Abstract
Background
Genome-wide association studies have identified polymorphisms linked to both smoking exposure and risk of lung cancer. The degree to which lung cancer risk is driven by increased smoking, genetics, or gene–environment interactions is not well understood.
Methods
We analyzed associations between 28 single nucleotide polymorphisms (SNPs) previously associated with smoking quantity and lung cancer in 7156 African-American females in the Women's Health Initiative (WHI), then analyzed main effects of top nominally significant SNPs and interactions between SNPs, cigarettes per day (CPD) and pack-years for lung cancer in an independent, multi-center case–control study of African-American females and males (1078 lung cancer cases and 822 controls).
Findings
Nine nominally significant SNPs for CPD in WHI were associated with incident lung cancer (corrected p-values from 0.027 to 6.09 × 10− 5). CPD was found to be a nominally significant effect modifier between SNP and lung cancer for six SNPs, including CHRNA5 rs2036527[A](betaSNP*CPD = − 0.017, p = 0.0061, corrected p = 0.054), which was associated with CPD in a previous genome-wide meta-analysis of African-Americans.
Interpretation
These results suggest that chromosome 15q25.1 variants are robustly associated with CPD and lung cancer in African-Americans and that the allelic dose effect of these polymorphisms on lung cancer risk is most pronounced in lighter smokers.
Keywords: African-Americans, Environment, Genetics, Lung Cancer, rs2036527, Single Nucleotide Polymorphisms, Smoking
Highlights
-
•
Genetic by environment (e.g., cigarettes/day, CPD) interactions for lung cancer are understudied in non-European ancestry populations.
-
•
We analyzed interactions between nominal smoking quantity SNPs (n = 7156 discovery sample) and CPD and risk of lung cancer (n = 1078 cases, n = 822 controls).
-
•
Six SNPs were effect modifiers of CPD for lung cancer, suggesting that the allelic dose effect is most pronounced in light smokers.
Lung cancer is the leading cause of cancer death, disproportionately affecting African-Americans. Prior studies have reported specific genetic markers linked to both smoking quantity and risk of lung cancer in multiple ethnic/racial groups. Investigators analyzed associations between 28 polymorphisms and average cigarettes smoked per day (CPD) in 7156 African-American females and examined interactions between the top polymorphisms and CPD in a cohort of African-American males and females (1078 lung cancer cases and 822 health control patients). The results suggested that six polymorphisms within one genomic region increased lung cancer risk in African-Americans, which was most pronounced in light smokers.
1. Introduction
It is well established that tobacco smoking is responsible for most of the attributable risk of lung cancer, which is the leading cause of cancer death in males and females in the U.S (U.S.-Department-of-Health-and-Human-Services, 2014). However, there is also growing evidence that genetic factors contribute to risk for developing lung cancer (Fisher, 1958a, Fisher, 1958b, Sullivan and Kendler, 1999, Li et al., 2003, Maes et al., 1999, Heath, 1990, Raaschou-Nielsen, 1960, Crumpacker et al., 1979, Eaves et al., 1993, Carmelli et al., 1992, Kaprio et al., 1984, Edwards et al., 1995, Hannah et al., 1985, Heath et al., 1993, Vink et al., 2005, Lessov et al., 2004, Broms et al., 2006). Genome-wide meta-analyses (GWAS) of linkage studies of smoking behaviors in European ancestry populations have identified three genomic regions with genome-wide suggestive or significant evidence for ever-smoking on chromosomes 5 (q33.1–5q35.2) and 17 (q24.3–q25.3) and maximum cigarettes smoked per day on chromosome 20 (q13.12–q13.32) (Han et al., 2010). Candidate gene association studies of smoking have had limited success in identifying replicable associations (Hirschhorn et al., 2002, Lohmueller et al., 2003, Munafo et al., 2004). Variation in the CYP2A6 locus, which plays the primary role in nicotine metabolism, has emerged as reliably influencing smoking behavior (Tyndale and Sellers, 2002) but slow metabolizing variants are uncommon in African-descent populations (Piliguian et al., 2014).
In GWAS, several single-nucleotide polymorphisms (SNPs) have been identified as being associated with lung cancer or smoking behavior. The rs1051730 SNP within the nicotinic acetylcholine receptor gene cluster (CHRNA5/CHRNA3/CHRNB4) cluster on chromosome 15q25.1 has been identified with being associated with both quantity of cigarettes smoked per day and lung cancer risk (Thorgeirsson et al., 2008). The relationship between CHRNA5-A3-B4 loci and smoking quantity has been replicated in European-ancestry smokers in large GWAS datasets including the European Network of Genomic and Genetic Epidemiology (ENGAGE) Consortium (Thorgeirsson et al., 2010). Moreover, a large (> 140,000 European-ancestry samples) GWAS by the Tobacco and Genetics Consortium confirmed an association between two SNPs in the CHRNA5/CHRNA3/CHRNB4 gene cluster with cigarettes per day (CPD) for rs1051730 (p = 2.8 × 10− 73) and rs1696998 (p = 5.6 × 10− 72) (Tobacco-and-Genetics-(TAG)-Consortium, 2010). Another meta-analysis (Saccone et al., 2010) reported that rs16969968, which is highly correlated with rs1051730, and rs588765 (separate loci within the chromosome 15q25.1 region) were both statistically-significantly associated with smoking quantity and lung cancer risk and chronic obstructive pulmonary disease (COPD) and emphysema after adjustment for average reported CPD. Given that smoking is causally related to both COPD/emphysema and lung cancer and that COPD/emphysema is considered a precursor condition to carcinoma of the lung (Etzel et al., 2008) and mediator of the relationship between smoking and lung cancer (Young et al., 2009), the mechanistic nature of the relationship between the chromosome 15q25.1 locus and risk of lung cancer, i.e., the degree to which smoking quantity is an effect-modifier, mediator or confound, remains a subject of ongoing investigation. In addition to chromosome 15q25.1, nine other regions have been associated with lung cancer susceptibility in European and Asian ancestry populations (Wang et al., 2015), though not all these regions have been replicated.
Exploration of genetic biomarkers for lung cancer risk is needed in non-European populations such as African-ancestry populations because of population differences in disease allele frequency, linkage disequilibrium patterns and phenotype prevalence (Rosenberg et al., 2010). African Americans, on average, initiate smoking later, smoke fewer CPD, yet are less likely to successfully quit smoking and have a higher risk of smoking-related lung cancer than many other populations (Haiman et al., 2006). Ethnic differences in clearance of metabolites have been shown to contribute to the observed differences in cigarette consumption across populations, mediated in part by variants in the cytochrome p450 2A6 (CYP2A6) gene, but these alleles are less common in individuals of African-descent which makes this locus less likely to be responsible for increased lung cancer risk (Benowitz et al., 2011, Mwenifumbo et al., 2007, Moolchan et al., 2003).
We seek to examine the hypothesis that the association between SNPs in this region and lung cancer is moderated by smoking quantity, through conducting a candidate gene based analysis in African-American WHI SNP Health Association Resource (SHARe) participants to identify nominal SNPs linked to lung cancer that are associated with smoking quantity, and then to conduct gene × smoking quantity interaction analyses in participants from a multicenter case–control study of lung cancer.
2. Material and Methods
2.1. Study Population
The WHI SHARe study population consisted of 7156 females, of which 7097 were healthy and 59 had been diagnosed with lung cancer for whom genotype, smoking variables and lung cancer status were available. Additional details about the methods of the larger WHI Study (Stefanick et al., 2003, Langer et al., 2003) and the WHI SHARe cohort (Langer et al., 2003, David et al., 2012) have been previously reported. The study population analyzed for lung cancer susceptibility were from a previously-described multicenter case–control study designed to include African-American lung cancer cases and controls from three collaborating institutions: University of California, San Francisco (UCSF) (447 cases & 453 controls); Wayne State University (WSU) (459 cases & 460 controls) and the MD Anderson Cancer Center (MDA) (479 cases & 376 controls) (Walsh et al., 2013). The final analytic sample from the multicenter case–control study included 1078 cases and 822 controls. Never smokers were excluded from interaction analyses.
The genotype data were generated from DNA extracted from whole blood samples and imputed using 1000 Genomes (Genomes Project et al., 2012), downloaded from dbGaP (dbGaP accession #phs000200.v10.p3.c1; protocol #4723). The data available on dbGaP contained imputed genotype information along with variables of interest such as age, CPD, and other covariates. This investigation was approved by the Stanford University Institutional Review Board (IRB) (Stanford IRB #27009) and the IRBs of the Women's Health Initiative and all participating institutions of the multi-center case–control study.
2.2. SNP Selection
Based upon the evidence from GWAS and fine-mapping studies of African-Americans, we selected 28 SNPs in the chromosome 15p12.33 and 15q25.1 regions reported in one or more studies of African-Americans to be associated with risk of lung cancer and/or smoking quantity (CHRNA3 rs1051730 (Amos et al., 2010), rs10519203 (Amos et al., 2010, Spitz et al., 2013), rs578776 (Hansen et al., 2010), rs4243084 (Walsh et al., 2012), and rs8029939 (Hansen et al., 2010); CHRNA5 rs11637635 (Hansen et al., 2010), rs169969968 (Walsh et al., 2012, Chen et al., 2012), rs17408276 (Hansen et al., 2010), rs17486278 (Walsh et al., 2013, Hansen et al., 2010), rs17486195 (Walsh et al., 2013), rs2036527 (David et al., 2012, Walsh et al., 2013, Walsh et al., 2012), rs564585 (Hansen et al., 2010), rs667282 (David et al., 2012, Amos et al., 2010), rs684513 (Amos et al., 2010), 7180002 (Walsh et al., 2013), and rs951255 (Hansen et al., 2010, Walsh et al., 2013); IREB2 rs17405217 (Walsh et al., 2013, Walsh et al., 2012); LOC123688 rs11852372 (Hansen et al., 2010), rs7164594 (Amos et al., 2010, Hansen et al., 2010), and rs7168796 (Hansen et al., 2010); RORA rs8031948 (Amos et al., 2010); and on 5p15.33, TERT rs2735940 (Walsh et al., 2013) and rs4635969 (Walsh et al., 2013). We also included SNPs associated with smoking quantity that achieved (CHRNA5 rs2036527) or approached genome-wide significance from the STOMP meta-GWAS (CHRNA5 rs667282, CHRNA3 rs938682, C1orf100 rs3101457, LOC503519 rs547843 and PSMA4 rs3813570) (David et al., 2012).
2.3. Statistical Analysis
2.3.1. Smoking Exposure Variables, Genotyping and Quality Control (QC) in WHI SHARe
All WHI SHARe samples were genotyped at the Fred Hutchinson Cancer Research Center using Affymetrix 6.0 arrays. SNPs with call-rates < 95%, < 1% minor allele frequency or significant (P < 10− 6) departure from Hardy–Weinberg equilibrium were excluded, as were individuals with excess autosomal heterozygosity, mismatch between reported and genetically determined sex, or first- or second-degree relatedness. Smokers were individuals reporting having smoked at least 100 cigarettes in their lifetime. Current smokers were individuals who reported smoking at their baseline assessment. Cigarettes per day, a categorical variable in WHI's dataset (< 1, 1–4, 5–14, 15–24, 25–34, 35–44, > 45), were converted to a quasi-continuous variable by using the midpoint of each level of the original variable as its continuous value, e.g., the 1–4 cigarette per day category would have a continuous value of 2.5. Women in the < 1 category were assigned 0 cigarettes per day and women who reported smoking > 45 were assigned 50. Pack-years were calculated as the number of cigarettes per day multiplied by the duration of a subject's smoking habit divided by 20 (David et al., 2012).
2.3.2. Analysis of WHI SHARe Samples
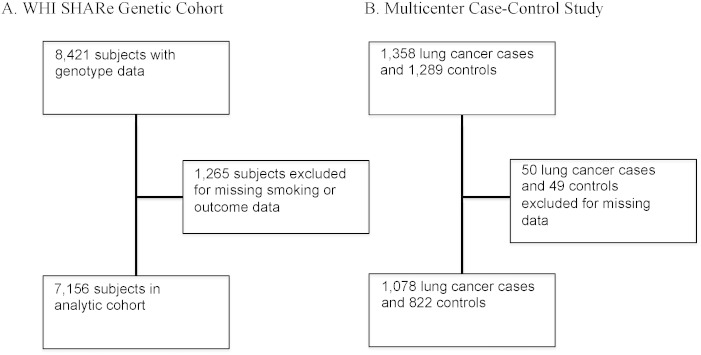
WHI genotype data were downloaded from dbGaP files. Prior to dbGaP deposition, principal components analysis and frappe analysis were performed for all genotyped SNPs first on all WHI samples and then towards African-American samples separately. Principal components analysis (PCA) was applied to African, European, Native American and East Asian separately for five iterations to remove self-identified African-American individuals with less than 20% African, resulting in an average of 80% African ancestry (David et al., 2012). Eigenstrat computes the eigenvectors (principle components) of a set of independent SNPs. These principal components were used as covariates in all phenotype association analyses (Price et al., 2006). Frappe is a model-based clustering program that estimates ancestry proportions, determining what percentage of the genome for any African-American individual is African and what percentage is European (Tang et al., 2005). Genome-wide Complex Trait Analysis (GCTA) was applied to the genotyped SNPs having a quality score greater than 0.98 in the 1000 Genome imputed data (Yang et al., 2011). SNPs with quality scores ≤ 0.98 were excluded (David et al., 2012). Fig. 1 provides details on the sample size adjustments for subjects removed for reasons of admixture, or missing phenotype data. From WHI SHARe, 1265 subjects out of 8421 genotyped subjects (15%) were excluded for missing smoking or outcomes data or because of non-African ancestry (defined above), of which 213 (2.5%) for non-African ancestry outliers.
Fig. 1.
Sample size flow chart.
Legend: Description of analytic sample with available genetic, smoking and lung cancer data for (A) Women's Health Initiative (WHI) Single Nucleotide Polymorphism Health Association Resource (SHARe) and (B) Multicenter Lung Cancer Case–Control Study. There were 8421 women in the downloaded dbGaP data for WHI SHARe (dbGaP accession #4723: Gene by environment interactions for lung cancer in cohort of African-American women in Women's Health Initiative), and 1265 were excluded for missing salient variables. The final WHI SHARe cohort included 7156 women with 59 cases of lung cancer and 29 cases of lung cancer mortality. There were 1,358 lung cancer cases and 1,289 controls in the multicenter case–control study of lung cancer. We excluded 50 lung cancer cases and 419 controls because of missing data, leaving 1,308 lung cancer cases and 1,241 controls as the final study population. When modeling interactions, never-smokers were excluded, leaving 1078 cases and 822 controls.
PCA was performed on the WHI genotype data and the top principal components were subsequently used as covariates to adjust for admixture. For each SNP we fit linear regression models with CPD as the dependent variable and SNP genotypes as the independent variable while adjusting for age and the first five principal components. All SNPs were coded using an additive risk scheme with the minor allele as the risk allele. Our results were consistent in a sensitivity analysis using only the first principal component and using the first 20 principal components.
As a secondary analysis, we refit the cigarette exposure models using pack-years as the dependent variable. We addressed multiple testing issues by adjusting the family-wise error rate using the Bonferroni method. The p-values from the models for the fit to WHI data were Bonferroni adjusted separately from the p-values resulting from the case–control data.
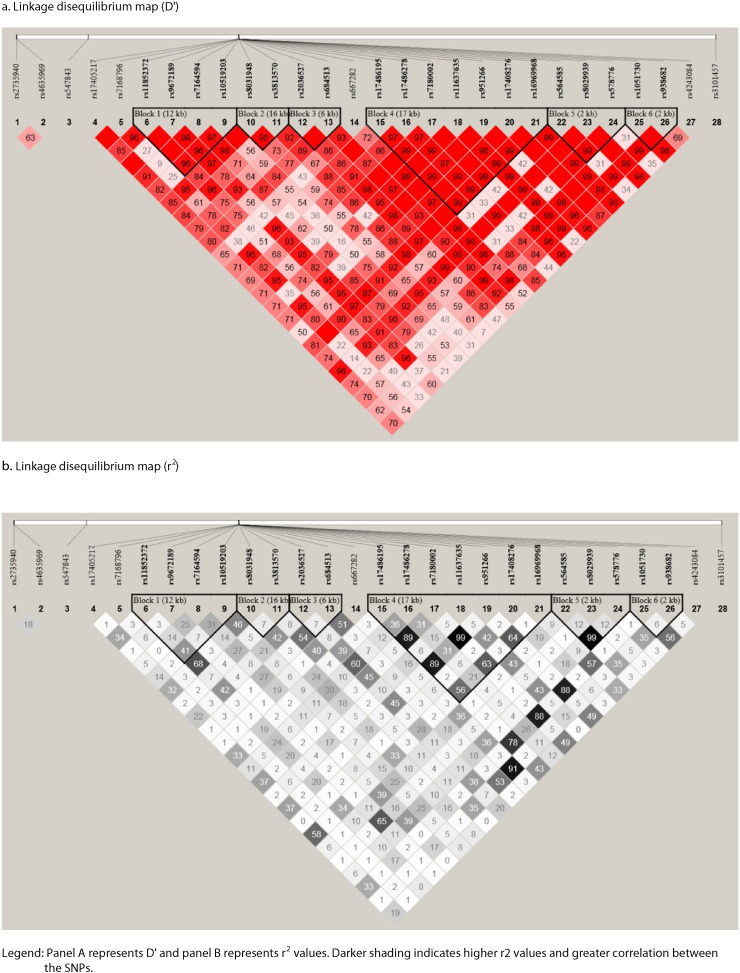
A haplotype map was generated including the top SNPs in order to determine D′ and r2 between the chromosome 15q25.1 SNPs associated with CPD to assess degree of linkage disequilibrium between potentially correlated loci in the same genomic region (Fig. 2).
Fig. 2.
Patterns of linkage disequilibrium in the chromosome 15q24–25.1 region.
Legend: Panel a represents D’ and panel b represents r2 values. Darker shading indicates higher r2 values and greater correlation between the SNPs.
2.3.3. Smoking Exposure, Lung Cancer Variables, Genotyping and QC in Multicenter Case–Control Study
UCSF samples were genotyped at the UCSF Genome Center, WSU samples were genotyped at the WSU Applied Genomics Technology Center, and MDA samples were genotyped at the MDA Cancer Center — all centers using the same Illumina Golden Gate Custom panel of 1536 SNPs (Walsh et al., 2013). Genotype reproducibility was verified with 7 duplicate samples, with concordance ranging from 99.93% to 100%. For all study sites, samples with genotyping call-rates less than 95% were excluded from analyses. SNPs with call-rates less than 95% in more than one study sites were excluded for analyses. To exclude poorly genotyped SNPs, any SNP with Hardy–Weinberg Equilibrium P value < 1.0 × 10− 4 in controls, stratified by site, was removed from analysis. All SNP quality control was carried out using Plink v1.07 (Purcell et al., 2007).
Cancer histology was determined using ICD-O codes abstracted from Surveillance Epidemiology and End Results (SEER) data from the California Cancer Registry (UCSF cases) or Detroit Cancer Registry (WSU cases). For MD-Anderson cases, histology was determined by extraction from medical records. The following ICD-O groupings were made: adenocarcinoma (ICD-O: 8140, 8230, 8250–8255, 8260, 8310, 8333, 8470, 8480, 8481, 8490, and 8550), squamous cell carcinoma (8052, 8070–8073, 8083, and 8084), and small cell carcinoma (8041–8045) (Walsh et al., 2013). Each of these diagnostic subtypes of lung cancer were pooled as a singular ‘lung cancer’ diagnosis for the purposes of the present investigation.
2.3.4. Analysis of Multicenter Case–Control Study Samples
Structure v2.3.1 was used to estimate percentage of membership in 3 distinct founder populations: sub-Saharan African, European, and East Asian, with East Asian ancestry as a proxy for American Indian descent (International HapMap C, 2003). Founder population allele frequencies were defined using SNP data from 102 unlinked (r2 < 0.20) ancestry informative markers, genotyped in 502 unrelated HapMap individuals (167 Yoruban Africans, 165 Europeans, 84 Chinese, and 86 Japanese). These same AIMs were genotyped in study participants for use with the Structure program (Walsh et al., 2013, Pritchard et al., 2000). PCA was performed on the multicenter case control genotype data and the top principal components were subsequently used as covariates to adjust for admixture. For the 9 available SNPs with the smallest p-values on the association between CPD and SNP, we fit logistic regression models where probability of lung cancer incidence was modeled with a logit link. These models were all adjusted for age, sex, study site, and percent African ancestry using first five principal components. As a secondary analysis, we refit the cigarette exposure model using pack-years in place of CPD. All models were refit with an interaction term between SNP and CPD or SNP and pack-years using the case–control data.
All SNP quality control was carried out using Plink v1.07 (Purcell et al., 2007). Haploview was used to generate haplotype maps (Fig. 1) (Barrett et al., 2005). WHI SHARe analyses were conducted in R 3.1 (R Foundation for Statistical Computing, Vienna, Austria) (R Foundation for Statistical Computing, 2013). The analyses on the samples from the multicenter case–control study were conducted in SAS v9. P-values < 0.05 were considered to be statistically significant.
3. Results
The WHI SHARe genetic cohort included N = 8421 female individuals. After excluding women without genotype data and women who were missing relevant variables, 7156 women remained (Fig. 1), of which 2765 (39%) were former smokers, 779 (11%) were current smokers and 3612 (50%) were never smokers. The mean age of participants was 61 years (standard deviation (SD) = 6.8) for healthy and 63.6 years (SD = 6.6) for lung cancer cases. The multicenter case–control study analytic cohort included N = 1900 female and male individuals, of which 934 (49%) were female, of which 751 (39%) were ever smokers, 778 (40.9%) were current smokers and 295 (15%) were former smokers. The mean age of participants was 62 years (standard deviation (SD) = 10) for healthy and 62 years (SD = 10) for lung cancer cases. Additional details including smoking history and demographics are described in Table 1 with extended descriptions published elsewhere (David et al., 2012, Walsh et al., 2013).
Table 1.
Participant characteristics.
| Total (%) |
Female (%) |
Male (%) |
Never smokers (%) |
Current smokers (%) |
Former smokers (%) | Age, mean (SD) | Age of onset, mean (SD) | CPD, mean (SD) |
|---|---|---|---|---|---|---|---|---|
| WHI SHARe | ||||||||
| Healthy | ||||||||
| N = 7097 (99.2%) | 7097 (100.0%) | – | 3,598 (50.7%) | 761 (10.7%) | 2,738 (38.6%) | 61.0 (6.8) | 20.9 (5.4) | 5.5 (8.5) |
| Lung cancer | ||||||||
| N = 59 (0.8%) | 59 (0.08%) | – | 14 (23.7%) | 18 (30.5%) | 27 (45.8%) | 63.6 (6.6) | 19.5 (4.1) | 14.4 (12.4) |
| UCSF, MD Anderson & Wayne State University Case–Control | ||||||||
| Healthy | ||||||||
| N = 822 (43.3%) | 404 (49.1%) | 418 (50.9%) | NA | 435 (52.9%) | 387 (47.1%) | 60.5 (10.2) | 16.9 (4.6) | 17.9 (13.9) |
| Lung cancer | ||||||||
| N = 1078 (56.7%) | 530 (49.2%) | 548 (50.8%) | NA | 660 (61.2%) | 418 (38.8%) | 63.0 (10.3) | 17.6 (4.9) | 20.2 (13.1) |
Legend: Abbreviations: WHI = Women's Health Initiative; SHARe = Single Nucleotide Polymorphism Health Association Resource; SD = standard deviation; % = percent healthy or lung cancer.
Note. The WHI study included only females. Never-smokers were not included in analyses of lung cancer outcomes.
The results of analyses of all included SNPs on CPD in the WHI SHARe sample are presented in Table 2. In the models with CPD as their outcome in the WHI SHARe cohort, 1 SNP was found to be associated with CPD following Bonferroni adjustment), rs1051730 (adjusted p = 0.027). For this SNP, each additional A allele increased subjects' expected number of CPD by 0.081 cigarettes. In addition, 10 other SNPs were nominally significant for association with CPD but not statistically significant following Bonferroni correction.
Table 2.
SNPs analyzed for association with cigarettes per day in WHI SHARe.
| SNP | Chromosome (base-pair) position | Nearby genes | Alleles | Coded Allele | Coded AF | ß (s.e.) | P-value | P-value (adjusted)* |
|---|---|---|---|---|---|---|---|---|
| rs1051730 | 15:78601997 | CHRNA3 | G/A | A | 0.13145 | 0.081 (0.029) | 0.00095 | 0.02669 |
| rs7180002 | 15:78581651 | CHRNA5 | A/T | T | 0.11941 | 0.072 (0.034) | 0.00307 | 0.08597 |
| rs951266 | 15:78586199 | CHRNA5 | G/A | A | 0.11944 | 0.069 (0.035) | 0.00383 | 0.10718 |
| rs2036527 | 15:78559273 | CHRNA5 | G/A | A | 0.22451 | 0.077 (0.029) | 0.00388 | 0.10872 |
| rs17486278 | 15:78575140 | CHRNA5 | A/C | C | 0.28691 | 0.072 (0.026) | 0.00492 | 0.13772 |
| rs16969968 | 15:78590583 | CHRNA5 | G/A | A | 0.08021 | 0.059 (0.036) | 0.00725 | 0.20295 |
| rs4243084 | 15:78619330 | CHRNA3 | G/C | C | 0.20197 | 0.072 (0.028) | 0.00745 | 0.20854 |
| rs17405217 | 15:78438807 | IREB2 | C/T | T | 0.09058 | 0.056 (0.033) | 0.01213 | 0.33967 |
| rs547843 | 15:26178900 | LOC503519 | C/G | G | 0.35954 | 0.068 (0.029) | 0.01740 | 0.48725 |
| rs938682 | 15:78604205 | CHRNA3 | A/G | G | 0.28587 | − 0.049 (0.025) | 0.02642 | 0.73971 |
| rs11852372 | 15:78509052 | HYKK | A/C | C | 0.16997 | 0.040 (0.029) | 0.03982 | 1 |
| rs478776 | 15:78596058 | CHRNA3 | A/G | G | 0.47358 | 0.029 (0.024) | 0.05376 | 1 |
| rs8031948 | 15:78523715 | HYKK | G/T | T | 0.18103 | 0.043 (0.028) | 0.05791 | 1 |
| rs10519203 | 15:78521704 | HYKK | A/G | G | 0.32025 | 0.042 (0.025) | 0.11335 | 1 |
| rs564585 | 15:78593885 | CHRNA3 | G/A | A | 0.47403 | 0.016 (0.026) | 0.12637 | 1 |
| rs17408276 | 15:78589276 | CHRNA5 | T/C | C | 0.1458 | − 0.062 (0.029) | 0.19065 | 1 |
| rs7164594 | 15:78510715 | HYKK | C/T | T | 0.40943 | − 0.020 (0.024) | 0.19103 | 1 |
| rs11637635 | 15:78584808 | CHRNA5 | G/A | A | 0.28379 | − 0.044 (0.026) | 0.19106 | 1 |
| rs3101457 | 1:244369912 | C1orf100 | A/G | G | 0.2478 | 0.072 (0.049) | 0.23023 | 1 |
| rs3813570 | 15:78540490 | PSMA4 | T/C | C | 0.27113 | − 0.027 (0.027) | 0.28473 | 1 |
| rs2735940 | 5:1296371 | TERT | G/A | A | 0.49586 | − 0.028 (0.033) | 0.32441 | 1 |
| rs17486195 | 15:78572855 | CHRNA5 | A/G | G | 0.13368 | 0.012 (0.029) | 0.33791 | 1 |
| rs8029939 | 15:78596007 | CHRNA3 | G/A | A | 0.12207 | 0.053 (0.035) | 0.34905 | 1 |
| rs4635969 | 5:1308437 | TERT | G/A | A | 0.31211 | 0.031 (0.025) | 0.40241 | 1 |
| rs667282 | 15:78571130 | CHRNA5 | T/C | C | 0.2945 | − 0.024 (0.026) | 0.48116 | 1 |
| rs7168796 | 15:78508152 | HYKK | T/C | C | 0.16283 | 0.048 (0.031) | 0.52338 | 1 |
| rs9672189 | 15:78509054 | HYKK | A/C | C | 0.15226 | 0.030 (0.028) | 0.69760 | 1 |
| rs684513 | 15:78566058 | CHRNA5 | C/G | G | 0.13145 | − 0.013 (0.031) | 0.89055 | 1 |
Legend: Abbreviations: AF, allele frequency; SNP, single-nucleotide polymorphism. Coded AF refers to the allele analyzed as the predictor allele; it is not necessarily the minor allele. All SNPs coded to NCBI Build 38/UCSC hg38 forward strand. *Bonferronni-adjusted P-value. Beta (ß) and standard error (s.e.) for change in CPD per increase in additive risk (increase in presence of coded allele.
Given the low number of lung cancer cases in the WHI sample (n = 86), we sought to conduct interaction analyses for the top 10 nominal SNPs for CPD from the WHI SHARe population in the multicenter lung cancer case–control study population described above. Genotype data were available for nine of these SNPs, with no data available for rs547843. The results for analyses of main effects of nominally significant WHI SHARe CPD SNPs on lung cancer in the multicenter case–control study are presented in Table 3. All nine SNPs were statistically significantly associated with lung cancer (Bonferroni-corrected P-values from 0.027 to 6.09 × 10− 5). There were six SNPs that demonstrated nominally significant interactions with CPD for risk of incident lung cancer (Table 4), two of which approached statistical significance after Bonferroni correction: rs2036527[A], beta = − 0.0171, p = 0.0549; rs7180002, beta = − 0.0205, p = 0.0576.
Table 3.
Associations between Nominal SNPs associated with cigarettes per day in WHI SHARe and lung cancer in African Americans in WHI SHARe and multicenter case–control study.
| Gene | SNP | ß (s.e.) | P | Adj. P⁎ |
|---|---|---|---|---|
| CHRNA5 | rs17486278 | 0.295 (0.061) | 1.66 × 10− 06 | 1.49 × 10− 05 |
| CHRNA5 | rs2036527 | 0.302 (0.067) | 6.77 × 10− 06 | 6.09 × 10− 5 |
| CHRNA3 | rs1051730 | 0.374 (0.085) | 1.18 × 10− 05 | 1.06 × 10− 04 |
| CHRNA5 | rs16969968 | 0.451 (0.115) | 5.43 × 10− 05 | 4.89 × 10− 04 |
| CHRNA5 | rs7180002 | 0.344 (0.088) | 9.57 × 10− 05 | 8.61 × 10− 4 |
| CHRNA3 | rs4243084 | 0.278 (0.071) | 9.86 × 10− 05 | 8.87 × 10− 04 |
| CHRNA5 | rs951266 | 0.333 (0.088) | 0.00015 | 0.001 |
| CHRNA3 | rs938682 | − 0.192 (0.065) | 0.00301 | 0.027 |
| IREB2 | rs17405217 | 0.339 (0.115) | 0.0032 | 0.029 |
Legend: SNP AFs in multicenter case–control reported elsewhere); (Walsh et al., 2013) Beta (ß) and standard error (s.e.) for change in CPD per increase in additive risk (increase in presence of coded allele) for WHI SHARe and for lung cancer risk in Multicenter Case–Control Study of Lung Cancer.
Bonferroni-adjusted p-value. The p-values were adjusted by a factor of 9. Although nominally associated with CPD in WHI SHARe, rs547843 was not genotyped in the Multi-Center Case–Control Study.
Table 4.
Interaction analyses for incident lung cancer.
| Pack-year × SNP interaction |
CPD × SNP interaction |
||||||
|---|---|---|---|---|---|---|---|
| rsID | Effect allele | ß (s.e.)SNP⁎PackYrs | PSNP⁎PackYears | Bonferroni P | ß (s.e.)SNP⁎CPD | PSNP⁎CPD | Bonferroni P |
| rs2036527 | A | − 0.0029 (0.004) | 0.4448 | 1 | − 0.0171 (0.006) | 0.0061 | 0.0549 |
| rs7180002 | T | − 0.0026 (0.005) | 0.5878 | 1 | − 0.0205 (0.008) | 0.0064 | 0.0576 |
| rs17486278 | C | − 0.0037 (0.003) | 0.2738 | 1 | − 0.0140 (0.006) | 0.0081 | 0.0729 |
| rs951266 | A | − 0.0022 (0.005) | 0.6585 | 1 | − 0.0197 (0.008) | 0.0089 | 0.0801 |
| rs1051730 | A | − 0.004 (0.005) | 0.4418 | 1 | − 0.0183 (0.007) | 0.0112 | 0.1008 |
| rs17405217 | T | < 0.001 (0.007) | 0.9546 | 1 | − 0.0251 (0.010) | 0.0136 | 0.1224 |
| rs16969968 | A | 0.0057 (0.007) | 0.4049 | 1 | − 0.018 (0.010) | 0.0754 | 0.6786 |
| rs4243084 | C | 0.0024 (0.004) | 0.5524 | 1 | − 0.008 (0.006) | 0.1959 | 1 |
| rs938682 | G | − 0.003 (0.004) | 0.4581 | 1 | − 0.0035 (0.006) | 0.5765 | 1 |
Legend: Abbreviations: AF = allele frequency; CPD = cigarettes per day; s.e. = standard error; SNP = single-nucleotide polymorphism.
Bonferroni-adjusted p-value. The p-values were adjusted by a factor of 9.
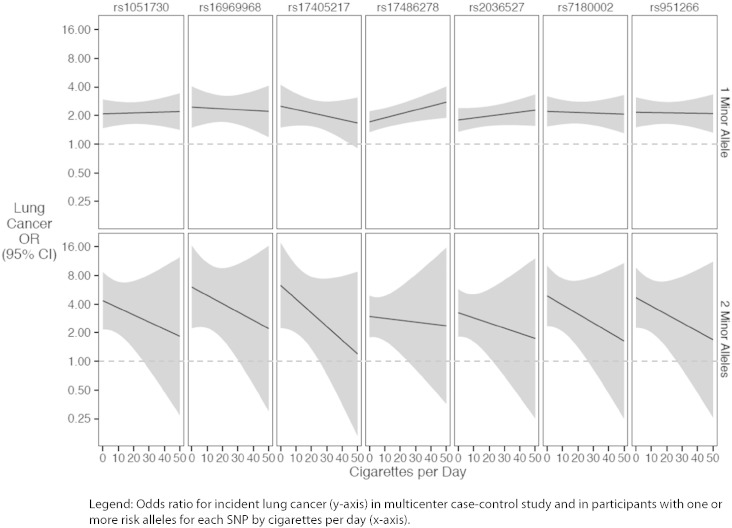
Fig. 3 illustrates the allele dose response relationships for each of these SNPs with incident lung cancer as estimated in the interaction models. The nature of each of these interactions was notable for a pattern suggesting – but not establishing, given that the interactions were not statistically significant after Bonferroni correction – stronger allele dose-responses for individuals who smoked fewer CPD. Odds ratios for lung cancer risk by average CPD are presented in Supplementary Materials.
Fig. 3.
Interaction between SNPs and cigarettes per day and risk of lung cancer.
Legend: Odds ratio for incident lung cancer (y-axis) in multicenter case–control study and in participants with one or more risk alleles for each SNP by cigarettes per day (x-axis).
4. Discussion
To our knowledge, there have been at least four fine-mapping case control studies of lung cancer in African Americans that have examined the chromosome 15q25.1 locus and additional loci on chromosomes 5p15.33 and 6p22.1–21.31 (Walsh et al., 2013, Amos et al., 2010, Spitz et al., 2013, Hansen et al., 2010). One study of 1058 cases and 1314 controls from the Detroit area Surveillance, Epidemiology, and End Results (SEER) registry found that SNP rs1051730 on chromosome 15q25.1 was associated with lung cancer in African-American ever-smokers (Schwartz et al., 2009) — findings similar to a larger study of European-ancestry cases (n = 1024) and controls (n = 32,244) (Thorgeirsson et al., 2008). Another study identified multiple SNPs and a haplotype within the chromosome 15q25.1 region with lung cancer in 448 African-American lung cancer cases and 611 controls, which suggests that SNPs in this region affecting expression of the alpha 5 (CHRNA5), alpha 3 (CHRNA3) and beta 4 (CHRNB4) nicotinic acetylcholine receptor genes may be independently associated with lung cancer (Amos et al., 2010, Hansen et al., 2010).
In a previously published meta-analysis using data from the WHI SHARe cohort (n = 8208) and twelve other study groups forming the Study of Tobacco Use in Minority Populations (STOMP) Genetics Consortium (N = 32,829 (David et al., 2012), rs2036527 was associated with CPD in African-Americans (David et al., 2012), which has subsequently been shown to portend increased potential benefit from smoking cessation treatment in treatment-seeking African-Americans smokers (Zhu et al., 2014). The s2036527 SNP is located in the 5′ distal enhancer region of the CHRNA5 gene, which forms a haplotype associated with increased CHRNA5 expression (Smith et al., 2011) and which has been associated with lung cancer in three fine-mapping studies of African Americans (Walsh et al., 2013, Amos et al., 2010, Hansen et al., 2010). However, mechanisms conferring increased risk for any of the highly correlated SNPs in this region are complex and may involve epigenetic effects on the CHRNB4 and TERT genes (Scherf et al., 2013) and decreased apoptosis in tumor cells resulting from over-expression of the PSMA4 gene, which has been suggested for rs2036527 (Liu et al., 2009).
The functional SNP rs16969968, which results in an amino acid change conferring increased alpha 5 nicotinic acetylcholine receptor expression (Bierut et al., 2008), was associated with CPD but was not reach nominal significance for effect modification of CPD for lung cancer. Rs1051730 was also associated with CPD. Although rs169969968 and rs1051730 are highly correlated with each other in European-ancestry populations (r2 = 1), they are less correlated in African-ancestry (r2 = 0.4) (HapMap3 Release 2 ASW). Both SNPs are rare or monomorphic in sub-Saharan Africans and African-Americans (David et al., 2012, Chen et al., 2012), and in the US-based WHI SHARe study population the minor alleles for both SNPs were relatively infrequent (mean allele frequencies for rs169969968 and rs1051730 are 0.08 and 0.13, respectively) compared to rs2036527 (MAF = 0.22) (Table 2). Our previous results indicated that rs2036527 was the primary index signal for CPD in African-Americans (David et al., 2012) after conditioning on this SNP. The STOMP meta-GWAS results suggested that the secondary SNP signals for CPD in African-Americans were likely the result of high linkage disequilibrium, indicating that the rs2036527 variant may be the most informative marker of heightened smoking exposure in African-Americans. Of note, neither rs16969968 nor rs1051730 approached genome-wide significance in the meta-analyses of over 32,000 African-Americans. Alternatively, less robust associations for these SNPs in African-Americans could also be influenced by reduced power resulting from diminished linkage disequilbrium smaller haplotype blocks and lower minor allele frequencies.
As mentioned above, only 11 of 29 SNPs tested were nominally associated with CPD. Lack of statistical significance for CPD for many of these variants could be explained either by small effect sizes for this phenotype, requiring larger sample sizes to detect and the possibility that some variants previously linked to lung cancer but not to CPD (e.g., HYKK rs11852372) (Fehringer et al., 2012) could operate through mechanisms other than increased smoking quantity to confer higher lung cancer susceptibility. The present study was unable to address this question.
Our hypothesis was that if polymorphisms associated with altered CHRNA3-A5-B4 expression concomitantly moderated both increased cigarette consumption and intrinsic risk for lung cancer, that gene by environment (smoking quantity) interactions would be additive. However, these data suggest a different scenario. The magnitude of increased risk associated with one or two risk alleles appeared to attenuate in individuals smoking more CPD, even though there was a positive relationship between CPD and risk of lung cancer. For example, individuals who smoked on average 5 CPD had 80% higher odds of developing lung cancer if they possessed one A risk allele for rs2036527 (odds ratio (OR) = 1.84; 95% confidence interval (CI) 1.41, 2.40), whereas they were 200% more likely to develop lung cancer if they possessed two A alleles (OR = 3.05, 95% CI: 1.78, 5.19). However, an individual who smoked 20 CPD was about twice as likely to develop lung cancer if they had one A allele (OR = 1.96, 95% CI: 1.56, 2.51) but slightly more than twice as likely to develop lung cancer if they had two risk alleles (OR = 2.53, 95% CI: 1.16, 5.62).
It is noteworthy that in our data, diminishing odds ratios for lung cancer with increasing smoking intensity were only observed for individuals with the highest genetic risk (i.e., possessing two risk alleles). Reduced effects of increased smoking intensity on lung cancer risk are a phenomenon that has been modeled in other studies (Lubin et al., 2007a), and in multiple tobacco-attributable cancers (Lubin et al., 2007b), that included studies of African-Americans. It has been shown that there is reduced exposure to N′-nitrosonornicotine (NNK) and its metabolites (NNAL) at higher levels of cotinine exposure (Lubin et al., 2007c, Lubin and Caporaso, 2006).
Mechanisms of increased lung cancer susceptibility independent of smoking quantity for individuals with chromosome 15q25.1 high-risk genotypes are not entirely clear. Multiple studies have demonstrated associations between SNPs in chromosome 15q25.1 and lung cancer in nonsmokers (Hung et al., 2008, Shiraishi et al., 2009). Functional studies have shown that a gene in this region (PSMA4) has been associated with cancer cell proliferation and apoptosis (Liu et al., 2009) and another gene (IREB2) (Fehringer et al., 2012) has been associated with lung cancer risk. A SNP (rs3813570) in the PSMA4 gene approached genome-wide significance in the STOMP Consortium meta-analysis and a variant in IREB2 (rs17405217) was associated with lung cancer risk in the present investigation and others (Walsh et al., 2013, Walsh et al., 2012). It is therefore possible that the high degree of linkage disequilibrium between chromosome 15q25.1 variants in cholinergic genes associated with increased smoking intensity and those in neighboring genes linked to tumor proliferation may result in frequent haplotypes capturing both heightened cigarette exposure and susceptibility to lung cancer pathogenesis. However, additional research is needed to confirm mechanisms of potential concomitant dual risk of exposure and cancer susceptibility.
It is worth noting that pack-years were not associated with the genotypes tested and this variable did not interact with genotype to predict lung cancer risk in either study population. While this may be the result of a combination of lack of sensitivity due to recall bias and sample size (Castaldi et al., 2011), a previously published study of smoking persistence in African-Americans examined pack-years of exposure, accounting for periods of non-smoking, and reported multiple statistically significant associations with SNPs in the 15q25.1 region (Hamidovic et al., 2011). Thus, it is not entirely clear why this measure of smoking exposure was not robust in our investigation. However, given pre-clinical evidence of much higher nicotine self-administration in CHRNA5 knockout mice (Fowler et al., 2011, Fowler et al., 2013, Fowler and Kenny, 2014), it is possible that humans with altered CHRNA5 expression possess a phenotype of markedly greater alveolar tobacco smoke exposure vis-à-vis increased smoking quantity, intensity and puff volume over a lifetime (Ware et al., 2012), which in turn portends greater overall exposure than can be captured with granularity using the measured phenotypes of pack-years or average CPD.
This study has some limitations. While we did have sufficient statistical power in an a priori power analysis to detect effects sizes of at least 2.1 (assuming power > .8, allele frequencies ≥ 0.15 and at least 85 lung cancer cases), in analyses not corrected for multiple comparison, adjusting for multiple comparisons may limited our ability to detect statistically-significant effects robust to multiple corrections. We did not have access to spirometry or diagnostic data for conditions such as COPD and emphysema to enable analyses of potential mediating effects of smoking quantity on the relationship between the CHRNA3-A5-B4 locus and lung cancer risk, which has been demonstrated in European-ancestry studies (El-Zein et al., 2012). Another limitation of the present investigation is that the WHI SHARe analyses did not include males. However, we previously published a genome-wide meta-analyses of smoking quantity that included both genders of African-Americans that confirmed rs2036527 in association with CPD after adjusting for gender (David et al., 2012), but nonetheless, we cannot rule out the possibility that other SNPs nominally associated with CPD in this investigation could be moderated by sex. In addition, we did not have information on type of cigarettes smoked such as menthol cigarettes, smoking topography or plasma cotinine levels. However, two earlier investigations of genetic predictors of lung cancer risk in African-Americans by Amos and colleagues did not show any differential effect of menthol on the relationship between SNP and lung cancer risk (Walsh et al., 2013, Amos et al., 2010).
5. Conclusions
Additional research is needed using larger sample sizes to conduct mediation analyses and including preclinical data to confirm that the increased lung cancer risk of associated SNPs is causally driven entirely by increased smoking intensity. However, our results add to the growing literature pointing towards rs2036527 as an informative polymorphism for smoking exposure and lung cancer risk in African-Americans, who may benefit from enhanced preventive interventions for smoking cessation treatment (Zhu et al., 2014) and genetically-informed lung cancer screening interventions (Young et al., 2012).
The following is the supplementary data related to this article.
Relationship between cigarettes smoked per day and risk of lung cancer in multicenter case–control study.
Role of the Funding Source
Funding — The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health (NIH), U.S. Department of Health and Human Services through contracts N01WH22110, 24152, 32100–2, 32105–6, 32108–9, 32111–13, 32115, 32118–32119, 32122, 42107–26, 42129–32, and 44221 (SPD, HH, KK, MD, LY, BAG, MS). Investigator support is also acknowledged from the NIH to R01CA14176, R01CA060691, N01PC35145, and contract HHSN261201000028C (AGS), and grants P30CA008748 (HF), U19CA148127 and R01CA141716 (CIA), R01ES06717 and R01CA52689 (MRW), R01CA55769 and R01CA127219 (MRS). The funder of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Declaration of Interests
SPD was a scientific advisor to and is a stockholder with BaseHealth. AWB is an associate with BioRealm.
Acknowledgments
SPD, CA, AW, MH and MD conceived study design, data and/or statistical analyses were conducted by HH, KK, KMW, CK, HT, MRS, AGS, ASW, JPQ, MRW. AGS was responsible for study design, data collection, analysis and data interpretation for the case–control study at Wayne State University. ASW was responsible for data collection and analysis for the case–control study at Wayne State University and review of the manuscript. Review of protocol and final manuscript was conducted by all other listed authors.
References
- Amos C.I., Gorlov I.P., Dong Q. Nicotinic acetylcholine receptor region on chromosome 15q25 and lung cancer risk among African Americans: a case-control study. J. Natl. Cancer Inst. 2010;102(15):1199–1205. doi: 10.1093/jnci/djq232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benowitz N.L., Dains K.M., Dempsey D., Wilson M., Jacob P. Racial differences in the relationship between number of cigarettes smoked and nicotine and carcinogen exposure. Nicotine Tob. Res. 2011;13(9):772–783. doi: 10.1093/ntr/ntr072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut L.J., Stitzel J.A., Wang J.C. Variants in nicotinic receptors and risk for nicotine dependence. Am. J. Psychiatry. 2008;165(9):1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms U., Silventoinen K., Madden P.A., Heath A.C., Kaprio J. Genetic architecture of smoking behavior: a study of Finnish adult twins. Twin Res. Hum. Genet. 2006;9(1):64–72. doi: 10.1375/183242706776403046. [DOI] [PubMed] [Google Scholar]
- Carmelli D., Swan G.E., Robinette D., Fabsitz R. Genetic influence on smoking–a study of male twins. N. Engl. J. Med. 1992;327(12):829–833. doi: 10.1056/NEJM199209173271201. [DOI] [PubMed] [Google Scholar]
- Castaldi P.J., Demeo D.L., Hersh C.P. Impact of non-linear smoking effects on the identification of gene-by-smoking interactions in COPD genetics studies. Thorax. 2011;66(10):903–909. doi: 10.1136/thx.2010.146118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.S., Saccone N.L., Culverhouse R.C. Smoking and genetic risk variation across populations of European, Asian, and African American ancestry - A meta-analysis of chromosome 15q25. Genet. Epidemiol. 2012;36(4):340–351. doi: 10.1002/gepi.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumpacker D.W., Cederlof R., Friberg L. A twin methodology for the study of genetic and environmental control of variation in human smoking behavior. Acta Genet. Med. Gemellol. 1979;28(3):173–195. doi: 10.1017/s0001566000009041. [DOI] [PubMed] [Google Scholar]
- David S.P., Hamidovic A., Chen G.K. Genome-wide meta-analyses of smoking behaviors in African Americans. Transl. Psychiatry. 2012;2 doi: 10.1038/tp.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves L.J., Silberg J.L., Hewitt J.K. Analyzing twin resemblance in multisymptom data: genetic applications of a latent class model for symptoms of conduct disorder in juvenile boys. Behav. Genet. 1993;23(1):5–19. doi: 10.1007/BF01067550. [DOI] [PubMed] [Google Scholar]
- Edwards K.L., Austin M.A., Jarvik G.P. Evidence for genetic influences on smoking in adult women twins. Clin. Genet. 1995;47(5):236–244. doi: 10.1111/j.1399-0004.1995.tb04303.x. [DOI] [PubMed] [Google Scholar]
- El-Zein R.A., Young R.P., Hopkins R.J., Etzel C.J. Genetic predisposition to chronic obstructive pulmonary disease and/or lung cancer: important considerations when evaluating risk. Cancer Prev. Res. (Phila.) 2012;5(4):522–527. doi: 10.1158/1940-6207.CAPR-12-0042. [DOI] [PubMed] [Google Scholar]
- Etzel C.J., Kachroo S., Liu M. Development and validation of a lung cancer risk prediction model for African-Americans. Cancer Prev. Res. (Phila.) 2008;1(4):255–265. doi: 10.1158/1940-6207.CAPR-08-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehringer G., Liu G., Pintilie M. Association of the 15q25 and 5p15 lung cancer susceptibility regions with gene expression in lung tumor tissue. Cancer Epidemiol. Biomark. Prev. 2012;21(7):1097–1104. doi: 10.1158/1055-9965.EPI-11-1123-T. [DOI] [PubMed] [Google Scholar]
- Fisher R.A. Cancer and moking. Nature. 1958;182(4635):596. doi: 10.1038/182596a0. [DOI] [PubMed] [Google Scholar]
- Fisher R.A. Lung cancer and cigarettes. Nature. 1958;182(4628):108. doi: 10.1038/182108a0. [DOI] [PubMed] [Google Scholar]
- Fowler C.D., Kenny P.J. Nicotine aversion: Neurobiological mechanisms and relevance to tobacco dependence vulnerability. Neuropharmacology. 2014;76(Pt B):533–544. doi: 10.1016/j.neuropharm.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler C.D., Lu Q., Johnson P.M., Marks M.J., Kenny P.J. Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature. 2011;471(7340):597–601. doi: 10.1038/nature09797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler C.D., Tuesta L., Kenny P.J. Role of alpha5* nicotinic acetylcholine receptors in the effects of acute and chronic nicotine treatment on brain reward function in mice. Psychopharmacology. 2013 doi: 10.1007/s00213-013-3235-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C., Abecasis G.R., Auton A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman C.A., Stram D.O., Wilkens L.R. Ethnic and racial differences in the smoking-related risk of lung cancer. N. Engl. J. Med. 2006;354(4):333–342. doi: 10.1056/NEJMoa033250. [DOI] [PubMed] [Google Scholar]
- Hamidovic A., Kasberger J.L., Young T.R. Genetic variability of smoking persistence in African Americans. Cancer Prev. Res. (Phila.) 2011;4(5):729–734. doi: 10.1158/1940-6207.CAPR-10-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Gelernter J., Luo X., Yang B.Z. Meta-analysis of 15 genome-wide linkage scans of smoking behavior. Biol. Psychiatry. 2010;67(1):12–19. doi: 10.1016/j.biopsych.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah M.C., Hopper J.L., Mathews J.D. Twin concordance for a binary trait. II. Nested analysis of ever-smoking and ex-smoking traits and unnested analysis of a "committed-smoking" trait. Am. J. Hum. Genet. 1985;37(1):153–165. [PMC free article] [PubMed] [Google Scholar]
- Hansen H.M., Xiao Y., Rice T. Fine mapping of chromosome 15q25.1 lung cancer susceptibility in African-Americans. Hum. Mol. Genet. 2010;19(18):3652–3661. doi: 10.1093/hmg/ddq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath A.C. Persist or quit? Testing for a genetic contribution to smoking persistence. Acta Genet. Med. Gemellol. 1990;39(4):447–458. doi: 10.1017/s0001566000003676. [DOI] [PubMed] [Google Scholar]
- Heath A.C., Cates R., Martin N.G. Genetic contribution to risk of smoking initiation: comparisons across birth cohorts and across cultures. J. Subst. Abus. 1993;5(3):221–246. doi: 10.1016/0899-3289(93)90065-j. [DOI] [PubMed] [Google Scholar]
- Hirschhorn J.N., Lohmueller K., Byrne E., Hirschhorn K. A comprehensive review of genetic association studies. Genet. Med. 2002;4(2):45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Hung R.J., McKay J.D., Gaborieau V. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- International HapMap C The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Kaprio J., Koskenvuo M., Langinvainio H. Finnish twins reared apart. IV: Smoking and drinking habits. A preliminary analysis of the effect of heredity and environment. Acta Genet. Med. Gemellol. 1984;33(3):425–433. doi: 10.1017/s0001566000005870. [DOI] [PubMed] [Google Scholar]
- Langer R.D., White E., Lewis C.E., Kotchen J.M., Hendrix S.L., Trevisan M. The Women's Health Initiative Observational Study: baseline characteristics of participants and reliability of baseline measures. Ann. Epidemiol. 2003;13(9 Suppl):S107–S121. doi: 10.1016/s1047-2797(03)00047-4. [DOI] [PubMed] [Google Scholar]
- Lessov C.N., Martin N.G., Statham D.J. Defining nicotine dependence for genetic research: evidence from Australian twins. Psychol. Med. 2004;34(5):865–879. doi: 10.1017/s0033291703001582. [DOI] [PubMed] [Google Scholar]
- Li M.D., Cheng R., Ma J.Z., Swan G.E. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98(1):23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu P., Wen W. Haplotype and cell proliferation analyses of candidate lung cancer susceptibility genes on chromosome 15q24-25.1. Cancer Res. 2009;69(19):7844–7850. doi: 10.1158/0008-5472.CAN-09-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller K.E., Pearce C.L., Pike M., Lander E.S., Hirschhorn J.N. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 2003;33(2):177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- Lubin J.H., Caporaso N.E. Cigarette smoking and lung cancer: modeling total exposure and intensity. Cancer Epidemiol. Biomark. Prev. 2006;15(3):517–523. doi: 10.1158/1055-9965.EPI-05-0863. [DOI] [PubMed] [Google Scholar]
- Lubin J.H., Caporaso N., Wichmann H.E., Schaffrath-Rosario A., Alavanja M.C. Cigarette smoking and lung cancer: modeling effect modification of total exposure and intensity. Epidemiology. 2007;18(5):639–648. doi: 10.1097/EDE.0b013e31812717fe. [DOI] [PubMed] [Google Scholar]
- Lubin J.H., Alavanja M.C., Caporaso N. Cigarette smoking and cancer risk: modeling total exposure and intensity. Am. J. Epidemiol. 2007;166(4):479–489. doi: 10.1093/aje/kwm089. [DOI] [PubMed] [Google Scholar]
- Lubin J.H., Caporaso N., Hatsukami D.K., Joseph A.M., Hecht S.S. The association of a tobacco-specific biomarker and cigarette consumption and its dependence on host characteristics. Cancer Epidemiol. Biomark. Prev. 2007;16(9):1852–1857. doi: 10.1158/1055-9965.EPI-07-0018. [DOI] [PubMed] [Google Scholar]
- Maes H.H., Woodard C.E., Murrelle L. Tobacco, alcohol and drug use in eight- to sixteen-year-old twins: the Virginia Twin Study of Adolescent Behavioral Development. J. Stud. Alcohol. 1999;60(3):293–305. doi: 10.15288/jsa.1999.60.293. [DOI] [PubMed] [Google Scholar]
- Moolchan E.T., Berlin I., Robinson M.L., Cadet J.L. Characteristics of African American teenage smokers who request cessation treatment: implications for addressing health disparities. Arch. Pediatr. Adolesc. Med. 2003;157(6):533–538. doi: 10.1001/archpedi.157.6.533. [DOI] [PubMed] [Google Scholar]
- Munafo M., Clark T., Johnstone E., Murphy M., Walton R. The genetic basis for smoking behavior: a systematic review and meta-analysis. Nicotine Tob. Res. 2004;6(4):583–597. doi: 10.1080/14622200410001734030. [DOI] [PubMed] [Google Scholar]
- Mwenifumbo J.C., Sellers E.M., Tyndale R.F. Nicotine metabolism and CYP2A6 activity in a population of black African descent: impact of gender and light smoking. Drug Alcohol Depend. 2007;89(1):24–33. doi: 10.1016/j.drugalcdep.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Piliguian M., Zhu A.Z., Zhou Q. Novel CYP2A6 variants identified in African Americans are associated with slow nicotine metabolism in vitro and in vivo. Pharmacogenet. Genomics. 2014;24(2):118–128. doi: 10.1097/FPC.0000000000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Foundation for Statistical Computing; Vienna, Austria: 2013. R: a language and environment for statistical computing. ( http://www.R-project.org/) [computer program] [Google Scholar]
- Raaschou-Nielsen E. Smoking habits in twins . Bull. 1960;7:82–88. [PubMed] [Google Scholar]
- Rosenberg N.A., Huang L., Jewett E.M., Szpiech Z.A., Jankovic I., Boehnke M. Genome-wide association studies in diverse populations. Nat. Rev. Genet. 2010;11(5):356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone N.L., Culverhouse R.C., Schwantes-An T.H. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6(8) doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf D.B., Sarkisyan N., Jacobsson H. Epigenetic screen identifies genotype-specific promoter DNA methylation and oncogenic potential of CHRNB4. Oncogene. 2013;32(28):3329–3338. doi: 10.1038/onc.2012.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz A.G., Cote M.L., Wenzlaff A.S., Land S., Amos C.I. Racial differences in the association between SNPs on 15q25.1, smoking behavior, and risk of non-small cell lung cancer. J. Thorac. Oncol. 2009;4(10):1195–1201. doi: 10.1097/JTO.0b013e3181b244ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi K., Kohno T., Kunitoh H. Contribution of nicotine acetylcholine receptor polymorphisms to lung cancer risk in a smoking-independent manner in the Japanese. Carcinogenesis. 2009;30(1):65–70. doi: 10.1093/carcin/bgn257. [DOI] [PubMed] [Google Scholar]
- Smith R.M., Alachkar H., Papp A.C. Nicotinic alpha5 receptor subunit mRNA expression is associated with distant 5' upstream polymorphisms. Eur. J. Hum. Genet. 2011;19(1):76–83. doi: 10.1038/ejhg.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz M.R., Amos C.I., Land S. Role of selected genetic variants in lung cancer risk in African Americans. J. Thorac. Oncol. 2013;8(4):391–397. doi: 10.1097/JTO.0b013e318283da29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanick M.L., Cochrane B.B., Hsia J., Barad D.H., Liu J.H., Johnson S.R. The Women's Health Initiative postmenopausal hormone trials: overview and baseline characteristics of participants. Ann. Epidemiol. 2003;13(9 Suppl):S78–S86. doi: 10.1016/s1047-2797(03)00045-0. [DOI] [PubMed] [Google Scholar]
- Sullivan P.F., Kendler K.S. The genetic epidemiology of smoking. Nicotine Tob. Res. 1999;1(Suppl. 2):S51–S57. doi: 10.1080/14622299050011811. (discussion S69-70) [DOI] [PubMed] [Google Scholar]
- Tang H., Peng J., Wang P., Risch N.J. Estimation of individual admixture: analytical and study design considerations. Genet. Epidemiol. 2005;28(4):289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson T.E., Geller F., Sulem P. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson T.E., Gudbjartsson D.F., Surakka I. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 2010;42(5):448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobacco-and-Genetics-(TAG)-Consortium Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 2010;42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyndale R.F., Sellers E.M. Genetic variation in CYP2A6-mediated nicotine metabolism alters smoking behavior. Ther. Drug Monit. 2002;24(1):163–171. doi: 10.1097/00007691-200202000-00026. [DOI] [PubMed] [Google Scholar]
- U.S.-Department-of-Health-and-Human-Services . The Health Consequences of Smoking-50 Years of Progress: A Report of the Surgeon General. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; Atlanta (GA): Atlanta, GA: 2014. The Health Consequences of Smoking-50 Years of Progress: A Report of the Surgeon General. [Google Scholar]
- Vink J.M., Willemsen G., Boomsma D.I. Heritability of smoking initiation and nicotine dependence. Behav. Genet. 2005;35(4):397–406. doi: 10.1007/s10519-004-1327-8. [DOI] [PubMed] [Google Scholar]
- Walsh K.M., Amos C.I., Wenzlaff A.S. Association study of nicotinic acetylcholine receptor genes identifies a novel lung cancer susceptibility locus near CHRNA1 in African-Americans. Oncotarget. 2012;3(11):1428–1438. doi: 10.18632/oncotarget.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K.M., Gorlov I.P., Hansen H.M. Fine-mapping of the 5p15.33, 6p22.1-p21.31, and 15q25.1 regions identifies functional and histology-specific lung cancer susceptibility loci in African-Americans. Cancer Epidemiol. Biomark. Prev. 2013;22(2):251–260. doi: 10.1158/1055-9965.EPI-12-1007-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wei Y., Gaborieau V. Deciphering associations for lung cancer risk through imputation and analysis of 12 316 cases and 16 831 controls. Eur. J. Hum. Genet. 2015 doi: 10.1038/ejhg.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware J.J., van den Bree M., Munafo M.R. From men to mice: CHRNA5/CHRNA3, smoking behavior and disease. Nicotine Tob. Res. 2012;14(11):1291–1299. doi: 10.1093/ntr/nts106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.P., Hopkins R.J., Christmas T., Black P.N., Metcalf P., Gamble G.D. COPD prevalence is increased in lung cancer, independent of age, sex and smoking history. Eur. Respir. J. 2009;34(2):380–386. doi: 10.1183/09031936.00144208. [DOI] [PubMed] [Google Scholar]
- Young R.P., Hopkins R.J., Gamble G.D. Clinical applications of gene-based risk prediction for lung cancer and the central role of chronic obstructive pulmonary disease. Front. Genet. 2012;3:210. doi: 10.3389/fgene.2012.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A.Z., Zhou Q., Cox L.S. Association of CHRNA5-A3-B4 SNP rs2036527 with smoking cessation therapy response in African-American smokers. Clin. Pharmacol. Ther. 2014;96(2):256–265. doi: 10.1038/clpt.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relationship between cigarettes smoked per day and risk of lung cancer in multicenter case–control study.