Abstract
Successful treatment of bacterial infections requires the timely administration of appropriate antimicrobial therapy. The failure to initiate the correct therapy in a timely fashion results in poor clinical outcomes, longer hospital stays, and higher medical costs. Current approaches to antibiotic susceptibility testing of cultured pathogens have key limitations ranging from long run times to dependence on prior knowledge of genetic mechanisms of resistance. We have developed a rapid antimicrobial susceptibility assay for Staphylococcus aureus based on bacterial cytological profiling (BCP), which uses quantitative fluorescence microscopy to measure antibiotic induced changes in cellular architecture. BCP discriminated between methicillin-susceptible (MSSA) and -resistant (MRSA) clinical isolates of S. aureus (n = 71) within 1–2 h with 100% accuracy. Similarly, BCP correctly distinguished daptomycin susceptible (DS) from daptomycin non-susceptible (DNS) S. aureus strains (n = 20) within 30 min. Among MRSA isolates, BCP further identified two classes of strains that differ in their susceptibility to specific combinations of beta-lactam antibiotics. BCP provides a rapid and flexible alternative to gene-based susceptibility testing methods for S. aureus, and should be readily adaptable to different antibiotics and bacterial species as new mechanisms of resistance or multidrug-resistant pathogens evolve and appear in mainstream clinical practice.
Keywords: Antibiotic resistance, Susceptibility tests, Staphylococcus aureus, Multidrug resistant bacteria
Highlights
-
•
Bacterial cytological profiling identifies antibiotic resistant S. aureus.
-
•
BCP predicts best treatment options for multidrug resistant MRSA.
-
•
Resistant strains are correctly identified within 1 h.
-
•
BCP does not require prior knowledge of resistance mechanism.
There is a great need for rapid antimicrobial susceptibility testing (AST) as it can dramatically improve clinical outcome for bacterial infections. Most currently proposed ASTs are dependent on knowledge of known resistance genes or based solely on growth/lysis. We have developed a new diagnostic method for rapidly determining antibiotic susceptibility of Staphylococcus aureus using quantitative fluorescence microscopy to measure antibiotic induced changes in cellular architecture. Our test has the potential to change the way antibiotic susceptibility testing is done in the future and is readily adaptable to different antibiotics and bacterial species regardless of the mechanisms of resistance.
1. Introduction
We are locked in an evolutionary race with bacteria that rapidly become resistant to each new antibiotic, necessitating the continued development of new treatments and diagnostic approaches. The aptly coined “ESKAPE” pathogens (Rice, 2008) — Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species can exhibit multi-resistance to all first line antibacterial drugs and pose significant clinical threats. Current therapeutic agents are losing effectiveness more rapidly than they are being replaced, and new drug discovery has stagnated due to high development costs coupled with a lower profit margin associated with antibiotics compared to other drug classes (Gwynn et al., 2010). Hence, in the recent decade there has been a push for the implementation of antimicrobial stewardship, a set of strategies to reduce the emergence and spread of resistant pathogens and to use existing antimicrobials more effectively. Rapid susceptibility testing plays a key role in antimicrobial stewardship by decreasing emergence of resistance and by reserving drugs of last resort to those cases in which they are warranted (Goff et al., 2012).
Traditional antimicrobial susceptibility tests include the disk diffusion test, E-test gradient diffusion test and broth dilution susceptibility tests. Although the tests are robust, they each rely on the growth of the bacteria to a dense culture while being exposed to the antibiotic panel. Consequently, these methods require 24–48 h to isolate the bacterium from the patient in pure culture and another 24–72 h to complete susceptibility testing (Goff et al., 2012). Due to the long time required to deliver test results compared to the tempo of infection, patients are often treated “empirically” with broader-spectrum regimens under the assumption that the infection could be drug-resistant.
To improve susceptibility testing times there are currently a few rapid platforms gaining acceptance in the clinical laboratory: polymerase chain reaction (PCR), poly-nucleic acid fluorescence in situ hybridization (PNA-FISH), and nanosphere hybridization. PCR method detects coding sequences of known resistance genes while PNA-FISH identifies species and/or resistance by hybridizing synthetic oligo-nucleotide fluorescence-labeled probes to species-specific ribosomal RNA and mRNA. Similarly, nanosphere hybridization uses nanoparticle probes to detect DNA, RNA or protein targets. Rapid susceptibility methods can improve patient outcomes and decrease cost of care. In a study comparing PCR to traditional methods, time required for identification of methicillin-susceptible S. aureus (MSSA) versus methicillin-resistant S. aureus (MRSA) was reduced by 1.7 days, cost per patient decreased from $69,737 to $48,350, and outcomes improved with 6.2 fewer days in the hospital and 20% lower mortality (Bauer et al., 2010). Unfortunately, both these current rapid methods depend on a priori knowledge of resistance genes.
S. aureus is a prevalent Gram-positive human pathogen that is a leading cause of bacteremia, pneumonia, skin/soft tissue infections and endocarditis worldwide (Tong et al., 2015). Asymptomatic colonization of up to 30% of healthy individuals allows for the continual transmission and proliferation of this pathogen (Wertheim et al., 2005). S. aureus is responsible for community-acquired (CA-) and hospital-acquired (HA-) infections in both healthy and immune-compromised individuals, and numerous lineages of MRSA including the USA300 clone have spread throughout the US and internationally (Mediavilla et al., 2012). Currently, rPCR is used by many clinical laboratories to detect the mecA gene, which confers beta-lactam resistance in S. aureus by encoding the low-affinity penicillin-binding protein, PBP2a (Pinho et al., 2001). Since MSSA lack mecA, detection of this gene is considered a gold standard for molecular identification of MRSA. However, occurrence of oxacillin-susceptible mecA + MRSA strains (Hososaka et al., 2007) and emergence of a mecA variant, mecC, which encodes a protein with < 63% AA identity to PBP2a (Laurent et al., 2012, García-Álvarez et al., 2011), highlight limitations in tests that detect only the presence of this gene.
In recent years, rapid phenotypic susceptibility assays for S. aureus have been proposed, but many of these assays are based solely on cell lysis and/or growth (Price et al., 2014, Kalashnikov et al., 2012, Choi et al., 2013, Kinnunen et al., 2012, Sinn et al., 2012). Recently, an imaging-based single-cell morphological analysis (SCMA) focused on early morphological changes in response to antibiotic exposure sought to determine susceptibility of five different bacterial pathogens to a variety of antibiotics within 3–4 h (Choi et al., 2014). Despite the impressive scale and scope of this study, S. aureus antibiotic susceptibility determination in this assay depended solely on cell division, similar to standard broth microdilution assays, and had an error rate of 5.4% compared to conventional testing.
Recently, we demonstrated the utility of bacterial cytological profiling (BCP) to determine specific antibacterial mechanisms of action (Lamsa et al., 2012, Nonejuie et al., 2013). BCP is based on the observation that treatment of a diverse range of bacterial species with different antibiotics yield unique and quantifiable changes in cytological profiles. BCP profiles are comprised of a multitude of parameters including DNA and cell size and shape and dye intensity on a single-cell level for hundreds of cells in a single run. Here, we applied BCP as a rapid method for determining the antibiotic susceptibility of S. aureus clinical isolates obtained from patient samples. With BCP, 100% (n = 30) of blind test isolates were correctly categorize as MRSA and MSSA within 1 h, and the MRSA strains were further subdivided into two groups that correlated with increased susceptibility to combinatorial drug therapy. Likewise, daptomycin susceptible (DS) and daptomycin non-susceptible (DNS) S. aureus strains (n = 20) were correctly classified after 30 min of antibiotic treatment. BCP provides a flexible alternative to current rapid susceptibility testing methods as it doesn't require prior knowledge of resistance genes and can be applied broadly to different antibiotics and species.
2. Materials and Methods
2.1. Strains and Culture Condition
The strains used in this study are listed in Table S3. MLST, spa type, and agr group were provided from their corresponding sources. MLST typing for cb strains were done by following published protocols (Ji, 2007). All cultures were grown in Muller-Hinton Broth (MHB) (HiMedia, Lot #140338) supplemented with 2% NaCl, 12 mg L− 1 Mg2 + and 25 mg L− 1 Ca2 + to ensure that the growth conditions and therefore the cytological profiles of the cells remained consistent for all antibiotics. The only exception was daptomycin, since this antibiotic requires 50 mg/L CaCl2 for activity. Antibiotics were prepared using the following concentrations and solvents: 10 mg mL− 1 cefoxitin (H2O, Fluka Analytical), 10 mg mL− 1 oxacillin (H2O, Fluka Analytical), 10 mg mL− 1 cephalexin (H2O, Sigma), 10 mg mL− 1 cefotaxime (H2O, MP Biomedical LLC), 10 mg mL− 1 meropenem (H2O, Sigma,), 5 mg mL− 1 daptomycin (DMSO, Cubist).
2.2. Minimal Inhibitory Concentration Determination and Synergy Assays
Minimum inhibitory concentration (MIC) data shown in Table S1 and Fig. 2, Fig. 3 were determined using microdilution method while synergy assays (Fig. 5b-c) were performed using checkerboard assays (CLSI, 2007, CLSI, 2012). In both cases, cells were grown in a culture tube to an OD600 of 0.35–0.6 and diluted to 5 × 107 CFU/mL (or 5 × 105 CFU/mL for daptomycin MIC) in cation-adjusted MHB. Cell solution was diluted 1:10 into a 96-well plate containing different concentrations of each antibiotic. Clinical isolates with an MIC for oxacillin higher than 4 μg/mL were categorized as MRSA, and used as the training set for the initial LDA experiments. MIC and synergy results were obtained after 24 h incubation at 30 °C. MIC values reported and synergy curve shown were calculated from the mean of triplicates (n = 3).
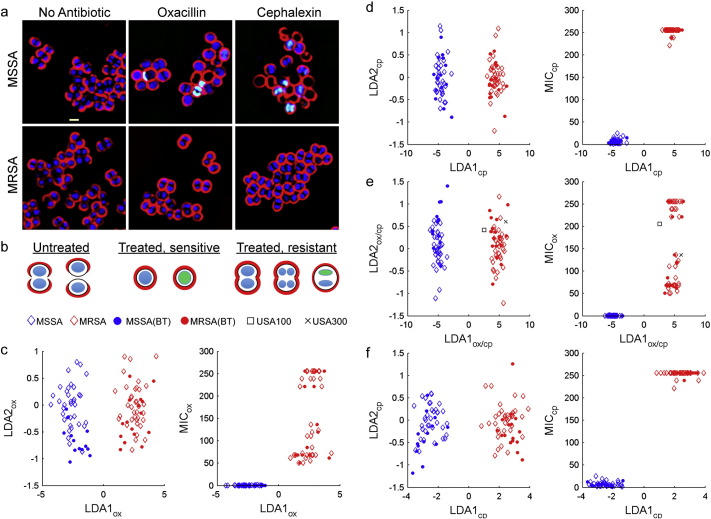
Fig. 2.
Discrimination of MRSA from MSSA with BCP. (a) Images of MRSA (ATCC 33591) and MSSA (ATCC 29213) after 2 h treatment either with no antibiotics, 10 μg/mL of oxacillin, or 20 μg/mL of cephalexin. In all the images, cell walls were stained with WGA-Cy5 (red), DNA was stained with DAPI (blue) and SYTOX Green (green). SYTOX Green is membrane impermeable and only stains permeabilized cells. (b) Cartoon showing the observed cellular phenotypes. (c–e) Linear discriminant analysis (LDA) plots were calculated from a cytological profile library consisting of 22 parameters measured for 71 isolates, 37 MRSA strains (red) and 36 MSSA strains (blue), treated with each antibiotic measured in triplicates. The mean of BCP profiles of each of the 71 strains (diamonds) is plotted after 2 h of treatment with (c) 10 μg/mL of oxacillin, (d) 20 μg/mL of cephalexin, (e) from the combined data from the measurements in (c) and (d). USA100 (N315; black square) and USA300 (TCH1516; black x) profiles were projected onto the MRSA/MSSA LDA plot. (f) LDA plots calculated from BCP profiles measured after 1 h treatment with 20 μg/mL of cephalexin. Blind test profiles (filled circles; n = 30) are overlaid on LDA plots of the training set
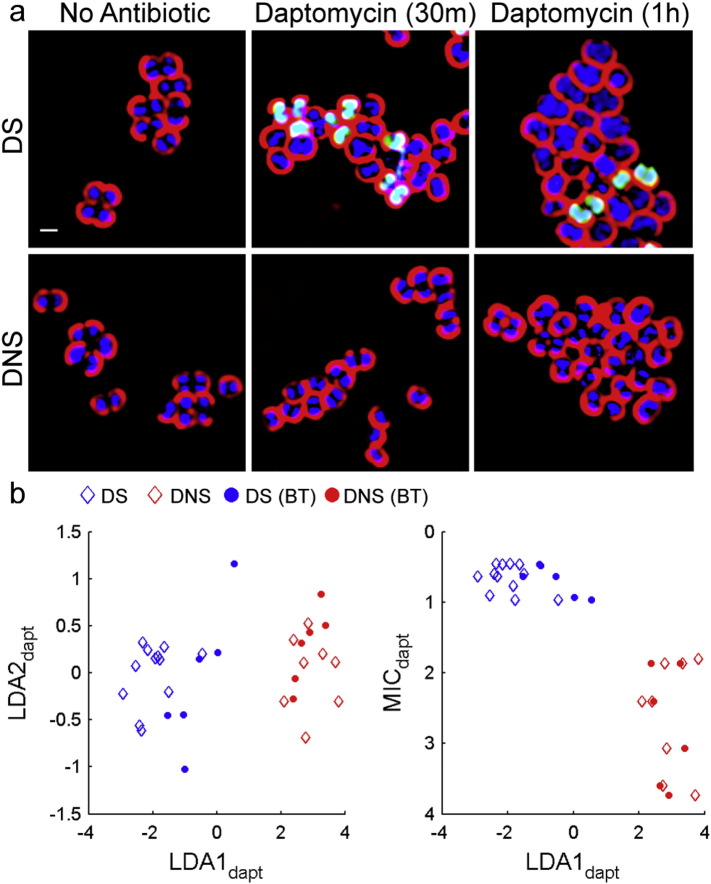
Fig. 3.
BCP determination of daptomycin susceptibility. (a) Daptomycin non-susceptible (DNS; gs5) and daptomycin susceptible (DS; ATCC29213) cell walls were stained with WGA-Cy5 (red) while the DNA was stained with DAPI (blue) and SYTOX Green (green). Images were taken after treatment with no antibiotics or 5 μg/mL of daptomycin for 30 min. (b) LDA plots were calculated from cytological profiles after 30 min of treatment with daptomycin where parameters were the mean of triplicates. The LDA plot consists of the mean cytological profiles 8 DNS strains (red) and 12 DS strains (blue), all of which forms the training set. Blind test profiles (filled circles; n = 12) are overlaid on the training set.
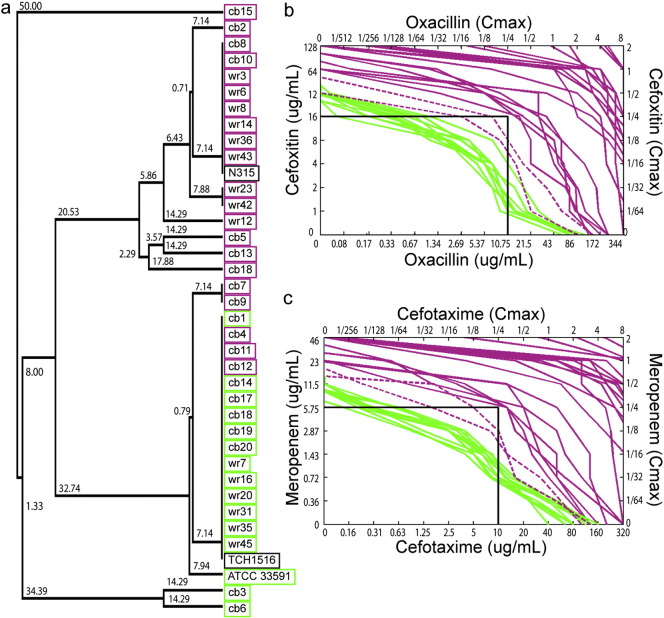
Fig. 5.
Genetic characterization and susceptibility to dual-beta lactam treatment for the two MRSA subgroups. (a) Dendrogram calculated using unweighted paired-group method with arithmetic mean (UPGMA) for the multilocus sequence typing (MLST) allelic profiles of MRSALL (purple) and MRSAHL (green). (b–c) Synergy assay curves. The position of each curve represents the combination of antibiotic concentrations that inhibits growth. Each curve represents the average of three independent synergy checkerboard assay for a single strain. There are 12 MRSA (green) strains that were not viable when treated with 1/4 Cmax of both antibiotics in combination; each of these was MRSAHL strains. There are 24 MRSA strains (purple) that were viable; each corresponds to MRSALL strains. The dotted purple curves represent two strains (wr12; cb15) that were viable at 1/4 Cmax of both antibiotics in at least one of the three independent synergy checkerboard assays.
2.3. Growth Curves
For growth curve determination, cells were grown to OD600 of 0.2. Cells were then either incubated with 10 μg mL− 1 oxacillin or with no antibiotic at 37 °C. OD600 readings were made every 30 min for 3 h. The exponential slope of the growth curve was used to determine doubling rate see in Table S3.
2.4. Fluorescence Microscopy
Exponential-phase cell cultures (OD600 ~ 0.2) were treated with antibiotics (10 μg mL− 1 oxacillin; 20 μg mL− 1 cephalexin; 5 μg mL− 1 daptomycin) and grown at 37 °C in a roller. Samples were collected for imaging every hour for 3 h (and at 30 min for daptomycin). Eight microliters of cells was added to 2 μL of dye mix consisting of 2.5 μM SYTOX Green, 10 μg mL− 1 DAPI and 20 μg mL− 1 of WGA-647 in 1 ×-Tbase and transferred to an agarose pad (10% MHB, 1% agarose). Exposure time of each wavelength was maintained constant for all images. Three images were taken for each strain. Microscopy was performed as previously described (Lamsa et al., 2012) except using a different set of dyes.
For time-lapse imaging, 8 μL of cell culture (OD600 ~ 0.2) was added to 2 μL of stain mix (10.25 μM SYTOX Green and 0.25 μg mL− 1 FM4-64) and transferred onto an agarose pad (10% MHB, 1% agarose, 5 μg mL− 1 oxacillin). The same field was imaged at 5 min intervals for 4 h.
2.5. Blind-test
In the first blind test, which consisted of MRSA and MSSA isolates, the test administrator reassigned thirty of the 71 isolates to BT 1–30. Similarly, in the second blind test which contained DNS and DS isolates, 12 of the 20 isolates were renamed BT 1–12. The tester and administrator were both unaware of what class (MSSA, MRSAHL, MRSALL; DNS, DS) each isolate belonged to.
2.6. Cytological Profiling
The threshold for the deconvolved images was adjusted on FIJI (ImageJ 2.0.0) in order to remove background and obtain shape edges. All SYTOX and DAPI intensity measurements were performed on nondeconvolved images. Cell measurements were made using CellProfiler 2.0 (Kamentsky et al., 2011). Each parameter is taken as the weighted average of three images. For each strain, three sets of profiles (each represented by three images), were obtained in three experiments on different days (n = 3).
2.7. Statistical Analysis
LDA analysis was completed using built-in MATLAB (R2013) function. LDA analysis was done on the triplicate data set (n = 3) of measured parameters (22 parameters per antibiotic) for each strain while only the average data set was graph on the LDA plots. Categorization of unknown isolates was determined based on a decision boundary of the LDA analysis. The decision boundary was defined as the mean distance between of the centroid of the two groups. Unweighted Pair Group Method with Arithmetic Mean (UPGMA) dendrogram based on the MLST of each strain was made using saureus.mlst.net online tree creation program. The UPGMA relationship is calculated based on pair-wise differences in the allelic profiles of the seven housekeeping genes.
3. Results
3.1. Rapid Antimicrobial Susceptibility Determination by BCP
To determine if BCP can distinguish between MSSA and MRSA, we created cytological profiles for 71 clinical S. aureus isolates (37 MRSA and 34 MSSA strains, with MRSA strains defined as having an MIC for oxacillin > 4 μg/mL). Isolates were cultured in MHB liquid media and treated with one of two beta-lactam antibiotics: oxacillin or cephalexin. After 1 or 2 h, cells were stained with fluorescent dyes and imaged (Fig. 1, Fig. 2a, b). For each bacterial cell, 22 different parameters were measured to create cytological profiles for each strain in the presence or absence of antibiotic treatment. We used linear discriminant analysis (LDA) to generate a linear classifier that could separate the profiles from this training set into two classes susceptible (S) and resistant (R) (Fig. 2c–f, diamond). Linear discriminant analysis is a supervised pattern recognition and machine learning statistical method used to find combinations of features which separates classes of objects. We input all the parameters measured into the LDA, and the algorithm identifies linear combinations of the parameters which maximize distances between groups and minimize distances within groups. The first LDA projection (LDA1) retains the most between-class variance in comparison to the second (LDA2). This analysis showed that better separation between S and R control strains was generated with cephalexin treatment (Fig. 2d, left) than oxacillin treatment (Fig. 2c, left). Aggregation of the profile data from cephalexin and oxacillin treatments into a single 44 parameter combined profile also generated excellent separation between S and R strains (Fig. 2e, left).
Fig. 1.
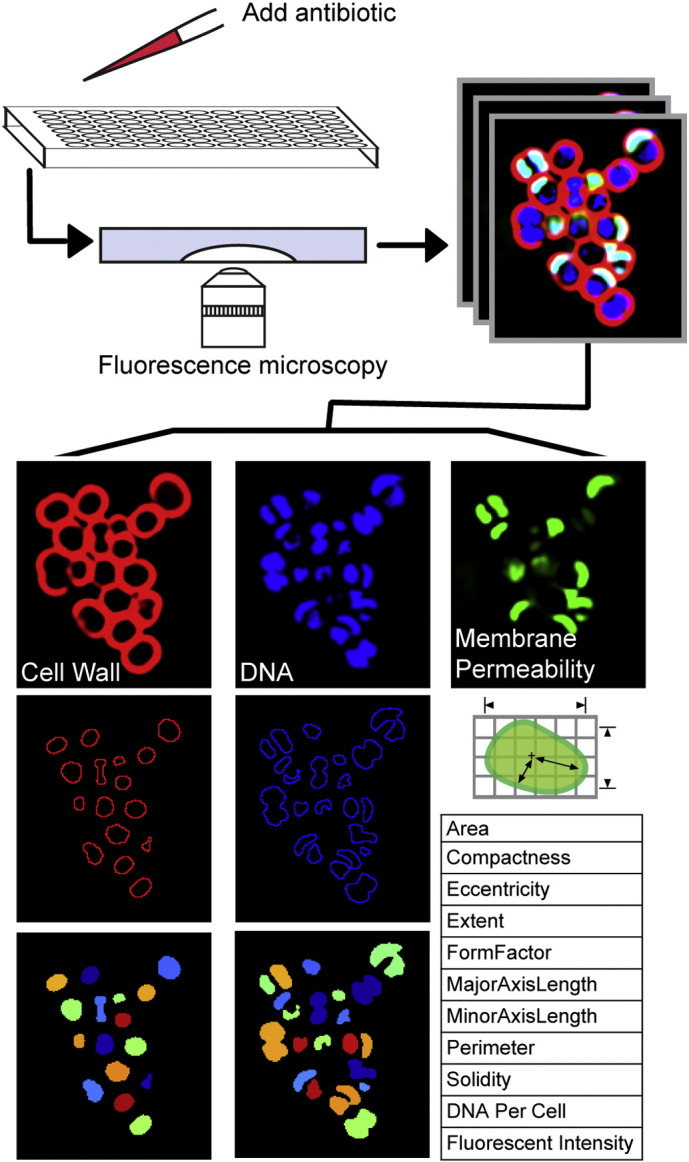
BCP methodology and analysis. Each antibiotic was added to exponentially growing cells and samples were collected for imaging hourly. Changes in cytological parameters were measured using CellProfiler. For each strain, three sets of profiles (each represented by three images), were obtained over three different days.
These results suggested that BCP can be applied to rapidly (within 2 h) determine S. aureus antibiotic susceptibility. We therefore performed a blinded experiment to test if BCP can accurately distinguish between MSSA and MRSA clinical isolates by plotting the profiles for 30 blinded isolates (filled circles) on the LDA graph using the linear classifier obtained from the LDA training set (Fig. 2c–f, left). For profiles derived from oxacillin or cephalexin treatment alone or in aggregate, we correctly categorized MSSA and MRSA strains with 100% (n = 71) accuracy after 2 h of antibiotic exposure. When the analysis was performed after only 1 h of antibiotic treatment, the separation between the two groups was somewhat reduced, but the accuracy of discrimination remained at 100% (Fig. 2f, left). LDA values strongly correlated with the MICs for cephalexin and oxacillin and correctly predicted susceptibility (Fig. 2c–f, right).
With high rates of vancomycin treatment failure (Levine, 2006, Charles et al., 2004, Stevens, 2006), daptomycin has emerged as an important treatment option for serious MRSA infections including bacteremia (Murray et al., 2013, Moore et al., 2012). Accordingly, we applied our method to daptomycin non-susceptible (DNS) and susceptible (DS) strains. Performing the same techniques described above but with daptomycin treatment, we observed distinct differences in bacterial cytological profiles between DS and DNS strains (n = 20) (Fig. 3a). LDA graphs showed that we could accurately discriminate between DS and DNS strains as early as 30 min post treatment in a double blinded test (n = 12) (Fig. 3b).
3.2. BCP Profiles can be Mined to Extract Additional Information and Detect Patterns
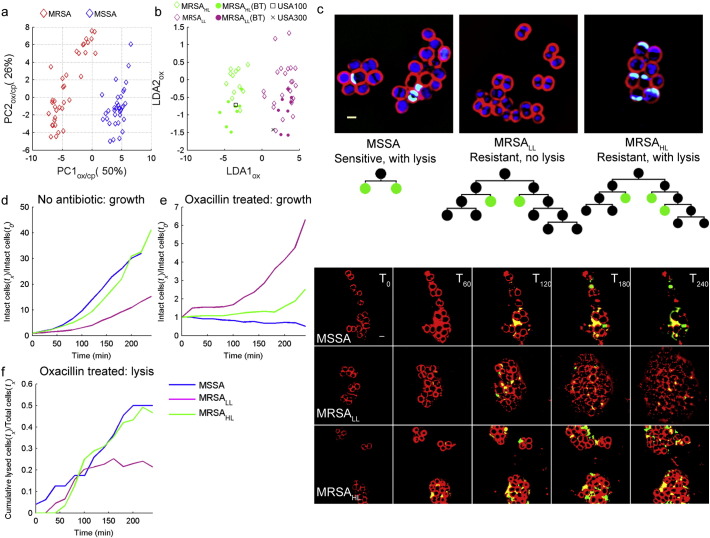
While cephalexin treatment alone was sufficient to distinguish between MSSA and MRSA strains, oxacillin profiles revealed strikingly different responses to antibiotic treatment among MRSA strains. Principal component analysis (PCA) of the combined profile data (aggregated profile of oxacillin and cephalexin treatment) showed two major clusters corresponding to the MSSA and MRSA groups (Fig. 4a), but large variance within principal component 2 (PC2) hinted at the existence of subgroups within the MRSA cohort. Further inspection of MRSA strains at opposing ends of this range revealed two distinctive phenotypes. LDA of the cytological profiles of oxacillin-treated MRSA strains in the top and bottom quartiles of PC2 was used to define the LDA space onto which all MRSA strains were plotted. This LDA parameter space separated MRSA into two groups with distinctive profiles (MRSALL, low lysis; MRSAHL, high lysis; Fig. 4b). When exposed to oxacillin, strains in the MRSALL (purple) group have an elliptical cell shape and low levels of cell lysis (average 3%, n = 24 strains), whereas MRSAHL strains (green) are circular with a high degree of lysis (average 20%, n = 12 strains). This result was unanticipated, as previous studies suggested that lysis alone is indicative of susceptibility (Matsuda et al., 1995, Best et al., 1974, Memmi et al., 2012, Kalashnikov et al., 2012); however, these clinical strains with high lysis possessed oxacillin MICs > 50 μg/mL.
Fig. 4.
Two classes of MRSA strains. (a) Principal component analysis (PCA) plot of training set data comprise of 37 MRSA (red) and 36 MSSA (blue) strains. (b) LDA plot showing the two classes of MRSA strains. The transformation matrix for the LDA plot was calculated from the cytological profiles of strains in the top and bottom quartiles of PC2 and used to plot 12 MRSAHL (green diamonds) and 24 MRSALL (purple diamonds) after 2 h of treatment with 10 μg/mL of oxacillin. Each data point represents the mean of triplicate cytological profiles for a different clinical isolate. The BCP profiles for the blind tests (filled circles), USA300 (TCH1516; black x) and USA100 (N315; black square) were overlaid on the training set (diamonds). (c) Time-lapse microscopy of MSSA (ATTC29213), MRSALL (ATCC33591), MRSAHL (wr7) was taken while strains were grown on agarose pads with 5 μg/mL oxacillin. (d–e) Comparison of the net growth rates of these three groups on agarose pads (d) without antibiotics and (e) with oxacillin. (f) Cumulative lysis of different strain types on agarose pad with oxacillin.
Time-lapse microscopy was performed to follow the progression of cytological profiles over time during treatment with oxacillin (Fig. 4c). The expected response to beta-lactam treatment is that MSSA strains halt growth and lyse while MRSA strains continue to grow with relatively little cell lysis. As expected upon oxacillin treatment, MSSA halted growth, became enlarged and began to lyse. In contrast, MRSALL strains exhibited an initial burst of lysis but then quickly recovered, proceeding to actively grow and divide. Interestingly, MRSAHL strains continued to undergo cycles of growth, division and lysis for the duration of the time-lapse experiment. Comparison of the growth rates of these three groups (MSSA, MRSALL, MRSAHL) on agarose pads without antibiotics revealed that MRSAHL and MSSA have comparable growth rates (generation time = 27 min) while MRSALL grew slower (generation time = 32 min) (Fig. 4d). With the addition of oxacillin, MSSA lysed, resulting in a negative growth rate while MRSALL grew faster than MRSAHL (Fig. 4e). In addition to a faster growth rate in the presence of oxacillin, MRSALL recovered after an initial burst of lysis, whereas MRSAHL continued to lyse throughout the assay (Fig. 4f).
These results suggest that MRSA strains are diverse in their phenotypic response to beta-lactam antibiotics. High lysis strains are better adapted for rapid growth in the absence of antibiotics, while low lysis strains are better adapted for growth in the presence of beta-lactams. The MRSALL fitness advantage over MRSAHL in the presence of antibiotics is reflected in higher MICs of low lysis strains compared to high lysis strains (Table S1). This result parallels observations that repeated selection for high level resistance to beta-lactams comes at a fitness cost to growth in the absence of antibiotic (Ender et al., 2004, Andersson and Levin, 1999). Although the majority of MRSALL strains have lower growth rates than MRSAHL when grown without antibiotic and higher growth rates than MRSAHL under antibiotic treatment, this trend did not apply to all the strains in our collection (Table S2). Such heterogeneity among MRSA strains explains why growth rate alone is insufficient to distinguish between MRSALL and MRSAHL. In contrast to growth rate measurements, BCP evaluates a multitude of parameters allowing rapid differentiation of MSSA and MRSA strains and the identification of subgroups within MRSA isolates.
Next we performed a genetic analysis of the S. aureus clinical isolates to determine if their cytological profiles correlated with specific genotypes. Specifically, we questioned if the two groups of MRSA identified as displaying distinct responses to beta-lactam antibiotics were genetically different. We classified our clinical isolates of S. aureus using a number of genetic markers including the accessory gene regulator agr locus, protein A, and a collection of seven housekeeping genes (Multi Locus Sequence Type, MLST) (Table S3). From the MLST, we constructed a dendrogram illustrating the relationship between all of the MRSA strains in our study (Fig. 5a). We found that all of the MRSAHL strains are closely related to each other (Fig. 5a, green) and to the community-acquired strain USA300. A majority of the MRSAHL belong to multilocus sequence type (MLST) ST8 (Table S3), a marker for USA300 lineage community-acquired MRSA in the U.S. (Nimmo, 2012). However, genotyping alone could not accurately predict whether strains would be low or high lysis. Three of the MRSALL strains were closely related to the community-acquired strain USA300, while the 9 MRSALL strains were more closely related to the hospital-acquired strain USA100 (Fig. 5a, purple). MRSALL isolates belonged to ST8 as well as ST5, a sequence type associated with hospital acquired (HA) MRSA (Gordon and Lowy, 2008, Takano et al., 2013). The three MRSALL strains with genetic identities close to USA300 have high MICs and most likely represent strains that have evolved through continued antibiotic selection to become similar to low lysis, highly antibiotic-resistant strains.
BCP profiles for USA300 (TCH1516) and USA100 (N315) showed that the strains correctly cluster with MRSA isolates (Fig. 2e), but they do not cluster into the same MRSA subgroups in the LDA analysis as they do on the dendrogram (Fig. 4b). N315, a pre-MRSA, exhibits a heterogeneous response to oxacillin exposure (Aiba et al., 2013, Ito et al., 1999). The fact that N315 shows genetic similarity to MRSALL and phenotypic similarity to MRSAHL demonstrates that genetic testing alone is insufficient for determining phenotypic response to antibiotic treatment. Because of the constant evolutionary pressures driving strain evolution, phenotypic assays such as BCP paired with molecular genetic tests can be used as a tool to track the evolution of antibiotic susceptibility patterns.
3.3. BCP Detects Groups that Respond to Different Potential Treatment Options
The existence of a MRSA group with high lysis in the presence of a low level (< 1/10th MIC) of oxacillin led us to explore the potential of eradicating the remaining population with a second antibiotic. Previous work showed that community-acquired strains of MRSA respond to dual beta-lactam therapy in vitro (Memmi et al., 2008). We thus tested each MRSA clinical isolate in synergy assays to determine if they were susceptible to physiologically relevant combinations of beta-lactams. We found that all of the MRSAHL strains responded synergistically to combinations of oxacillin + cefoxitin (Fig. 5b) or cefotaxime + meropenem (Fig. 5c). The MRSAHL strains (green) did not grow when treated with both antibiotics at concentrations below 1/4 Cmax (maximum IV concentration), modeling physiologically achievable antibiotic concentrations (ADD-Vantage®, 2015, PFIZER, 2009, NOVATION, 2014). Strains that did not grow at or below this cutoff are candidate responders to dual antibiotic combination therapy. Testing of the remaining strains (MRSALL, purple) revealed either no synergy or synergy at concentrations that were too high to be physiologically achievable. Finally, two MRSALL strains (dotted purple) that bordered the cutoff were not reproducibly inhibited below 1/4 Cmax. Together these results show that BCP can detect two distinct subgroups of MRSA and predict which strains will be susceptible to combinations of beta-lactam antibiotics.
4. Discussion
Our study demonstrates that applications of BCP extend beyond determination of antibiotic mechanism of action to determination of antibiotic susceptibility and identification of optimally effective combinatorial therapies. This single cell phenotype-driven approach provides a rapid susceptibility test that can be performed in parallel with traditional MIC testing, but which delivers a result within 1–2 h, rather than 1–2 days. The test is precise, yielding no major or minor errors in our blinded tests. Rather than relying on a single parameter (such as growth or permeability), BCP as implemented here assesses 22 different parameters that together capture the wide array of affects antibiotics have on bacterial cells, including changes in cell length, width, permeability, and in chromosome number, compactness and shape (Lamsa et al., 2012, Nonejuie et al., 2013). This likely explains why BCP is more accurate (as well as more rapid) than other proposed phenotype-driven tests (Choi et al., 2014, Ligozzi et al., 2002). Indeed, phenotypic methods that rely only on lysis (Price et al., 2014, Kalashnikov et al., 2012, Choi et al., 2013) would have miscategorized the MRSAHL strains as MSSA, whereas methods that rely only on growth would have failed to identify the two classes of MRSA strains that differ in their susceptibility to combinatorial drug therapies.
Our mechanistic studies have demonstrated that BCP works for all antibiotics, natural products and antimicrobial peptides tested so far, and for all species tested (S. aureus, Escherichia coli, Bacillus subtilis, E. faecium, Enterococcus faecalis, Streptococcus pneumoniae and A. baumannii) (Lamsa et al., 2012, Nonejuie et al., 2013, Sakoulas et al., 2015a, Sakoulas et al., 2015b, Lin et al., 2015, Hindler et al., 2015, Werth et al., 2014). A major strength of BCP is that, in contrast to the rapid gene and genome-based susceptibility tests, it does not rely on prior knowledge of the mechanism of antibiotic resistance or prior identification of all possible genes and mutations that confer resistance. This is critical, even for drugs that bind well-defined targets, such as the ribosome, since the number of mutations that confer drug resistance in the clinic continues to expand (Wilson, 2014). It is even more critical for drugs such as daptomycin, for which several different mutations confer various levels of resistance (Diaz et al., 2014, Mishra et al., 2014, Bayer et al., 2015, Bayer et al., 2013, Berti et al., 2015). The reliance of BCP on phenotype rather than genotype makes it highly adaptable, allowing future applications to new drug resistant pathogens and new antibiotics. BCP can be readily optimized to provide a more accurate test for different species or antibiotics. For example, the susceptibility test reported here relies on a different combination of fluorescent stains than used in our previous BCP assays.
BCP provides a robust and rapid technique that is readily adapted to identify opportunities for antibiotic treatment synergy. Such synergies often derive from the additive effects of multiple genetic factors, making it difficult or impossible to detect them using a gene-based technique. This was found to be the case for the beta-lactam plus daptomycin synergy in E. faecium, which depends on multiple PBPs (Sakoulas et al., 2015a) and for daptomycin resistance in S. aureus and E. faecium for which multiple mutations contribute to resistance (Bayer et al., 2013) by modifying either the cell membrane or the cell wall (Humphries et al., 2013). Hence we propose that BCP could be used to guide treatment options including the usage of dual beta-lactam treatment or the substitution or addition of different class antibiotics to lower the chances of treatment failure. Identification of synergistic combinations in vitro followed by appropriate clinical correlation studies to determine the efficacy in vivo allows for the assessment of new treatment options using existing antibiotics.
The cytological library we have developed provides a versatile reference data set for the analysis of S. aureus strains that can be expanded to include new data from reference strains as well as new strains as they evolve. Indeed, realizing the clinical potential of BCP as an antimicrobial susceptibility test will require that we expand our current reference data set to include additional clinical isolates to generate a more robust training set that includes strains from more diverse locations. Currently we have tested culture samples, in the future we would like to expand our method to test directly from patient samples for a variety of bacterial pathogens and antibiotics. Additionally we expect that BCP could work for determining susceptibility in mixed cultures by categorizing the species according to cytological variation. Rapid, mass spectrometry based methods for species identification might also be applied, which would help identify the appropriate species-specific BCP reference library to determine susceptibility.
5. Conclusions
The objective of this study was to apply BCP as a rapid susceptibility test for S. aureus. We demonstrated that BCP could be used to distinguish MRSA from MSSA and DNS from DS S. aureus for a set of clinical isolates on a timescale competitive with existing rapid susceptibility tests. BCP provided information beyond susceptibility testing and identified two subgroups of MRSA that responded differently to oxacillin and to specific combinations of antibiotics. These subgroups were not predicted based on the presence of commonly assessed genetic markers (mecA), highlighting the potential power of phenotypic cell based assays to rapidly identify the best treatment options. Because cytological profiles consist of tens of parameters, one could retrospectively mine them for parameters that correlate with factors such as new sequence types and groups corresponding to treatment failure. For every new strain tested and added to the library, BCP data can be coupled with susceptibility information, sequence and strain type, and clinical data, such as infection type and treatment outcome. Since sequence typing of different isolates is routinely performed for phylogenetic and population-based analysis, BCP profiles that are obtained during susceptibility determination can complement future epidemiological studies by serving as an early indicator of emerging trends that require more public health attention. Combining sequence and BCP data will also allow us to better understand the cellular consequences of the genetic changes that occur as bacterial pathogens adapt to the clinic and acquire resistance to new antimicrobial agents.
Funding Sources
This research was supported by the National Institute of Health AI113295 (to JP and KP).
Conflict of Interest Statement
JP and KP have an equity interest in Linnaeus Bioscience, Inc. (La Jolla, CA). The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. US Patent application #13/816,471 has been filed related to this topic. Linnaeus Bioscience, Inc. has licensed the rights to US Patent application #13/816,471 from UCSD. GS has received consulting and speaking honoraria fees from Merck and Actives Pharmaceuticals and research grant funds from Actives Pharmaceuticals. VN has received consulting fees from Cidara Therapeutics and Loxbridge Research, and has laboratory sponsored research agreements with Roche, InhibRx and Altermune Technologies.
Author Contributions
GS, JP and KP each contributed to the concept design, data interpretation and writing of the paper. VN provided general counsel and contributed to the writing of the paper. DQ contributed to the performance of the experiments, analysis of the data and writing of the paper.
Acknowledgments
We would like to thank Warren E. Rose (University of Wisconsin — Madison) for providing the S. aureus strains used in this project. Special thanks to Poochit Nonejuie and Anne Lamsa for many discussions and guidance.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.01.020.
Appendix A. Supplementary Data
Supplementary material 1
Supplementary material 2
References
- ADD-Vantage® . Sandoz Inc.; NJ: 2015. Oxacillin Sodium ADD-Vantage® Princeton. [Google Scholar]
- Aiba Y., Katayama Y., Hishinuma T., Murakami-Kuroda H., Cui L., Hiramatsu K. Mutation of RNA polymerase β-subunit gene promotes heterogeneous-to-homogeneous conversion of β-lactam resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2013;57:4861–4871. doi: 10.1128/AAC.00720-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson D.I., Levin B.R. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 1999;2:489–493. doi: 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- Bauer K.A., West J.E., Balada-Llasat J.M., Pancholi P., Stevenson K.B., Goff D.A. An antimicrobial stewardship program's impact with rapid polymerase chain reaction methicillin-resistant Staphylococcus aureus/S. aureus blood culture test in patients with S. aureus bacteremia. Clin. Infect. Dis. 2010;51:1074–1080. doi: 10.1086/656623. [DOI] [PubMed] [Google Scholar]
- Bayer A.S., Mishra N.N., Chen L., Kreiswirth B.N., Rubio A., Yang S.J. Frequency and distribution of single-nucleotide polymorphisms within mprF in methicillin-resistant Staphylococcus aureus clinical isolates and their role in cross-resistance to daptomycin and host defense antimicrobial peptides. Antimicrob. Agents Chemother. 2015;59:4930–4937. doi: 10.1128/AAC.00970-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer A.S., Schneider T., Sahl H.G. Mechanisms of daptomycin resistance in Staphylococcus aureus: role of the cell membrane and cell wall. Ann. N. Y. Acad. Sci. 2013;1277:139–158. doi: 10.1111/j.1749-6632.2012.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti A.D., Baines S.L., Howden B.P., Sakoulas G., Nizet V., Proctor R.A., Rose W.E. Heterogeneity of genetic pathways toward daptomycin nonsusceptibility in Staphylococcus aureus determined by adjunctive antibiotics. Antimicrob. Agents Chemother. 2015;59:2799–2806. doi: 10.1128/AAC.04990-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best G.K., Best N.H., Koval A.V. Evidence for participation of autolysins in bactericidal action of oxacillin on Staphylococcus aureus. Antimicrob. Agents Chemother. 1974;6:825–830. doi: 10.1128/aac.6.6.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles P.G., Ward P.B., Johnson P.D., Howden B.P., Grayson M.L. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clin. Infect. Dis. 2004;38:448–451. doi: 10.1086/381093. [DOI] [PubMed] [Google Scholar]
- Choi J., Jung Y.G., Kim J., Kim S., Jung Y., Na H., Kwon S. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip. 2013;13:280–287. doi: 10.1039/c2lc41055a. [DOI] [PubMed] [Google Scholar]
- Choi J., Yoo J., Lee M., Kim E.G., Lee J.S., Lee S., Joo S., Song S.H., Kim E.C., Lee J.C., Kim H.C., Jung Y.G., Kwon S. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3009650. (267ra174) [DOI] [PubMed] [Google Scholar]
- CLSI . CLSI document M100-S17. Clinical and Laboratory Standards Institute; Wayne, PA: 2007. Performance standards for antimicrobial susceptibility testing; seventeenth informational supplement. [Google Scholar]
- CLSI . CLSI document M07-A9. Ninth ed. Clinical and Laboratory Standards Institute; Wayne, PA: 2012. Method for dilution antimicrobial susceptibility test for bacteria that grow aerobically; approved standard. [Google Scholar]
- Diaz L., Tran T.T., Munita J.M., Miller W.R., Rincon S., Carvajal L.P., Wollam A., Reyes J., Panesso D., Rojas N.L., Shamoo Y., Murray B.E., Weinstock G.M., Arias C.A. Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob. Agents Chemother. 2014;58:4527–4534. doi: 10.1128/AAC.02686-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender M., Mccallum N., Adhikari R., Berger-Bächi B. Fitness cost of SCCmec and methicillin resistance levels in Staphylococcus aureus. Antimicrob. Agents Chemother. 2004;48:2295–2297. doi: 10.1128/AAC.48.6.2295-2297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Álvarez L., Holden M.T., Lindsay H., Webb C.R., Brown D.F., Curran M.D., Walpole E., Brooks K., Pickard D.J., Teale C., Parkhill J., Bentley S.D., Edwards G.F., Girvan E.K., Kearns A.M., Pichon B., Hill R.L., Larsen A.R., Skov R.L., Peacock S.J., Maskell D.J., Holmes M.A. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect. Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff D.A., Jankowski C., Tenover F.C. Using rapid diagnostic tests to optimize antimicrobial selection in antimicrobial stewardship programs. Pharmacotherapy. 2012;32:677–687. doi: 10.1002/j.1875-9114.2012.01137.x. [DOI] [PubMed] [Google Scholar]
- Gordon R.J., Lowy F.D. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 2008;46(Suppl. 5):S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwynn M.N., Portnoy A., Rittenhouse S.F., Payne D.J. Challenges of antibacterial discovery revisited. Ann. N. Y. Acad. Sci. 2010;1213:5–19. doi: 10.1111/j.1749-6632.2010.05828.x. [DOI] [PubMed] [Google Scholar]
- Hindler J.A., Wong-Beringer A., Charlton C.L., Miller S.A., Kelesidis T., Carvalho M., Sakoulas G., Nonejuie P., Pogliano J., Nizet V., Humphries R. In vitro activity of daptomycin in combination with β-lactams, gentamicin, rifampin, and tigecycline against daptomycin-nonsusceptible enterococci. Antimicrob. Agents Chemother. 2015;59:4279–4288. doi: 10.1128/AAC.05077-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hososaka Y., Hanaki H., Endo H., Suzuki Y., Nagasawa Z., Otsuka Y., Nakae T., Sunakawa K. Characterization of oxacillin-susceptible mecA-positive Staphylococcus aureus: a new type of MRSA. J. Infect. Chemother. 2007;13:79–86. doi: 10.1007/s10156-006-0502-7. [DOI] [PubMed] [Google Scholar]
- Humphries R.M., Pollett S., Sakoulas G. A current perspective on daptomycin for the clinical microbiologist. Clin. Microbiol. Rev. 2013;26:759–780. doi: 10.1128/CMR.00030-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Katayama Y., Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y. Methods in Molecular Biology. Humana Press; Totowa, NJ: 2007. Methicillin-resistant Staphylococcus aureus (MRSA) protocols. [DOI] [PubMed] [Google Scholar]
- Kalashnikov M., Lee J.C., Campbell J., Sharon A., Sauer-Budge A.F. A microfluidic platform for rapid, stress-induced antibiotic susceptibility testing of Staphylococcus aureus. Lab Chip. 2012;12:4523–4532. doi: 10.1039/c2lc40531h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamentsky L., Jones T.R., Fraser A., Bray M.A., Logan D.J., Madden K.L., Ljosa V., Rueden C., Eliceiri K.W., Carpenter A.E. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnunen P., Mcnaughton B.H., Albertson T., Sinn I., Mofakham S., Elbez R., Newton D.W., Hunt A., Kopelman R. Self-assembled magnetic bead biosensor for measuring bacterial growth and antimicrobial susceptibility testing. Small. 2012;8:2477–2482. doi: 10.1002/smll.201200110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamsa A., Liu W.T., Dorrestein P.C., Pogliano K. The Bacillus subtilis cannibalism toxin SDP collapses the proton motive force and induces autolysis. Mol. Microbiol. 2012;84:486–500. doi: 10.1111/j.1365-2958.2012.08038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent F., Chardon H., Haenni M., Bes M., Reverdy M.E., Madec J.Y., Lagier E., Vandenesch F., Tristan A. MRSA harboring mecA variant gene mecC, France. Emerg. Infect. Dis. 2012;18:1465–1467. doi: 10.3201/eid1809.111920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine D.P. Vancomycin: a history. Clin. Infect. Dis. 2006;42(Suppl. 1):S5–12. doi: 10.1086/491709. [DOI] [PubMed] [Google Scholar]
- Ligozzi M., Bernini C., Bonora M.G., De Fatima M., Zuliani J., Fontana R. Evaluation of the VITEK 2 system for identification and antimicrobial susceptibility testing of medically relevant gram-positive cocci. J. Clin. Microbiol. 2002;40:1681–1686. doi: 10.1128/JCM.40.5.1681-1686.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Nonejuie P., Munguia J., Hollands A., Olson J., Dam Q., Kumaraswamy M., Rivera H., Corriden R., Rohde M., Hensler M.E., Burkart M.D., Pogliano J., Sakoulas G., Nizet V. Azithromycin synergizes with cationic antimicrobial peptides to exert bactericidal and therapeutic activity against highly multidrug-resistant gram-negative bacterial pathogens. EBioMedicine. 2015;2:690–698. doi: 10.1016/j.ebiom.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K., Nakamura K., Adachi Y., Inoue M., Kawakami M. Autolysis of methicillin-resistant Staphylococcus aureus is involved in synergism between imipenem and cefotiam. Antimicrob. Agents Chemother. 1995;39:2631–2634. doi: 10.1128/aac.39.12.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mediavilla J.R., Chen L., Mathema B., Kreiswirth B.N. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA) Curr. Opin. Microbiol. 2012;15:588–595. doi: 10.1016/j.mib.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Memmi G., Filipe S.R., Pinho M.G., Fu Z., Cheung A. Staphylococcus aureus PBP4 is essential for beta-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob. Agents Chemother. 2008;52:3955–3966. doi: 10.1128/AAC.00049-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memmi G., Nair D.R., Cheung A. Role of ArlRS in autolysis in methicillin-sensitive and methicillin-resistant Staphylococcus aureus strains. J. Bacteriol. 2012;194:759–767. doi: 10.1128/JB.06261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N.N., Bayer A.S., Weidenmaier C., Grau T., Wanner S., Stefani S., Cafiso V., Bertuccio T., Yeaman M.R., Nast C.C., Yang S.J. Phenotypic and genotypic characterization of daptomycin-resistant methicillin-resistant Staphylococcus aureus strains: relative roles of mprF and dlt operons. PLoS One. 2014;9 doi: 10.1371/journal.pone.0107426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C.L., Osaki-Kiyan P., Haque N.Z., Perri M.B., Donabedian S., Zervos M.J. Daptomycin versus vancomycin for bloodstream infections due to methicillin-resistant Staphylococcus aureus with a high vancomycin minimum inhibitory concentration: a case-control study. Clin. Infect. Dis. 2012;54:51–58. doi: 10.1093/cid/cir764. [DOI] [PubMed] [Google Scholar]
- Murray K.P., Zhao J.J., Davis S.L., Kullar R., Kaye K.S., Lephart P., Rybak M.J. Early use of daptomycin versus vancomycin for methicillin-resistant Staphylococcus aureus bacteremia with vancomycin minimum inhibitory concentration > 1 mg/L: a matched cohort study. Clin. Infect. Dis. 2013;56:1562–1569. doi: 10.1093/cid/cit112. [DOI] [PubMed] [Google Scholar]
- Nimmo G.R. USA300 abroad: global spread of a virulent strain of community-associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2012;18:725–734. doi: 10.1111/j.1469-0691.2012.03822.x. [DOI] [PubMed] [Google Scholar]
- Nonejuie P., Burkart M., Pogliano K., Pogliano J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl. Acad. Sci. 2013;110:16169–16174. doi: 10.1073/pnas.1311066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOVATION . Irving; Tx: 2014. Meropenem. [Google Scholar]
- PFIZER . NY; New York: 2009. Cefotaxime. [Google Scholar]
- Pinho M.G., Filipe S.R., De Lencastre H., Tomasz A. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J. Bacteriol. 2001;183:6525–6531. doi: 10.1128/JB.183.22.6525-6531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C.S., Kon S.E., Metzger S. Rapid antibiotic susceptibility phenotypic characterization of Staphylococcus aureus using automated microscopy of small numbers of cells. J. Microbiol. Methods. 2014;98:50–58. doi: 10.1016/j.mimet.2013.12.021. [DOI] [PubMed] [Google Scholar]
- Rice L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 2008;197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- Sakoulas G., Kumaraswamy M., Nonejuie P., Werth B., Rybak M.J., Pogliano J., Rice L.B., Nizet V. Differential effects of penicillin binding protein deletion on the susceptibility of Enterococcus faecium to cationic peptide antibiotics. Antimicrob. Agents Chemother. 2015 doi: 10.1128/AAC.00486-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoulas G., Nonejuie P., Kullar R., Pogliano J., Rybak M.J., Nizet V. Examining the use of ceftaroline in the treatment of Streptococcus pneumoniae meningitis with reference to human cathelicidin LL-37. Antimicrob. Agents Chemother. 2015;59:2428–2431. doi: 10.1128/AAC.04965-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinn I., Albertson T., Kinnunen P., Breslauer D.N., Mcnaughton B.H., Burns M.A., Kopelman R. Asynchronous magnetic bead rotation microviscometer for rapid, sensitive, and label-free studies of bacterial growth and drug sensitivity. Anal. Chem. 2012;84:5250–5256. doi: 10.1021/ac300128p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D.L. The role of vancomycin in the treatment paradigm. Clin. Infect. Dis. 2006;42(Suppl. 1):S51–S57. doi: 10.1086/491714. [DOI] [PubMed] [Google Scholar]
- Takano T., Hung W.C., Shibuya M., Higuchi W., Iwao Y., Nishiyama A., Reva I., Khokhlova O.E., Yabe S., Ozaki K., Takano M., Yamamoto T. A new local variant (ST764) of the globally disseminated ST5 lineage of hospital-associated methicillin-resistant Staphylococcus aureus (MRSA) carrying the virulence determinants of community-associated MRSA. Antimicrob. Agents Chemother. 2013;57:1589–1595. doi: 10.1128/AAC.01147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S.Y., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werth B.J., Steed M.E., Ireland C.E., Tran T.T., Nonejuie P., Murray B.E., Rose W.E., Sakoulas G., Pogliano J., Arias C.A., Rybak M.J. Defining daptomycin resistance prevention exposures in vancomycin-resistant Enterococcus faecium and E. faecalis. Antimicrob. Agents Chemother. 2014;58:5253–5261. doi: 10.1128/AAC.00098-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim H.F., Melles D.C., Vos M.C., Van Leeuwen W., Van Belkum A., Verbrugh H.A., Nouwen J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- Wilson D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014;12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2