Abstract
Integrins are bidirectional, allosteric transmembrane receptors that play a central role in hemostasis and arterial thrombosis. Using cryo-electron microscopy, multireference single-particle reconstruction methods, and statistics-based computational fitting approaches, we determined three-dimensional structures of human integrin αIIbβ3 embedded in a lipid bilayer (nanodiscs) while bound to domains of the cytosolic regulator talin and to extracellular ligands. We also determined the conformations of integrin in solution by itself to localize the membrane and the talin-binding site. To our knowledge, our data provide unprecedented three-dimensional information about the conformational states of intact, full-length integrin within membrane bilayers under near-physiological conditions and in the presence of cytosolic activators and extracellular ligands. We show that αIIbβ3 integrins exist in a conformational equilibrium clustered around four main states. These conformations range from a compact bent nodule to two partially extended intermediate conformers and finally to a fully upright state. In the presence of nanodiscs and the two ligands, the equilibrium is significantly shifted toward the upright conformation. In this conformation, the receptor extends ∼20 nm upward from the membrane. There are no observable contacts between the two subunits other than those in the headpiece near the ligand-binding pocket, and the α- and β-subunits are well separated with their cytoplasmic tails ∼8 nm apart. Our results indicate that extension of the ectodomain is possible without separating the legs or extending the hybrid domain, and that the ligand-binding pocket is not occluded by the membrane in any conformations of the equilibrium. Further, they suggest that integrin activation may be influenced by equilibrium shifts.
Introduction
Integrin receptors transmit bidirectional signals across the plasma membrane, thereby modulating signals that regulate hemostasis, proliferation, differentiation, migration, and cell death. These receptors are composed of a noncovalent mix of heterodimer transmembrane α- and β-subunits, each of which possesses a single transmembrane and cytoplasmic tail domain (1). They are expressed on the cell surface and are believed to be in either a low- or high-affinity state with respect to extracellular ligand binding. Integrins require an activation step that switches them from the low-affinity state to the high-affinity state (2, 3, 4, 5, 6). This activation triggers extracellular ligand binding and initiates adhesion. Upon activation, integrins aggregate, recruit integrin-associated proteins, and form small clusters. The low- and high-affinity states are crucial for regulating cell adhesion, particularly in the case of platelets, which must circulate freely in a nonadherent state until they are stimulated by agonists at the site of a wound. Such stimulation turns on the fibrinogen binding function of the αIIbβ3 integrin, leading to platelet aggregation and the formation of a thrombus that halts the loss of blood.
Much of the current thought regarding the conformational rearrangements in integrin that lead to activation is inferred from crystal structures of the β3 integrin ectodomains (7, 8, 9, 10, 11, 12). All of these structures show that the receptor adopts a bent conformation in which the headpiece containing the ligand-binding site is pointed in the same direction as the tail, presumably toward the membrane. This conformation is generally assumed to be the low-affinity state, in part because the ligand-binding pocket would be too close to the membrane to be accessible to extracellular ligands. Negative-stain reconstructions in the absence of membrane show that bent αVβ3 integrins are capable of binding relatively bulky fibronectin fragments (13), but it is unclear whether the presence of the membrane would prevent binding by steric hindrance. A recent three-dimensional (3D) reconstruction of manually selected, dehydrated, and negatively stained integrins in lipid bilayer nanodiscs (14) shows a bent conformation somewhat similar to the crystal structures but with the ligand-binding domain pointing away from the membrane.
Negative-stain-based, two-dimensional (2D) electron microscopy (EM) studies of integrin samples (4, 11, 15, 16, 17, 18) indicate that integrin can also adopt an upright conformation, which is generally assumed to correspond to the high-affinity state (19). 2D projection images of upright conformations show the α- and β-legs both separated and closed together, although the β-leg is mostly poorly resolved. In the crystal structures and the negative-stain reconstruction of the bent integrin (13, 14), the legs are closed together. However, because 3D reconstructions of upright integrins were not obtained, it is unclear whether other conformations of integrin exist or whether 2D projection images that appear to be upright with closed legs are projections of the open-leg upright conformation captured in different orientations.
To adopt the upright conformation with separated legs, the hybrid domain, which connects the β-head with the β-leg, must swing out from its position in the bent crystal structure conformation. Such movement of the hybrid domain was observed within a crystal lattice when RGD peptide was soaked into crystals of the integrin αIIbβ3 headpiece (12). Talin binds to the cytoplasmic side of the β integrin subunit (20), and binding of talin domains to cytoplasmic fragments of β integrin leads to dissociation of integrin αIIbβ3 transmembrane segments (21, 22). A separation of the α- and β-legs in response to talin binding was demonstrated in vivo for αLβ2 integrins (23). However, this dissociation effect of talin has not been demonstrated conclusively in the context of the membrane-embedded full-length αIIbβ3 integrin molecule.
In this study we investigated the conformations of full-length integrin in the activated state, induced by the presence of cytosolic activators and extracellular ligands. Samples were analyzed in a fully hydrated environment and while embedded in a membrane, using cryo-EM and single-particle reconstruction techniques. The use of nanodiscs (nanometer-scale phospholipid bilayer membrane islands (18, 24, 25)) in conjunction with specialized assembly preparation protocols ensured that only single integrin molecules would be incorporated into each ∼10-nm-diameter nanodisc, thus avoiding clustering effects. We determined the 3D conformations of single, full-length, fully hydrated integrin embedded in nanodiscs with bound talin head domains and RGD peptides. Native gels confirmed the integration of single heterodimers of integrin in the nanodisc under the conditions used for imaging. In addition, we determined the conformations of full-length integrin samples in solution without activators, ligands, or nanodiscs to allow localization of components and to compare conformations in the high- and low-affinity states. In all of the samples, αIIbβ3 exists in an equilibrium anchored at four main conformational states: a compact nodule that is similar to the bent conformer of the αIIbβ3 integrin observed in its crystal structure and in negative stain (14), a conformation that is clearly identifiable as being upright with well separated lower legs, and two intermediate conformations that are consistent with the closed lower legs but exhibit different degrees of extension and opening of the hybrid domain. In all of the membrane-embedded conformations, including that of the compact nodule, the ligand-binding pocket of the integrin headpiece is pointing away from the membrane. For the experimental conditions employed here, the 3D data analysis indicates that intact human integrins αIIbβ3 exist in a continuous conformational equilibrium between the two extremes (i.e., the bent and upright conformations) and that this equilibrium is significantly shifted toward the upright conformation in the presence of cytosolic activators, extracellular ligands, and nanodiscs. We show that extension of the ectodomain is possible without separating the legs or extending the hybrid domain, and that the ligand-binding pocket is not occluded by the membrane in any of the conformations in the equilibrium landscape.
Materials and Methods
Purification of αIIbβ3 integrins
We purified αIIbβ3 integrins from human platelets (26, 27) and characterized their biochemical activities as previously described (28). Briefly, washed, outdated platelets were lysed in a 20 mM Tris buffer, pH adjusted to 7.4 by addition of HCl (Tris-HCl buffer, containing 150 mM NaCl, 1% Triton X-100, 5 mM phenylmethylsulfonyl fluoride, 1 mM CaCl2, and 10 μM leupeptin) overnight at 4°C. The lysate was centrifuged at 50,000 × g and passed over Con-A-Sepharose (Amersham Pharmacia Biotech) at 4°C. Proteins were eluted with buffer A (20 mM Tris buffer, pH 7.0, 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 0.1% Triton X-100, and 0.05% NaN3) containing 200 mM methyl α-D-mannopyranoside (Sigma) and 10 μM leupeptin (Sigma). The flow-through fractions were pooled after three passes over a KYGRGDS-Sepharose column. The flow-through fraction was then run over a heparin-agarose column at 4°C, further purified by gel size-exclusion chromatography on a Sephacryl S300 HR, and dialyzed against buffer A. The bound fraction was eluted from the KYGRGDS-Sepharose column using soluble RGD peptide dialyzed against buffer A. The flow-through fraction and the eluted bound fraction were treated as independent biochemical preparations during the analysis process. The purity and integrity of each sample were assessed by SDS-PAGE. For the studies presented here, samples were freshly prepared and experiments were performed at physiological levels of Ca2+ and Mg2+ without Mn2+.
Talin, RGD, and lipid nanodiscs
The F3 domain of talin (GSGVSFFLVK EKMKGKNKLV PRLLGITKES VMRVDEKTKE VIQEWSLTNI KRWAASPKSF TLDFGDYQDG YYSVQTTEGE QIAQLIAGYI DIILKKKS, pI = 9.46, 11.2 KD) was provided by Drs. K.L. Wegener and I.D. Campbell (Oxford University). The full-length head domain of talin was provided by Drs. B. Patel and D.R. Critchley (University of Leicester). RGD peptide was generated as Ac-CGSGRGDF-amide in-house. Preformed phospholipid MSP1/DPPC nanodiscs were kindly provided by Dr. Mark A. McLean (University of Illinois) and generated as previously described (24).
Incorporation of integrin into nanodiscs
To form nanodisc-integrin complexes, 0.25 μM of integrin was incubated at room temperature for 15–20 min with 3.5 μM of preformed MSP1/DPPC nanodiscs in 40 mM Tris-HCl buffer, 1 mM MgCl2, and 1 mM CaCl2 in the presence of 11 μM of the talin F3 domain or full-length talin head domain, and 800 μM of RGD peptide, with both ligands at saturating conditions. After incubation, the samples were analyzed with the use of native alkaline electrophoresis 4–20% Tris-Glycine PAGE (Invitrogen). Reaction products were detected by Coomassie staining and were directly processed for cryo-EM. For protein fluorescein modification, RGD or F3 peptides were incubated with two molar excess of fluorescein-maleimide in 20 mM HEPES, 50 mM NaCl, pH 8.0, overnight at 4°C.
Cryo-EM
Samples (4 μL) from the reaction mixtures described above were applied to glow-discharged Quantifoil R 1.2/1.3 holey carbon film on a 400 mesh copper transmission electron microscopy grid (Quantifoil Micro Tools) and manually plunge-frozen in liquid-nitrogen-cooled liquefied ethane. Images of all samples suspended over holes were acquired under low-dose conditions using either a Tecnai F20 G2 Twin transmission electron microscope (FEI) equipped with a field emission gun operated at 200 kV and a nominal magnification of 67,000×, or a Tecnai T12 Twin transmission electron microscope (FEI) equipped with a Lab6 filament (Denka, Denki Kagaku Kogyo Kabushiki Kaisha) at 120 kV at a nominal magnification of 62,000×. Micrographs were recorded with both microscopes on Kodak ISO-163 plates (Eastman Kodak) with a total dose of 20–50 e−/Å2, with a defocus of 1.5–2.5 μm on the T12, and 2.5–3.5 μm on the F20. The micrographs were developed in a full-strength Kodak D19 developer, digitized using an SCAI scanner (Intergraph) at 7 μm raster, and compressed to a final pixel size of 0.6 nm. To allow cross-validation of results, nine different biochemical preparations (four with nanodiscs, talin, and RGD, and five without) were separately imaged and analyzed.
Image preprocessing
A total of 48,479 particles were interactively selected from 954 micrographs using the boxer routine from the EMAN package (29) (Fig. S1 in the Supporting Material). Phase correction for the contrast transfer function was applied for all images using ctfit from EMAN. An amplitude correction was applied during the iterative refinement. The low-resolution part of a curve used for correction was obtained by simultaneously fitting the contrast transfer function of several sets of particles. For final runs, a spherically averaged structure factor amplitude curve was computed from the respective fitted models, which was then used to construct the high-resolution part of the correction curve. No B-factor sharpening was applied to the reconstructions. To elucidate the composition of the conformations in the sample, a reference-free 2D alignment (30) was run for each of the nine data sets. Consistent with previous observations in negatively stained samples, the resulting class averages indicated the existence of at least two different conformations, one globular and one extended to different degrees, with some class averages showing clearly separated legs.
Initial iterative sorting procedure
Initial models for sorting the data into different conformations were constructed according to the results of the reference-free alignment results. We used the bent conformation of the crystal structure and constructed a rough model of an upright conformation. Both models were low-pass filtered to 6-nm resolution and then used in a multireference alignment scheme (31) as implemented in EMAN2 (32) for all nine data sets independently. The sorted data were then separated for further processing. Each sorted set was subjected to iterative refinement as implemented in EMAN2 until convergence was reached (eight to 25 iterations). The refined reconstructions were then used as input for a new iteration of the sorting process. The process was iterated until no more changes were obtained in the sorting (three to five iterations) of each independent set. Reconstruction from class averages was achieved using the gridding algorithm implemented in SPARX (33, 34).
Model dependence testing and refinement of sorted groups
Model dependence is a major indicator for inhomogeneity within data sets. We devised a particularly stringent test for model dependence that is very efficient at detecting mixtures of conformations (35). The procedure is based on deliberately biasing the starting model of the structure refinement step of the reconstruction procedure by swapping the references after sorting. For example, to run this test on data sorted between bent and upright conformations, the upright model is used as a starting model for refinement with the data sorted into the bent conformation. If there is a substantial contamination of conformations that are in between the upright and bent conformations within that sorted subset, the reconstruction will depend significantly on the starting model and the resulting reconstruction will be less bent. We performed this procedure independently for each of the nine data sets (Fig. S2). In addition to the swapped reference, we also ran the procedure with a model constructed from the reference-free class averages of the initial data set and the common-lines procedure implemented in EMAN2. A second level of model dependence testing is based on cross-validation of results between the independent nine data sets. Different preparations of the same conditions should yield the same subdivision between conformers and should also yield similar reconstructions for each class. If significant model dependence is detected by either method in one of the sorted subsets, the new model from the biased run is added as a reference for the sorting procedure and a new round of iterative sorting is initiated (see supplement in Xu et al. (35)). The success of the procedure depends to some extent on the criterion used for comparing the reconstructions. If the criterion is too tight, convergence will not be achieved. If the criterion is too loose, sorted subsets will not be optimal. With four classes, considering that the similarity test was passed if the Fourier shell correlation 0.5 cutoff was at 4.5 nm or better gave the best results over all independent data sets.
Final refinement
Subdivision into four classes yielded optimal results in terms of model independence over all nine data sets both within the data sets and by cross-validation. Attempts to further split the data (for example, to detect talin domains in the intermediate states) did not yield stable class separations. To obtain the highest possible quality and resolution, we performed reference-based iterative refinement using the consensus references for the four conformations on the respective data subsets once we had confirmed model independence. Independent data sets for the same conditions were combined for this purpose. Convergence was achieved within eight to 25 iterations. Only the upright and bent conformations showed an improvement in resolution at this stage, indicating that the two intermediate classes were significantly more variable than those two. After final convergence was reached, we performed one round of sorting to compile the percentages of the four conformers. We ran this analysis independently on the original independent data sets to estimate the standard deviations of these percentages, which, with 1–2% (Table 1), indicated a high level of consistency for all four reconstructions under all conditions. Iteration of the sorting procedure inherently breaks the assumption of complete independence for the refinement, which is a requirement for commonly used resolution-determination schemes (36). Because of the lack of independence, splitting the data after band-limited refinement was a more adequate method for estimating resolution in this study (36). The resolution determined with this procedure was between 3 and 4.5 nm for the reconstructions.
Table 1.
Statistical Analysis of the Conformational Equilibrium Distribution of Integrins
| Bent (%) | First Intermediate (%) | Second Intermediate (%) | Upright (%) | |
|---|---|---|---|---|
| Integrin | 13.27 ± 1.95 | 20.89 ± 1.10 | 37.63 ± 1.25 | 28.21 ± 1.85 |
| Integrin, N, T, R | 6.13 ± 1.37 | 17.43 ± 0.68 | 33.11 ± 0.85 | 43.33 ± 0.88 |
Statistical analysis of integrins and integrins embedded in nanodiscs (N) and in the presence of talin (T) and RGD (R). The statistics were compiled from a structural analysis of five independent preparations in the absence of nanodiscs and four independent preparations with integrins embedded in nanodiscs (a total of 48,479 particles). Standard deviations for each average are indicated. The different conditions were deliberately chosen to account for variations in sample preparation, to ensure that the results would be independent of those variations. For example, one data set was obtained from samples that were fresh right after purification and another was obtained after the samples were frozen, stored at −80°C, and then thawed before preparation for cryo-EM.
Docking and model building
We used difference mapping between reconstructions of integrins with and without nanodiscs to locate the densities corresponding to the nanodiscs. Density corresponding to the talin head domain was also identified by difference mapping, in this case between integrin in nanodiscs with the talin F3 domain and full-length talin head present. Only the reconstruction of the upright conformation yielded a significant difference peak. We used a combination of normal-mode-based (37) and statistics-based (38) docking to fit integrin αIIbβ3 domains extracted from crystal structures (11, 39, 40, 41, 42, 43) into the reconstructions. The use of statistics-based docking allows one to objectively estimate the docking precision even at low resolution (44) and to identify regions of the structure with inherent flexibility. The analysis indicates that the atoms can be reproducibly localized in the density with a root-mean-square deviation of ∼0.6 nm for the model overall and ∼0.7 nm for the distance between the centers of the α- and β-legs at the membrane interface, a small fraction of the respective intermodel root mean-square deviations. For the other conformers, rigid-body docking of larger domains was used and further refinement of relative positioning was not pursued.
For both intermediate conformations, the quality and resolution of the maps indicated that they represented mixtures of conformations. Neither of the two reconstructions was of sufficient quality to allow reliable docking of individual domains according to the statistical analysis. Thus, the headpiece and the leg domain from the crystal structure were docked as rigid bodies into the first intermediate conformation. Breaking the structure up into smaller units for docking did not yield statistically significant improvements. For the second intermediate conformation, the structure needed to be further modified to fit the density. The headpiece and α-leg were taken from the model of the upright conformation and docked as rigid bodies. The lower β-leg was attached to the calf-2 domain using the contacts observed in the crystal structure. The remaining I-EGF domains were modeled into the density using connectivity and density constraints. All correlation values and statistical analyses were calculated with a conservative 45-Å-resolution cutoff. Data and models were deposited in the Electron Microscopy Data Bank and the Protein Data Bank.
Results and Discussion
Single integrin αIIbβ3 incorporated into lipid bilayer nanodiscs
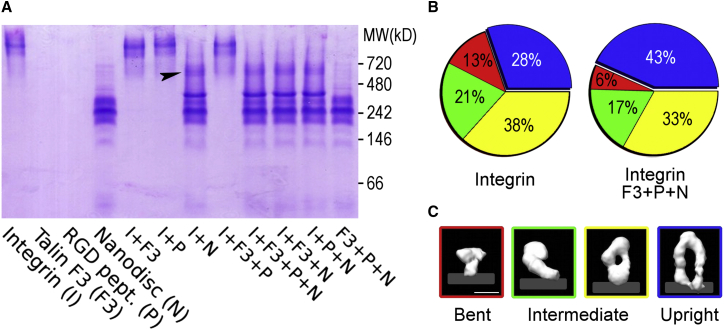
We determined 3D structures of full-length integrin in a fully hydrated environment within a membrane bilayer while it was bound to cytosolic and extracellular ligands. Nanodiscs (nanometer-scale, protein-supported phospholipid bilayer discs (24)) were used as a model membrane. They recapitulate the properties of biological membranes more closely than liposomes (45), and the small size (11 nm × 15 nm) of the oblate nanodiscs (25) ensures that under defined biochemical conditions, only single integrin molecules are incorporated, eliminating the possibility that interactions between integrin molecules (e.g., clustering) could affect the results of the study. Ye et al. (18) previously demonstrated the physiologically relevant functionality of nanodisc-embedded integrins by measuring the binding of the activation-specific antibody PAC1 and demonstrating its ability to bind fibrin. Here, we incorporated purified, intact αIIbβ3 integrins into nanodisc phospholipid bilayers in the presence of talin head domains and saturating concentrations of RGD peptides. The incorporation of αIIbβ3 into the nanodisc membranes in physiologic buffer was followed in native gels via the large shift in mobility of the integrin, indicating nearly quantitative incorporation of single integrins into the nanodiscs (Fig. 1 A).
Figure 1.
Integrin exists in a conformational equilibrium of four major conformers. (A) Nondenaturating native gel electrophoresis was used to assess the integration of integrin into nanodiscs. When integrin and nanodiscs were incubated together, the integrin band spatially relocated on the gel (arrowhead) without splitting. (B) Distribution of integrin conformers in the absence and presence of nanodiscs, talin head domain, and RGD peptide. In both conditions, four conformations are observed: bent (red), first intermediate (green), second intermediate (yellow), and upright (blue). The standard deviation for all percentages is between 1% and 2% (Table 1). (C) Surface representations of the 3D reconstructions corresponding to the colors in the pie charts are shown. The contour level was chosen to account for integrin’s molecular weight. All four images are at the same magnification. The light-gray strip indicates the approximate membrane position as suggested by reconstructions of integrins bound to nanodiscs. Scale bar, 10 nm. To see this figure in color, go online.
Imaging of fully hydrated, membrane-embedded integrin αIIbβ3 by cryo-EM
The αIIbβ3 integrins and integrin-nanodisc assemblies were imaged in a fully hydrated state in physiological buffers by cryo-EM. This procedure contrasts with most other EM studies on integrins, which used either negative staining (4, 11, 13, 14, 15, 16, 17, 18, 46, 47, 48) or rotary shadowing (49, 50). In rotary shadowing, heavy metal is evaporated onto surface-adsorbed molecules, whereas in negative-staining approaches, samples are embedded in a heavy-metal stain, absorbed on a carbon film support, and dehydrated. It was shown that this type of preparation technique can introduce serious structural distortions (e.g., flattening) into nanodisc-embedded integrins (18), raising the possibility that other studies of integrins based on negative staining might suffer from similar artifacts, and highlighting the importance of obtaining structures under more native conditions. Furthermore, the majority of the studies mentioned above were carried out using 2D projection images, which introduces ambiguities into the observed conformations, for example, concerning the degree of leg separation (Fig. S4). Therefore, to obtain accurate representations and percentages of integrin conformations, a complete 3D analysis of cryo-EM data is necessary.
The results of our study also differ from those of Adair and Yeager (51), who provided the only other 3D structure of integrin αIIbβ3 determined by cryo-EM. Their study, which was done in the absence of a phospholipid membrane, showed αIIbβ3 integrin in a single conformation. However, the image reconstruction scheme employed in that study assumed the existence of a single conformation, and thus all images were mapped onto that single conformer. Indeed, it has been suggested (4) that the structure reported by Adair and Yeager is an average of bent and extended conformations.
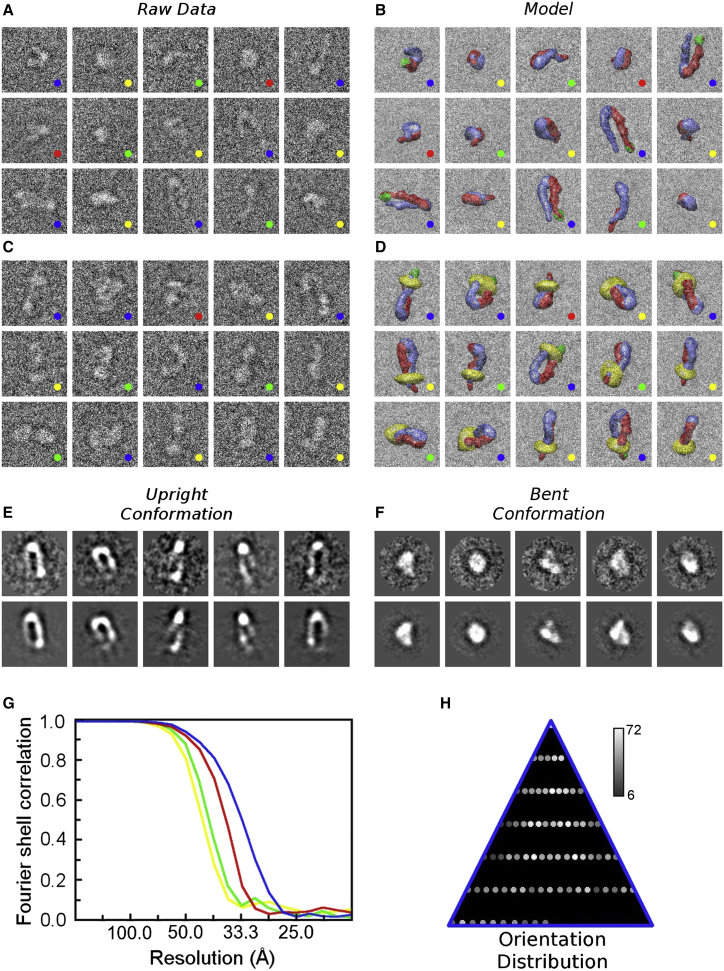
Here, we sorted individual conformations using unbiased iterative, multireference, single-particle 3D reconstruction techniques (31, 35). Image processing was run independently for four conditions, each of which was duplicated with two separate biochemical integrin preparations. For each of these, either two or three independent reconstructions were generated, further facilitating cross-validation of the results and resolution (36), leading to a total of nine independent runs. In total, nearly 50,000 images of individual particles, selected from more than 950 micrographs, were used for the analysis (Fig. 2). For each reconstruction, we ensured the absence of model bias by using multiple starting models as well as cross-validation approaches (35).
Figure 2.
Representative cryo-EM data and data-quality indicators for integrin samples. (A) Representative gallery of particles picked from unprocessed micrographs of samples with integrin. These particles represent random projections of all four conformations with defined Euler angles with respect to the 3D reconstructions. The conformations are color coded as in Fig. 1, B and C: upright (blue), bent (red), first intermediate (green), and second intermediate (yellow). The width of the boxes is 44 nm (applies to entire figure). (B) Surface representations of models of the corresponding integrin conformations in matching orientations (using the defined Euler angles) of images in (A). Blue, α-subunit; red, β-subunit; green, talin; yellow, nanodisc. (C) Representative gallery of particles picked from unprocessed micrographs of samples with integrin, talin F3, RGD peptide, and nanodisc. As in (A), these particles represent all four conformations. (D) Surface representations of models of the corresponding integrin conformations in matching orientations (using the defined Euler angles) of images in (C). (E and F) Five representative class averages (top row) drawn from the angular classification (see H) and corresponding reconstruction projections (bottom row) for the upright (E) and bent (F) conformations generated from the same data set that was used to generate (C), but after iterative multireference classification. (G) Fourier shell correlation curves for the 3D reconstructions of integrin conformations in the presence of nanodiscs, talin head domain, and RGD peptide. The 0.5 cutoff criterion indicates resolutions between 4.5 and 3 nm. The color code of the curves corresponds to that used in Fig. 1, B and C. (H) Distribution of integrin particles in the upright conformation on the sample support grid with an angular sampling of 15°. The number of particles corresponding to a particular orientation is proportional to the brightness of the spots (see bar). The distribution indicates that the particles are nearly randomly distributed. The distributions of Euler angles for the other data sets display similarly uniform coverage. To see this figure in color, go online.
Integrin αIIbβ3 exists in a continuous conformational equilibrium clustered around four main states
We identified four main conformational groups of αIIbβ3 and determined their structures in three dimensions (Fig. 1). These included i) a compact nodule that is similar to the compact conformer observed in the crystal structure (11) and to the negative-stain-based reconstruction of nanodisc-embedded integrin (14), ii and iii) two intermediate conformations that have closed lower legs but exhibit different degrees of straightening and opening of the hybrid domain, and iv) a conformation that is clearly identifiable as being upright with well separated legs. A resolution analysis indicates that each of these groups represents a cluster of conformers around the main conformation. Structurally, the upright conformation (iv) is the best defined, followed by the compact nodule (i), and the intermediate conformations (ii and iii) are the least well defined.
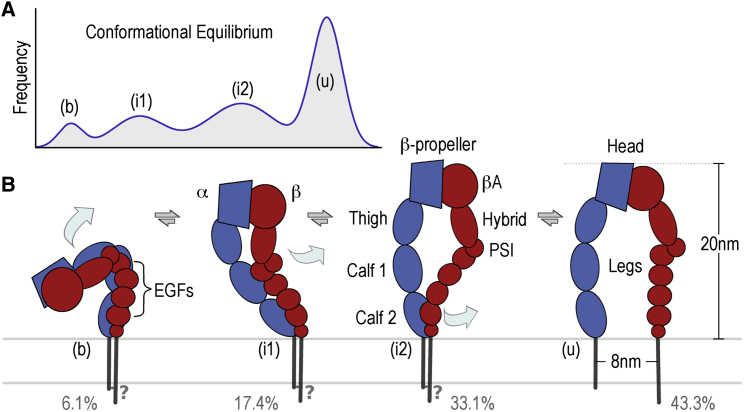
We compared the distributions of the four conformers of αIIbβ3 in different data sets by tallying the fraction of integrin particles observed for each of the conformers. Integrin by itself is distributed across the four conformations, ranging from 13% in the compact nodule to 21% in the first intermediate, 38% in the second intermediate, and 28% in the fully upright stance. Addition of talin head domains, RGD peptide, and nanodiscs caused a large shift in the distribution, with only 6% for the compact nodule, 17% and 33% for the intermediates, and 43% for the upright conformation (Fig. 1 B; Table 1).
Locations of nanodiscs and talin head domains in the reconstructions
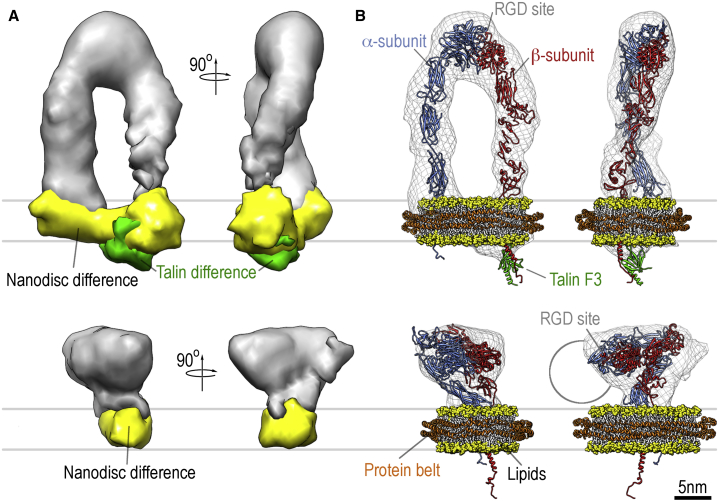
We employed difference mapping, modeling, and density-docking methods to further interpret the reconstructions in light of the available structures, and to provide structural models for the four conformational groups (Figs. 3, 4, and S3). Difference mapping between reconstructions obtained from conditions with and without nanodiscs allowed us to deduce the location of the nanodiscs (Figs. 3 A and S5). This location unequivocally labels the position of the transmembrane portion of integrin within the respective densities and thus allows the model of the ectodomain to be anchored in the density. Difference mapping between reconstructions of the upright stance with the full-length talin head domain and reconstructions with the much smaller talin F3 domain allowed us to locate extra density corresponding to talin (Fig. 3 A). All of the difference maps were cross-validated using reconstructions from independent biochemical preparations. The talin head domain is attached to the cytoplasmic tail of β3 (39) and is visible in our reconstruction near one of the separated legs, identifying the cytoplasmic side of the β-subunit in the density (Fig. 3 A).
Figure 3.
Molecular model of extended upright (upper panel) and bent (lower panel) conformations of integrin in lipid bilayers. (A) The gray surfaces represent the reconstructions of integrin in the absence of nanodisc and talin. The yellow surfaces represent the density attributable to nanodiscs by difference mapping, and the green density in the upright conformation (upper panel) represents the density for the talin head domain obtained by difference mapping. No extra density for bound talin head domain could be observed for the bent conformation. (B) The bilayer of the nanodisc is shown in space-filling representation. The protein belts of the nanodisc (orange), integrin (red and blue), and talin F3 (green) are shown in cartoon representation. The density map is shown in chicken-wire representation for reference. The distance from the surface of the nanodisc to the ligand-binding site (RGD site) in the head domain of the upright conformation is ∼20 nm. The approximate size of the integrin-binding domain of fibronectin (13) is indicated as a circle in the side view of the bent conformation. To see this figure in color, go online.
Figure 4.
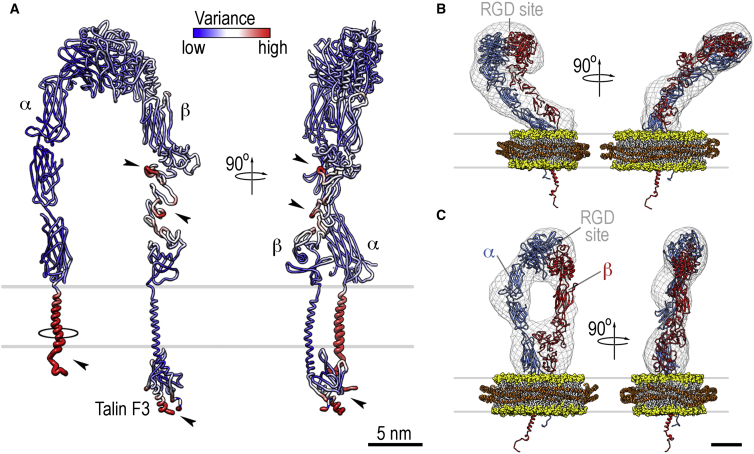
Variance of the upright model and molecular models of intermediate conformations of integrin in lipid bilayers. (A) Variance from the docking analysis mapped onto the model for the upright conformation with bound talin F3. The largest variances are seen in the cytoplasmic domains, the transmembrane helix of the α-subunit, and at the top in the I-EGF domains of the β-leg (all marked by arrowheads). The largest observed deviation of the α-subunit transmembrane helix that is still compatible with the docking is shown as a black circle. (B and C) Models of the two intermediate conformations. The lipid bilayer of the nanodisc is shown in space-filling representation, with the headgroups in yellow and the hydrophobic tails in white. The protein belts of the nanodisc (orange) and integrin (red and blue) are shown in cartoon representation. The density maps are shown in chicken-wire representation for reference. Scale bar, 5 nm. To see this figure in color, go online.
Quantitative evaluation of the reconstructions using statistics-based fitting combined with difference mapping
Using the information derived from the difference mapping to place the transmembrane portion of the structures within the reconstructions, we created models of the membrane-embedded conformations of integrin based on available atomic structures derived from x-ray crystallography and NMR (11, 39, 40, 41, 42, 43) using statistics-based docking (52) with cross-validation (53). In our modeling strategy we used a conservative approach to ensure that the known atomic structures were subjected to the minimum number of changes necessary to explain the density. Although additional changes cannot be ruled out, this strategy was shown to minimize the dangers of overfitting at low resolution (54). For the bent conformation, no changes from the original crystal structure of the full-length ectodomain (9) were supported by the statistics. To model the first intermediate conformers, a hinge was introduced at the bend of the crystal structure, and the angle between the two was refined using the density as a constraint. No further modifications were supported by the statistics. In addition to a similar hinge-based extension of the second intermediate conformer, opening of the hybrid domain using the crystal structure of legless αIIbβ3 containing the PSI/Hybrid/βA and propeller domains (41) as a template was supported by the statistics. An additional separation of the lower legs was supported for the upright conformation.
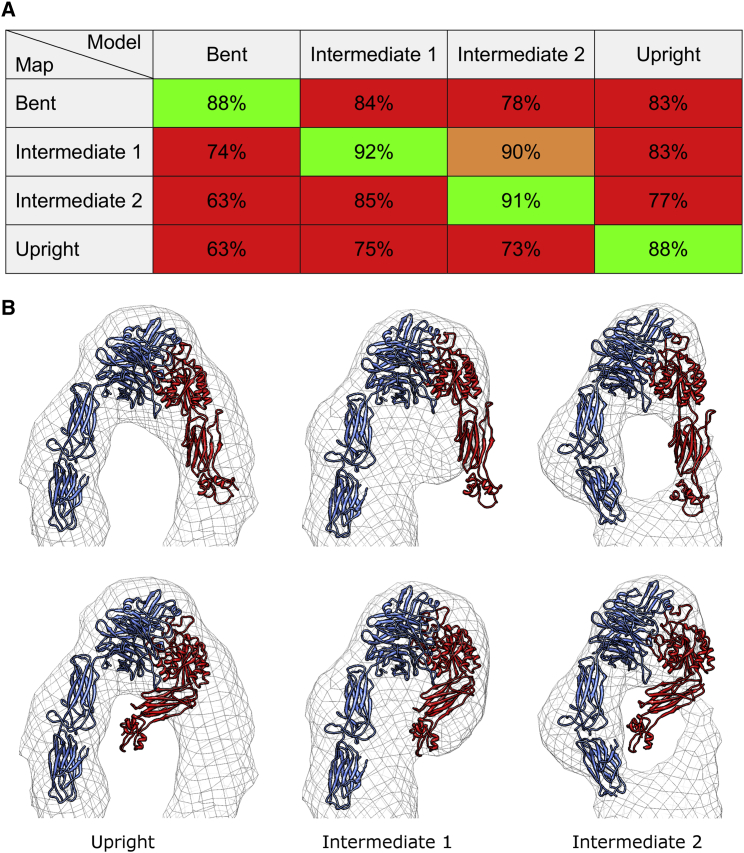
The statistical formalism of the docking method (53) allows to test whether the introduced changes lead to a significant improvement in the fits. For example, using the open hybrid-domain conformation from the legless αIIbβ3 crystal structure (41) instead of the closed hybrid-domain configuration of the full-length ectodomain structure (9) for the second intermediate conformer leads to an improvement of the correlation from 85% to 91% (Fig. 5). The statistical analysis indicates that this improvement is significant at a confidence level of 99.99% (α = 0.0001; α is the chosen threshold value for experimental p-values to be considered significant, and the smaller the value of α, the more stringent are the confidence intervals). A statistical test for placement of the β-subunit near the extra talin density in the upright conformation (88.2% correlation) versus the α-subunit (83.3%) favors the β-subunit as significantly better at α = 0.0001.
Figure 5.
Statistical evaluation of the differences in conformation. (A) Color-coded table showing the values of correlations between the models and the respective reconstructions (maps). All correlation values were calculated using a conservative resolution cutoff of 45 Å. The cells are color coded according to the confidence level at which the null hypothesis that the structure fits the density as well as the best fit (green) can be rejected. The red cells can be rejected at α = 0.0001 (confidence level 99.99%) and the orange cell can be rejected at α = 0.0005 (99.95%). (B) Illustration of discrimination between different degrees of swing-out of the hybrid domain for the upright and two intermediate conformations. In each case, the state of the hybrid domain can be unequivocally determined by the visual fit to the density, consistent with the results of the statistical analysis. To see this figure in color, go online.
The fully extended conformation of integrin αIIbβ3 shows a clear separation between α- and β-legs
To our knowledge, our model for the upright conformation is the first molecular-resolution 3D structure of a complete, upright integrin embedded in a phospholipid bilayer in a fully hydrated environment. All other models of upright integrin conformations reported in the literature were based on 2D projection data, and most were obtained in the absence of membrane and in negative stain. In the 3D model of the upright stance presented here, there are no observable contacts between the two subunits other than those in the β3A domain and the propeller domain of αIIb, which come together to form the ligand-binding site. The integrin extends ∼20 nm upward from the membrane. The β3 hybrid domain is swung out, being similar to the configuration in the crystal structure of legless αIIbβ3 (41). According to the docking statistics, the most flexible region in the ectodomain of the upright stance is found within the I-EGF leg region of β3 (Fig. 4 A). In the crystal structure of αvβ3, many of the residues that show high flexibility in our model are not resolved (9). Most 2D EM images of upright integrins in the absence of membrane show the legs of the β-subunit to be poorly resolved, and electron tomographic analyses of αIIbβ3 integrin in negative stain (46) as well as random-conical-tilt, negative-stain reconstructions of αVβ3 integrin (55) show the β-leg in many different conformations. In contrast, the 3D reconstructions of membrane-embedded upright integrin presented here show a very clearly defined β-leg, indicating that membrane embedding promotes stabilization of the lower legs. Consistently, our 3D reconstructions of the upright stance in the absence of membrane and ligands are significantly less well defined in the β-leg region. In the membrane-embedded structure, the transmembrane domains and cytoplasmic tails are well separated with a distance of 8 (± 0.7) nm.
The compact conformation of integrin αIIbβ3 allows access to the RGD-binding site
The compact nodular conformation of αIIbβ3 observed here allows the crystal structure of αIIbβ3 to be fitted without modification (Fig. 3 B). Although the difference density corresponding to the nanodisc clearly shows its location, the fact that integrin in its compact conformation can freely move within the lipid bilayer (14) leads to a washing out of the nanodisc density due to averaging over a multitude of orientations. The fit of the crystal structure can be visually improved by a slight opening of the head and leg domains, consistent with the random-conical-tilt reconstructions of manually selected, negatively stained bent αIIbβ3 integrins in nanodiscs (14), but the improvement in fit is not statistically significant at our resolution. The correlation between the crystal structure and the reconstruction of the bent conformer is 88.2%. For the model derived from the negative-stain reconstruction (14), the correlation is 87.6%. The 0.6% difference is not statistically significant, even if α is raised to 0.005. To distinguish between the two possibilities, higher-resolution data are required. The ligand-binding site is accessible to ligands in both models and is not obscured by the membrane (Fig. 3 B). In both cases, there is enough space available to bind fibronectin, as shown for the bent conformation of αVβ3 integrins in the absence of nanodiscs (13).
Intermediate conformations of integrin αIIbβ3 show different degrees of extension and hybrid-domain swing
For the first intermediate state (ii), the difference density attributable to the nanodisc (Fig. S5) indicates that integrin in this conformation is somewhat tilted with respect to the membrane rather than optimally extended. The fact that not all of the nanodisc is resolved indicates that there is some freedom of movement in the nanodisc for this conformation as well. To derive a structural model, the head of the crystal structure needs to be moved into a different position than in the bent conformation, but all contacts and structural relationships between the legs and within the head do not need to be altered from those in the crystal structure (Fig. 4 B). For the second intermediate conformer (iii), the difference density for the nanodisc is also washed out, indicating some freedom of movement (Fig. S5). Docking experiments show that this reconstruction is most consistent with a configuration in which the contacts in the lower legs are still similar to those in the crystal structure but the hybrid domain is swung out to a similar position as the one we observe in the upright conformation (Fig. 5 B).
Conclusions
Our study shows that αIIbβ3 integrins and integrins embedded in nanodiscs exist in an equilibrium of states even while they are bound to cytosolic and extracellular ligands. These conformations range from a compact nodular structure similar to the crystal structures of ectodomains to a fully upright conformation and two intermediate states (Fig. 6). The addition of talin head domains, RGD peptide, and phospholipid bilayer to the system significantly shifts the conformational equilibrium toward the upright conformation.
Figure 6.
Schematic model for integrin conformational equilibrium. (A) Schematic distribution landscape of conformational equilibrium. The bent (b) and upright (u) conformations have the narrowest peaks, indicating the smallest amount of variability in their vicinity. The two intermediate peaks (i1 and i2) are much broader, indicating a larger variability. (B) Schematic overview of key conformers in the integrin αIIbβ3 conformational equilibrium. Transitions include an opening between the head and leg domains from the bent conformation (b) so that the legs can untwist and straighten out to form the first intermediate conformer (i1). A swing-out of the hybrid domain in the β-subunit converts the first intermediate conformer (i1) into the second intermediate conformer (i2). Separation of the legs in the membrane then converts the second intermediate conformer into the fully extended upright conformation (u). To see this figure in color, go online.
NMR structure determination of the αIIbβ3 integrin single-pass transmembrane segments showed that they associate closely in membrane environments (40). Mutations that lead to dissociation of the transmembrane segments and thus to leg separation have shown profound effects on the ligand affinity of integrins (56, 57, 58). Binding of talin domains to the cytoplasmic tail of β3 (21, 39) was shown to trigger dissociation of isolated transmembrane segments (22) or cytoplasmic tails (59), and separation of the α- and β-legs of αLβ2 integrins in response to talin binding was demonstrated (23). Thus, there is strong evidence that disruption of the transmembrane segments induces integrin activation and that talin triggers this transition. Here, we show that the talin head domain binds to the upright conformation of integrin while it is embedded in the lipid bilayer nanodisc, and that the two integrin subunits are well separated on the cytosolic side in this configuration.
The existence of multiple conformations of integrin has implications for the activation mechanism and suggests that integrin activation may involve equilibrium shifts. Our results indicate that the ligand-binding site is not obscured by the membrane for any of the conformational states in equilibrium, and that extension of the ectodomain is possible without separating the legs or the hybrid domain. Although our resolution of the bent conformation is too low to determine whether modification of the crystal structure is appropriate, the orientation of the integrin density with respect to the membrane clearly shows that the ligand-binding site is not obstructed even for natural ligands such as fibronectin (Fig. 3 B, lower panel), consistent with the negative-stain reconstruction of the bent conformation in the membrane (14) and the capability of bent αVβ3 integrin to bind fibronectin (13). Because the distance between the legs is so large (8 ± 0.7 nm), even the relatively low resolution of our reconstructions is fully sufficient to determine whether the legs are separated or not.
This study demonstrates that a simplified reconstituted system containing intact full-length integrins, phospholipid bilayers, and physiologically relevant cytosolic and extracellular integrin ligands is amenable to structural analysis in three dimensions in its native environment. The use of cryo-EM and multireference, single-particle analysis with this system circumvents the artifacts that are potentially associated with negative staining or rotary shadowing, the use of truncated constructs, analysis of 2D projections, undetected conformational mixtures, or crystal lattices. Despite the relatively low resolution of this study, however, careful statistical analysis and cross-validation allow us to draw reliable structural conclusions that are not attainable by any other means. This opens the avenue to detailed structural investigations of the integrin inside-out and outside-in activation mechanisms, and the role of additional integrin ligands such as kindlin or larger fibronectin fragments. Through controlled size variations of the nanodisc system, it will also be possible to investigate integrin clusters of controlled size to unravel the effect of clustering on integrin activation.
Author Contributions
J.W.S., N.V., and D.H. designed the study. E.K. and M.S. performed the research. N.V. and X.-P.X. analyzed the data. N.V. and D.H. prepared the figures and wrote the manuscript.
Acknowledgments
We thank Mrs. Larnele Hazelwood for EM sample preparation and screening, Mrs. Sheryl Harvey for purification and characterization of the integrin samples, Dr. Robert Jeng for supplemental biochemical analysis, Drs. Mark Ginsberg and Bill Weis for comments on the manuscript, and Drs. Mark A. McLean and Stephen G. Sligar for providing the nanodisc sample.
This work was supported by grants from the National Institutes of Health (R01 HL072862 to J.W.S., R01 CA179087 to N.V., and Program Project grant P01 GM098412 to D.H.).
Editor: Andreas Engel.
Footnotes
Eldar Kim’s present address is MSM Protein Technologies, Inc., Medford, Massachusetts.
Five figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)00091-6.
Contributor Information
Niels Volkmann, Email: niels@burnham.org.
Dorit Hanein, Email: dorit@burnham.org.
Supporting Material
References
- 1.Hynes R.O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 2.Banno A., Ginsberg M.H. Integrin activation. Biochem. Soc. Trans. 2008;36:229–234. doi: 10.1042/BST0360229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carman C.V., Springer T.A. Integrin avidity regulation: are changes in affinity and conformation underemphasized? Curr. Opin. Cell Biol. 2003;15:547–556. doi: 10.1016/j.ceb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Luo B.H., Carman C.V., Springer T.A. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouaouina M., Harburger D.S., Calderwood D.A. Talin and signaling through integrins. Methods Mol. Biol. 2012;757:325–347. doi: 10.1007/978-1-61779-166-6_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye F., Kim C., Ginsberg M.H. Reconstruction of integrin activation. Blood. 2012;119:26–33. doi: 10.1182/blood-2011-04-292128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong J.-P., Mahalingham B., Arnaout M.A. Crystal structure of the complete integrin alphaVbeta3 ectodomain plus an alpha/beta transmembrane fragment. J. Cell Biol. 2009;186:589–600. doi: 10.1083/jcb.200905085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong J.-P., Stehle T., Arnaout M.A. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 9.Xiong J.P., Stehle T., Arnaout M.A. Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J., Luo B.-H., Springer T.A. The structure of a receptor with two associating transmembrane domains on the cell surface: integrin alphaIIbbeta3. Mol. Cell. 2009;34:234–249. doi: 10.1016/j.molcel.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J., Luo B.-H., Springer T.A. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell. 2008;32:849–861. doi: 10.1016/j.molcel.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J., Zhu J., Springer T.A. Complete integrin headpiece opening in eight steps. J. Cell Biol. 2013;201:1053–1068. doi: 10.1083/jcb.201212037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adair B.D., Xiong J.P., Yeager M. Three-dimensional EM structure of the ectodomain of integrin alphaVbeta3 in a complex with fibronectin. J. Cell Biol. 2005;168:1109–1118. doi: 10.1083/jcb.200410068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi W.-S., Rice W.J., Coller B.S. Three-dimensional reconstruction of intact human integrin αIIbβ3: new implications for activation-dependent ligand binding. Blood. 2013;122:4165–4171. doi: 10.1182/blood-2013-04-499194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishida N., Xie C., Springer T.A. Activation of leukocyte beta2 integrins by conversion from bent to extended conformations. Immunity. 2006;25:583–594. doi: 10.1016/j.immuni.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 16.Takagi J., Petre B.M., Springer T.A. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 17.Xie C., Zhu J., Springer T.A. Structure of an integrin with an alphaI domain, complement receptor type 4. EMBO J. 2010;29:666–679. doi: 10.1038/emboj.2009.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye F., Hu G., Ginsberg M.H. Recreation of the terminal events in physiological integrin activation. J. Cell Biol. 2010;188:157–173. doi: 10.1083/jcb.200908045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liddington R.C. Structural aspects of integrins. Adv. Exp. Med. Biol. 2014;819:111–126. doi: 10.1007/978-94-017-9153-3_8. [DOI] [PubMed] [Google Scholar]
- 20.Calderwood D.A., Zent R., Ginsberg M.H. The talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem. 1999;274:28071–28074. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- 21.Wegener K.L., Partridge A.W., Campbell I.D. Structural basis of integrin activation by talin. Cell. 2007;128:171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 22.Kim C., Lau T.L., Ginsberg M.H. Interactions of platelet integrin alphaIIb and beta3 transmembrane domains in mammalian cell membranes and their role in integrin activation. Blood. 2009;113:4747–4753. doi: 10.1182/blood-2008-10-186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M., Carman C.V., Springer T.A. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 24.Denisov I.G., Grinkova Y.V., Sligar S.G. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 25.Xu X.-P., Zhai D., Hanein D. Three-dimensional structure of Bax-mediated pores in membrane bilayers. Cell Death Dis. 2013;4:e683. doi: 10.1038/cddis.2013.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fitzgerald L.A., Leung B., Phillips D.R. A method for purifying the platelet membrane glycoprotein IIb-IIIa complex. Anal. Biochem. 1985;151:169–177. doi: 10.1016/0003-2697(85)90067-3. [DOI] [PubMed] [Google Scholar]
- 27.Kouns W.C., Hadvary P., Steiner B. Conformational modulation of purified glycoprotein (GP) IIb-IIIa allows proteolytic generation of active fragments from either active or inactive GPIIb-IIIa. J. Biol. Chem. 1992;267:18844–18851. [PubMed] [Google Scholar]
- 28.Yan B., Hu D.D., Smith J.W. Probing chemical and conformational differences in the resting and active conformers of platelet integrin alpha(IIb)beta(3) J. Biol. Chem. 2000;275:7249–7260. doi: 10.1074/jbc.275.10.7249. [DOI] [PubMed] [Google Scholar]
- 29.Ludtke S.J., Baldwin P.R., Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 30.Yang Z., Fang J., Penczek P.A. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20:237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spahn C.M., Penczek P.A. Exploring conformational modes of macromolecular assemblies by multiparticle cryo-EM. Curr. Opin. Struct. Biol. 2009;19:623–631. doi: 10.1016/j.sbi.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang G., Peng L., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Hohn M., Tang G., Ludtke S.J. SPARX, a new environment for cryo-EM image processing. J. Struct. Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Penczek P.A., Renka R., Schomberg H. Gridding-based direct Fourier inversion of the three-dimensional ray transform. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 2004;21:499–509. doi: 10.1364/josaa.21.000499. [DOI] [PubMed] [Google Scholar]
- 35.Xu X.-P., Rouiller I., Volkmann N. Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 2012;31:236–247. doi: 10.1038/emboj.2011.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheres S.H., Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tama F., Miyashita O., Brooks C.L., 3rd Flexible multi-scale fitting of atomic structures into low-resolution electron density maps with elastic network normal mode analysis. J. Mol. Biol. 2004;337:985–999. doi: 10.1016/j.jmb.2004.01.048. [DOI] [PubMed] [Google Scholar]
- 38.Volkmann N., Hanein D. Docking of atomic models into reconstructions from electron microscopy. Methods Enzymol. 2003;374:204–225. doi: 10.1016/S0076-6879(03)74010-5. [DOI] [PubMed] [Google Scholar]
- 39.Anthis N.J., Wegener K.L., Campbell I.D. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 2009;28:3623–3632. doi: 10.1038/emboj.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau T.-L., Kim C., Ulmer T.S. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009;28:1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi M., Foo S.Y., Lescar J. A structural hypothesis for the transition between bent and extended conformations of the leukocyte beta2 integrins. J. Biol. Chem. 2007;282:30198–30206. doi: 10.1074/jbc.M701670200. [DOI] [PubMed] [Google Scholar]
- 42.Shi M., Sundramurthy K., Lescar J. The crystal structure of the plexin-semaphorin-integrin domain/hybrid domain/I-EGF1 segment from the human integrin beta2 subunit at 1.8-A resolution. J. Biol. Chem. 2005;280:30586–30593. doi: 10.1074/jbc.M502525200. [DOI] [PubMed] [Google Scholar]
- 43.Xiao T., Takagi J., Springer T.A. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu X.-P., Volkmann N. Validation methods for low-resolution fitting of atomic structures to electron microscopy data. Arch. Biochem. Biophys. 2015;581:49–53. doi: 10.1016/j.abb.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw A.W., McLean M.A., Sligar S.G. Phospholipid phase transitions in homogeneous nanometer scale bilayer discs. FEBS Lett. 2004;556:260–264. doi: 10.1016/s0014-5793(03)01400-5. [DOI] [PubMed] [Google Scholar]
- 46.Iwasaki K., Mitsuoka K., Yamada T. Electron tomography reveals diverse conformations of integrin alphaIIbbeta3 in the active state. J. Struct. Biol. 2005;150:259–267. doi: 10.1016/j.jsb.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 47.Takagi J., Strokovich K., Walz T. Structure of integrin alpha5beta1 in complex with fibronectin. EMBO J. 2003;22:4607–4615. doi: 10.1093/emboj/cdg445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu J., Choi W.-S., Coller B.S. Structure-guided design of a high-affinity platelet integrin αIIbβ3 receptor antagonist that disrupts Mg2+ binding to the MIDAS. Sci. Transl. Med. 2012;4:125ra32. doi: 10.1126/scitranslmed.3003576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weisel J.W., Nagaswami C., Bennett J.S. Examination of the platelet membrane glycoprotein IIb-IIIa complex and its interaction with fibrinogen and other ligands by electron microscopy. J. Biol. Chem. 1992;267:16637–16643. [PubMed] [Google Scholar]
- 50.Litvinov R.I., Nagaswami C., Weisel J.W. Functional and structural correlations of individual alphaIIbbeta3 molecules. Blood. 2004;104:3979–3985. doi: 10.1182/blood-2004-04-1411. [DOI] [PubMed] [Google Scholar]
- 51.Adair B.D., Yeager M. Three-dimensional model of the human platelet integrin alpha IIbbeta 3 based on electron cryomicroscopy and x-ray crystallography. Proc. Natl. Acad. Sci. USA. 2002;99:14059–14064. doi: 10.1073/pnas.212498199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Volkmann N., Hanein D. Quantitative fitting of atomic models into observed densities derived by electron microscopy. J. Struct. Biol. 1999;125:176–184. doi: 10.1006/jsbi.1998.4074. [DOI] [PubMed] [Google Scholar]
- 53.Volkmann N. Confidence intervals for fitting of atomic models into low-resolution densities. Acta Crystallogr. D Biol. Crystallogr. 2009;65:679–689. doi: 10.1107/S0907444909012876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Volkmann N. The joys and perils of flexible fitting. Adv. Exp. Med. Biol. 2014;805:137–155. doi: 10.1007/978-3-319-02970-2_6. [DOI] [PubMed] [Google Scholar]
- 55.Veesler D., Cupelli K., Johnson J.E. Single-particle EM reveals plasticity of interactions between the adenovirus penton base and integrin αVβ3. Proc. Natl. Acad. Sci. USA. 2014;111:8815–8819. doi: 10.1073/pnas.1404575111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo B.-H., Carman C.V., Springer T.A. Disrupting integrin transmembrane domain heterodimerization increases ligand binding affinity, not valency or clustering. Proc. Natl. Acad. Sci. USA. 2005;102:3679–3684. doi: 10.1073/pnas.0409440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Melker A.A., Kramer D., Sonnenberg A. The two phenylalanines in the GFFKR motif of the integrin alpha6A subunit are essential for heterodimerization. Biochem. J. 1997;328:529–537. doi: 10.1042/bj3280529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hughes P.E., Diaz-Gonzalez F., Ginsberg M.H. Breaking the integrin hinge. A defined structural constraint regulates integrin signaling. J. Biol. Chem. 1996;271:6571–6574. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- 59.Vinogradova O., Velyvis A., Qin J. A structural mechanism of integrin alpha(IIb)beta(3) “inside-out” activation as regulated by its cytoplasmic face. Cell. 2002;110:587–597. doi: 10.1016/s0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.