |
| 4 | MarR (G81fs) | 63.98 (24.51) | 0.21 (0.12) | 2.62 (0.62) |
| 5 | SoxR (G121D) | 1.23 (0.11) | 12.19 (3.22) | 2.29 (0.48) |
| 6 | MarR (Q42fs) | 35.74 (8.98) | 0.33 (0.13) | 2.16 (0.39) |
| 12 | MarR (N126fs) | 36.36 (6.25) | 0.311 (0.12) | 2.22 (0.28) |
| 13 | SoxR (G121D) | 1.73 (0.54) | 10.55 (4.06) | 2.51 (0.88) |
| 14 | MarR (C47STOP) | 63.91 (7.21) | 0.23 (0.09) | 1.39 (0.25) |
| 20 | MarR (K62fs) | 66.41 (22.37) | 0.21 (0.07) | 2.88 (1.02) |
| 24 | MarR (C47STOP) | 28.22 (4.81) | 0.34 (0.14) | 2.30 (0.99) |
| 25 | MarR (V65fs) | 46.27 (14.21) | 0.24 (0.09) | 1.75 (0.37) |
| 27 | MarR (V63fs) | 9.20 (2.90) | 0.32 (0.16) | 2.21 (0.64) |
| 28 | MarR (V21D) | 79.73 (1.22) | 0.29 (0.15) | 2.86 (0.65) |
fs, frameshift; STOP, stop codon.
Mutation detected in the MarR binding site (22 bp before the GTG start codon).
Inactivation of MarR increases the expression of marA, and with increased marA expression, the transcription of the pump genes acrAB and tolC is amplified, with consequently increased efflux of ciprofloxacin (16). To verify that MarR mutations caused a loss of repressor activity, we measured the expression levels of marA by RT-qPCR (Table 2) and found increased expression levels in the 11 strains containing MarR mutations. Similarly, expression of soxS is regulated by soxR, and increased soxS expression also leads to increases in acrAB transcript levels (17). We found 10- to 12-fold overexpression of soxS in the two strains with SoxR mutations (Table 2), thus establishing that in these mutants, SoxR was constitutively active. Finally, to establish whether the acrAB pump genes were overexpressed, we measured transcript levels of acrA in all 43 mutant strains by RT-qPCR. Eleven of the 13 MarR/SoxR mutant strains had 2- to 4-fold increases in the expression level of acrA, while the remaining 2 showed 1.39- to 1.75-fold increases (Table 2). acrA expression levels were not increased in any of the other mutants.
We also measured the relative expression levels of other efflux pumps that might contribute to ciprofloxacin resistance (Table 3). The ydhE efflux pump was overexpressed 2- to 3-fold in 6 mutants, and the mdtEF pump was overexpressed at least 2-fold in 9 mutants, in both cases independently of mutations in MarR or SoxR. Mutant 9-2 had a 2-fold increase in the expression level of acrE, and mutant 3-2, in addition to increased ydhE expression, also had a 2-fold increase in the expression level of tolC. Among the 9 mutants with overexpression of the mdtEF genes, mutants 15 and 28 showed 2-fold increases in the expression levels of the pump regulator evgA, the response regulator of the EvgAS two-component system (18). A 2- to 3-fold increase in the evgA expression level was also seen in mutants 2 and 4 containing MarR mutations and lacking overexpression of the mdtEF pump. None of the strains with overexpression of the mdtEF pump contained mutations in the 798-bp-long promoter region of gadE, another mdtEF regulator (19, 20). No changes in mdfA or in the fis and dsrA (21) regulators of MdtEF expression were detected.
TABLE 3.
Relative expression levels of other efflux pumps and regulatorsa
| Strain | Mean fold change relative to J53(pMG252) (SEM) |
||||||
|---|---|---|---|---|---|---|---|
| mdfA | ydhE | acrE | tolC | mdtE | mdtF | evgA | |
| 2-1 | 0.87 (0.25) | 2.58 (0.82) | 1.15 (0.44) | 1.37 (0.15) | 0.59 (0.19) | 0.84 (0.30) | |
| 2-2 | 0.88 (0.23) | 3.15 (0.93) | 1.35 (0.54) | 1.29 (0.26) | 0.62 (0.12) | 0.78 (0.18) | |
| 3-1 | 1.23 (0.35) | 2.70 (0.31) | 0.89 (0.43) | 1.18 (0.14) | 0.32 (0.08) | 0.52 (0.05) | |
| 3-2 | 0.65 (0.05) | 2.17 (0.47) | 1.43 (0.59) | 2.02 (0.38) | 1.18 (0.53) | 1.46 (0.68) | |
| 6-1 | 0.73 (0.29) | 2.12 (0.42) | 1.18 (0.34) | 1.03 (0.11) | 0.82 (0.29) | 1.06 (0.32) | |
| 8-1 | 1.23 (0.42) | 2.10 (0.30) | 0.98 (0.25) | 0.86 (0.23) | 0.35 (0.08) | 0.61 (0.22) | |
| 9-2 | 1.13 (0.31) | 1.84 (0.79) | 2.01 (0.80) | 1.56 (0.25) | 0.94 (0.15) | 1.34 (0.18) | |
| 2 | 1.34 (0.36) | 0.94 (0.18) | 1.03 (0.24) | 2.04 (0.29) | |||
| 4 | 1.70 (0.55) | 1.71 (0.22) | 1.40 (0.44) | 2.40 (0.73) | |||
| 12 | 0.92 (0.25) | 2.11 (0.39) | 2.78 (1.50) | 1.25 (0.30) | |||
| 13 | 1.61 (0.34) | 2.10 (0.82) | 3.76 (2.24) | 1.44 (0.27) | |||
| 15 | 0.78 (0.13) | 2.68 (0.71) | 1.66 (0.84) | 2.10 (0.45) | |||
| 21 | 0.55 (0.11) | 2.10 (0.56) | 1.52 (0.42) | 1.14 (0.22) | |||
| 24 | 1.52 (0.91) | 2.78 (0.88) | 2.01 (0.46) | 1.33 (0.31) | |||
| 25 | 0.83 (0.08) | 2.43 (1.03) | 2.22 (0.48) | 1.42 (0.15) | |||
| 26 | 0.57 (0.29) | 3.04 (1.28) | 1.58 (0.47) | 1.37 (0.26) | |||
| 28 | 1.52 (0.60) | 2.50 (0.56) | 2.38 (0.80) | 2.44 (0.40) | |||
| 30 | 0.73 (0.50) | 6.96 (2.71) | 5.49 (2.23) | 0.96 (0.44) | |||
At least three different assays with three different RNA extractions were performed for each gene tested.
LPS defects.
Since a change in pump substrate specificity could conceivably account for the combination of decreased ciprofloxacin but increased novobiocin susceptibility, we sequenced genes for efflux pumps, acrAB, acrEF, acrD, acrS, mdtEF, and ydhE, but found no alterations. A clue to novobiocin hypersensitivity came from whole-genome sequencing, which disclosed a mutation in rfaE. RfaE participates in the biosynthesis of the ADP-l-glycero-d-manno-heptose precursors used in the assembly of inner core LPS. E. coli mutants lacking heptose in the LPS core display a variety of phenotypes due to the reduced stability of the outer membrane, including hypersensitivity to hydrophobic antibiotics such as novobiocin (22). Further sequencing (see Table 5) revealed two additional mutants with defects in rfaE and two strains with mutations in rfaD. All of the rfaD and rdfE mutants were novobiocin hypersusceptible, but we were unable to detect a defect in six other novobiocin-hypersusceptible mutants despite sequencing 18 genes involved in LPS biosynthesis.
TABLE 5.
Other mutations detected in J53(pMG252) Cipr strains
| Protein | Mutation detected | J53(pMG252) Cipr strain(s) |
|---|---|---|
| RfaE | ΔIS10 | 16 |
| H356L | 20 | |
| M217fs | 21 | |
| RfaD | ΔIS10 | 18, 19 |
| IcdA | ΔIS10 | 17 |
| RpoB | R1246C | 9 |
| GudD | R213fs | 8-1 |
| YgeG | Y62H | 2-2 |
| YoaA | F308LLLDdel | 5-2 |
| pYejAa | −4 G→A | 5-1, 5-2 |
| pSdhCa | −156 C→T | 8-2 |
| Protein of unknown function under GenBank accession no. EIE38512 | G27fs | 2-1, 2-2, 3-2, 8-1, 8-2, 10-1, 10-2 |
Promoter changes.
Porin expression.
The major outer membrane proteins of E. coli, OmpF and OmpC, also have a role in susceptibility since they provide channels for ciprofloxacin entry and, when downregulated, decrease ciprofloxacin susceptibility (23, 24). Expression of the ompC and ompF porin genes was evaluated in all the mutants (Table 4). ompF expression was reduced 25% or more in 30 of the 43 mutants, while ompC expression was reduced by the same amount in 15 mutants. All but one of the mutants with marR or soxR defects had decreased ompF expression, averaging a decrement of >80%. In 9 of the 12 novobiocin-hypersusceptible mutants, ompF expression was reduced by at least 25%, averaging a decrease of 68%.
TABLE 4.
Relative expression levels of porins
| Strain | Gene or promoter affecteda | Mean fold change relative to J53(pMG252) (SEM) |
|
|---|---|---|---|
| ompC | ompF | ||
| J53(pMG252) | None | 1.0 | 1.0 |
| J53(pMG252) Cipr mutants | |||
| 2 | marR | 1.688 (0.575) | 0.240 (0.044) |
| 3 | marR | 1.612 (0.452) | 0.168 (0.081) |
| 4 | marR | 1.681 (0.406) | 0.166 (0.051) |
| 5 | soxR | 1.059 (0.291) | 0.243 (0.107) |
| 6 | marR | 1.078 (0.189) | 0.127 (0.036) |
| 7 | — | 2.381 (0.140) | 1.312 (0.120) |
| 8 | qnrA ↑ | 2.702 (0.474) | 0.900 (0.303) |
| 9 | rpoB | 1.181 (0.182) | 0.234 (0.006) |
| 10 | — | 0.624 (0.213) | 0.782 (0.061) |
| 11 | — | 1.574 (0.468) | 0.240 (0.027) |
| 12 | marR, mdtE ↑ | 1.379 (0.279) | 0.199 (0.088) |
| 13 | soxR, mdtE ↑ | 1.298 (0.458) | 0.281 (0.100) |
| 14 | marR | 0.873 (0.206) | 0.134 (0.015) |
| 15 | mdtE ↑ | 1.074 (0.088) | 0.342 (0030) |
| 16 | rfaE | 0.972 (0.113) | 0.271 (0.019) |
| 17 | icdA | 0.907 (0.158) | 1.287 (0.443) |
| 18 | rfaD | 0.700 (0.075) | 0.274 (0.036) |
| 19 | rfaD, qnrA ↑ | 0.862 (0.052) | 0.350 (0.042) |
| 20 | marR, rfaE | 0.831 (0.392) | 0.127 (0.044) |
| 21 | rfaE, mdtE ↑ | 0.781 (0.326) | 0.295 (0.030) |
| 22 | — | 1.191 (0.112) | 1.278 (0.239) |
| 23 | — | 0.465 (0.129) | 0.632 (0.072) |
| 24 | marR, mdtE ↑ | 1.756 (0.663) | 0.206 (0.016) |
| 25 | marR, mdtE ↑ | 1.525 (0.290) | 0.202 (0.067) |
| 26 | mdtE ↑ | 0.494 (0.106) | 0.934 (0.326) |
| 27 | marR | 1.862 (1.214) | 0.994 (0.432) |
| 28 | marR, mdtE ↑ | 0.879 (0.280) | 0.156 (0.027) |
| 29 | — | 1.984 (0.426) | 0.413 (0.082) |
| 30 | mdtE ↑ | 5.367 (1.882) | 0.529 (0.026) |
| 2-1 | ydhE ↑ | 0.757 (0.207) | 1.091 (0.268) |
| 2-2 | ygeG, ydhE ↑ | 0.572 (0.109) | 1.064 (0.126) |
| 3-1 | ydhE ↑ | 0.372 (0.053) | 0.951 (0.278) |
| 3-2 | tolC ↑, ydhE ↑ | 0.575 (0.264) | 0.515 (0.081) |
| 4-1 | — | 0.404 (0.026) | 0.631 (0.147) |
| 5-1 | pyejA | 0.764 (0.110) | 0.886 (0.221) |
| 5-2 | pyejA, yoaA | 0.375 (0.088) | 1.087 (0.353) |
| 6-1 | ydhE ↑ | 0.313 (0.070) | 0.801 (0.074) |
| 8-1 | gudD, ydhE ↑ | 0.271 (0.040) | 0.458 (0.131) |
| 8-2 | psdhC, qnrA ↑ | 0.642 (0.251) | 0.425 (0.133) |
| 9-1 | acrE ↑ | 0.329 (0.018) | 0.707 (0.183) |
| 9-2 | — | 0.382 (0.038) | 1.146 (0.365) |
| 10-1 | — | 0.278 (0.082) | 0.351 (0.055) |
| 10-2 | — | 0.211 (0.030) | 0.679 (0.216) |
—, no mutation was detected. The up arrow indicates increased.
Other chromosomal mutations.
Mutations in eight other genes were detected in the remaining mutants but have not been proven to be responsible for the observed decrease in ciprofloxacin susceptibility (Table 5). IcdA is isocitrate dehydrogenase, a component of the tricarboxylic acid cycle. Defects in icdA have been associated with decreased nalidixic acid and ciprofloxacin susceptibilities via metabolic acrA activation (25), but acrA expression was not increased in our mutant. RpoB is the β-subunit of RNA polymerase. A mutation in the α-subunit (but not the β-subunit) has been associated with decreased ciprofloxacin susceptibility in Salmonella enterica (26). GudD is glucarate dehydrogenase. YgeG is a putative secretion chaperone and a remnant of a presumably nonfunctional pathogenicity island. YoaA is a protein of unknown function with a nucleoside triphosphate hydrolysis domain. YejA is a component of a periplasmic peptide ABC transporter. SdhC is a membrane protein component of succinate:quinone oxidoreductase. The alterations in YejA and SdhC were located in their promoter regions. The mutant promoter from SdhC had 40% reduced activity when cloned upstream of lacZ in plasmid pRS415. In the same way, the mutant promoter from YejA showed 20% reduced activity. Finally, seven mutants obtained in the same experiment were defective in a 43-amino-acid protein of unknown function encoded by several E. coli genomes (GenBank accession number EIE38512). Further studies are needed to evaluate the role of these genes in resistance.
Changes in plasmid pMG252.
Expression of qnrA is not regulated in pMG252, but the gene is surrounded by two copies of ISCR1 with the potential for transposition to the chromosome or another site on the plasmid. qnrA expression was measured in all the mutants by RT-qPCR. Its expression levels were 2.4-fold higher in mutant 8, 2.2-fold higher in mutant 8-2, and 3.6-fold higher in mutant 19. No mutations were detected in the corresponding qnrA promoter regions. Mutants 8, 8-2, and 19 also had 1.9-fold, 3.1-fold, and 2.4-fold increases in qnrA copy numbers without a change in copy numbers for sul1 or blaFOX-5, two other resistance genes on pMG252, indicating that the qnrA copy number increase was gene specific. When pMG252 was displaced from J53(pMG252) mutants 8, 8-2, and 19 by selection with tetracycline for the entry of plasmid R692, belonging to the same Inc group as pMG252, ciprofloxacin susceptibility returned to the baseline value, supporting that a change in pMG252 is responsible for enhanced resistance. When plasmid pMG252 was outcrossed from these strains to E. coli C600, the level of ciprofloxacin resistance was 2- to 4-fold higher than that with unmodified pMG252. Furthermore, the copy number and expression level of qnrA in C600(pMG252) from mutants 8, 8-2, and 19 were also increased >3-fold. Although the plasmids have not been mapped, these changes suggest that qnrA duplication on pMG252 is the mechanism for decreased ciprofloxacin susceptibility in J53(pMG252) Cipr mutants 8, 8-2, and 19.
Unexplained mutants.
We have not yet been able to detect a chromosomal or plasmid alteration in 10 of the 43 mutants despite whole-genome sequencing. In particular, the entirety of the topoisomerase genes gyrA, gyrB, parC, and parE was examined in these strains, without discovering mutations within or outside the known QRDR.
Low-level ciprofloxacin resistance mutants of E. coli J53 without qnrA.
To explore whether J53 without qnrA exposed to low concentrations of ciprofloxacin produced the same variety of mutations as seen with J53(pMG252), selection was performed with 0.01 to 0.16 μg/ml ciprofloxacin. Mutations in the QRDR of gyrA were present in 0 of 12 mutants selected with 0.01 or 0.02 μg/ml, 3 of 6 mutants selected with 0.04 μg/ml, and all 7 mutants selected with 0.08 or 0.16 μg/ml, including 1 with an amber mutation at serine 83 in GyrA. Such a mutant was reported previously (4) and is viable because of a coincident suppressor mutation (our unpublished observations). Nineteen mutants were analyzed in detail (Table 6). Ciprofloxacin MICs of the mutants varied from 0.032 to 1 μg/ml, and nalidixic acid MICs varied from 1 to 2,048 μg/ml, with high MICs resulting from gyrA mutations. Four strains were hypersensitive to novobiocin, while four had 2- to 4-fold increases in novobiocin resistance. Changes in susceptibility to ampicillin, chloramphenicol, erythromycin, and trimethoprim of 4-fold or higher were noted for some strains, as shown in Table 6. In 2 of the 4 novobiocin-hypersensitive strains, defects in core lipopolysaccharide biosynthesis were identified (I12L in lpcA and an IS10 insertion in rfaE). Five strains had mutations that increased efflux pump expression: 3 with R20C in SoxR and 14- to 26-fold increases in soxS expression, 1 in MarR with K61stop and a 65-fold increase in marA expression, and 1 with a 2-fold increase in mdfA expression. Alterations in other genes, including gyrB, parC, and other genes of lipopolysaccharide biosynthesis, and increased expression of the pump gene mdtE or ydhE were not detected. In 7 mutants with 4-fold increases in ciprofloxacin MICs, no mutation was found, indicating that the spectrum of low-level resistance mechanisms extends beyond those detected here.
TABLE 6.
Characterization, susceptibility, and porin gene expression of E. coli J53 ciprofloxacin-resistant mutants
| Strain | Ciprofloxacin concn for selection (μg/ml) | Mutation detected | Mean fold change relative to J53 (SEM)a |
MIC (μg/ml)b |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ompC | ompF | Cip | Nal | Novo | Amp | Chl | Ery | Trim | |||
| J53 | 1 | 1 | 0.016 | 8 | 640 | 8 | 8 | 128 | 0.25 | ||
| Cipr mutants | |||||||||||
| 7 | 0.01 | mdfA ↑ | 0.979 (0.132) | 2.107 (0.195) | 0.064 | 8 | 640 | 8 | 8 | 128 | 0.25 |
| 8 | 0.01 | 0.765 (0.272) | 1.326 (0.070) | 0.064 | 8 | 640 | 8 | 8 | 64 | 0.25 | |
| 9 | 0.01 | RfaE | 0.310 (0.082) | 0.213 (0.036) | 0.032 | 1 | 20 | 8 | 2 | 8 | 0.125 |
| 10 | 0.01 | 0.606 (0.240) | 1.370 (0.188) | 0.064 | 8 | 640 | 8 | 8 | 128 | 0.25 | |
| 11 | 0.01 | 0.627 (0.141) | 1.170 (0.011) | 0.064 | 8 | 640 | 8 | 8 | 128 | 0.5 | |
| 12 | 0.01 | 0.851 (0.138) | 0.231 (0.033) | 0.064 | 8 | 640 | 16 | 16 | 128 | 0.25 | |
| 13 | 0.02 | LpcA | 0.166 (0.053) | 0.362 (0.016) | 0.064 | 8 | 40 | 8 | 2 | 8 | 0.125 |
| 14 | 0.02 | GyrA D391Ac | 0.534 (0.198) | 0.900 (0.080) | 0.064 | 1 | 20 | 8 | 4 | 32 | 0.25 |
| 15 | 0.02 | 0.479 (0.035) | 0.348 (0.050) | 0.064 | 32 | 640 | 16 | 8 | 128 | 4 | |
| 16 | 0.02 | SoxR | 0.326 (0.141) | 0.481 (0.165) | 0.125 | 32 | 1280 | 16 | 32 | 128 | 2 |
| 17 | 0.02 | SoxR | 0.531 (0.192) | 0.529 (0.137) | 0.125 | 8 | 1280 | 16 | 32 | 128 | 2 |
| 18 | 0.02 | 0.497 (0.106) | 0.252 (0.013) | 0.064 | 1 | 80 | 16 | 32 | 64 | 0.25 | |
| 19 | 0.04 | MarR | 0.474 (0.474) | 0.091 (0.011) | 0.25 | 32 | 1280 | 32 | 32 | 128 | 1 |
| 20 | 0.04 | SoxR | 0.312 (0.100) | 0.268 (0.125) | 0.25 | 32 | 2560 | 32 | 32 | 256 | 2 |
| 21 | 0.04 | 0.406 (0.051) | 2.355 (0.270) | 0.064 | 8 | 640 | 16 | 8 | 128 | 2 | |
| 24 | 0.04 | GyrA G81D | 0.571 (0.288) | 0.380 (0.023) | 0.25 | 32 | 640 | 8 | 16 | 128 | 0.25 |
| 25 | 0.08 | GyrA S83L | 0.986 (0.284) | 1.726 (0.147) | 0.5 | 2048 | 640 | 8 | 8 | 128 | 0.5 |
| 26 | 0.08 | GyrA S83L | 0.847 (0.157) | 1.504 (0.355) | 1 | 2048 | 320 | 8 | 8 | 128 | 0.25 |
| 27 | 0.08 | GyrA S83amberd | 0.531 (0.216) | 0.423 (0.065) | 0.5 | 128 | 640 | 8 | 8 | 128 | 0.25 |
Values in boldface type differ by 50% or more from values for porin expression in the J53 parent.
MIC values in boldface type differ from values for the parent by at least 4-fold. Abbreviations: Cip, ciprofloxacin; Nal, nalidixic acid; Novo, novobiocin; Amp, ampicillin; Chl, chloramphenicol; Ery, erythromycin; Trim, trimethoprim.
Mutation outside the quinolone resistance-determining region.
Strain contains an amber suppressor.
DISCUSSION
The rarity of gyrA mutations in E. coli J53 carrying qnrA has been reported by several groups (4, 27). This result was initially unexpected because when qnr is added to a strain with a ciprofloxacin-resistant gyrA mutation, resistances are additive (28, 29), and both gyrA mutations and qnr genes are commonly found together in E. coli and other enterobacterial clinical isolates (30–32). Some combinations of qnr and gyrA mutants have a measureable decrease in growth rate (33), but the effect of the common GyrA S83L mutation and qnrA was an insignificant 1%, especially compared to the average 9% lower growth rate of the nalidixic acid-hypersusceptible mutants that we selected than of others (our unpublished observations), making fitness cost as an explanation for the lack of gyrA mutations unlikely. Mutations in gyrA were also rare in J53 carrying plasmids with enzymatic ciprofloxacin resistance [aac(6′)-Ib-cr] or a ciprofloxacin efflux pump (qepA), so the lack of gyrA mutations is not qnr specific. Furthermore, we have been unable to obtain gyrA mutants with secondary selection of higher ciprofloxacin resistance from J53(pMG252) Cipr mutant 3 with a marR defect or Cipr mutant 15 with defects in rfaE and increased mdtE expression. Mutant selection in an E. coli strain carrying qnr thus provides a way to explore the range of nongyrase mutants that decrease ciprofloxacin susceptibility.
Mutations that are selected from J53(pMG252) with decreased ciprofloxacin susceptibility are mainly those that activate efflux pumps or alter LPS core biosynthesis. Mutations in MarR or SoxR that augment the expression of the major AcrAB/TolC pump were the most common, but increased expression of the MdtEF/TolC, YdhE, or AcrE efflux pump was found in other mutants. AcrAB is well known for ciprofloxacin efflux (34), and the potential for MdtEF (35), YdhE (36, 37), and AcrE (38) to efflux ciprofloxacin was also reported previously. Mutations upregulating drug efflux may have a fitness cost (39) but are relevant to resistance evolution because of their pleiotropic effects on susceptibility to other agents and potential for occurrence at a rate higher than that required for specific changes in the QRDR of gyrA. Gyrase mutations that provide ciprofloxacin resistance are relatively rare events because a change to an alternate amino acid at a relatively small number of localized sites in DNA gyrase is required. Mutations in pump regulatory proteins like MarR are more common because insertion, deletions, and substitutions at many sites can block translation. Gyrase mutations provide higher-level resistance than pump or porin changes, but Qnr, by competing with ciprofloxacin for its target (40); AAC(6′)-Ib-cr, by modifying the drug (7); and QepA, by enhancing ciprofloxacin efflux (8), all decrease the effective concentration of ciprofloxacin so that more common if less potent mutations at sites other than gyrA can predominate.
Measuring susceptibility to nalidixic acid and novobiocin as well as ciprofloxacin proved to be a useful phenotypic screening test. Enhanced resistance to all three agents suggested efflux pump activation, while unchanged nalidixic acid susceptibility and novobiocin hypersusceptibility suggested a defect in the core LPS pathway. Defects in many genes have been reported to cause novobiocin hypersusceptibility, including cls (41), djlA (42), dnaK and dnaJ (43), mukB (44), and surA (45), but especially genes of LPS biosynthesis (46, 47). We detected defects in rfaD and rfaE after discovering that 12 of the 43 J53(pMG252) ciprofloxacin-resistant mutants were novobiocin hypersusceptible and also found an lpcA mutant among low-level-ciprofloxacin-resistant mutants of J53 without qnrA. Increased novobiocin susceptibility combined with decreased quinolone susceptibility has been reported for E. coli mutants selected with tigecycline that had defects in the core LPS genes lpcA, rfaC, rfaE, and rfaF (48). Defects in LPS reduce the outer membrane barrier to novobiocin entry (49). Why ciprofloxacin susceptibility decreases instead relates to porin regulation.
OmpF is the major channel for ciprofloxacin entry in E. coli, with OmpC playing a lesser role (49, 50). Decreased expression levels of both ompF and ompC were detected in many of the ciprofloxacin-resistant mutants. These genes are transcriptionally regulated by the two-component systems OmpR-EnvZ (51) and CpxA-CpxR (52) in response to effects on the physical properties of the cell membrane due to such conditions as temperature, osmolarity, pH, and chemical stress (53). Each porin is also subject to posttranscriptional control by the small regulatory RNA molecules micF and micC that downregulate OmpF and OmpC expression (54, 55). Both MarA and SoxS activate micF production, explaining the decrease in ompF expression in MarR or SoxR mutants. We hypothesize that membrane stress from the mutant cell wall decreases porin expression in LPS mutants and explains their decreased ciprofloxacin susceptibility. These changes in pump and porin expression also affect susceptibility to other antimicrobials, as is evident from changes in ampicillin, chloramphenicol, erythromycin, and trimethoprim susceptibilities with mutations providing low-level changes in ciprofloxacin susceptibility in E. coli J53 (Table 6).
In three strains, a change in the expression of qnrA1 itself in pMG252 seemed to be responsible for enhanced resistance. qnrA1 is bracketed by two copies of ISCR1 (6), which has the potential to transpose to other sites on pMG252 or the E. coli chromosome. The increased copy number and expression of qnrA in these mutants as well as their transconjugants suggest that an increased gene dosage from replication on pMG252 is the mechanism for their reduced ciprofloxacin susceptibility.
Whether the other gene alterations that we discovered (Table 5) contribute to ciprofloxacin resistance requires further study. Quinolones are mutagenic and activate the cell's SOS system. At least 12 of the analyzed mutants had more than a single defect detected, so random mutations unrelated to ciprofloxacin resistance need to be excluded.
Low-level ciprofloxacin resistance can thus arise by many mechanisms, some of which increase or decrease susceptibility to chemically unrelated antibiotics. By decreasing the effective selecting ciprofloxacin concentration, qnr and other PMQR genes facilitate the selection of alterations in pumps, porins, and LPS, with multiple consequences for antimicrobial susceptibility.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02167-15.
REFERENCES
- 1.Martínez-Martínez L, Pascual A, Jacoby GA. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 2.Hooper DC. 2003. Mechanisms of quinolone resistance, p 41–67. In Hooper DC, Rubinstein E (ed), Quinolone antimicrobial agents, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 3.Fabrega A, Madurga S, Giralt E, Vila J. 2009. Mechanism of action of and resistance to quinolones. Microb Biotechnol 2:40–61. doi: 10.1111/j.1751-7915.2008.00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesaro A, Bettoni RR, Lascols C, Merens A, Soussy CJ, Cambau E. 2008. Low selection of topoisomerase mutants from strains of Escherichia coli harbouring plasmid-borne qnr genes. J Antimicrob Chemother 61:1007–1015. doi: 10.1093/jac/dkn077. [DOI] [PubMed] [Google Scholar]
- 5.Jacoby GA, Han P. 1996. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol 34:908–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran JH, Jacoby GA. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc Natl Acad Sci U S A 99:5638–5642. doi: 10.1073/pnas.082092899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med 12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 8.Périchon B, Courvalin P, Galimand M. 2007. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob Agents Chemother 51:2464–2469. doi: 10.1128/AAC.00143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hedges RW. 1974. R factors from Providence. J Gen Microbiol 81:171–181. doi: 10.1099/00221287-81-1-171. [DOI] [PubMed] [Google Scholar]
- 10.CLSI. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 12.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 13.Drlica K. 2003. The mutant selection window and antimicrobial resistance. J Antimicrob Chemother 52:11–17. doi: 10.1093/jac/dkg269. [DOI] [PubMed] [Google Scholar]
- 14.McHugh GL, Swartz MN. 1977. Elimination of plasmids from several bacterial species by novobiocin. Antimicrob Agents Chemother 12:423–426. doi: 10.1128/AAC.12.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duval V, McMurry LM, Foster K, Head JF, Levy SB. 2013. Mutational analysis of the multiple-antibiotic resistance regulator MarR reveals a ligand binding pocket at the interface between the dimerization and DNA binding domains. J Bacteriol 195:3341–3351. doi: 10.1128/JB.02224-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alekshun MN, Levy SB. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol 7:410–413. doi: 10.1016/S0966-842X(99)01589-9. [DOI] [PubMed] [Google Scholar]
- 17.Koutsolioutsou A, Pena-Llopis S, Demple B. 2005. Constitutive soxR mutations contribute to multiple-antibiotic resistance in clinical Escherichia coli isolates. Antimicrob Agents Chemother 49:2746–2752. doi: 10.1128/AAC.49.7.2746-2752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eguchi Y, Oshima T, Mori H, Aono R, Yamamoto K, Ishihama A, Utsumi R. 2003. Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology 149:2819–2828. doi: 10.1099/mic.0.26460-0. [DOI] [PubMed] [Google Scholar]
- 19.Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. 2005. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol Microbiol 55:1113–1126. [DOI] [PubMed] [Google Scholar]
- 20.Deng Z, Shan Y, Pan Q, Gao X, Yan A. 2013. Anaerobic expression of the gadE-mdtEF multidrug efflux operon is primarily regulated by the two-component system ArcBA through antagonizing the H-NS mediated repression. Front Microbiol 4:194. doi: 10.3389/fmicb.2013.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishino K, Yamasaki S, Hayashi-Nishino M, Yamaguchi A. 2011. Effect of overexpression of small non-coding DsrA RNA on multidrug efflux in Escherichia coli. J Antimicrob Chemother 66:291–296. doi: 10.1093/jac/dkq420. [DOI] [PubMed] [Google Scholar]
- 22.Valvano MA, Messner P, Kosma P. 2002. Novel pathways for biosynthesis of nucleotide-activated glycero-manno-heptose precursors of bacterial glycoproteins and cell surface polysaccharides. Microbiology 148:1979–1989. doi: 10.1099/00221287-148-7-1979. [DOI] [PubMed] [Google Scholar]
- 23.Hirai K, Aoyama H, Suzue S, Irikura T, Iyobe S, Mitsuhashi S. 1986. Isolation and characterization of norfloxacin-resistant mutants of Escherichia coli K-12. Antimicrob Agents Chemother 30:248–253. doi: 10.1128/AAC.30.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapman JS, Georgopapadakou NH. 1988. Routes of quinolone permeation in Escherichia coli. Antimicrob Agents Chemother 32:438–442. doi: 10.1128/AAC.32.4.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helling RB, Janes BK, Kimball H, Tran T, Bundesmann M, Check P, Phelan D, Miller C. 2002. Toxic waste disposal in Escherichia coli. J Bacteriol 184:3699–3703. doi: 10.1128/JB.184.13.3699-3703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webber MA, Whitehead RN, Mount M, Loman NJ, Pallen MJ, Piddock LJ. 2015. Parallel evolutionary pathways to antibiotic resistance selected by biocide exposure. J Antimicrob Chemother 70:2241–2248. doi: 10.1093/jac/dkv109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goto K, Kawamura K, Arakawa Y. 2015. Contribution of QnrA, plasmid-mediated quinolone resistance peptide, to survival of Escherichia coli exposed to lethal ciprofloxacin concentration. Jpn J Infect Dis 68:196–202. doi: 10.7883/yoken.JJID.2014.153. [DOI] [PubMed] [Google Scholar]
- 28.Martínez-Martínez L, Pascual A, García I, Tran J, Jacoby GA. 2003. Interaction of plasmid and host quinolone resistance. J Antimicrob Chemother 51:1037–1039. doi: 10.1093/jac/dkg157. [DOI] [PubMed] [Google Scholar]
- 29.Briales A, Rodriguez-Martinez JM, Velasco C, Diaz de Alba P, Dominguez-Herrera J, Pachon J, Pascual A. 2011. In vitro effect of qnrA1, qnrB1, and qnrS1 genes on fluoroquinolone activity against isogenic Escherichia coli isolates with mutations in gyrA and parC. Antimicrob Agents Chemother 55:1266–1269. doi: 10.1128/AAC.00927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lascols C, Robert J, Cattoir V, Bébéar C, Cavallo JD, Podglajen I, Ploy MC, Bonnet R, Soussy CJ, Cambau E. 2007. Type II topoisomerase mutations in clinical isolates of Enterobacter cloacae and other enterobacterial species harbouring the qnrA gene. Int J Antimicrob Agents 29:402–409. doi: 10.1016/j.ijantimicag.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Yang J, Luo Y, Li J, Ma Y, Hu C, Jin S, Ye L, Cui S. 2010. Characterization of clinical Escherichia coli isolates from China containing transferable quinolone resistance determinants. J Antimicrob Chemother 65:453–459. doi: 10.1093/jac/dkp478. [DOI] [PubMed] [Google Scholar]
- 32.Ruiz E, Saenz Y, Zarazaga M, Rocha-Gracia R, Martinez-Martinez L, Arlet G, Torres C. 2012. qnr, aac(6′)-Ib-cr and qepA genes in Escherichia coli and Klebsiella spp.: genetic environments and plasmid and chromosomal location. J Antimicrob Chemother 67:886–897. doi: 10.1093/jac/dkr548. [DOI] [PubMed] [Google Scholar]
- 33.Machuca J, Briales A, Labrador G, Diaz-de-Alba P, Lopez-Rojas R, Docobo-Perez F, Martinez-Martinez L, Rodriguez-Bano J, Pachon ME, Pascual A, Rodriguez-Martinez JM. 2014. Interplay between plasmid-mediated and chromosomal-mediated fluoroquinolone resistance and bacterial fitness in Escherichia coli. J Antimicrob Chemother 69:3203–3215. doi: 10.1093/jac/dku308. [DOI] [PubMed] [Google Scholar]
- 34.Swick MC, Morgan-Linnell SK, Carlson KM, Zechiedrich L. 2011. Expression of multidrug efflux pump genes acrAB-tolC, mdfA, and norE in Escherichia coli clinical isolates as a function of fluoroquinolone and multidrug resistance. Antimicrob Agents Chemother 55:921–924. doi: 10.1128/AAC.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohnert JA, Schuster S, Fahnrich E, Trittler R, Kern WV. 2007. Altered spectrum of multidrug resistance associated with a single point mutation in the Escherichia coli RND-type MDR efflux pump YhiV (MdtF). J Antimicrob Chemother 59:1216–1222. doi: 10.1093/jac/dkl426. [DOI] [PubMed] [Google Scholar]
- 36.Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, Tsuchiya T. 1998. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother 42:1778–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang S, Clayton SR, Zechiedrich EL. 2003. Relative contributions of the AcrAB, MdfA and NorE efflux pumps to quinolone resistance in Escherichia coli. J Antimicrob Chemother 51:545–556. doi: 10.1093/jac/dkg126. [DOI] [PubMed] [Google Scholar]
- 38.Jellen-Ritter AS, Kern WV. 2001. Enhanced expression of the multidrug efflux pumps AcrAB and AcrEF associated with insertion element transposition in Escherichia coli mutants selected with a fluoroquinolone. Antimicrob Agents Chemother 45:1467–1472. doi: 10.1128/AAC.45.5.1467-1472.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcusson LL, Frimodt-Moller N, Hughes D. 2009. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog 5:e1000541. doi: 10.1371/journal.ppat.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim E, Chen C, Braun M, Kim HY, Okumura R, Wang Y, Jacoby GA, Hooper DC. 2015. Interactions between QnrB, QnrB mutants, and DNA gyrase. Antimicrob Agents Chemother 59:5413–5419. doi: 10.1128/AAC.00771-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tropp BE, Ragolia L, Xia W, Dowhan W, Milkman R, Rudd KE, Ivanisevic R, Savic DJ. 1995. Identity of the Escherichia coli cls and nov genes. J Bacteriol 177:5155–5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernard S, Clarke DJ, Chen MX, Holland IB, Jacq A. 1998. Increased sensitivity of E. coli to novobiocin, EDTA and the anticalmodulin drug W7 following overproduction of DjlA requires a functional transmembrane domain. Mol Gen Genet 259:645–655. doi: 10.1007/PL00008629. [DOI] [PubMed] [Google Scholar]
- 43.Modrzewska M, Karpinski P, Grudniak A, Wolska KI. 2002. Effect of null mutations in dnaK and dnaJ genes on conjugational DNA transfer, proteolysis and novobiocin susceptibility of Escherichia coli. Acta Microbiol Pol 51:217–224. [PubMed] [Google Scholar]
- 44.Adachi S, Hiraga S. 2003. Mutants suppressing novobiocin hypersensitivity of a mukB null mutation. J Bacteriol 185:3690–3695. doi: 10.1128/JB.185.13.3690-3695.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong M, Ferrell B, Lu W, Chai Q, Wei Y. 2013. Insights into the function and structural flexibility of the periplasmic molecular chaperone SurA. J Bacteriol 195:1061–1067. doi: 10.1128/JB.01143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamaki S, Sato T, Matsuhashi M. 1971. Role of lipopolysaccharides in antibiotic resistance and bacteriophage adsorption of Escherichia coli K-12. J Bacteriol 105:968–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pazoles CJ, Kulpa CF Jr. 1977. Biosynthesis and structure of lipopolysaccharide in an outer membrane-defective mutant of Escherichia coli J5. Biochim Biophys Acta 466:160–175. doi: 10.1016/0005-2736(77)90216-4. [DOI] [PubMed] [Google Scholar]
- 48.Linkevicius M, Sandegren L, Andersson DI. 2013. Mechanisms and fitness costs of tigecycline resistance in Escherichia coli. J Antimicrob Chemother 68:2809–2819. doi: 10.1093/jac/dkt263. [DOI] [PubMed] [Google Scholar]
- 49.Delcour AH. 2009. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huguet A, Pensec J, Soumet C. 2013. Resistance in Escherichia coli: variable contribution of efflux pumps with respect to different fluoroquinolones. J Appl Microbiol 114:1294–1299. doi: 10.1111/jam.12156. [DOI] [PubMed] [Google Scholar]
- 51.Forst S, Delgado J, Inouye M. 1989. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc Natl Acad Sci U S A 86:6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Batchelor E, Walthers D, Kenney LJ, Goulian M. 2005. The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins ompF and ompC. J Bacteriol 187:5723–5731. doi: 10.1128/JB.187.16.5723-5731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clarke EJ, Voigt CA. 2011. Characterization of combinatorial patterns generated by multiple two-component sensors in E. coli that respond to many stimuli. Biotechnol Bioeng 108:666–675. doi: 10.1002/bit.22966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chou JH, Greenberg JT, Demple B. 1993. Posttranscriptional repression of Escherichia coli OmpF protein in response to redox stress: positive control of the micF antisense RNA by the soxRS locus. J Bacteriol 175:1026–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen S, Zhang A, Blyn LB, Storz G. 2004. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J Bacteriol 186:6689–6697. doi: 10.1128/JB.186.20.6689-6697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jacoby GA, Strahilevitz J, Hooper DC. 2014. Plasmid-mediated quinolone resistance. Microbiol Spectr 2:PLAS-0006-2013. doi: 10.1128/microbiolspec.PLAS-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


