Abstract
Nutritionally variant streptococci (NVS) are fastidious Gram-positive cocci comprised of the species Abiotrophia defectiva, Granulicatella adiacens, and Granulicatella elegans. NVS are an important cause of bacteremia and infective endocarditis (IE) associated with significant morbidity and mortality. Antimicrobial susceptibility testing (AST) was performed for 14 antimicrobials using the broth microdilution MIC method described in the Clinical and Laboratory Standards Institute (CLSI) M45 guideline. A total of 132 clinical NVS blood isolates collected from 2008 to 2014 were tested. Species level identification of NVS isolates was achieved by 16S rRNA gene sequencing and/or matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). Ninety isolates were identified as G. adiacens, 37 as A. defectiva, and 5 as G. elegans. All isolates were susceptible to vancomycin (MIC90 = 1 μg/ml), and none displayed high-level resistance to aminoglycosides. G. adiacens was considerably more susceptible to penicillin than A. defectiva (38.9% versus 10.8% of isolates susceptible) but was less susceptible to cephalosporins than was A. defectiva (43.3% versus 100% of isolates susceptible to ceftriaxone). Several isolates were resistant to levofloxacin (6%), erythromycin (51%), and clindamycin (10%). The MIC90 for daptomycin was ≥4 μg/ml for G. adiacens and A. defectiva. G. elegans isolates were 100% susceptible to all antimicrobials tested, with the exception of erythromycin, to which only 20% were susceptible. This study provides antimicrobial susceptibility data for a recent collection of NVS and demonstrates important NVS species-related differences with respect to susceptibility to penicillin, cephalosporins, carbapenems, and daptomycin. Species-level identification of NVS organisms when susceptibility testing is not readily available may aid in treatment decisions.
INTRODUCTION
The nutritionally variant streptococci (NVS) are fastidious Gram-positive bacteria, requiring either l-cysteine- or pyridoxal-supplemented medium to support growth (1, 2). Chromosomal DNA-DNA hybridization in 1989 (3) and 16S rRNA gene sequencing data in 1995 and 2000 (4, 5) resulted in the reclassification of NVS isolates into two genera, Abiotrophia and Granulicatella, which comprise four recognized species: Abiotrophia defectiva, Granulicatella adiacens, Granulicatella elegans, and Granulicatella balaenopterae. A fifth species, Granulicatella para-adiacens, has been proposed but not formally recognized (6).
These bacteria, with the exception of G. balaenopterae (isolated from the minke whale [7]), are normal constituents of the human oropharyngeal, gastrointestinal, and urogenital flora (1). The NVS have been implicated in cases of bacteremia/septicemia and account for an estimated 4 to 8% of all cases of infective endocarditis (IE) due to viridans group streptococcus-like organisms (8–11). The NVS have also been associated with other invasive infections, including abscess, wound infections, and meningitis (1, 12). The incidence of IE caused by NVS may be underreported due to the fastidious nature of these organisms and the challenge of recovering isolates from clinical specimens (i.e., “culture-negative IE”) (11, 13–16). Nevertheless, retrospective case review has demonstrated that cases of NVS IE are associated with higher rates of complications, including relapse, bacteriologic failure, embolization, and death, than IE caused by other streptococci (17).
Given the fastidious nature of the organisms, antimicrobial susceptibility testing (AST) is challenging for clinical laboratories. This fact, combined with the limited clinical data and high rate of complications, has led the American Heart Association (AHA) and British Society for Antimicrobial Chemotherapy (BSAC) to suggest NVS IE be treated following the guidelines put forth for enterococcal IE, that is to say, with a combination of benzylpenicillin or ampicillin plus gentamicin for a duration of 4 to 6 weeks (18, 19). In contrast to enterococcal IE, vancomycin can be used alone (without gentamicin) for 6 weeks for patients with penicillin allergy (18, 19).
Little clinical data support the AHA/BSAC regimen recommended for treatment of NVS IE. A recent retrospective case review of eight patients in Taiwan documented successful outcomes for all the patients, although for 2 patients, antimicrobial regimens were adjusted to include vancomycin, teicoplanin, or cefotaxime due to poor initial responses (i.e., continued fever) (16). In addition, several case reports describe outcomes varying from clinical cure to death following treatment of NVS IE with benzylpenicillin plus gentamicin (20–22). Few data exist regarding the in vitro susceptibility of NVS to antimicrobials typically used for treatment. The Clinical and Laboratory Standards Institute (CLSI) MIC breakpoints for NVS (23) are derived primarily from two small studies performed over a decade ago (39 isolates in 2000 [24] and 20 isolates in 2001 [25]), in addition to data adapted from Streptococcus spp. (23). Data are also lacking for newer antimicrobials (e.g., daptomycin and linezolid) that have come to market since these reports were published.
We performed AST for 132 clinical NVS isolates recovered from blood cultures collected between 2008 and 2014 at Los Angeles hospitals. We confirm and expand upon important species-related differences among the NVS with respect to susceptibility to several antimicrobials, including penicillin, cephalosporins, carbapenems, and daptomycin. Additionally, we examine the benefit of using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) for species-level identification of NVS as a possible aid for treatment decisions in those situations where AST is not readily available.
MATERIALS AND METHODS
Bacterial isolates.
From July 2011 through August 2014, the University of California, Los Angeles, CA (UCLA), clinical microbiology laboratory identified 21 blood culture isolates from 19 patients at UCLA as “NVS,” “A. defectiva,” or “Granulicatella spp.” Additionally, from January 2008 to August 2014, the UCLA clinical microbiology laboratory received 161 blood culture isolates (corresponding to 154 patients) identified as “NVS,” “A. defectiva,” or “Granulicatella spp.” from various referring hospitals in Los Angeles County for AST. The isolates were stocked at −70°C in brucella broth with 15% glycerol (BD Diagnostics, Sparks, MD).
A total of 146 isolates from 145 patients were recovered and included for species level identification by MALDI-TOF MS. In brief, all isolates from the frozen stock cultures were subcultured twice onto BBL chocolate II agar (BD Diagnostic Systems) and incubated at 35°C in a humidified atmosphere with 5% CO2. Isolates were identified using a Vitek MS (bioMérieux, Craponne, France) at a 99.9% confidence value. Isolates not confirmed as belonging to one of the NVS species were excluded from further study (data not shown).
For the first 20 NVS isolates encountered, Vitek MS species level identifications were confirmed by 16S rRNA gene sequencing. Genomic DNA was isolated using the EZ1 DNA Tissue kit with the BioRobot EZ1 (Qiagen, Valencia, CA), and the 16S rRNA gene was amplified and sequenced using the Applied Biosystems MicroSeq 500 16S rRNA gene PCR and sequencing kits (Life Technologies, Carlsbad, CA) with the ABI Prism 3130xL genetic analyzer (Life Technologies) according to the manufacturer's protocols. Species identification was determined by querying 16S rRNA gene sequences against the MicroSeq ID rDNA 500 Library (v2.0; Life Technologies) and/or confirmed by nucleotide BLAST (BLASTN) search. Isolates that were ≥99% homologous to the reference rRNA gene sequence of A. defectiva, G. adiacens, or G. elegans were identified as such (26).
AST by BMD.
Cefotaxime, ceftaroline, ceftriaxone, clindamycin, daptomycin, erythromycin, gentamicin, levofloxacin, linezolid, imipenem, meropenem, penicillin, streptomycin, and vancomycin were tested by broth microdilution (BMD) according to CLSI standards (23, 27). Panels were prepared in house using cation-adjusted Mueller-Hinton broth (CAMHB) (Difco, BD Diagnostics) supplemented with 2.5% lysed horse blood (LHB) (Remel, Thermo Fisher Scientific, Lenexa, KS) and 1 μg/ml pyridoxal hydrochloride (Sigma-Aldrich, St. Louis, MO) (23, 27). For daptomycin, the test medium was supplemented with calcium to a final concentration of 50 μg/ml. Antimicrobials were purchased from Sigma-Aldrich, with the exception of daptomycin (Merck & Co.), ceftaroline (Allergan), and linezolid (United States Pharmacopeia, Rockville, MD). MICs were interpreted using the CLSI M45 breakpoints for Abiotrophia spp. and Granulicatella spp. when available (23). For quality control, CLSI-recommended Streptococcus pneumoniae ATCC 49619 (23) and several in-house-selected strains were tested.
To evaluate the potential effect of pyridoxal and the reproducibility of daptomycin MICs, daptomycin testing was repeated at an outside reference laboratory (Laboratory Specialists, Inc.) for 5 A. defectiva and 5 G. adiacens isolates with elevated daptomycin MICs (2 to >4 μg/ml). In addition, daptomycin MICs were measured with Staphylococcus aureus ATCC 29213 in CAMHB, CAMHB plus LHB, and CAMHB plus LHB plus 1 μg/ml pyridoxal (all supplemented with 50 μg/ml calcium).
Statistical analysis.
Where indicated, susceptibility data were compared by using chi-square and Fisher's exact tests in Prism 4.0 (GraphPad Software, La Jolla, CA). P values of <0.05 were considered significant. Tentative epidemiological cutoff values (ECVs) were calculated for all antimicrobials, using ECOFFinder (28), which applies the iterative statistical method of calculating the ECV. ECVs are intended to distinguish between organisms without and with phenotypically expressed resistance mechanisms for a species and a drug in a defined test system (i.e., BMD in this study). The ECVs defined here are tentative, as they are based on MICs obtained from a single laboratory. ECVs were not determined for antimicrobials if the modal MIC for a given species was less than or equal to the lowest concentration of the antimicrobial tested.
RESULTS
Identification by MALDI-TOF MS.
We validated the Vitek MS against 16S rRNA gene sequencing as a means to identify NVS, using a subset of 20 clinical isolates (11 G. adiacens, 3 A. defectiva, and all 6 G. elegans isolates). Similar to results obtained by Ratcliffe et al. (29), our Vitek MS identifications matched those of sequencing for all 20 isolates. Therefore, we used the Vitek MS for species level identification of the remainder of the NVS isolates (and exclusion of non-NVS isolates). Among the 146 isolates selected, Vitek MS identified 135 NVS (91 G. adiacens, 38 A. defectiva, and 6 G. elegans). The 11 isolates that were not identified as one of the three NVS species were excluded from further study. They were identified by 16S rRNA gene sequencing as Streptococcus mitis group (n = 5), Streptococcus agninosus group (n = 5), and Propionibacterium granulosum (n = 1). Incidentally, 5 of them (all S. mitis group) did not grow on sheep's blood agar (data not shown), which is presumably why these “nutritionally variant” viridans group streptococci were originally misidentified as Abiotrophia or Granulicatella spp.
AST by BMD.
AST results for 132 of 135 NVS isolates were evaluable. Three isolates (one of each species) were eliminated due to poor or no growth during AST. The MIC range, MIC50, and MIC90 and the percentage of isolates susceptible to each antimicrobial agent tested are listed in Table 1. Of note, MIC and susceptibility data for each of the antimicrobials tested did not change significantly based on the year the NVS isolates was recovered (P > 0.05) (data not shown).
Beta-lactams.
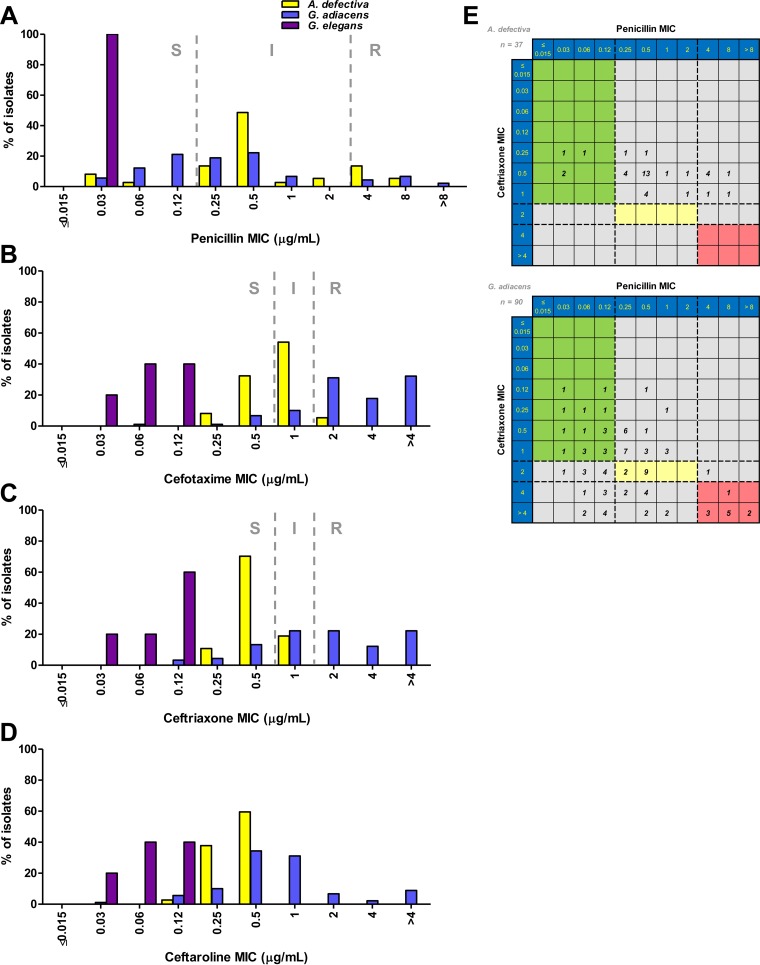
Overall 33% of the 132 NVS isolates were susceptible to penicillin. A greater percentage of isolates of G. adiacens than of A. defectiva were susceptible to penicillin (38.9% versus 10.8%) (Table 1 and Fig. 1A); however, penicillin MIC90 values were 4 μg/ml for both of the species. Compared to penicillin, NVS were more susceptible to the third-generation cephalosporins cefotaxime (43.2% susceptible) and ceftriaxone (61.4% susceptible), and again, species-related differences for these agents were noted (Fig. 1B and C). Interestingly, A. defectiva was more susceptible than G. adiacens to the third-generation cephalosporins (94.6% versus 18.9% for cefotaxime and 100% versus 43.3% for ceftriaxone). MIC90 values for both cefotaxime and ceftriaxone were 1 μg/ml for A. defectiva and >4 μg/ml for G. adiacens (Table 1). Overall, 48.6% of G. adiacens isolates that were susceptible to penicillin were also susceptible to ceftriaxone compared to 100% of A. defectiva isolates (Fig. 1E).
TABLE 1.
Antimicrobial susceptibility testing data by broth microdilution
| Drug and isolatec | MIC (μg/ml) |
MIC interpretation (%)a |
Tentative ECV (μg/ml)b | ||||
|---|---|---|---|---|---|---|---|
| Range | MIC50 | MIC90 | S | I | R | ||
| Penicillin | |||||||
| All isolates | 0.03 to >8 | 0.5 | 4 | 33.3 | 52.3 | 14.4 | 4 |
| A. defectiva | 0.03 to 8 | 0.5 | 4 | 10.8 | 70.3 | 18.9 | 1 |
| G. adiacens | 0.03 to >8 | 0.25 | 4 | 38.9 | 47.8 | 13.3 | 2 |
| G. elegans | 0.03 | 0.03 | 0.03 | 100 | 0 | 0 | ND |
| Ceftriaxone | |||||||
| All isolates | 0.03 to >4 | 1 | >4 | 61.4 | 15.1 | 23.5 | >4 |
| A. defectiva | 0.25 to 1 | 0.5 | 1 | 100 | 0 | 0 | 1 |
| G. adiacens | 0.12 to >4 | 2 | >4 | 43.3 | 22.2 | 34.5 | >4 |
| G. elegans | 0.03 to 0.12 | 0.12 | 0.12 | 100 | 0 | 0 | ND |
| Cefotaxime | |||||||
| All isolates | 0.03 to >4 | 2 | >4 | 43.2 | 22.7 | 34.1 | >4 |
| A. defectiva | 0.25 to 2 | 1 | 1 | 94.6 | 5.4 | 0 | 2 |
| G. adiacens | 0.06 to >4 | 4 | >4 | 18.9 | 31.1 | 50.0 | >4 |
| G. elegans | 0.03 to 0.12 | 0.06 | 0.12 | 100 | 0 | 0 | ND |
| Ceftaroline | |||||||
| All isolates | 0.03 to >4 | 0.5 | 2 | − | − | − | 2 |
| A. defectiva | 0.12 to 0.5 | 0.5 | 0.5 | − | − | − | 0.5 |
| G. adiacens | 0.03 to >4 | 0.5 | 4 | − | − | − | 4 |
| G. elegans | 0.03 to 0.12 | 0.06 | 0.12 | − | − | − | ND |
| Imipenem | |||||||
| All isolates | ≤0.06 to 0.25 | ≤0.06 | 0.12 | 100 | 0 | 0 | 0.125 |
| A. defectiva | ≤0.06 to 0.25 | 0.12 | 0.25 | 100 | 0 | 0 | 0.25 |
| G. adiacens | ≤0.06 to 0.25 | ≤0.06 | 0.12 | 100 | 0 | 0 | ND |
| G. elegans | ≤0.06 | ≤0.06 | ≤0.06 | 100 | 0 | 0 | ND |
| Meropenem | |||||||
| All isolates | ≤0.06 to 0.5 | 0.12 | 0.5 | 100 | 0 | 0 | 0.5 |
| A. defectiva | 0.25 to 0.5 | 0.5 | 0.5 | 100 | 0 | 0 | 0.5 |
| G. adiacens | ≤0.06 to 0.5 | 0.12 | 0.5 | 100 | 0 | 0 | 0.5 |
| G. elegans | ≤0.06 | ≤0.06 | ≤0.06 | 100 | 0 | 0 | ND |
| Vancomycin | |||||||
| All isolates | 0.25 to 1 | 0.5 | 1 | 100 | − | − | 1 |
| A. defectiva | 0.25 to 0.5 | 0.5 | 0.5 | 100 | − | − | 0.5 |
| G. adiacens | 0.25 to 1 | 0.5 | 1 | 100 | − | − | 1 |
| G. elegans | 0.5 | 0.5 | 0.5 | 100 | − | − | ND |
| Daptomycin | |||||||
| All isolates | 0.25 to >4 | 4 | >4 | − | − | − | >4 |
| A. defectiva | 1 to >4 | 2 | 4 | − | − | − | 4 |
| G. adiacens | 0.25 to >4 | 4 | >4 | − | − | − | >4 |
| G. elegans | 0.25 to 0.5 | 0.5 | 0.5 | − | − | − | ND |
| Linezolid | |||||||
| All isolates | 0.5 to 2 | 1 | 2 | − | − | − | 2 |
| A. defectiva | 1 to 2 | 1 | 1 | − | − | − | 2 |
| G. adiacens | 0.5 to 2 | 1 | 2 | − | − | − | 4 |
| G. elegans | 0.5 to 1 | 0.5 | 1 | − | − | − | ND |
| Levofloxacin | |||||||
| All isolates | ≤0.5 to >8 | ≤0.5 | 1 | 93.9 | 0 | 6.1 | ND |
| A. defectiva | ≤0.5 | ≤0.5 | ≤0.5 | 100 | 0 | 0 | ND |
| G. adiacens | ≤0.5 to >8 | 1 | 2 | 91.9 | 0 | 8.9 | ND |
| G. elegans | ≤0.5 to 1 | ≤0.5 | 1 | 100 | 0 | 0 | ND |
| Clindamycin | |||||||
| All isolates | ≤0.25 to >1 | ≤0.25 | 0.5 | 87.1 | 3.0 | 9.9 | ND |
| A. defectiva | ≤0.25 to >1 | ≤0.25 | ≤0.25 | 91.9 | 5.4 | 2.7 | ND |
| G. adiacens | ≤0.25 to >1 | ≤0.25 | >1 | 84.5 | 2.2 | 13.3 | ND |
| G. elegans | ≤0.25 | ≤0.25 | ≤0.25 | 100 | 0 | 0 | ND |
| Erythromycin | |||||||
| All isolates | ≤0.25 to >1 | 1 | >1 | 49.2 | 0 | 50.8 | ND |
| A. defectiva | ≤0.25 to >1 | >1 | >1 | 45.9 | 0 | 54.1 | ND |
| G. adiacens | ≤0.25 to >1 | ≤0.25 | >1 | 52.2 | 0 | 47.8 | ND |
| G. elegans | ≤0.25 to >1 | >1 | >1 | 20.0 | 0 | 80.0 | ND |
| Gentamicin | |||||||
| All isolates | ≤1 to 4 | ≤1 | 2 | − | − | − | ND |
| A. defectiva | ≤1 to 4 | ≤1 | 2 | − | − | − | ND |
| G. adiacens | ≤1 to 4 | ≤1 | 2 | − | − | − | ND |
| G. elegans | ≤1 | ≤1 | ≤1 | − | − | − | ND |
| Streptomycin | |||||||
| All isolates | ≤1 to 4 | ≤1 | 2 | − | − | − | ND |
| A. defectiva | ≤1 to 4 | ≤1 | 2 | − | − | − | ND |
| G. adiacens | ≤1 to 4 | 2 | 2 | − | − | − | ND |
| G. elegans | ≤1 | ≤1 | ≤1 | − | − | − | ND |
Interpreted according to criteria listed in CLSI M45 when available; S, susceptible; I, intermediate; R, resistant; −, no breakpoint available.
Tentative ECVs were calculated for all antimicrobials using ECOFFinder (28); ECVs were not determined (ND) for antimicrobials if the modal MIC for a given species was less than or equal to the lowest concentration of the antimicrobial tested.
All isolates, n = 132; A. defectiva, n = 37; G. adiacens, n = 90; G. elegans, n = 5.
FIG 1.
(A to D) A. defectiva, G. adiacens, and G. elegans MIC distributions for penicillin (A), cefotaxime (B), ceftriaxone (C), and ceftaroline (D). CLSI breakpoints (dashed lines) are shown. S, susceptible; I, intermediate; R, resistant. (E) Scatter plot comparison of ceftriaxone (y axis) and penicillin (x axis) MIC values for A. defectiva (top) and G. adiacens (bottom). The number of isolates with the given combination of MIC values is indicated in each square. Ceftriaxone and penicillin breakpoints (dashed lines) are plotted on either axis. The colored areas indicate where both MIC values are susceptible (green), intermediate (yellow), or resistant (red).
We also evaluated NVS susceptibility to the advanced-generation cephalosporin ceftaroline (Table 1 and Fig. 1D). The MIC90 was lower for ceftaroline than for either cefotaxime or ceftriaxone (2 versus >4 μg/ml) when analyzing all isolates and differed for A. defectiva (0.5 μg/ml) and G. adiacens (4 μg/ml) (Table 1). Of note, 51.6% (16 of 31 isolates) of G. adiacens isolates resistant to ceftriaxone (MIC ≥ 4 μg/ml) had ceftaroline MICs of ≤1 μg/ml. Additionally, 32.3% (10 of 31 isolates) of G. adiacens isolates resistant to ceftriaxone (MIC ≥ 4 μg/ml) had ceftaroline MICs of ≥4 μg/ml. These 10 isolates were also resistant to penicillin (MIC ≥ 4 μg/ml).
Although only a small number of G. elegans isolates (n = 5) were evaluated, all were susceptible to penicillin, cefotaxime, and ceftriaxone and had lower individual MIC and MIC90 values for these agents, and also for ceftaroline, than A. defectiva and G. adiacens (Table 1 and Fig. 1).
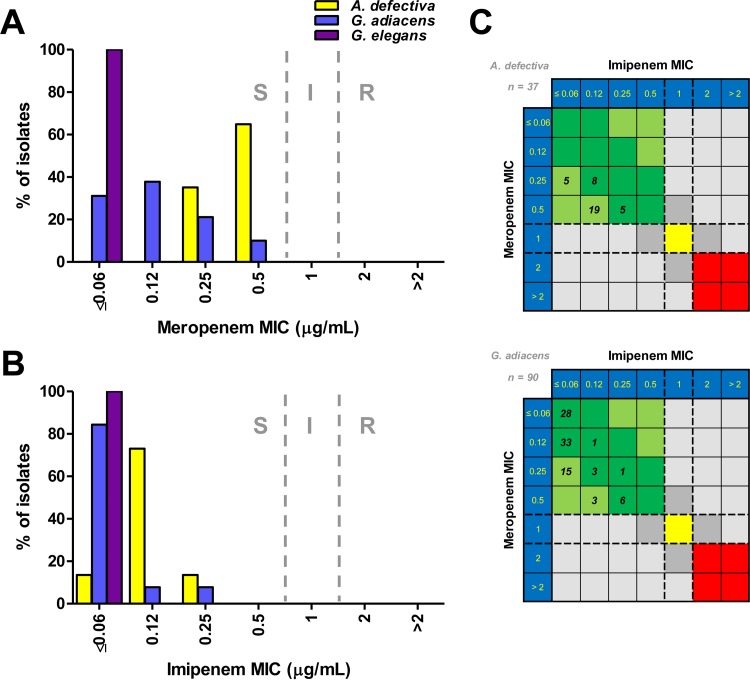
In contrast to the other beta-lactam antimicrobials tested (penicillin, cefotaxime, and ceftriaxone), resistance to meropenem or imipenem was not observed (Table 1). All 5 G. elegans isolates demonstrated MICs of ≤0.06 μg/ml for both meropenem and imipenem (Fig. 2A and B). The meropenem MIC50 (but not MIC90) was higher for A. defectiva than for G. adiacens, and both the imipenem MIC50 and MIC90 were higher for A. defectiva than for G. adiacens (Table 1). Furthermore, meropenem MICs were generally 2-fold higher than imipenem MICs for both species, although 31.1% (28 of 90) of G. adiacens isolates had MIC values of ≤0.06 for both carbapenems (Fig. 2C).
FIG 2.
(A and B) A. defectiva, G. adiacens, and G. elegans MIC distributions for meropenem (A) and imipenem (B). CLSI breakpoints (dashed lines) are shown. S, susceptible; I, intermediate; R, resistant. (C) Scatter plot comparison of meropenem (y axis) and imipenem (x axis) MIC values for A. defectiva (top) and G. adiacens (bottom). The number of isolates with the given combination of MIC values is indicated in each square. Meropenem and imipenem breakpoints (dashed lines) are plotted on either axis. The colored areas indicate where both MIC values are susceptible (green), intermediate (yellow), or resistant (red).
Non-beta-lactams.
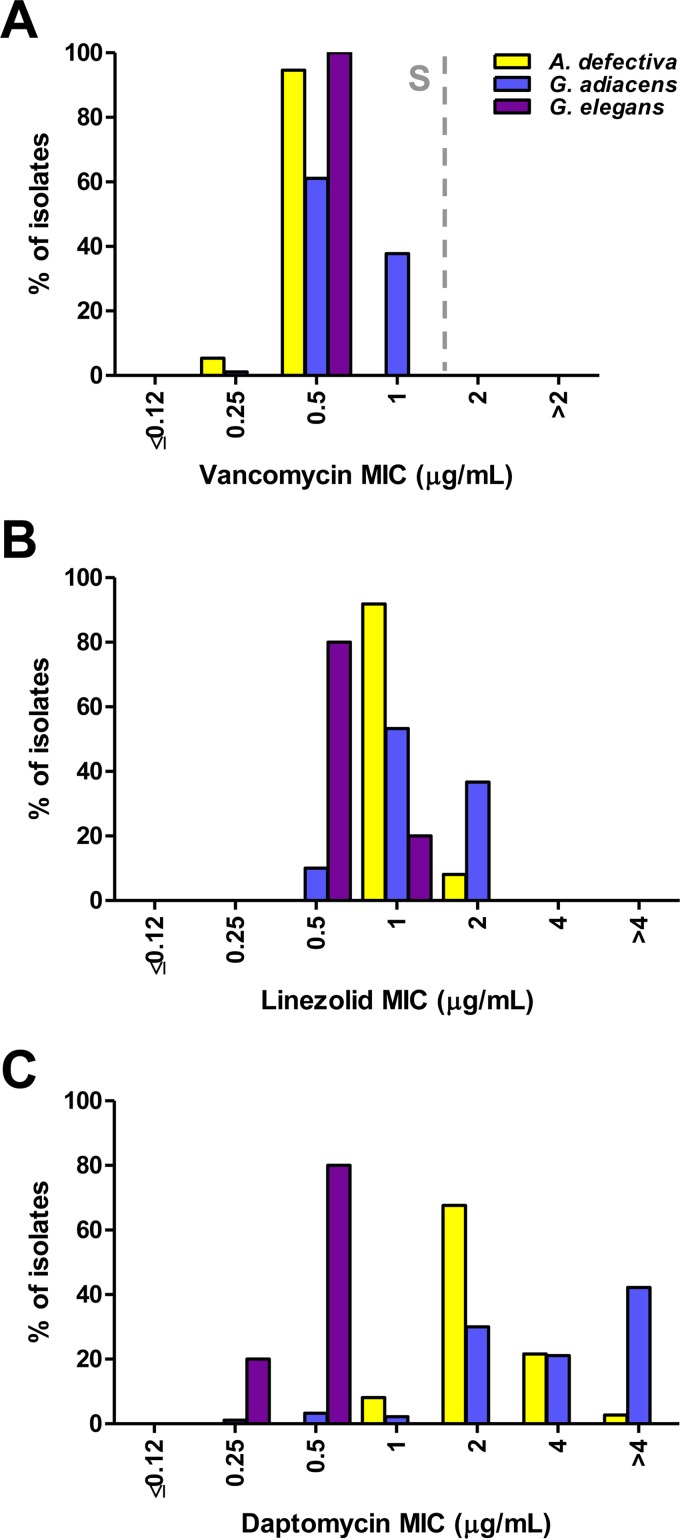
Gentamicin and streptomycin MICs were ≤4 μg/ml for all isolates, indicating a lack of high-level aminoglycoside resistance (Table 1). All the isolates were susceptible to vancomycin at ≤1 μg/ml (Table 1 and Fig. 3A).
FIG 3.
A. defectiva, G. adiacens, and G. elegans MIC distributions for vancomycin (A), linezolid (B), and daptomycin (C). The vancomycin CLSI breakpoint (dashed line) is shown. S, susceptible.
Individual linezolid MIC values were generally lower for G. elegans than for A. defectiva and G. adiacens (Fig. 3B), and the linezolid MIC90 was slightly higher for G. adiacens than for A. defectiva and G. elegans (2 versus 1 μg/ml) (Table 1). The daptomycin MIC90s for A. defectiva and G. adiacens were >4 and 4 μg/ml, respectively, whereas the MIC90 for G. elegans was only 0.5 μg/ml (Fig. 3C). These elevated MICs were confirmed, for a subset of 10 isolates, at an outside reference laboratory.
For levofloxacin, A. defectiva had a lower MIC90 value (≤0.5 μg/ml) than G. adiacens (2 μg/ml), and 8.9% (8 of 90 isolates) of G. adiacens isolates were resistant to levofloxacin (MIC ≥ 8 μg/ml) (Table 1 and Fig. 4A).
FIG 4.
(A to C) A. defectiva, G. adiacens, and G. elegans MIC distributions for levofloxacin (A), erythromycin (B), and clindamycin (C). CLSI breakpoints (dashed lines) are shown. S, susceptible; I, intermediate; R, resistant. (D) Scatter plot comparison of erythromycin (y axis) and clindamycin (x axis) MIC values for all NVS (left), A. defectiva (left middle), G. adiacens (right middle), and G. elegans (right). The number of isolates with the given combination of MIC values is indicated in each square. Erythromycin and clindamycin breakpoints (dashed lines) are plotted on either axis. The colored areas indicate where both MIC values are susceptible (green), intermediate (yellow), or resistant (red).
Erythromycin MIC distributions were bimodal for A. defectiva, G. adiacens, and G. elegans, with similar percentages of resistant isolates for A. defectiva and G. adiacens (Table 1 and Fig. 4B). Clindamycin MIC50 values were equivalent for all three species; however, the MIC90 was higher for G. adiacens, which can be attributed to a higher percentage of resistant isolates (13.3% versus 2.7% and 0% for A. defectiva and G. elegans, respectively) (Table 1 and Fig. 4C). Interestingly, one isolate (G. adiacens) was clindamycin resistant (MIC ≥ 1 μg/ml) but susceptible to erythromycin (MIC ≤ 0.25 μg/ml), which was confirmed on repeat testing (Fig. 4D).
DISCUSSION
Historically, combination therapy with penicillin and gentamicin (or vancomycin alone) has been used for the treatment of NVS IE. However, anecdotal evidence of resistance to penicillin and concern for possible emerging resistance to other agents led us to examine susceptibilities among several old and newer agents in a large contemporary collection of clinical NVS isolates. We did not observe evidence of high-level aminoglycoside resistance among the 132 NVS isolates tested. Additionally, and in line with previous studies (24, 25, 30), we did not find any evidence of vancomycin resistance, and vancomycin MICs for all isolates were ≤1 μg/ml. Nevertheless, the MIC90 for vancomycin was 2-fold higher for G. adiacens than for A. defectiva or G. elegans, which was consistent with the observations by Tuohy et al. (24).
Only 33% of the isolates examined in this study were susceptible to penicillin and 14% were resistant (MIC ≥ 4 μg/ml); the remaining 53% of isolates had penicillin MICs in the intermediate category (0.25 to 2 μg/ml). This penicillin resistance rate is substantially higher than the 5 to 8% resistance rates reported in two earlier studies (24, 25). The penicillin MIC90 for all NVS isolates was also substantially higher (4 μg/ml) than previously reported (0.5 to 2 μg/ml) (24, 25). At the species level, G. adiacens was considerably more susceptible to penicillin than was A. defectiva (38.9% versus 10.8%; P = 0.008). Tuohy et al. observed a similar pattern when examining 27 G. adiacens and 12 A. defectiva isolates (55% versus 8% susceptibility) (24). In addition, we observed that G. elegans isolates were all highly susceptible to penicillin (MIC = 0.03 μg/ml for all 5 isolates), in contrast to the other NVS species. Importantly, AST results for G. elegans have not been reported previously.
In addition to the species-related differences that were noted for penicillin susceptibility, Tuohy et al. also observed a similar but inverse relationship with respect to susceptibility to ceftriaxone (24). They determined that 83% of A. defectiva isolates compared to 63% of G. adiacens isolates were susceptible to ceftriaxone at a MIC of ≤0.5 μg/ml. Although we used a higher susceptibility breakpoint for ceftriaxone (≤1 μg/ml, according to CLSI M45 [23]), we noted a similar pattern of ceftriaxone susceptibility for A. defectiva and G. adiacens (100% versus 43%; P < 0.00001). Reanalysis of our data using the MIC breakpoint of ≤0.5 μg/ml, as in the previous report, demonstrated that the percentage of A. defectiva isolates susceptible to ceftriaxone is nearly the same (81%). However, ceftriaxone susceptibility for G. adiacens in our collection of isolates is substantially lower (21%) than that reported by Tuohy et al. (63%) (24). Additionally, all five of the G. elegans isolates were susceptible to ceftriaxone (MIC, 0.03 to 0.12 μg/ml). The reason for these differences is not immediately clear; however, similar to streptococci, it may be related to genetic differences (including pbp gene mosaicism) in one or more of the penicillin-binding proteins (PBPs) for A. defectiva and Granulicatella spp.
Although the AHA/BSAC recommendations do not endorse combination therapy with cephalosporins for NVS IE (18, 19), the European Society of Cardiology (ESC) guidelines differ slightly, as they recommend 6 weeks of benzylpenicillin, ceftriaxone, or vancomycin, combined with an aminoglycoside for at least the first 2 weeks (31). Thus, the observation of these important species-related differences with respect to penicillin and ceftriaxone that were first identified by Tuohy et al. (24) and expanded upon here may be important in certain clinical settings. For instance, a species level identification of A. defectiva may suggest ceftriaxone treatment is appropriate, as all the isolates tested were susceptible to the agent, whereas if G. adiacens is reported, an alternative agent, such as vancomycin, should be considered unless susceptibility can be confirmed.
We also evaluated the susceptibility of the isolates to meropenem and imipenem. We did not observe species-related differences for meropenem MIC90 values, similar to Tuohy et al. (24). In contrast, we observed at least 2-fold-higher imipenem MIC90 values for A. defectiva than for G. adiacens and G. elegans; however, all the isolates were susceptible to both antimicrobials. Additionally, imipenem MIC values were approximately 2-fold lower than meropenem MIC values for the majority of individual isolates. This is an interesting finding that has also been observed in Lactobacillus spp. and may be related to PBP differences (23).
While we did not find evidence of vancomycin resistance, we found high MICs of the lipopeptide daptomycin. In particular, we observed a striking difference in daptomycin MICs between our study and that of Piper et al., who evaluated a small number of NVS isolates (32). We noted substantially higher MICs for NVS isolates as a whole and also highlight species-related differences. It is not clear from the other study's methods (32) if the CAMHB growth medium used for testing was supplemented with pyridoxal, as recommended by the CLSI M45 (23). Nonetheless, we found that the addition of 1 μg/ml pyridoxal to the test medium did not impact daptomycin MIC results when testing the quality control strain S. pneumoniae ATCC 49619 or S. aureus ATCC 29213. Our data indicate that a substantial percentage of NVS isolates (89.4%) would be considered nonsusceptible to daptomycin according to the CLSI breakpoint established for viridans group streptococci (MIC ≤ 1 μg/ml) (33). The MIC distribution pattern differences between G. adiacens and A. defectiva support inherent resistance to daptomycin, which may be related to differences in cell membrane composition. In contrast, MIC90 values for linezolid were 2-fold higher for G. adiacens than for A. defectiva and G. elegans (2 versus 1 μg/ml). To our knowledge, this is the first report of MIC distribution data for linezolid with NVS (CLSI has not established breakpoints for NVS with the antimicrobial). Nevertheless, results from the LEADER surveillance program demonstrate that several groups of Gram-positive organisms, including Enterococcus and viridans group streptococci, share a linezolid MIC90 (1 μg/ml) (34). Furthermore, our data indicate that all NVS isolates would be considered susceptible according to the linezolid susceptible breakpoint (MIC ≤ 2 μg/ml) used for viridans group streptococci (33).
As with the other antimicrobials tested, we also noted species-related differences with respect to levofloxacin susceptibility. Similar to the results of Tuohy et al., the MIC90 was higher for G. adiacens than for A. defectiva (24). We also showed that the MIC90 was higher for G. adiacens than for G. elegans, and 8 G. adiacens isolates were levofloxacin resistant. Resistance to levofloxacin among NVS has been reported only once, in a G. elegans isolate from a patient with neutropenic fever and bacteremia who had previously received levofloxacin prophylactic therapy (25). We were unable to determine if antibiotic pressure played a role in the development of resistance to levofloxacin in the patients in our study. Furthermore, the mechanism of resistance is not known in the previously reported case and the cases here, although point mutations in parC and/or gyrA have been described for other Gram-positive organisms (35).
In agreement with previous reports (25, 36), we observed a bimodal pattern for the distribution of individual erythromycin MICs with respect to NVS organisms as a group. Here, we extend this analysis to show that all three species exhibit this bimodal pattern with similar frequencies of resistant isolates. Interestingly, 22% of erythromycin-resistant G. adiacens isolates were also resistant to clindamycin compared to only 5% and 0% of erythromycin-resistant A. defectiva and G. elegans isolates. Overall, 18% of erythromycin-resistant NVS isolates demonstrated resistance to clindamycin (MIC ≥ 1 μg/ml), which was in contrast to a related observation by Liao et al. (54% of azithromycin-resistant [MIC ≥ 1 μg/ml] NVS isolates) (30). This pattern is suggestive of the constitutive macrolide-lincosamide-streptogramin B (cMLSB) phenotype encoded by erm (erythromycin ribosomal methylase) genes (37). The presence of erm(B) in A. defectiva, G. adiacens, and G. elegans isolates resistant to both erythromycin and clindamycin has been described previously (36, 38). Poyart et al. demonstrated that the erm(B) gene was harbored within Tn3872, a Tn916-derivitive transposon common to macrolide-resistant S. pneumoniae isolates (39), in a clinical isolate of A. defectiva resistant to erythromycin and clindamycin (38). Zheng et al. did not perform a characterization in as much depth but did note that the three NVS isolates (G. adiacens and G. elegans) positive for erm(B) were also positive for the tetracycline resistance gene, tet(M), which is frequently found with erm(B) on the same transposon (36). Interestingly, the same authors found no evidence for the inducible MLSB (iMLSB) phenotype among their five NVS isolates (G. adiacens and A. defectiva) resistant to erythromycin but susceptible to clindamycin (36). Rather, these five isolates, which were negative for erm(A) and erm(B), were all positive for the efflux gene, mef(A) (36). Nevertheless, the fastidious nature and specific growth requirements of NVS have precluded proper evaluation of the iMLSB phenotype by either disk diffusion (D-zone test) or BMD. Finally, we also found one G. adiacens isolate that was clindamycin resistant but susceptible to erythromycin, a phenotype that has been infrequently described in Streptococcus spp. (40–42).
Overall, our data confirm and expand upon NVS species-related differences in susceptibility to several antimicrobials that were first noted by Tuohy et al. for a much smaller collection of isolates (24). Interestingly these observations are in contrast to those of Liao et al., who observed no difference in MIC distributions for A. defectiva and G. adiacens in 2004 (30). However, it is not clear whether the CAMHB testing medium used by Liao et al. (30) was supplemented with pyridoxal as recommended by the CLSI (23). Nevertheless, our data reveal that generally (with the exception of cephalosporins and carbapenems) G. adiacens has higher MIC values than both A. defectiva and G. elegans.
The species-related differences in antimicrobial susceptibility observed here raise a number of important questions for both laboratorians and clinicians regarding identification, AST, and perhaps treatment strategies for NVS. Specifically, it should be recognized that significant differences were observed in the tentative beta-lactam ECVs for A. defectiva and G. adiacens and possibly even G. elegans—such differences may cause different clinical responses to therapy, although there is a paucity of data by which to evaluate this. In addition, there are few data regarding treatment strategies for NVS IE and particularly for other invasive infections caused by NVS. As mentioned above, the ESC, but not the AHA or BSAC, supports cephalosporin therapy for NVS IE. Although it is not entirely clear what evidence was used to support this recommendation, the data reported here may provide an additional rationale to formally evaluate other antimicrobials (e.g., ceftriaxone for A. defectiva or G. elegans infections) in animal models and/or small prospective clinical trials. Finally, we reaffirm and strongly advocate the benefits of using MALDI-TOF MS for species level identification of NVS (as well as preventing the misidentification of nutritionally variant viridans group streptococci as NVS) and feel that its use in conjunction with present and future data could aid in treatment decisions in those areas or situations where appropriate AST is not readily available.
ACKNOWLEDGMENTS
We thank Karina Hernandez, Myra Maldonado, Cinthia Flores, Tom Garcia, Shelley Miller, Nico Magnano, Farzaneh Sooudipour, Ian McHardy, and Max Wu (UCLA) for invaluable technical support; Laura Koeth (Laboratory Specialists, Inc.) for confirmatory testing of daptomycin; John Turnidge (University of Adelaide) for assistance with determination of epidemiological cutoff values; Jenny Brook (UCLA) for statistical consultation; Marty Cohen, Ellen Kato, Kevin Ward, and Omai Garner (UCLA) for helpful discussions; and Nagendra Mishra and Arnold Bayer (Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center) for helpful discussions.
Funding Statement
This work was supported by a UCLA Department of Pathology and Laboratory Medicine Translational Research Fund Award to R.M.H. and M.O.A. Statistical consultation was supported by an NIH/National Center for Advancing Translational Science (NCATS) grant to UCLA CTSI (UL1TR000124).
REFERENCES
- 1.Ruoff KL. 1991. Nutritionally variant streptococci. Clin Microbiol Rev 4:184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frenkel A, Hirsch W. 1961. Spontaneous development of L forms of streptococci requiring secretions of other bacteria or sulphydryl compounds for normal growth. Nature 191:728–730. doi: 10.1038/191728a0. [DOI] [PubMed] [Google Scholar]
- 3.Bouvet A, Grimont F, Grimont PAD. 1989. Streptococcus defectivus sp. nov. and Streptococcus adjacens sp. nov., Nutritionally variant streptococci from human clinical specimens. Int J Syst Bacteriol 39:290–294. doi: 10.1099/00207713-39-3-290. [DOI] [Google Scholar]
- 4.Kawamura Y, Hou XG, Sultana F, Liu S, Yamamoto H, Ezaki T. 1995. Transfer of Streptococcus adjacens and Streptococcus defectivus to Abiotrophia gen. nov. as Abiotrophia adiacens comb. nov. and Abiotrophia defectiva comb. nov., respectively. Int J Syst Bacteriol 45:798–803. doi: 10.1099/00207713-45-4-798. [DOI] [PubMed] [Google Scholar]
- 5.Collins MD, Lawson PA. 2000. The genus Abiotrophia (Kawamura et al.) is not monophyletic: proposal of Granulicatella gen. nov., Granulicatella adiacens comb. nov., Granulicatella elegans comb. nov. and Granulicatella balaenopterae comb. nov. Int J Syst Evol Microbiol 50:365–369. doi: 10.1099/00207713-50-1-365. [DOI] [PubMed] [Google Scholar]
- 6.Kanamoto T, Sato S, Inoue M. 2000. Genetic heterogeneities and phenotypic characteristics of strains of the genus Abiotrophia and proposal of Abiotrophia para-adiacens sp. nov. J Clin Microbiol 38:492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawson PA, Foster G, Falsen E, Sjoden B, Collins MD. 1999. Abiotrophia balaenopterae sp. nov., isolated from the minke whale (Balaenoptera acutorostrata). Int J Syst Bacteriol 49:503–506. doi: 10.1099/00207713-49-2-503. [DOI] [PubMed] [Google Scholar]
- 8.Hase R, Otsuka Y, Yoshida K, Hosokawa N. 2015. Profile of infective endocarditis at a tertiary-care hospital in Japan over a 14-year period: characteristics, outcome and predictors for in-hospital mortality. Int J Infect Dis 33:62–66. doi: 10.1016/j.ijid.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Roberts RB, Krieger AG, Schiller NL, Gross KC. 1979. Viridans streptococcal endocarditis: the role of various species, including pyridoxal-dependent streptococci. Rev Infect Dis 1:955–966. doi: 10.1093/clinids/1.6.955. [DOI] [PubMed] [Google Scholar]
- 10.Hoen B, Alla F, Selton-Suty C, Beguinot I, Bouvet A, Briancon S, Casalta JP, Danchin N, Delahaye F, Etienne J, Le Moing V, Leport C, Mainardi JL, Ruimy R, Vandenesch F. 2002. Changing profile of infective endocarditis: results of a 1-year survey in France. JAMA 288:75–81. doi: 10.1001/jama.288.1.75. [DOI] [PubMed] [Google Scholar]
- 11.Brouqui P, Raoult D. 2001. Endocarditis due to rare and fastidious bacteria. Clin Microbiol Rev 14:177–207. doi: 10.1128/CMR.14.1.177-207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen JJ, Facklam RR. 2001. Granulicatella and Abiotrophia species from human clinical specimens. J Clin Microbiol 39:3520–3523. doi: 10.1128/JCM.39.10.3520-3523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koh YR, Yi J, Kim HH, Chang CL, Kim SY. 2014. Discrepant satellitism for identification of Granulicatella adiacens isolates. Ann Lab Med 34:174–176. doi: 10.3343/alm.2014.34.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casalta JP, Habib G, La Scola B, Drancourt M, Caus T, Raoult D. 2002. Molecular diagnosis of Granulicatella elegans on the cardiac valve of a patient with culture-negative endocarditis. J Clin Microbiol 40:1845–1847. doi: 10.1128/JCM.40.5.1845-1847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houpikian P, Raoult D. 2005. Blood culture-negative endocarditis in a reference center: etiologic diagnosis of 348 cases. Medicine 84:162–173. doi: 10.1097/01.md.0000165658.82869.17. [DOI] [PubMed] [Google Scholar]
- 16.Lin CH, Hsu RB. 2007. Infective endocarditis caused by nutritionally variant streptococci. Am J Med Sci 334:235–239. doi: 10.1097/MAJ.0b013e3180a6eeab. [DOI] [PubMed] [Google Scholar]
- 17.Stein DS, Nelson KE. 1987. Endocarditis due to nutritionally deficient streptococci: therapeutic dilemma. Rev Infect Dis 9:908–916. doi: 10.1093/clinids/9.5.908. [DOI] [PubMed] [Google Scholar]
- 18.Baddour LM, Wilson WR, Bayer AS, Fowler VG Jr, Bolger AF, Levison ME, Ferrieri P, Gerber MA, Tani LY, Gewitz MH, Tong DC, Steckelberg JM, Baltimore RS, Shulman ST, Burns JC, Falace DA, Newburger JW, Pallasch TJ, Takahashi M, Taubert KA, Committee on Rheumatic Fever Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, Councils on Clinical Cardiology, Stroke, Cardiovascular Surgery and Anesthesia, American Heart Association, Infectious Diseases Society of Amrrica. 2005. Infective endocarditis: diagnosis, antimicrobial therapy, and management of complications: a statement for healthcare professionals from the Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, and the Councils on Clinical Cardiology, Stroke, and Cardiovascular Surgery and Anesthesia, American Heart Association: endorsed by the Infectious Diseases Society of America. Circulation 111:e394–e434. doi: 10.1161/CIRCULATIONAHA.105.165564. [DOI] [PubMed] [Google Scholar]
- 19.Gould FK, Denning DW, Elliott TS, Foweraker J, Perry JD, Prendergast BD, Sandoe JA, Spry MJ, Watkin RW, Working Party of the British Society for Antimicrobial Chemotherapy. 2012. Guidelines for the diagnosis and antibiotic treatment of endocarditis in adults: a report of the Working Party of the British Society for Antimicrobial Chemotherapy. J Antimicrob Chemother 67:269–289. doi: 10.1093/jac/dkr450. [DOI] [PubMed] [Google Scholar]
- 20.Giuliano S, Caccese R, Carfagna P, Vena A, Falcone M, Venditti M. 2012. Endocarditis caused by nutritionally variant streptococci: a case report and literature review. Infez Med 20:67–74. [PubMed] [Google Scholar]
- 21.Ramos JN, dos Santos LS, Vidal LM, Pereira PM, Salgado AA, Fortes CQ, Vieira VV, Mattos-Guaraldi AL, Junior RH, Damasco PV. 2014. A case report and literature overview: Abiotrophia defectiva aortic valve endocarditis in developing countries. Infection 42:579–584. doi: 10.1007/s15010-014-0595-3. [DOI] [PubMed] [Google Scholar]
- 22.Adam EL, Siciliano RF, Gualandro DM, Calderaro D, Issa VS, Rossi F, Caramelli B, Mansur AJ, Strabelli TM. 2015. Case series of infective endocarditis caused by Granulicatella species. Int J Infect Dis 31:56–58. doi: 10.1016/j.ijid.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 23.CLSI. 2015. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria, 3rd ed, CLSI guideline M45 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 24.Tuohy MJ, Procop GW, Washington JA. 2000. Antimicrobial susceptibility of Abiotrophia adiacens and Abiotrophia defectiva. Diagn Microbiol Infect Dis 38:189–191. doi: 10.1016/S0732-8893(00)00194-2. [DOI] [PubMed] [Google Scholar]
- 25.Murray CK, Walter EA, Crawford S, McElmeel ML, Jorgensen JH. 2001. Abiotrophia bacteremia in a patient with neutropenic fever and antimicrobial susceptibility testing of Abiotrophia isolates. Clin Infect Dis 32:E140–E142. doi: 10.1086/320150. [DOI] [PubMed] [Google Scholar]
- 26.CLSI. 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved standard, CLSI document MM18-A. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 27.CLSI. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 10th ed, CLSI document M07-A10 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 28.Turnidge J, Kahlmeter G, Kronvall G. 2006. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin Microbiol Infect 12:418–425. doi: 10.1111/j.1469-0691.2006.01377.x. [DOI] [PubMed] [Google Scholar]
- 29.Ratcliffe P, Fang H, Thidholm E, Borang S, Westling K, Ozenci V. 2013. Comparison of MALDI-TOF MS and VITEK 2 system for laboratory diagnosis of Granulicatella and Abiotrophia species causing invasive infections. Diagn Microbiol Infect Dis 77:216–219. doi: 10.1016/j.diagmicrobio.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Liao CH, Teng LJ, Hsueh PR, Chen YC, Huang LM, Chang SC, Ho SW. 2004. Nutritionally variant streptococcal infections at a University Hospital in Taiwan: disease emergence and high prevalence of beta-lactam and macrolide resistance. Clin Infect Dis 38:452–455. doi: 10.1086/381098. [DOI] [PubMed] [Google Scholar]
- 31.Habib G, Hoen B, Tornos P, Thuny F, Prendergast B, Vilacosta I, Moreillon P, de Jesus Antunes M, Thilen U, Lekakis J, Lengyel M, Muller L, Naber CK, Nihoyannopoulos P, Moritz A, Zamorano JL, ESC Committee for Practice Guidelines. 2009. Guidelines on the prevention, diagnosis, and treatment of infective endocarditis (new version 2009): the Task Force on the Prevention, Diagnosis, and Treatment of Infective Endocarditis of the European Society of Cardiology (ESC). Endorsed by the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and the International Society of Chemotherapy (ISC) for Infection and Cancer. Eur Heart J 30:2369–2413. doi: 10.1093/eurheartj/ehp285. [DOI] [PubMed] [Google Scholar]
- 32.Piper KE, Steckelberg JM, Patel R. 2005. In vitro activity of daptomycin against clinical isolates of Gram-positive bacteria. J Infect Chemother 11:207–209. doi: 10.1007/s10156-005-0395-X. [DOI] [PubMed] [Google Scholar]
- 33.CLSI. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement, CLSI document M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 34.Flamm RK, Mendes RE, Hogan PA, Ross JE, Farrell DJ, Jones RN. 2015. In vitro activity of linezolid as assessed through the 2013 LEADER surveillance program. Diagn Microbiol Infect Dis 81:283–289. doi: 10.1016/j.diagmicrobio.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 35.Jacoby GA. 2005. Mechanisms of resistance to quinolones. Clin Infect Dis 41(Suppl 2):S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 36.Zheng X, Freeman AF, Villafranca J, Shortridge D, Beyer J, Kabat W, Dembkowski K, Shulman ST. 2004. Antimicrobial susceptibilities of invasive pediatric Abiotrophia and Granulicatella isolates. J Clin Microbiol 42:4323–4326. doi: 10.1128/JCM.42.9.4323-4326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leclercq R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis 34:482–492. doi: 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- 38.Poyart C, Quesne G, Acar P, Berche P, Trieu-Cuot P. 2000. Characterization of the Tn916-like transposon Tn3872 in a strain of abiotrophia defectiva (Streptococcus defectivus) causing sequential episodes of endocarditis in a child. Antimicrob Agents Chemother 44:790–793. doi: 10.1128/AAC.44.3.790-793.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDougal LK, Tenover FC, Lee LN, Rasheed JK, Patterson JE, Jorgensen JH, LeBlanc DJ. 1998. Detection of Tn917-like sequences within a Tn916-like conjugative transposon (Tn3872) in erythromycin-resistant isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 42:2312–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malbruny B, Werno AM, Anderson TP, Murdoch DR, Leclercq R. 2004. A new phenotype of resistance to lincosamide and streptogramin A-type antibiotics in Streptococcus agalactiae in New Zealand. J Antimicrob Chemother 54:1040–1044. doi: 10.1093/jac/dkh493. [DOI] [PubMed] [Google Scholar]
- 41.Montagnani F, Zanchi A, Stolzuoli L, Croci L, Cellesi C. 2007. Clindamycin-resistant Streptococcus pneumoniae. Emerg Infect Dis 13:801–802. doi: 10.3201/eid1305.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merino Diaz L, Torres Sanchez MJ, Aznar Martin J. 2008. Prevalence and mechanisms of erythromycin and clindamycin resistance in clinical isolates of beta-haemolytic streptococci of Lancefield groups A, B, C and G in Seville, Spain. Clin Microbiol Infect 14:85–87. doi: 10.1111/j.1469-0691.2007.01881.x. [DOI] [PubMed] [Google Scholar]