Abstract
In multicellular organisms, single-fluorophore imaging is obstructed by high background. To achieve a signal/noise ratio conducive to single-molecule imaging, we adapted reflected light-sheet microscopy (RLSM) to image highly opaque late-stage Drosophila embryos. Alignment steps were modified by means of commercially available microprisms attached to standard coverslips. We imaged a member of the septate-junction complex that was used to outline the three-dimensional epidermal structures of Drosophila embryos. Furthermore, we show freely diffusing single 10 kDa Dextran molecules conjugated to one to two Alexa647 dyes inside living embryos. We demonstrate that Dextran diffuses quickly (∼6.4 μm2/s) in free space and obeys directional movement within the epidermal tissue (∼0.1 μm2/s). Our single-particle-tracking results are supplemented by imaging the endosomal marker Rab5-GFP and by earlier reports on the spreading of morphogens and vesicles in multicellular organisms. The single-molecule results suggest that RLSM will be helpful in studying single molecules or complexes in multicellular organisms.
Introduction
The high autofluorescence background present in living animals usually obstructs single-molecule imaging with wide-field microscopy. For the purpose of reducing this background, several techniques have been developed that are able to excavate the signal from dim light sources. With its excellent signal/noise ratio (SNR), total-internal-reflection microscopy is widely used for single-molecule studies in vitro and to observe membrane-associated events at the single-molecule level in vivo (1, 2). Although out-of-focus signal is greatly reduced by the selective excitation of only a few hundred nanometers, total-internal-reflection microscopy is intrinsically restricted to image regions near a refractive index mismatch. Highly inclined laminated microscopy was developed for three-dimensional (3D) single-molecule imaging within single cells and has been shown to be able to detect single membrane proteins in living animals by virtue of optimized protein labeling (3). Difficulties arise, however, when an attempt is made to image dense regions where the increased highly inclined laminated microscopy light-sheet thickness lowers the SNR (4).
We believe that the development of novel microscopic techniques with optimized detection and illumination efficiency will bring new information about the connection between stochastic single-molecule behavior and multicellular pattern formation. Moreover, studying stochastic events in naturally embedded cells that have previously only been studied in cell culture will bring us closer to authentic observations.
Originally developed for imaging morphogenetic processes on a multicellular scale (5), light-sheet microscopy (LSM) offers the most promising addition to single-molecule imaging techniques (6, 7, 8, 9, 10). Several groups have been able to demonstrate protein binding to DNA in the nucleus of salivary gland cells (7) and stem cell spheroids (6). Samples are optically sectioned via sheet-like excitation volumes parallel to the focal plane of the detection objective. The orthogonal arrangement of the illumination and detection objectives restricts the use to low to medium numerical-aperture objectives that result in a poor photon transmission efficiency. Maximizing photon yield is, however, crucially needed for single-molecule imaging.
We adapted the principles of reflected LSM (RLSM) as introduced by Gebhardt et al. (8) and recently implemented as single-objective RLSM (or single-objective selective-plane illumination microscopy) by Galland et al. (10). RLSM uses a small reflective element to circumvent the spatial limitations imposed by conventional LSM. The excitation light beam is deflected shortly after the illumination objective onto the focal plane of a detection objective.
In this report, we show that the alignment can be readily performed with commercially available microprisms that are attached to standard microscope coverslips (custom made by Optikron, Jena, Germany). Thus, samples can be placed directly onto the disposable platform without time-consuming handling or preparation. In fact, we reconstructed the 3D epidermal structure and, moreover, were able to observe single Dextran-Alexa647 molecules in the surrounding of the epidermis of living and highly opaque late-stage Drosophila embryos.
Materials and Methods
Optical setup
Two laser lines were collinearly aligned via mirror and dichroic beam splitter (488 nm, 50 mW 488-50 Sapphire, Coherent, Santa Clara, CA; 642 nm, 140 mW LuxX642, Scienta Omicron, Taunusstein, Germany). Laser lines were modulated by an acoustooptical tunable filter (AOTF.4C-VIS-TN, Optoelectronics, Hawthorne, CA) within a few microseconds and triggered with the camera TTL exposure output signal. The beam was collimated after the optical fiber by a reflective collimator (RC04FC-P01, Thorlabs, Newton, NJ) to a beam diameter of 4 mm. A cylindrical 5× Keplerian beam telescope expanded the beam diameter (cylindrical achromatic doublets 50/250 mm; ACY254-050-A/ACY254-250-A, Thorlabs). A spherical aperture (SM1D12C, Thorlabs) further controlled the final beam diameter (4–12 mm) and, hence, the light-sheet thickness. A single cylindrical lens (150 mm cylindrical achromatic doublets, ACY254-150-A, Thorlabs) conjugated the back focal plane of a 20×, 0.95 NA water-immersion objective (working distance 2 mm; XLUMPLFL20XWIR, Olympus, Center Valley, PA) to define the light-sheet width (Fig. 1). The entire optical illumination block was moved with an XYZ manual translation stage (PT3/M, Thorlabs) on a custom-made breadboard that was attached to a commercial inverted Zeiss microscope (Axiovert Observer D1.m, Carl Zeiss, Oberkochen, Germany).
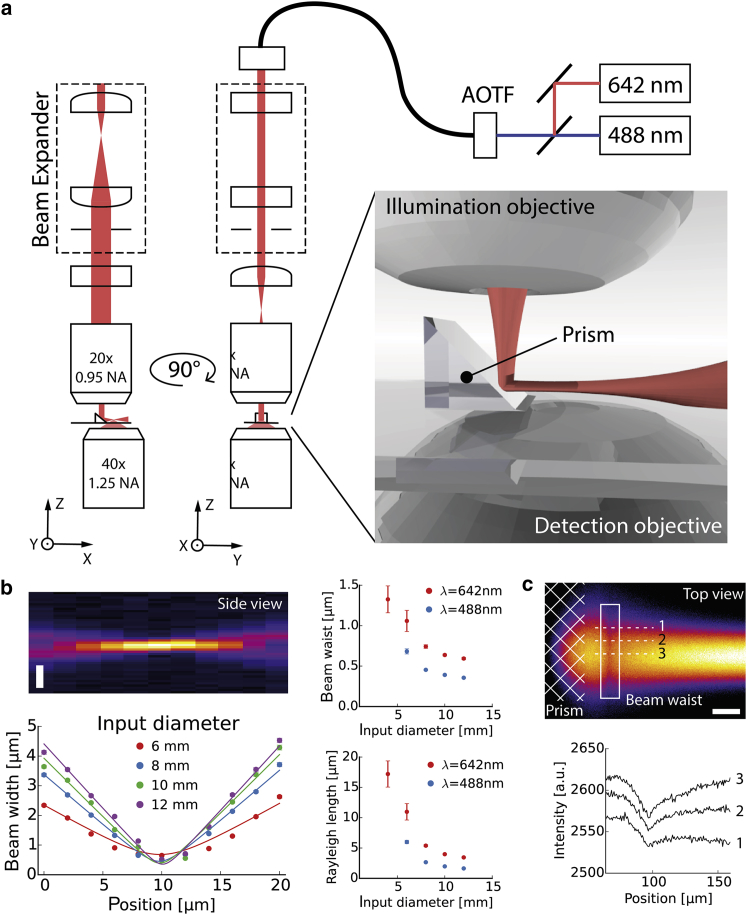
Figure 1.
Optical setup. (a) Schematic depicting the optical setup of our reflected-light-sheet microscope (RLSM). The 642 nm and 488 nm laser lines are modulated via AOTF and coupled into the optical system. A cylindrical beam expander forms an elliptical beam profile that is dynamically controlled by the beam input diameter. The third cylindrical lens conjugates the back focal plane of the illumination objective. The light sheet is reflected via a microprism onto the focal plane of a high-NA detection objective. The setup is illustrated from two different directions to highlight the elliptical beam geometry. (b) Raw data for the measured light-sheet profile along the propagation direction every 2 μm without reflection at 488 nm. Vertical scale bar, 2 μm. Gaussian beam propagation and fits are shown for 488 nm and four different beam input diameters. Beam waist and Rayleigh length were extracted from fits for both wavelengths. (c) Imaging of the light sheet after reflection of the microprism in Cy5 in water. Line profiles are shown for three different positions along the light-sheet width. Scale bar, 40 μm.
The coverslip (no. 1.5, 22 × 60 mm, Menzel, Berlin, Germany) with custom-attached 500 × 500 × 500-μm micro prism (Optikron) was placed under the microscope to reflect the illumination beam onto the focal plane of a high-NA water-immersion objective (40×, 1.25 NA, CFI Apochromat 40× WI Lambda-S, Nikon, Tokyo, Japan). Collected fluorescent signal was cleaned by a double-color emission filter for 642 nm and 488 nm (ZET488/640, AHF Analysentechnik, Tubingen, Germany) and projected onto the electron-multiplying charge-coupled device camera (Cascade II 512, Photometrics, Tucson, AZ) after further 2.5× magnification (2.5× C-Mount Adaptor, Carl Zeiss).
Light-sheet characterization
Light-sheet profiles were obtained by imaging the beam without reflection by the microprism. By moving along the propagation direction of the illumination beam in a stepwise manner, cross sections at every position were extracted and fitted to , where b is the lateral shift, d the baseline, w the width of the Gaussian beam, a the amplitude, and x the position. The extracted beam widths at different z-positions were then fitted to Gaussian beam propagation. Beam waist and Rayleigh length were extracted with and , where b is the lateral shift, z the propagation position, λ the wavelength, and w the beam waist, respectively. All fits were performed with a nonlinear least-square algorithm implemented in SciPy (11).
3D volume imaging
Drosophila embryos endogenously expressing green-fluorescent-protein (GFP)-tagged neuroglian were raised at 25°C and prepared according to established protocols. Briefly, embryos were picked at embryonic stages 14–15, dechorionated for 5 min with 50% household bleach, washed with H2O, and placed on a fresh grape juice agar stripe. Single embryos were placed on a coverslip with their anterior or posterior site pointing toward the capillary holder. A mounted 180 μm capillary (TSP180350, Optronis, Kehl, Germany) with heptane glue was then slowly moved toward the embryo and lifted after a few seconds of drying time. A motorized linear actuator (z steps of 0.5 μm; Z625B, Thorlabs) controlled the custom-made capillary holder along the z-direction. The camera exposure time was set to 50 ms with 2500 Gain. The power of the 488 nm illumination laser was kept at 130 μW (∼130 W/cm2). Images were analyzed and visualized with Fiji (ImageJ) and the plugin Volume Viewer.
In vivo single-molecule experiments
In typical injection experiments, embryos are dechorionated and covered with halocarbon oil to prevent dehydration while supplying sufficient oxygen. To accommodate the use of the water-dipping illumination objective, the injection protocol (12) was modified as follows: pasiflora1Δ embryos (13) expressing GFP-tagged neuroglian were picked at the syncytial blastoderm stage, placed on a double-sided tape, and desiccated for 10 min. Embryos were covered with halocarbon oil to avoid further desiccation and injected with a solution of 1 μg/μL 10 kDa Dextran-Alexa Fluor 647 in PBS (1× phosphate-buffered saline, Life Technology, Carlsbad, CA). After the injection, the embryos were placed in a dark and humid chamber to incubate for ∼20 h until they had reached embryonic stages 14-15. Embryos were carefully washed with n-heptane in a collection vial to remove residual halocarbon oil, rinsed three times with PBTw buffer (phosphate-buffered saline with 0.1% Tween 20), and further processed as described in the previous section. Under wide-field microscopy, the gut morphology served as a marker for the final staging (stages 15 to early 16) just before the single-molecule experiments were conducted. Normally developed embryos were closely positioned with their anterior-posterior axis parallel to the microprism without overlapping with each other. All steps of this procedure, except for the imaging, were carried out at 18°C. The following fly strains were obtained from published sources: pasiflora1Δ (13), GFP-trap NrgG00305 (14), paired-Gal4, and UAS-Rab5-GFP (Bloomington Drosophila Stock Center, Bloomington, IN). All strains were raised at 25°C.
Camera settings were chosen to optimize image quality (50 ms exposure time, 3800 gain, 1× binning, frame shift with 10 MHz read-out speed). The laser intensity was set to 60 μW (∼60 W/cm2) at 488 nm and 1.4 mW (∼1.4 kW/cm2) at 642 nm. Other single-molecule studies have used comparable laser power densities (8).
Single-particle detection and tracking
Inspired by image correlation spectroscopy (15), we propose a Gaussian window filter in the frequency space to simplify the automatic spot detection. The filtering removed the immobile fraction and bleaching kinetics. For this purpose, image stacks were fast-Fourier transformed (FFT), multiplied with the transmission window, , where f is the frequency and cut the cut-off frequency (0.08 s−1), and transformed back into real space. First, the FFT filter was benchmarked with tracks obtained from beads with 46 nm diameter (FluoSpheres 660/680, Thermo Scientific, Waltham, MA) diffusing in water. Settings were chosen to obtain similar raw images as acquired during Drosophila imaging (50 ms exposure time, 0 gain, 1× binning, frame shift with 10 MHz read-out speed). All the spots and tracks were then extracted from image sequences with TrackMate (16).
Next, the cumulative distribution of jump distances extracted from Drosophila embryos was fitted to four different models (Eqs. 1 and 2) with incremental species components (6) and compared using residuals between predicted and actual data:
| (1) |
| (2) |
where Di and Ai are the diffusion coefficient and fraction of diffusive species, respectively. Mean-square displacement (MSD) curves for directional movement were fitted to the anomalous diffusion model
| (3) |
where D is the diffusion coefficient and α is the exponent accounting for anomalous diffusion. All steps were processed with Python 2.7 and the SciPy packages for optimization, data handling, and visualization (11).
Results and Discussion
Optical setup
The assembly of our optical setup with opposed detection and illumination objective was realized with a few custom-made and commercial parts on an inverted microscope. An autonomous illumination module with lasers, fast AOTF, and single-mode optical fiber was assembled to easily connect the light source with our optical block. We further used achromatic lenses to maintain constant dimensions of the beam input diameter across different illumination wavelengths. A maximal beam input diameter of ∼12 mm was obtained with the 5× beam expander and the calibrated spherical aperture. A 150 mm achromatic cylindrical lens controlled the light-sheet width by conjugating the back focal plane of the illumination objective (Fig. 1 a).
We then sought to quantify the geometrical properties of the excitation light sheet with different beam input diameters. Therefore, we measured the light sheet’s cross-section by projecting it without reflection onto the focal plane of the detection objective in water. By moving the detection objective across the beam waist in a stepwise fashion, we determined the beam widths at 11 different propagation depths. The reconstructed profile was fitted to Gaussian beam propagation to obtain the beam waist for different beam input diameters (Fig. 1 b). In all our single-molecule experiments, we used a 642 nm laser line for excitation, which results in lower autofluorescence from the biological sample, and the 488 nm laser line to localize the epidermal structure. The beam input diameter was adjusted to 7 mm to create a final light sheet of ∼1 μm thickness (∼11 μm Rayleigh length) at 642 nm and ∼0.6 μm (∼8 μm Rayleigh length) at 488 nm wavelength. Furthermore, we visualized the light-sheet profile after reflection of the microprism by illuminating 500 nM Atto488 and Cy5 in water. By estimating the light-sheet waist position, we found that along the detection objective’s focal plane, a total light-sheet tilt of <∼1.5° was achieved (Fig. 1 c).
3D imaging capabilities
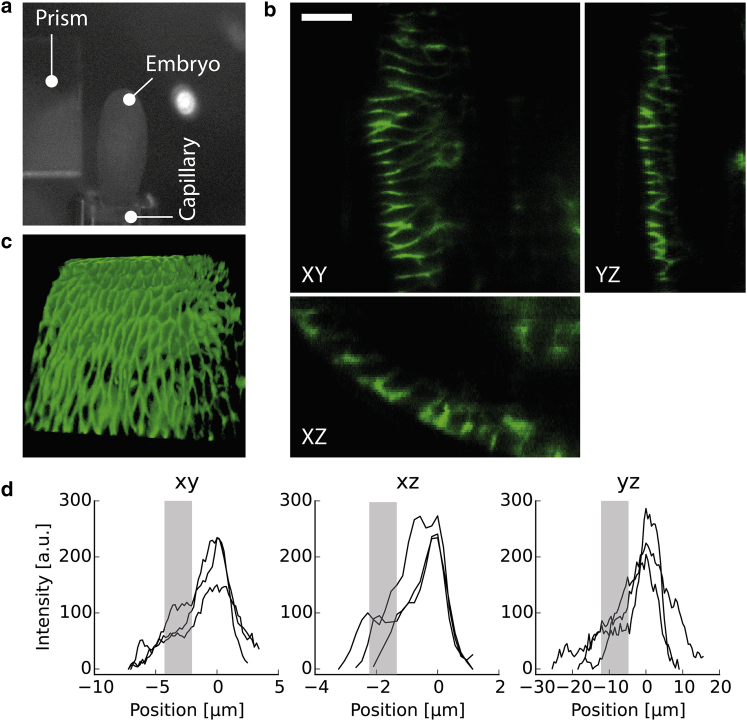
To investigate the ability of 3D sectioning with our optical setup, we positioned late-stage Drosophila embryos expressing GFP-tagged neuroglian (NrgGFP) on the tip of a microscale capillary with 180 μm inner diameter (Fig. 2 a). Mounting the embryo with common heptane glue, as used during live imaging of Drosophila with a confocal microscope on the glass capillary rendered a fast, robust, and user-friendly method of micrometer positioning. We implemented a linear actuator that offered a resolution of 0.5 μm in the z-direction, which was sufficient to obtain image stacks of the epidermal structure (Fig. 2).
Figure 2.
Volume imaging. (a) Positioning of Drosophila embryo with a capillary in front of the microprism’s reflective surface. A motorized linear actuator moved the capillary and the embryo in the z-direction. (b) Orthogonal views of images acquired with 0.5 μm resolution along the z-direction. Scale bar, 10 μm. NrgGFP accumulates at the apicolateral site of epithelial cells to hinder free diffusion along the paracellular space. (c) Reconstructed 3D volume of the epidermal structure. Single cells were readily resolved with RLSM. The ∼1 μm light-sheet thickness for optimal single-molecule imaging is accompanied by a limited region of homogeneous illumination (Fig. 1, b and c). Thus, we restricted the 3D volume imaging on the outer epidermal layer. (d) Line profiles along the cell-cell junction reveal an ∼2-fold increase in the NrgGFP signal at the apicolateral site (0 μm) compared to what we found at the basolateral site (gray bar) in all three directions of space.
It is important to note that the penetration depth is limited by light scattering as experienced with Gaussian-based light-sheet techniques. Furthermore, the light-sheet thickness of ∼1 μm provided a good SNR for single molecule imaging, but reduced the applicability on the global embryonic scale. Therefore, we primarily focused on the outer epidermal structure to test the volume-imaging capabilities of our RLSM implementation (17).
As a member of the membrane-associated septate junction (SJ) complex, neuroglian is strongly expressed and readily detected with 13 μW (∼130 W/cm2) at an excitation wavelength of 488 nm. As expected, NrgGFP accumulated at the apicolateral site where the properly formed SJ complex impedes free diffusion along the paracellular space (Fig. 2, b and c) (18). We further analyzed the data by measuring the intensity profile along the lateral membrane of some cells. Background-corrected intensity profiles (Fig. 2 d) show intensity peaks (200–300 counts) at 0 μm with lower-intensity plateaus (gray bar, 50–100 counts). The roughly twofold increase of NrgGFP signal toward the apicolateral site holds true for all three spatial projections, which indicates homogeneous illumination within the region of interest.
Single-molecule imaging in vivo
In recent years, single-molecule imaging has been successfully applied to a wide array of biological specimens ranging from the nuclei of single eukaryotic cells (6, 7, 8, 10) to membrane proteins in Caenorhabditis elegans (3). To test whether we could detect freely diffusing single molecules in the highly opaque Drosophila embryo (17), we decided to follow known paths and introduced a solution of 10 kDa Dextran as a single-molecule probe, since Dextran is being widely used for transportation studies in the biological context (13, 19, 20).
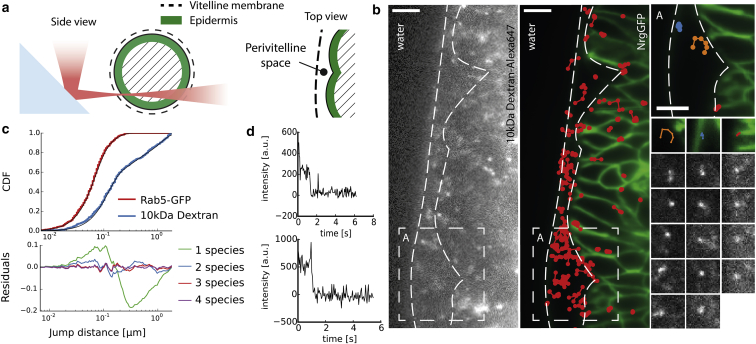
We first confirmed that single molecules of Dextran-Alexa647 are indeed detectable in living Drosophila embryos, and in a second step, we addressed questions regarding the diffusive behavior of Dextran on the multicellular scale. To this end, syncytial blastoderm embryos were collected and immediately injected with 10 kDa Dextran-Alexa647 and placed in a dark humid chamber. Embryos were staged (15 h after egg laying) and individually placed in H2O with their anterior-posterior axis parallel to the microprism’s reflective surface (Fig. 3 a). 3D mounting was unpractical for this purpose, because high-speed image acquisition is essential for single-molecule imaging in solution. As schematically depicted in Fig. 3 a, the light-sheet waist was positioned in the lower-left quadrant, since we expected that optical aberrations along the illumination as well as the emission path would worsen the SNR when imaging in deeper tissue (17).
Figure 3.
Single-molecule detection and tracking. (a) Illustration of the excitation path and cross section of the embryo at the region of interest. Imaging at the lower left quadrant of the embryo reduced optical aberrations. (b) Single raw image of 10 kDa Dextran-Alexa647 and the temporal average of NrgGFP with superimposed trajectories. Scale bar, 10 μm. Subregion and tracks show examples for fast (left), medium (center), and slow (right) particles. Scale bar, 5 μm. (c) Cumulative distribution function (CDF) of jump distances pooled from three different embryos and a three-component fitted model. Additionally, the CDF and a fitted curve (two-component model) are shown for Rab5-GFP particles found in the Drosophila embryo. (d) Example of Alexa647 bleaching steps found during the first seconds of imaging with the 642 nm laser line.
Because morphogenetic movement was negligible during the timescale of imaging, the GFP and Alexa647 channels could be acquired consecutively. Again, NrgGFP (488 nm laser line for excitation with ∼60 W/cm2) was used to adjust the light sheet and the focal position of the detection objective. Due to the interdependence of SJ proteins for localization, NrgGFP is now evenly distributed along the lateral membrane in the SJ mutant pasiflora1Δ (13) and therefore clearly outlines the epidermal structure (Fig. 3 b, center; Movie S1 in the Supporting Material).
In the Alexa647 channel, immobilized intensity spots that showed bleaching steps during the first few seconds of acquisition confirmed that we were indeed detecting single molecules in living Drosophila embryos (one to two dyes per Dextran according to the manufacturer). Particle locations of bleaching-step trajectories were found manually and their intensity was corrected for background signal. A locally dependent emission justifies the uneven level of signal counts for spatially separated fluorophores (Fig. 3 d).
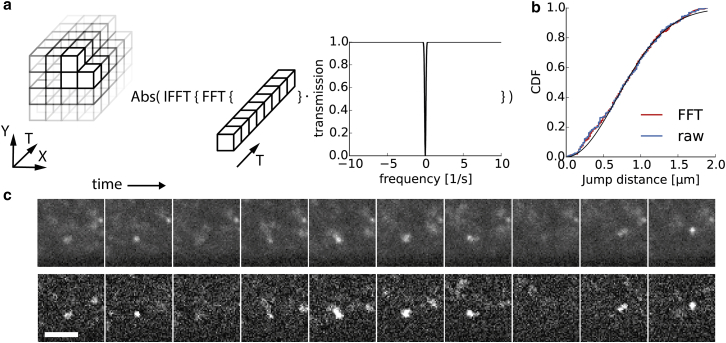
For single-particle tracking, the immobile fraction and low-frequency bleaching kinetics were removed via FFT filtering (Fig. 4; Movie S2). Before applying the FFT filter to the biological data, we imaged and tracked 46-nm-diameter beads with and without filtered image sequences to exclude possible artifacts introduced by the proposed temporal filter. As seen in Fig. 4 b, no apparent difference between the cumulative distributions of the two data sets could be observed. Fitting the jump-distance cumulative distribution to a single-component model led to the same diffusion coefficient for both data sets (Draw = 4.62 ± 0.04 μm2/s, Dfiltered = 4.63 ± 0.03 μm2/s).
Figure 4.
FFT filtering. (a) Every pixel in the image stack was independently processed using the temporal FFT filter. A Gaussian window was applied to remove immobile fraction and bleaching kinetics to improve the automatic spot detection. (b) CDF of freely diffusing 46-nm-diameter beads before and after FFT filtering. (c) Sequences of single frames of freely diffusing 10 kDa Dextran-Alexa647 in Drosophila embryos are shown for unfiltered (upper row) and filtered (lower row) movies. Scale bar, 5 μm. To see this figure in color, go online.
Single particles in Drosophila embryos were then detected and tracked with TrackMate after FFT filtering. Particles were excluded from further analysis when exceeding a certain size (diameter >1.84 μm) or tracks being outside of the expected lifetime range (3 frames ≤ included tracks ≤ 30 frames). The cumulative jump-distance distribution for all tracks was fitted to models consisting of 1, 2, 3, or 4 diffusive components. We found that our data were best described by a three-component model without overfitting, as indicated by the calculated residuals (Fig. 3 c; A1 = 0.56 with D1 = 0.068 ± 0.001 μm2/s, A2 = 0.16 with D2 = 0.520 ± 0.032 μm2/s, and A3 = 0.28 with D3 = 6.404 ± 0.072 μm2/s).
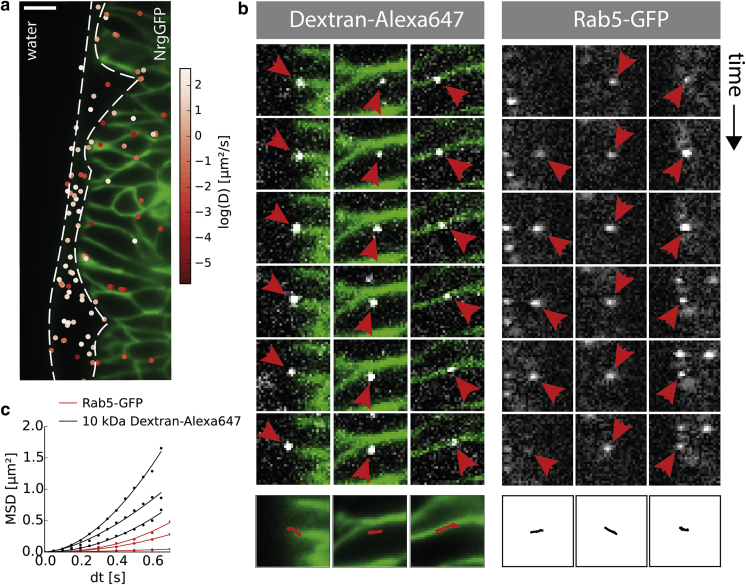
To investigate the spatial distribution of fast-, medium-, and slow-diffusing components, we calculated the apparent diffusion coefficient for a time lag of 50 ms, averaged over all equally spaced time lags in each track, and superimposed the color-coded mean track position onto the temporal averaged NrgGFP image stack. Diffusion coefficients are plotted on a log scale to account for the wide range of values (Fig. 5 a). It appears that fast tracks (∼6.4 μm2/s) occur in the free space between the vitelline membrane and the epidermis. A similar diffusion coefficient of 7 μm2/s was reported for a 23 kDa morphogen in the extracellular space of living zebrafish embryos by fluorescence correlation spectroscopy (21).
Figure 5.
Spatial heterogeneity of diffusion. (a) Superimposed mean track position with color-coded apparent diffusion coefficient. The apparent diffusion coefficient was calculated for all equally spaced time lags of 50 ms along entire individual trajectories. Fast diffusion coefficients are mainly found in the perivitelline space. In contrast, slow particles are mostly found in the paracellular space or at the cell cortex of epidermal cells. Scale bar, 10 μm. (b) Six representative time-lapse image sequences for particles associated with Dextran-Alexa647 (left three columns, green, NrgGFP) and Rab5-GFP (right three columns), with directional movement indicated by arrowheads. (c) Tracks were extracted and MSD curves reveal an anomalous diffusion coefficient of α > 1.
In contrast, slower particles were mostly found in the epidermal structure and seemed to obey directional movement, challenging the Brownian motion model. Clearly visible and long-living candidates were manually extracted and tracked, and the MSD for every track was fitted to the anomalous diffusion model (Fig. 5, b and c) (22, 23). We found that slow particles move in a directed manner (anomalous diffusion coefficient α > 1) along the cell cortex as well as the paracellular space (D1 = 0.87 ± 0.03 μm2/s1.81, α1 = 1.81 ± 0.05; D2 = 0.37 ± 0.02 μm2/s1.97, α2 = 1.97 ± 0.09; D3 = 0.47 ± 0.03 μm2/s1.57, α3 = 1.57 ± 0.08). The slow directional movement of particles suggests active vesicle trafficking as a possible explanation. This interpretation is supported by a study that proposed dynamin-dependent vesicle transportation of morphogens (D = 0.10 μm2/s) during wing development in Drosophila (24). In addition, previous experimental data showed that ∼80% of Dextran-positive structures colocalize with the GFP-tagged endosomal marker Rab5 during cellularization of early Drosophila embryos (19).
To further test our theory about endosomal trafficking, we expressed Rab5-GFP and acquired image stacks of Drosophila. Before analysis, the image sequence was postprocessed according to the proposed FFT filter with the same frequency transmission window as used for the single-molecule tracking. We found that the transportation direction appeared to align perpendicular to the anterior-posterior embryonic axis. The same preferred directionality could be observed for the Dextran-Alexa647 particles. Moreover, similar diffusion coefficients found by the cumulative distribution (Fig. 3 c; A1 = 0.78 with D1 = 0.0112 ± 0.0001 μm2/s and A2 = 0.22 with D2 = 0.052 ± 0.002 μm2/s) and the MSDs (Fig. 5 b; D1 = 0.270 ± 0.019 μm2/s2.341, α1 = 2.341 ± 0.138; D2 = 0.148 ± 0.005 μm2/s2.108, α2 = 2.108 ± 0.007; D3 = 0.020 ± 0.001 μm2/s1.578, α3 = 1.578 ± 0.046) support our interpretation. The key problem with this comparison of fitted diffusion coefficients is due to the strong influence of the particle’s transportation direction with respect to the imaging plane. Particles that translate along the light sheet will exhibit a higher diffusion coefficient than particles traversing across it.
Conclusions
We report on an optical setup for imaging single molecules to entire cellular structures in 3D of living late-stage Drosophila embryos. Methodologically, we show that by adapting RLSM, single-molecule imaging is readily achievable even within the highly opaque Drosophila embryo. Using commercially available microprisms offers a user-friendly and fast way to obtain the reflective surface that is necessary for RLSM. Our setup could easily be supplemented with oil-immersion detection objectives that would, however, limit the accessible imaging depth. In principle, the same optical approach could be used for single-cell studies when grown in 3D cell culture. However, it is important to note here that RLSM techniques are currently constrained to a few micrometers above the coverslip surface and consequently make the relative positioning of mirror and sample of central importance (8, 10).
We believe that the RLSM implementation presented here will not only offer an alternative to high-resolution particle-imaging techniques, e.g., spinning disk microscopy, but most importantly will provide a way to study the fundamental level of stochastic single-molecule dynamics from the multicellular perspective. In addition, more relevant data for cell biology will be generated, since the principles of LSM give the additional advantage of reduced photobleaching and phototoxic effects (25).
Author Contributions
F.G. and D.B. constructed the optical setup and F.G. analyzed the data. F.G. and M.D. performed the experiments. F.G., M.D., C.J., U.G., and D.B. contributed to designing the experiments, discussion, and writing the manuscript.
Acknowledgments
We thank the Fabrication team of the Physics Department and Gene Center of Ludwig Maximilians Universität München, and P. Bandilla, M. Hanf, and C. Ludwig for their technical and experimental expertise.
F.G. is supported by a Deutsche Forschungsgemeinschaft Fellowship through the Graduate School of Quantitative Biosciences Munich (QBM). Financial support from SFB 1032 Project A4 (D.B.), from the Alexander von Humboldt-Professorship by the Bundesministerium für Bildung und Forschung (U.G.) is acknowledged.
Editor: David Wolf.
Footnotes
Two movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)00042-4.
Supporting Material
Freely diffusing 10 kDa Dextran-Alexa647 molecules superimposed onto the NrgGFP channel. The Dextran-Alexa647 channel was post-processed by the temporal FFT filter. Scale bar, 10 μm.
Image sequences are shown before (left) and after (right) post-processing with the proposed FFT filter. Post-processing simplified the automatic spot detection in order to extract the diffusion coefficients. Scale bar, 10 μm.
References
- 1.Li G.-W., Xie X.S. Central dogma at the single-molecule level in living cells. Nature. 2011;475:308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ulbrich M.H., Isacoff E.Y. Subunit counting in membrane-bound proteins. Nat. Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhan H., Stanciauskas R., Pinaud F. In vivo single-molecule imaging identifies altered dynamics of calcium channels in dystrophin-mutant C. elegans. Nat. Commun. 2014;5:4974. doi: 10.1038/ncomms5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tokunaga M., Imamoto N., Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 5.Huisken J., Swoger J., Stelzer E.H. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 6.Chen B.C., Legant W.R., Betzig E. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ritter J.G., Veith R., Kubitscheck U. Light sheet microscopy for single molecule tracking in living tissue. PLoS One. 2010;5:e11639. doi: 10.1371/journal.pone.0011639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gebhardt J.C.M., Suter D.M., Xie X.S. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods. 2013;10:421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedrich M., Nozadze R., Harms G.S. Detection of single quantum dots in model organisms with sheet illumination microscopy. Biochem. Biophys. Res. Commun. 2009;390:722–727. doi: 10.1016/j.bbrc.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 10.Galland R., Grenci G., Sibarita J.-B. 3D high- and super-resolution imaging using single-objective SPIM. Nat. Methods. 2015;12:641–644. doi: 10.1038/nmeth.3402. [DOI] [PubMed] [Google Scholar]
- 11.Jones, E., T. Oliphant, and P. Peterson. 2007. SciPy: Open source scientific tools for Python.http://www.scipy.org.
- 12.Iordanou E., Chandran R.R., Jiang L. RNAi interference by dsRNA injection into Drosophila embryos. J. Vis. Exp. 2011;50:2795. doi: 10.3791/2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deligiannaki M., Casper A.L., Gaul U. Pasiflora proteins are novel core components of the septate junction. Development. 2015;142:3046–3057. doi: 10.1242/dev.119412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morin X., Daneman R., Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potvin-Trottier L., Chen L., Wiseman P.W. A nu-space for image correlation spectroscopy: characterization and application to measure protein transport in live cells. New J. Phys. 2013;15:085006–085021. doi: 10.1088/1367-2630/15/8/085006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schindelin J., Arganda-Carreras I., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller P.J., Schmidt A.D., Stelzer E.H.K. Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nat. Methods. 2010;7:637–642. doi: 10.1038/nmeth.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genova J.L., Fehon R.G. Neuroglian, Gliotactin, and the Na+/K+ ATPase are essential for septate junction function in Drosophila. J. Cell Biol. 2003;161:979–989. doi: 10.1083/jcb.200212054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabrowski P., Necakov A.S., De Renzis S. Tubular endocytosis drives remodelling of the apical surface during epithelial morphogenesis in Drosophila. Nat. Commun. 2013;4:2244. doi: 10.1038/ncomms3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pluen A., Netti P.A., Berk D.A. Diffusion of macromolecules in agarose gels: comparison of linear and globular configurations. Biophys. J. 1999;77:542–552. doi: 10.1016/S0006-3495(99)76911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu S.R., Burkhardt M., Brand M. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature. 2009;461:533–536. doi: 10.1038/nature08391. [DOI] [PubMed] [Google Scholar]
- 22.Metzler R., Klafter J. The random walk’s guide to anomalous diffusion: a fractional dynamics approach. Phys. Rep. 2000;339:1–77. [Google Scholar]
- 23.Reverey J.F., Jeon J.-H., Selhuber-Unkel C. Superdiffusion dominates intracellular particle motion in the supercrowded cytoplasm of pathogenic Acanthamoeba castellanii. Sci. Rep. 2015;5:11690. doi: 10.1038/srep11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kicheva A., Pantazis P., González-Gaitán M. Kinetics of morphogen gradient formation. Science. 2007;315:521–525. doi: 10.1126/science.1135774. [DOI] [PubMed] [Google Scholar]
- 25.Pampaloni F., Chang B.-J., Stelzer E.H.K. Light sheet-based fluorescence microscopy (LSFM) for the quantitative imaging of cells and tissues. Cell Tissue Res. 2015;360:129–141. doi: 10.1007/s00441-015-2144-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Freely diffusing 10 kDa Dextran-Alexa647 molecules superimposed onto the NrgGFP channel. The Dextran-Alexa647 channel was post-processed by the temporal FFT filter. Scale bar, 10 μm.
Image sequences are shown before (left) and after (right) post-processing with the proposed FFT filter. Post-processing simplified the automatic spot detection in order to extract the diffusion coefficients. Scale bar, 10 μm.