Abstract
Background
Antibiotic resistance is rising in important bacterial pathogens. Phage therapy (PT), the use of bacterial viruses infecting the pathogen in a species-specific way, is a potential alternative.
Method
T4-like coliphages or a commercial Russian coliphage product or placebo was orally given over 4 days to Bangladeshi children hospitalized with acute bacterial diarrhea. Safety of oral phage was assessed clinically and by functional tests; coliphage and Escherichia coli titers and enteropathogens were determined in stool and quantitative diarrhea parameters (stool output, stool frequency) were measured. Stool microbiota was studied by 16S rRNA gene sequencing; the genomes of four fecal Streptococcus isolates were sequenced.
Findings
No adverse events attributable to oral phage application were observed (primary safety outcome). Fecal coliphage was increased in treated over control children, but the titers did not show substantial intestinal phage replication (secondary microbiology outcome). 60% of the children suffered from a microbiologically proven E. coli diarrhea; the most frequent diagnosis was ETEC infections. Bacterial co-pathogens were also detected. Half of the patients contained phage-susceptible E. coli colonies in the stool. E. coli represented less than 5% of fecal bacteria. Stool ETEC titers showed only a short-lived peak and were otherwise close to the replication threshold determined for T4 phage in vitro. An interim analysis after the enrollment of 120 patients showed no amelioration in quantitative diarrhea parameter by PT over standard care (tertiary clinical outcome). Stool microbiota was characterized by an overgrowth with Streptococcus belonging to the Streptococcus gallolyticus and Streptococcus salivarius species groups, their abundance correlated with quantitative diarrhea outcome, but genome sequencing did not identify virulence genes.
Interpretation
Oral coliphages showed a safe gut transit in children, but failed to achieve intestinal amplification and to improve diarrhea outcome, possibly due to insufficient phage coverage and too low E. coli pathogen titers requiring higher oral phage doses. More knowledge is needed on in vivo phage–bacterium interaction and the role of E. coli in childhood diarrhea for successful PT.
Funding
The study was supported by a grant from Nestlé Nutrition and Nestlé Health Science. The trial was registered with Identifier NCT00937274 at ClinicalTrials.gov.
Abbreviations: Cfu, colony forming unit; ETEC, enterotoxigenic E. coli; EPEC, enteropathogenic E. coli; EAEC, enteroaggregative E. coli; M, ColiProteus phage cocktail from Microgen; ORS, oral rehydration solution; P, placebo; pfu, plaque forming unit; PT, phage therapy; qPCR, quantitative polymerase chain reaction; RCT, randomized controlled trial; T, T4 phage cocktail from NRC
Keywords: Bacteriophages, Diarrhea, Escherichia coli, Streptococcus, Bifidobacterium, Children, Bangladesh
Highlights
-
•
Coliphages given orally to children with bacterial diarrhea appeared in the stool, but did not improve clinical outcome.
-
•
In microbiologically diagnosed E. coli diarrhea, pathogen titers were close to the replication threshold of coliphages.
-
•
Acute bacterial diarrhea displayed a marked dysbiosis with fecal streptococci that stabilized with recovery from diarrhea.
Antibiotic resistance of bacterial infections reached alarming levels. Phage therapy is a potential alternative antimicrobial. We demonstrated that two different oral phage preparations did not improve acute bacterial diarrhea in children from Bangladesh. We observed fecal excretion of the oral phage, but no major phage amplification in the gut. E. coli pathogen levels were low and the fecal microbiota showed a transient overgrowth with streptococci. Future phage trials should first verify the titer and association of the targeted pathogen with the disease.
1. Introduction
In view of the growing threat of antibiotic resistance, WHO has warned for a return into a pre-antibiotic era (Martinez, 2012). Alternatives to antibiotics are thus urgently needed (Piddock, 2012, Stanton, 2013). Phage therapy (PT), the use of bacterial viruses (phages) against pathogens, is a potentially attractive option for the prevention and treatment of some bacterial infections (Sulakvelidze et al., 2001, Brüssow, 2005, Brüssow, 2012). Indeed, a large, randomized and placebo-controlled trial conducted in children from Tbilisi/Republic of Georgia during the 1960s described prevention of Shigella dysentery and Escherichia coli diarrhea with orally applied Shigella phages (Sulakvelidze et al., 2001). PT was practiced in Poland in patients with antibiotic-resistant bacterial infections on a case-by-case basis, apparently with high success (Międzybrodzki et al., 2012). However, randomized controlled trials (RCTs) in humans are needed to rationally assess the potential of PT. So far, a single small RCT documented an effect in carefully selected patients suffering from otitis externa (“swimmer's ear”) (Wright et al., 2009).
E. coli is an important cause of diarrhea in children from developing countries (Qadri et al., 2005), resistant against many antibiotics (Jiang et al., 2002) while efficient E. coli diarrhea vaccines are not yet available (Ahmed et al., 2013). Other than zinc and oral rehydration solution, no specific, effective, safe and affordable treatment is available to reduce the severity or duration of illness caused by this bacterial agent. We hypothesize that oral administration of coliphage will be effective in reducing severity of diarrhea in children with proven E. coli induced diarrhea. Therefore a RCT of PT with two different coliphage cocktails for which safe use was demonstrated in healthy adults (Sarker et al., 2012, McCallin et al., 2013) was performed in children with E. coli diarrhea. The primary endpoint was the safety of oral coliphage in children infected with E. coli, the secondary endpoint was the titration of fecal coliphage and E. coli pathogen to assess in vivo lytic phage activity and the tertiary endpoint was the impact of oral phage on quantitative diarrhea parameters. Oral coliphage reached the intestine, but did not achieve treatment effects over placebo most likely because intestinal E. coli titers were low and close to the replication threshold of coliphages. Microbiota analysis revealed a marked outgrowth of fecal streptococci during the acute phase of diarrhea.
2. Methods
2.1. Participants and Study Design
A prospective, single center, randomized, placebo-controlled, parallel group clinical trial was undertaken to assess the safety and efficacy of T4-like phage cocktail compared to Microgen ColiProteus phage cocktail or placebo in 6–24-month-old male (to obtain stool without urine contamination) children presenting with acute onset of dehydrating diarrhea of less than 48 h duration. The study took place at the Dhaka Hospital of the International Centre for Diarrheal Disease Research, Bangladesh (icddr,b) between June 2009 and September 2011. The trial was approved by the Research Review Committee and Ethical Review Committee of icddr,b (protocol #2008-062) and registered with the Identifier NCT00937274 at the ClinicalTrials.gov site. The study was performed according to Good Clinical Practice and the Declaration of Helsinki (http://www.who.int/bulletin/archives/79(4)373.pdf). Written informed consent was obtained from parents or legal guardians of the child before study enrollment. The T4 phage cocktail contained eleven T4-like phages (AB2, 4, 6, 11, 46, 50, 55; JS34, 37, 98, D1.4) (Bourdin et al., 2014a, Bourdin et al., 2014b). The composition of the Microgen ColiProteus phage cocktail was described previously by metagenome sequencing and electron microscopy (McCallin et al., 2013). The placebo was reduced osmolarity oral rehydration solution (ORS) supplemented with zinc, the standard treatment for uncomplicated watery diarrhea at icddr,b. The primary outcome of the safety part of the study was the acceptability of the phage products by the patients as determined by clinical observation and liver, renal and hematology tests described previously (Sarker et al., 2012, McCallin et al., 2013). The secondary endpoint was the determination of coliphage and total E. coli titers and their phage susceptibility in stool and the clearance of ETEC from stool. The tertiary outcome was changes in total stool output (expressed as g/kg of body weight per day), frequency of stool (number/day) and ORS need for rehydration (in ml/kg body weight per day) from randomization to resolution of illness.
Children with at least four liquid stools during the previous 24 h, with some degree of dehydration (WHO methodology) were considered for study enrollment after obtaining informed consent.
2.2. Randomization
A random permuted block design (with block size of 6) (to equalize the number of patients in the two treatment groups after short intervals) was used for allocation of patients to the study interventions, namely the two active products, i.e. T4 coliphage cocktail, Microgen phage cocktail in ORS or placebo (0.9% NaCl) at a 1:1:1 ratio. The icddr,b hospital pharmacist, not associated with the study in any other way, prepared the products and handed it over to the study nurse unaware of the product for dispensing to the patients following a computer-generated list of random numbers developed by an independent biostatistician not associated with the study. Code envelopes were kept by the sponsor and by the investigator for un-blinding in emergency situations and after the blind review meeting. The subjects, principal investigator, hospital staff and laboratory personnel were masked to the treatment assignments. After mixing phage or placebo with ORS powders in 30 ml mineral water (Vittel mineral water, pH 7.6, bicarbonate 384 mg/l) and a food dye, the interventions products were indistinguishable. The study products were given 3 times per day (8 am, noon, 4 pm) for four days.
2.3. Power Calculation
We estimated the sample size based on stool output. In a previous trial at icddr,b in children hospitalized with untreated ETEC and EPEC diarrhea we observed a stool output of 176 + 100 ml/kg body weight in a 4-day period (Casswall et al., 2000). With an anticipated 30% reduction in stool volume, we estimated a sample size of 71 at 5% significance level and 80% power. To adjust for a 5% drop-out rate, we decided to enroll 225 children into the trial. Note that the interim analysis was done with 120 patients, 53% of the planned enrollment.
2.4. Procedures
Following confirmation of the absence of rotavirus in stool by ELISA and Vibrio by dark field microcopy, and sending stool samples for further microbiological assessments, i.e. Salmonella, Shigella, Aeromonas and Campylobacter according to standard procedures at icddr,b (Harris et al., 2008) and for identification of ETEC, EPEC or EAEC by multiplex PCR assay using specific primers as described (Svenungsson et al., 2000), the enrolled children were randomized to intervention. Children with systemic infection, invasive diarrhea, severe acute malnutrition (weight for height z score < − 3SD of WHO median or presence of edema), significant medical abnormalities and who had received or needed antibiotic treatment were excluded. Initial correction of dehydration was performed using hypo-osmolar ORS solution or initial intravenous rehydration solution (for children with severe dehydration) followed by ORS solution, equal to the amount of abnormal (watery or liquid) stool loss, until diarrhea resolves.
Children whose fecal culture identified other bacterial enteric pathogens or no E. coli pathogen remained in the study and their data were used for Intention-to-Treat (ITT) analysis. Sub-group analysis was done for confirmed E. coli infection. Freshly passed stool was obtained daily at about 4 pm (the time was kept fixed as the first dose was fed exactly at 4 pm) for quantitative colony counts of E. coli on EMB and MacConkey agar plates, total bacterial count by qPCR (Nadkarni et al., 2002) and microbiota analysis. Three putative E. coli colonies from day 1 stool agar plates were tested for sensitivity to the applied phage cocktails at NRC, Switzerland.
The nursing staff recorded the frequency, consistency and volume of stool and urine passed every six hours (to a sensitivity of 1 g). This was achieved through a combination of placing the subject on a cholera cot and collecting urine in a pediatric urine collection bag. The data collected from this procedure was also used to calculate amounts of ORS solution required to account for fluid losses due to diarrhea. Stool frequency was counted by the study nurse and consistency of every stool passed was recorded. The passage of the last abnormal (watery) stool prior to formed or soft or no stool produced during two consecutive 6-hour periods were used to estimate the duration of diarrhea. Feeding appropriate for age was introduced early as ad libitum breastfeeding (for breast fed children) or formula feeding (for non-breast fed children) or standardized hospital diet to provide approximately 100 kcal per kg body weight per day.
If diarrhea continued beyond five days, treatment was judged a failure and the child was transferred to the general ward of the hospital for further management following the hospital's standard protocol. Resolution of illness was defined as passage of formed or soft stools during two consecutive six-hour periods. Patient follow-up visit occurred 21 days after first dose.
Safety evaluation of patients included, in addition to clinical observation, chemistry tests which comprised liver and renal function assays and hematology tests that were done at the admission and discharge from the hospital as described previously (Sarker et al., 2012, McCallin et al., 2013).
2.5. Phage Detection
One gram of stool sample was resuspended in TS (8.5 g NaCl and 1 g tryptone/l) to a final volume of 5 ml and centrifuged for 15 min at 14,500 × g. The cleared supernatant was filtered through a Millex AP20 prefilter followed by a 0.45 μm pore size Minisart filter. The presence of phages was determined on two indicator strains: E. coli strain WG5 obtained from J. Jofre, University of Barcelona (Spain), a common indicator strain used in phage ecology studies (Guzmán et al., 2008), and E. coli strain K803, a K-12 derivative lacking prophage lambda, obtained from E. Kutter, Evergreen College, Olympia, WA, USA. This strain lacks restriction-modification genes and contains only the core part of the lipopolysaccharide making it susceptible to a wide range of coliphages (Chibani-Chennoufi et al., 2004). The strains were propagated in Hershey broth supplemented with casein-derived amino acids (15 g/l). Phages were enumerated using the double layer plaque assay, with top agar at 7.5 g/l on a bottom layer of 15 g agar/l in broth (Chibani-Chennoufi et al., 2004). Phage dilutions were prepared in TS: 100 μl of stool filtrate and 100 μl of an exponentially growing bacterial culture were mixed together, incubated at room temperature for 15 min and used to inoculate 3 ml of top agar. Plates were incubated over-night at 37 °C and phage plaques were counted. Phage titer was calculated for plaque forming units (pfu) per g stool.
2.6. Stool Microbiota Analysis
Total DNA was extracted using the QIAamp DNA Stool Mini Kit (QIAGEN), following the manufacturer's instructions, except for the addition of a series of mechanical disruption steps (11 × 45 s) using a FastPrep apparatus and Lysing Matrix B tubes (MP Biochemicals) as described (Junick and Blaut, 2012).
The 16S variable region V1 to V3 was PCR amplified and sequenced on Roche 454 GS-FLX-Titanium Sequencer as described (Sanchez et al., 2014) Raw sequence data were deposited in the GenBank Short Read Archive (Accession number: SRP067629) and analyzed using Mothur v.1.33.0 and QIIME v.1.8 software packages (Schloss et al., 2009, Caporaso et al., 2010). Pyrosequencing reads were denoised with the Mothur implementation of PyroNoise (Quince et al., 2009) according to the 454 SOP (Schloss and Westcott, 2011). Chimeras were identified using usearch61 in QIIME (Edgar et al., 2011). The sequences were then trimmed as described in the Mothur 454 SOP in order to keep sequences overlapping the same 16S region. OTUs de novo picking at 97% identity was performed using uclust in QIIME (Edgar, 2010). Taxonomy assignment of OTU representative sequences used the RDP Classifier in QIIME with confidence threshold of 0.6 on the Greengenes reference database v.13.8(Simeoni et al., 2015). At species level, the taxonomic assignment was determined by BLAST (Altschul et al., 1990) using the website http://blast.ncbi.nlm.nih.gov with a 97% identity threshold. Diversity analyses were performed in QIIME, Mothur and Calypso (http://bioinfo.qimr.edu.au/calypso/). Dirichlet Multinomial Mixture analysis was performed in Mothur as described (Ding and Schloss, 2014). Figures were created with GraphPad Prism version 6.07 for Windows (GraphPad Software, La Jolla California USA) and Microsoft Excel 2010.
2.7. Genome Sequencing
Individual bacterial colonies were isolated from feces on a Streptococcus-enriching agar, characterized as streptococci by API fermentation gallery (API 20 STREP, bioMérieux, France) and four colonies identified as Streptococcus by 16S rDNA sequencing, three derived from different diarrhea patients and one derived from a local age-matched control child, underwent whole genome sequencing.
Genomic DNA was extracted from mid-exponential cultures using Gentra DNA Purgene kit (Qiagen). 10 Kb libraries were prepared following Pacific Biosciences (PacBio) protocol. Sequencing was performed on the PacBio RS platform using C2/C2 chemistry and four single-molecule-real-time (SMRT) cells were used per strain with a 120 min collection protocol. The sub-reads were de novo assembled using the PacBio Hierarchical Genome Assembly Process (HGAP)/Quiver software package (Chin et al., 2013). The HGAP workflow consists of three steps: Preassembly to generate long and highly accurate sequences; assembly of the long and highly accurate sequences using Celera assembler and finally, Consensus Polishing using the Quiver algorithm to derive a highly accurate consensus for the final assembly.
The strains were assembled into one to three contigs. The results of the sequencing and assemblies are summarized in Suppl. Table 1. The genomes were annotated using the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) and have been deposited at GenBank (NCBI) under accession numbers CP0113688 and CP013689 for isolates S1 and S5 (1 contig) and LPVQ00000000 and LPVR00000000 for isolates S3 and S6 (2–3 contigs). Plasmid DNA was not sequenced by our approach, but plasmids were not detected in the sequenced strains.
A global alignment of chromosomes was plotted using MUMMER package (nucmer parameters: –maxmatch-c 20-g 0-b 1-l 20–noextend). The KSNP phylogenetic tree was built with KSNP3 tool (Gardner and Hall, 2013) with the following parameters: –k 13–ML.
2.8. Genomic Safety Analysis
In order to reduce the complexity, sequence reads for the four streptococcal genomes showing 100% sequence identity were removed from the total reads to generate a set of non-redundant (nr) reads. This set of reads was investigated for virulence factors in three ways. First, the reads were mapped against closely related streptococci with proven (Streptococcus gallolyticus) and suspected (Streptococcus lutetiensis) virulence genes. Matches were manually investigated by additional BlastX searches to verify the annotations from the deposited and the newly sequenced streptococci. Second, the nr reads were mapped against all genes present in an in-house Database for Undesirable Genes described previously (Denou et al., 2009). Third, the nr reads were screened against two public databases compiling antibiotic resistance genes (ARDB) (Liu and Pop, 2009) and bacterial virulence factor genes (VFDB) (Chen et al., 2012). Only hits having two or more reads aligned and covering more than 30% of the gene were considered as positive hits and were further investigated.
2.9. Statistical Analysis
Outcome variables, e.g., stool output (g/kg × d), and ORS Intake (ml/kg × d) were compared among the groups using one-way analysis of variance (ANOVA) and between the groups using student's t test. Confounders, e.g., age, duration of diarrhea before hospitalization, and nutritional status were adjusted using multivariate analysis (multiple regression or logistic regression as appropriate). The data with skewed distribution were normalized by log transformation, and the t test was applied on transformed data. The Wilcoxon's rank test was used for skewed data that cannot be normalized by transformation.
3. Results
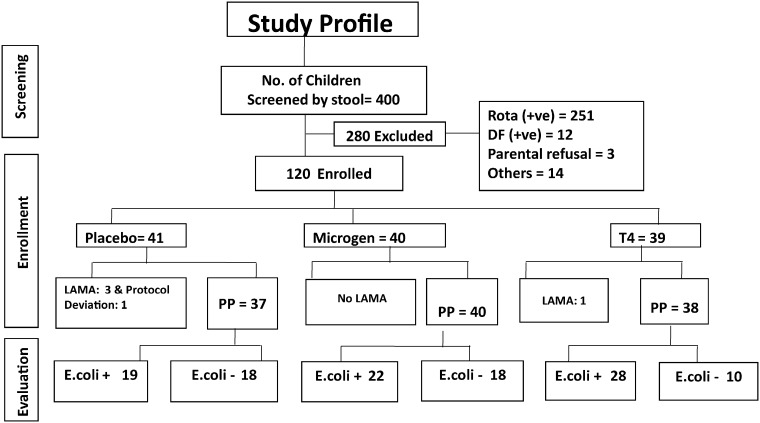
Four hundred children hospitalized with acute watery diarrhea were screened for this study; 280 were excluded mainly for rotavirus (n = 251) or Vibrio diagnosis (n = 12); 120 fulfilled the entrance criteria and were randomized: 41 received placebo (saline) with standard treatment (ORS plus zinc) (P), 40 received orally 1.4 × 109 pfu of the Microgen ColiProteus (McCallin et al., 2013) cocktail (M) and 39 children received 3.6 × 108 pfu of the Nestlé Research Centre (NRC) T4-like coliphage cocktail (T) in addition to standard treatment (Sarker et al., 2012, Bourdin et al., 2014a, Bourdin et al., 2014b). Fig. 1 shows the study flow chart.
Fig. 1.
Study flow chart.
The numbers indicate the children with diarrhea who were screened, enrolled and evaluated. Rota(+ ve), patients with rotavirus positive stool in ELISA; DF(+ ve), patients with V. cholerae positive stool in dark field microscopy; others, patients meeting other exclusion criteria; Microgen, commercial ColiProteus phage cocktail from Microgen, Russia; T4, phage cocktail consisting of T4-like phages from NRC; LAMA, left the study against medical advice; PP, per protocol, E. coli +, patient displaying a pathogenic E. coli in the stool; E. coli-patient without a pathogenic E. coli in the stool.
3.1. Safety Analysis
Per protocol a safety analysis was done after enrollment of 75 patients. No observations indicative of a Jarisch–Herxheimer reaction (Yang et al., 2010) were seen (Table 1). Liver, kidney and hematological functions were normal when using standard tests (Sarker et al., 2012, McCallin et al., 2013). One T4 phage and one placebo recipient developed generalized tonic–clonic convulsion a few hours after the initial product application, which was attributed by the clinician to hyponatremia and acidosis associated with diarrhea.
Table 1.
Physical examinations in children hospitalized with diarrhea at admission and after four days of treatment with the allotted phage or placebo product.
| Treatment |
T4-phage |
Microgen phage |
Placebo |
|||
|---|---|---|---|---|---|---|
| Time |
Adm |
Day 4 |
Adm |
Day 4 |
Adm |
Day 4 |
| n | 25 | 23 | 26 | 20 | 27 | 23 |
| Age, month | 10 ± 3.2 (8.8–11.3) | 12 ± 4.1 (10.4–13.6) | 13 ± 4.8 (11.2–14.8) | |||
| Weight, kg | 8.1 ± 1.3 (7.6–8.6) | 8.0 ± 1.3 (7.5–8.5) | 8.4 ± 1.4 (7.9–8.9) | 8.3 ± 1.5 (7.6–9.0) | 8.4 ± 1.5 (7.8–9.0) | 8.2 ± 1.7 (7.5–8.9) |
| Temp, °C | 36.9 ± 0.5 (36.7–37.1) | 36.7 ± 0.5 (36.5–36.9) | 36.7 ± 0.6 (36.5–36.7) | 36.5 ± 0.4 (36.3–36.7) | 36.9 ± 0.8 (36.6–37.2) | 36.8 ± 0.7 (36.5–37.1) |
| Pulse/min | 119 ± 7 (116–122) | 118 ± 4 (116–120) | 121 ± 5 (119–123) | 118 ± 5 (116–120) | 120 ± 6 (118–122) | 119 ± 5 (117–121) |
| Respiration/min | 33 ± 6 (31–35) | 31 ± 2 (30–32) | 32 ± 5 (30–34) | 30 ± 3 (29–31) | 34 ± 3 (33–35) | 32 ± 2 (31–33) |
| BP systolic (mmHg) | 89 ± 6 (87–91) | 86 ± 5 (84–88) | 89 ± 6 (87–91) | 87 ± 5 (85–89) | 88 ± 5 (86–90) | 87 ± 5 (85–89) |
| BP diastolic | 54 ± 5 (52–56) | 53 ± 4 (51–55) | 56 ± 6 (54–58) | 52 ± 3 (51–53) | 57 ± 8 (54–60) | 55 ± 4 (53–57) |
n, number of children per investigated group. Values are means ± standard deviations (95% CI).
No significant differences were seen between clinical parameters at admission and 4 days of phage or placebo treatment in diarrhea patients in paired t-tests. However, T4-phage treated children were significantly younger than placebo recipients (P = 0.01 in t-test), but did not differ from Microgen phage treated children.
3.2. Lack of Clinical Efficacy of Oral Phage
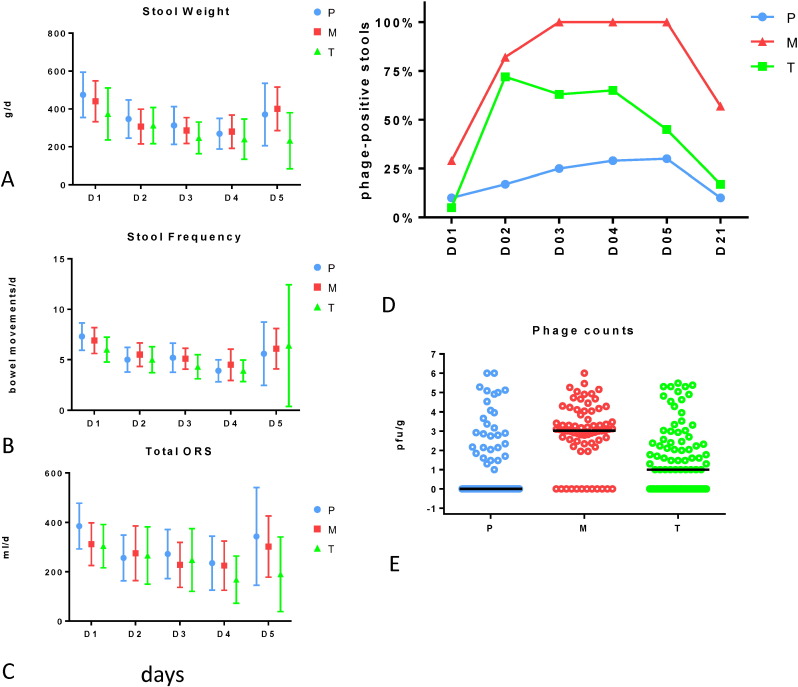
At enrollment, children in the three groups did not differ for prior duration of diarrhea, stool frequency, or vomiting (Table 2). At an interim analysis with 120 enrolled and 115 evaluated children, no significant difference was seen between the groups when evaluating quantitative diarrhea criteria (stool frequency, stool weight, oral rehydration solution needed to correct dehydration) (Fig. 2A–C) (P > 0.1 for all comparisons, t test). In the acute phase, we observed lower means for stool weight in T compared to M; T also happened to have a higher percentage of E. coli-positive cases than M and P (Fig. 1). However, even when limited to proven E. coli infections, oral phage had no significant impact on quantitative diarrhea criteria (Table 3). Four days after admission, 50% of the children were discharged from the hospital (no difference between the groups). The increase in quantitative diarrhea values seen at day 5 in Fig. 2 reflects therefore enrichment for children with more severe diarrhea. At day six, 93, 88, and 84% of patients treated with T, M or P, respectively, had recovered from diarrhea (difference not significant, t test).
Table 2.
Diarrhea patients' characteristics before first dose of allotted treatment (baseline).
| Group | Number of patients | Admission weight kg | Diarrhea duration before 1st dose (hours) | Stool frequency/day before 1st dose | No of vomiting/day before 1st dose |
|---|---|---|---|---|---|
| Placebo | 41 | 8.70 ± 1.7 (8.2–9.2) | 32.1 ± 10.3 (29.0–35.2) | 16.0 ± 7.1 (13.8–18.2) | 5.9 ± 4.7 (4.5–7.3) |
| Microgen phages | 40 | 8.43 ± 1.4 (8.0–8.9) | 33.1 ± 10.4 (32.5–33.7) | 14.6 ± 6.5 (12.6–16.6) | 6.4 ± 5.7 (4.6–8.2) |
| T4-like phages | 39 | 8.36 ± 1.3 (8.0–8.8) | 32.6 ± 10.2 (29.4–35.8) | 16.0 ± 7.6 (13.6–18.4) | 7.7 ± 7.2 (5.4–10.0) |
Values are means ± standard deviation (numbers in parenthesis denote 95% CI).
Two-tailed P values in unpaired Student t-tests > 0.19 for all comparisons.
Fig. 2.
Oral phages reach the intestine in children hospitalized with acute bacterial diarrhea, but show no impact on quantitative clinical diarrhea outcomes.
Left: Mean and standard deviation for stool weight (measured without urine contamination) in g/day (A), stool frequency per day (B), and need for oral rehydration solution in ml/day to correct dehydration (C) for the specified day of hospitalization in children enrolled into the three treatment groups identified by the color code. No significant difference was detected between the treatment groups.
Right, D: Prevalence of phage positive stools (≥ 10 pfu/g stool on E. coli indicator strain K-12) at the indicated time points (D01–05: days 1 to 5 of hospitalization; D21: re-convalescent visit 21 days after hospital admission) in the three treatment groups; green: NRC T4-like phage cocktail (T), red: Russian Microgen phage cocktail (M), blue: placebo (P).
E: Titer distribution and median fecal phage titer in log10 pfu/g stool for the indicated treatment groups.
Table 3.
Quantitative diarrhea parameters in patients with microbiologically confirmed E. coli diarrhea treated with Microgen or T4-like phage cocktail or placebo.
| Confirmed E. coli infections (n = 69) | Day 1 | Day 2 | Day 3 | Day 4 |
|---|---|---|---|---|
| Stool frequency: | ||||
| Placebo (n = 19) | 7.5 ± 3.4 (6.0–9.0) | 6.0 ± 3.1 (4.6–7.4) | 6.3 ± 4.5 (4.3–8.3) | 4.8 ± 3.9 (3.0–6.6) |
| Microgen (n = 22) | 6.3 ± 4.1 (4.6–8.0) | 5.3 ± 2.9 (4.1–6.5) | 4.2 ± 2.5⁎ (3.2–5.2) | 4.6 ± 5.1 (2.5–6.7) |
| T4-like phage (n = 28) | 6.6 ± 4.1 (5.1–8.1) | 5.8 ± 4.3 (4.2–7.4) | 5.0 ± 3.9 (3.6–6.4) | 4.3 ± 3.2 (3.1–5.5) |
| Stool weight: | ||||
| Placebo (n = 19) | 514 ± 444 (314–714) | 422 ± 360 (260–584) | 350 ± 352 (192–508) | 316 ± 280 (190–442) |
| Microgen (n = 22) | 433 ± 265 (322–544) | 276 ± 196 (194–358) | 278 ± 213 (189–367) | 293 ± 279 (176–410) |
| T4-like phage (n = 28) | 406 ± 480 (228–584) | 344 ± 321 (225–463) | 280 ± 284 (175–385) | 280 ± 345 (152–408) |
| ORS need: | ||||
| Placebo (n = 19) | 393 ± 261 (276–510) | 341 ± 341 (188–494) | 333 ± 319 (190–476) | 283 ± 373 (115–451) |
| Microgen (n = 22) | 328 ± 220 (236–420) | 252 ± 294 (129–375) | 192 ± 256 (85–299) | 216 ± 280 (99–333) |
| T4-like phage (n = 28) | 330 ± 289 (223–437) | 281 ± 368 (137–425) | 293 ± 438 (131–455) | 201 ± 312 (85–317) |
Values are means ± standard deviation (95% CI, confidence interval).
Days 1–4 of treatment.
Stool frequency: number of bowel movements per day for the indicated day of hospitalization.
Stool weight: stool weight in g per patient and the indicated 24 h period.
ORS: volume of ORS in ml needed per indicated day to correct dehydration.
n = number of patients per specified group.
P = 0.07 in unpaired Student t-test, for all other comparisons P > 0.1.
3.3. Detection of Fecal Phage
Oral Microgen, and to a lesser extent the T4 phage cocktail, resulted in a significantly higher fecal phage prevalence and titer than seen in placebo recipients likely producing endogenous phage (Chibani-Chennoufi et al., 2004) (Fig. 2 D, E). No patient showed higher fecal output than oral input. Electron microscopy done for the first 56 patients with sensitivity to detect 106 viral particles per g stool (Goldsmith and Miller, 2009) demonstrated only few phage particles in three patients excluding substantial in vivo replication of oral phage (data not shown).
Fecal coliphage titers did not differ significantly between patients with diagnosed ETEC or EPEC/EAEC infection or patients lacking a diarrheagenic E. coli (Suppl. Fig. 1A). In addition, fecal phage titers were not higher in patients harboring a phage-sensitive E. coli colony in the stool (50% of all patients) compared to patients lacking such colonies (three random colonies were tested per patient, Suppl. Fig. 1B) suggesting passive transit of oral phage through the gut.
In the plaque assay T4-like and T7-like coliphages can be differentiated by plaque size (Suppl. Fig. 2A–C). If a differential survival or replication occurred for these phages in the gut, one would expect a shift in relative proportion from the 1:1 ratio seen for T4 and T7 phages of the M cocktail (McCallin et al., 2013) in the stool of recipients of this cocktail. No such shift was observed (Suppl. Fig. 2D).
3.4. Etiology of Diarrhea
Stool series from the first 56 patients underwent detailed clinical microbiology analysis (Harris et al., 2008, Svenungsson et al., 2000): 77% yielded an E. coli pathogen by culture and subsequent pathotype analysis (ETEC: 23; EAEC: 10; enteropathogenic (EPEC): 2; mixed E. coli: 8 patients).
The presence of E. coli virulence genes was corroborated for these 56 patients by PCR (Vidal et al., 2005) on whole stool DNA. ETEC-specific genes were detected in 28 (23 elt+ est+; 3 elt+; 2 est+); EPEC in 5 (4 eae+ bfp+; 1 eae+); EAEC (daaD+) in 7 patients for at least 2 successive stool samples. Stool samples were also cultured for other bacterial pathogens (Harris et al., 2008). Campylobacter jejuni, Campylobacter coli, Aeromonas, Vibrio cholerae, Salmonella, and Shigella flexneri were detected in 9, 2, 2, 2, 1 and 1 patients, respectively. All but two also harbored an E. coli pathogen. Electron microscopy of putative viral pellets from these 56 stools identified one patient infected with rotavirus, which apparently escaped ELISA detection of group A rotavirus.
3.5. Fecal E. coli Titers
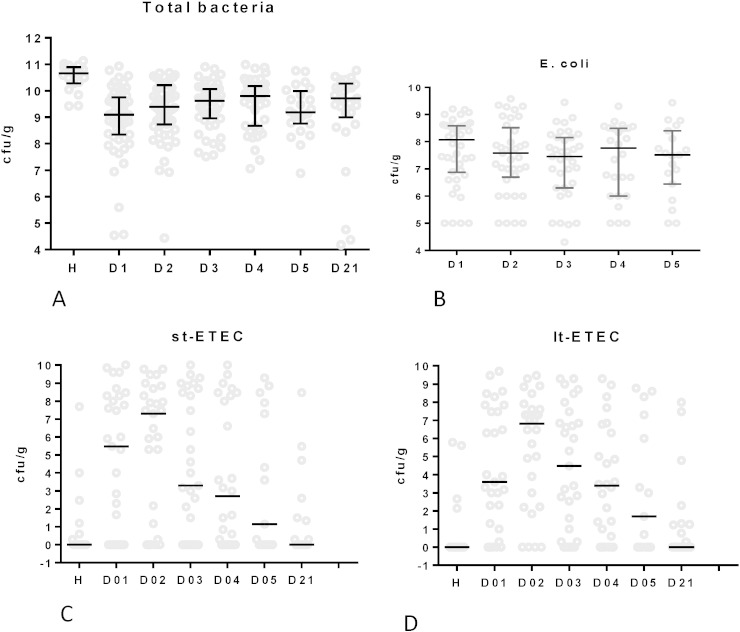
Total fecal bacterial counts as determined by real time PCR (Nadkarni et al., 2002) were significantly lower in patients than in controls (P < 0.001 in Dumm test) (Fig. 3A). Viable E. coli colony counts represented only 5% of all fecal bacteria in diarrhea patients (Fig. 3B) and were at most 10-fold higher in E. coli as compared to non-E. coli diarrhea patients (median viable stool E. coli titers were 5 × 108 cfu/g stool for ETEC, 2 × 108 for EAEC/EPEC patients and 5 × 107 for patients without E. coli diarrhea diagnosis).
Fig. 3.
Total bacteria, E. coli and enterotoxigenic E. coli titer development in serial stool samples of hospitalized children during an episode of acute diarrhea.
A. Median titer with interquartile range for total stool bacteria determined by real time qPCR with universal bacterial 16S rDNA primers; expressed as log10 cfu/g stool equivalents for healthy local control children (H) and diarrhea patients at the indicated day after hospital admission. Only H is significantly different from the other time points (Dumm's multiple comparisons tests).
B. Median with interquartile range of viable E. coli counts on MacConkey agar determined as log10 cfu/g fresh stool (ordinate) for the indicated day of hospitalization of diarrhea patients (no significant difference).
C and D. Titer distribution and median titers for heat-stable (st-ETEC, C) and heat-labile (lt-ETEC, D) enterotoxin-carrying bacteria in the stool of diarrhea patients hospitalized with a microbiologically confirmed ETEC infection. Titers determined by real time PCR are for the indicated days of hospitalization and are compared to healthy control children (H). The titers are expressed as log10 median titers in ETEC cfu equivalents.
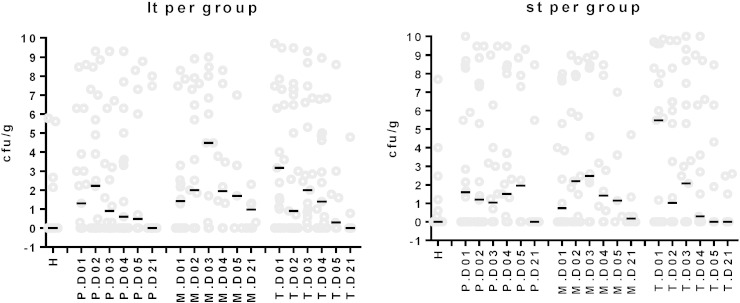
The titer of heat-stabile (Fig. 3C) and heat-labile (Fig. 3D) enterotoxin genes were investigated over the course of diarrhea by real time PCR (Guion et al., 2008). By calibration against an elt+ est+ E. coli strain of known titer, the data were expressed as cfu ETEC per g stool. Median titers were low, but the titers were widely spread. When limited to microbiologically confirmed ETEC infections, peak median titers of 3 × 107 ETEC cfu/g stool were observed on day 2 of hospitalization with rapid decline over the next days (Fig. 3C, D). Most children with microbiologically defined EPEC or EAEC infection and healthy controls had enterotoxin gene counts below the detection limit of the test, confirming the specificity of the test. No significant decrease was seen for ETEC titers as assessed by real time PCR of heat-labile and heat-stable enterotoxin genes in phage-treated patients compared to the placebo recipients (P > 0.1, t test) (Fig. 4).
Fig. 4.
No oral phage treatment effect on fecal ETEC titers.
Titer distribution and median titers for heat-labile (lt, left) and heat-stable (st, right) enterotoxin gene-carrying ETEC bacteria in the stool of diarrhea patients treated with placebo (P), Microgen phage (M) or NRC T4 phage (T). Titers were determined by real time PCR for the indicated day of hospitalization (D01 to D05) and the return visit (D21). Titers in healthy control children (H) are given for comparison. The titers are expressed as log10 titers in ETEC cfu equivalents.
3.6. Fecal Microbiota Composition by 16S rDNA Sequencing: Transient Streptococcus Overgrowth
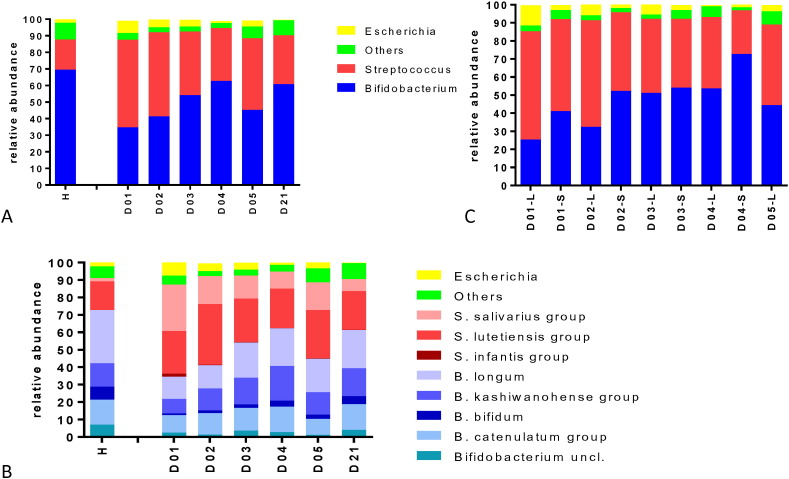
We determined the microbiota composition in daily collected stool samples for the first 56 patients enrolled into the trial by 454 sequencing of PCR-amplified 16S rDNA using universal bacterial primers (Sanchez et al., 2014) and compared it with the microbiota composition in stools sampled at the same time period from 20 age-matched healthy control children (median age 11 months) living in the same city under comparable socioeconomic conditions. During the first day of hospitalization, fecal Streptococcus represented a significantly higher abundance in diarrhea patients than in healthy controls (P = 0.001, Fisher's exact test); Escherichia was higher in patients than in controls, but not significantly so (P = 0.06) (Fig. 5A). Control children showed a higher stool abundance of Bifidobacterium than diarrhea patients in the acute phase (P = 0.003). Over the next days of hospitalization, a gradual increase in Bifidobacterium and a concomitant decrease in Streptococcus abundance were observed (Fig. 5A, P = 0.008). This shift from Streptococcus to Bifidobacterium during the hospital stay was seen whether the sequence analysis was based on mean (Fig. 5A) or median abundance (Fig. 6A–C) or whether the analysis was based on the V123 (all presented data in the current report) or on the V456 regions (data not shown) of the 16S rRNA gene. When further differentiated for the diarrhea pathogen, the decrease in fecal Streptococcus abundance during hospitalization was clearer for patients with EPEC and EAEC infections or diarrhea without E. coli pathogen than for ETEC patients (Fig. 6D, E). Subjects with elevated Escherichia abundance had ETEC, but not EPEC/EAEC infections (Fig. 6C, F). Children with non-E. coli diarrhea (Fig. 6F) and healthy controls (Fig. 6C) showed with few exceptions < 1% Escherichia abundance.
Fig. 5.
Fecal microbiota analysis by 16S rRNA gene sequencing in serial stool samples from children hospitalized with acute bacterial diarrhea.
A. Bacterial community structure profiles for fecal samples from 20 healthy control children (H) and 56 diarrhea patients at the indicated day of hospitalization (D01–D21). The panel gives the mean relative abundance in percent for the bacterial genera identified by the color code at the right.
B. Bacterial community structure at species level. Healthy controls (H) and diarrhea patients at the specified day of hospitalization are displayed for relative abundance at species level defined by the color code at the right. S. salivarius and S. vestibularis are members of the S. salivarius group. S. lutetiensis, S. infantarius, S. equinus, S. pasteurianus, S. gallolyticus, S. macedonicus, S. alactolyticus and S. porcorum are members of the S. gallolyticus group. B. catenulatum, B. pseudocatenulatum, B. angulatum, B. ruminantium, B. merycicum, B. adolescentis, B. dentium, and B. stercoris are members of the B. catenulatum group. B. kashiwanohense and B. breve are members of the B. kashiwanohense group.
C. Bacterial community structure for 56 diarrhea patients that were hospitalized for 4 day (S—short stay) or 5 days (L—long stay) at the specified day of hospitalization. The panel gives the mean relative abundance in percent for the bacterial genera identified by the same color code as in panel A.
Fig. 6.
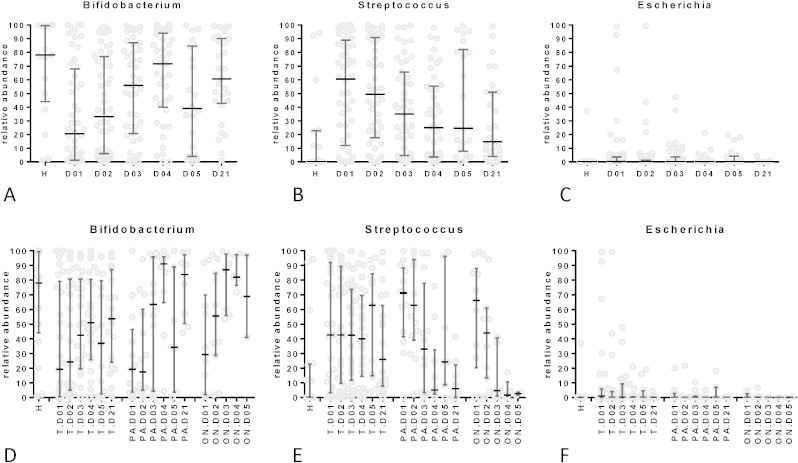
Succession of the three major bacterial genera in the gut microbiota during recovery from diarrhea.
Median percentage with interquartile range and distribution for the individual patients with respect to Bifidobacterium (A and D), Streptococcus (B and E) and Escherichia (D and F) contribution to the stool microbiota at the indicated day of hospitalization. In A to C data are presented for all investigated diarrhea patients; in D–F, patients were separated with respect to etiology (ON: other bacterial pathogens than E. coli and no pathogens detected; PA: EPEC and EAEC; T: ETEC infections) compared to healthy local controls (H).
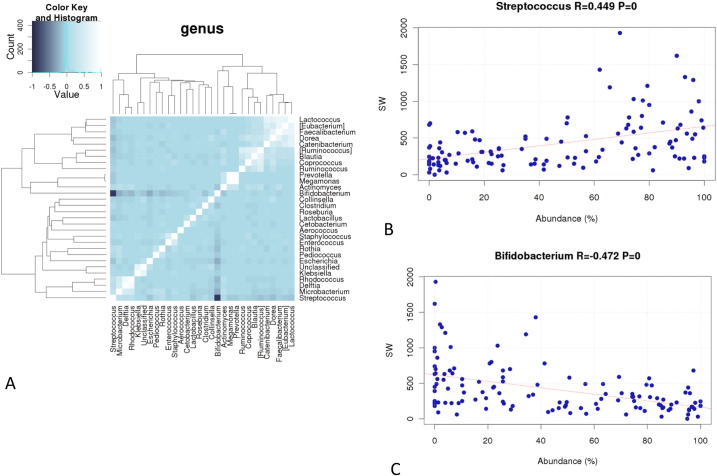
The heat map of correlation between bacterial genera showed a strong anti-correlation between Bifidobacterium and Streptococcus while only a weak anti-correlation between Bifidobacterium and Escherichia was observed (Fig. 7A). No significant difference was observed for microbiota composition and temporal development of the stool microbiota between placebo and phage recipients when all patients were considered (data not shown). Even when limited to patients with proven ETEC infection, only a difference of borderline statistical difference was seen for microbiota composition (not involving Escherichia) between placebo and T, but not M phage recipients (Suppl. Table 2).
Fig. 7.
Anti-correlation between Streptococcus and Bifidobacterium abundance in the feces and their association with stool output in diarrhea.
Heat map of correlation between the indicated bacterial genera identified in the stool of diarrhea patients (A). The strength for correlation and anti-correlation is given in the color key at the top left.
Correlation between percentage of Streptococcus (B) or Bifidobacterium (C) in fecal microbiota (abscissa) and stool weight in g/day (ordinate) for patients hospitalized with ETEC diarrhea. Correlation coefficients and their P value are given at the top of the panels.
When the 16S rRNA gene sequence data were analyzed at the species level (Fig. 5B), fecal streptococci (van den Bogert et al., 2013) were categorized into two groups, namely the S. gallolyticus/lutetiensis (Schlegel et al., 2003) and the Streptococcus salivarius groups, with very few S. infantis calls. These were the same streptococcal species which we found, albeit with different proportions, in healthy control children. Likewise, the same Bifidobacterium groups, namely B. longum, B. kashiwanohense, B. bifidum and B. catenulatum group, were detected both in diarrhea patients and healthy controls (Fig. 5B).
3.7. Streptococcal Abundance Correlates With Diarrhea Symptoms
Two observations associated Streptococcus abundance with diarrhea symptoms in the enrolled patients. First, an increase in Streptococcus abundance was observed in patients at day 5 of hospitalization (Fig. 5A). Since 50% of the initially enrolled patients were discharged from the hospital at day 4, these subjects represented more severely affected patients as also seen for quantitative diarrhea parameters at day 5 (Fig. 2C). Interestingly, a fecal microbiota difference was already observed in the early phase of hospitalization between patients later staying for 4 (short hospitalization) or 5 days (long hospitalization) in the hospital: children with an earlier diarrhea resolution also showed an earlier normalization of the Bifidobacterium/Streptococcus ratio and a lower Escherichia percentage than children with a later diarrhea resolution (day 5 or later) (Fig. 5C).
Second, when we plotted the diarrhea stool output of a patient during hospitalization against the Streptococcus abundance in the stool microbiota, we observed a positive correlation. This correlation was significant when all patients, only ETEC patients (Fig. 7B) or diarrhea patients without an E. coli infection were analyzed. Notably, Bifidobacterium stool abundance showed a significant negative correlation with diarrhea stool output (Fig. 7C). Escherichia abundance did not correlate with stool output (data not shown).
3.8. Streptococcal Genomics
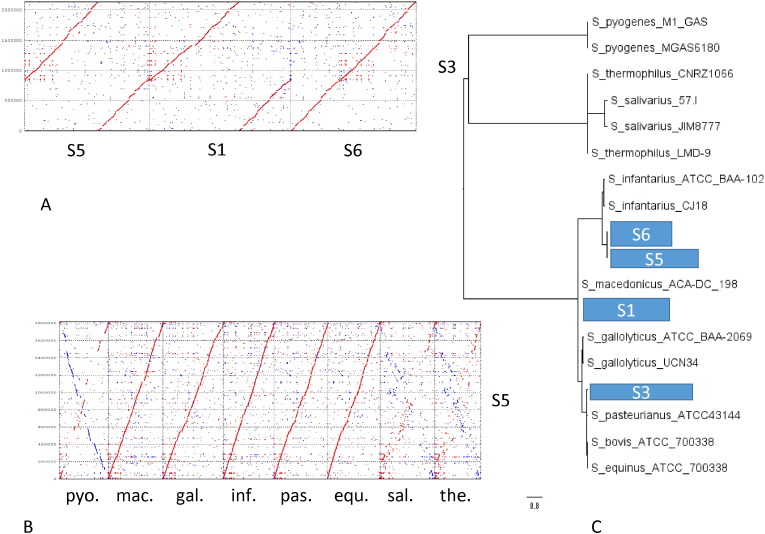
Streptococci were isolated from the stools of three diarrhea patients and identified by API fermentation gallery and 16S rDNA sequencing as S. gallolyticus (strains S1, S3) or S. infantarius (strain S5). Their genome length was 1.8 to 2.05 Mb and represented by a single (S1, S5) or 2 contigs (S3) (Suppl. Table 1). Their genomes were closely related to each other as well as to the sequenced Streptococcus strain S6 isolated from a healthy control child (Fig. 8A). These genomes were closely related to sequenced strains of the S. bovis/S. equinus species complex to which also belong food-associated species (S. pasteurianus, S. macedonicus), commensal species (S. infantarius, S. equinus) and facultative pathogens (S. gallolyticus) (Fig. 8B). By phylogenetic tree analysis based on single nucleotide polymorphisms of whole sequenced genomes (Gardner and Hall, 2013) S5 and S6 were placed close to S. infantarius, S1 associated with S. macedonicus, S3 with S. gallolyticus (Fig. 8C). More distantly related streptococci belonged to the S. salivarius complex comprising oral commensals (S. salivarius) and dairy strains (S. thermophilus) and S. pyogenes, a human pathogen (Fig. 8B, C).
Fig. 8.
Genome comparisons for the isolated and sequenced fecal streptococci.
A: Dotplot alignment of the genome from S. gallolyticus fecal isolate S3 (y-axis) against S. infantarius isolate S5, S. galloyticus isolate S1, and S. infantarius isolate S6 (x-axis) obtained from three different patients and one control (S6) at the DNA sequence level.
B: Dotplot alignment of the genome from the fecal S. infantarius isolate S5 (y-axis) against streptococcal strains of the database at the DNA sequence level. The following genomes were chosen for comparison (from left to right on the x-axis): S. pyogenes (human pathogen), S. macedonicus (dairy strain), S. gallolyticus (human pathogen), S. infantarius (dairy strain), S. pasteurianus (human gut commensal), S. equinus (horse gut commensal), S. salivarius (oral commensal), and S. thermophilus (dairy strain) (from left to right).
C: Phylogenetic tree analysis based on single nucleotide polymorphisms of the indicated sequenced genomes with the kSNPv2 software (Gardner and Hall, 2013). The position of the four sequenced fecal streptococci is indicated by the boxes S1, 3, 4, and 5 next to representative streptococci marked with their identifier.
The S1 chromosome is with a length of 2′052 kb 298 kb shorter than the clinical S. gallolyticus isolate UCN34 isolated from an endocarditis patient (Danne et al., 2011). When using BlastN 80% of S1 chromosome aligns to UCN34 chromosome with an identity of 80% bp identity or more. On the dotplot (Suppl. Fig. 4A), we can see 2 large loci that are absent from S1 chromosome. Locus 1 is approximately 47 kb long (56 prophage genes), and Locus 2 is approximately 60 kb long (71 genes) and comprises the pilus genes which were biologically proven virulence genes of this endocarditis isolate (Danne et al., 2011). Locus 3 is specific to S1 chromosome and is 53.5 kb long.
Our isolates were also closely related to S. lutetiensis strain 0033 (Suppl. Fig. 4B) identified in Chinese children hospitalized with diarrhea for which no known enteropathogen could be identified (Jin et al., 2013). The Chinese researchers proposed numerous virulence factors in their isolate, some were shared by our isolates (Suppl. Table 3). However, upon reinvestigation, alternative annotations were found for them that question their role as virulence genes.
3.9. Fecal Microbiota Association With Diarrhea
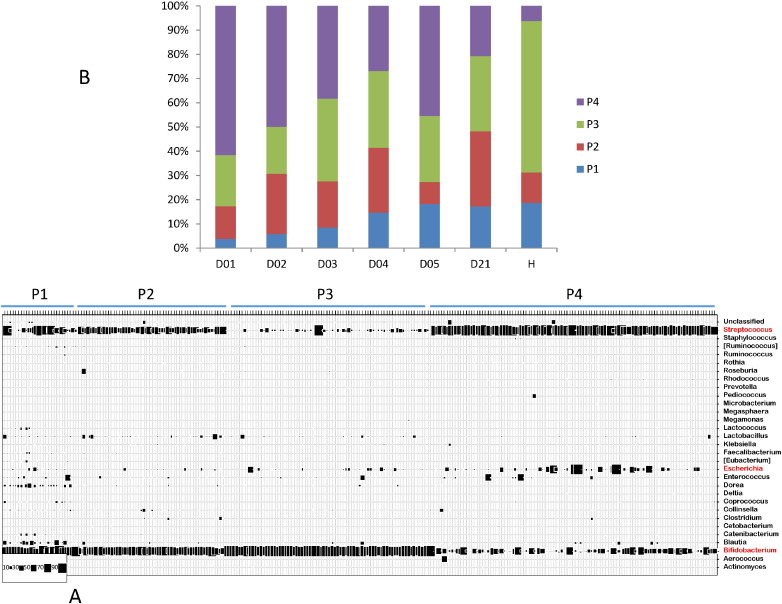
When the three major fecal bacterial genera were analyzed for the transition period from acute diarrhea into recovery, neither Escherichia nor Streptococcus showed a clear association with this transition. We found low abundance of Escherichia in the acute phase of EPEC/EAEC diarrhea patients and a high Streptococcus persistence into the recovery phase in ETEC patients. A high Streptococcus abundance was also seen in some healthy controls (Suppl. Fig. 4). The Dirichlet Multinomial Mixture modeling (Holmes et al., 2012) classified the investigated stool samples in four distinct partitions (“fecal community types”) displayed as a bubble plot in Fig. 9A and described in Suppl. Table 4. The genera Bifidobacterium and Streptococcus are by far the most important contributors to the differences in community structures of the partitions (37 and 31%, respectively), followed by Escherichia and Dorea (4% each) (Suppl. Table 5). Partition 3, which is characterized by a very high abundance of Bifidobacterium (mean = 90.6%) and a low abundance of Streptococcus (4%), was the dominant community type in control children. This community type was lower in diarrhea patients during the acute phase with a trend for increase with recovery (Fig. 9B). Partition 4, which is the most variable partition (Suppl. Table 4) is characterized by a high abundance of Streptococcus (68.5%) combined with either elevated amounts of Escherichia (4.9%) or a reduced amount of Bifidobacterium (16.0%). Compared to controls partition 4 was increased in all diarrhea patients over the hospitalization period (Fig. 9B), and this was independent of diarrhea etiology (data not shown). Partitions 2 and 1 showed medium Bifidobacterium (58.2 and 46.6%, respectively), elevated Streptococcus (34.4 and 16.0%), and small, but widely distributed Dorea (in partition 1 = 7.7%) and very low Escherichia (0.8 and 2.3%, respectively) abundance — this combined constellation was found with comparable abundance in both diarrhea patients and controls while partition P1 on its own increased with recovery from diarrhea (Fig. 9B). Notably, partition 4 gradually decreased with recovery from diarrhea (Fig. 9B), as if fecal streptococci develop a pathological potential only in the presence of Escherichia or if not counter-acted by bifidobacteria.
Fig. 9.
Fecal Community Type analysis in the diarrhea patients and healthy controls.
A. A Dirichlet Multinomial Mixture modeling defined four partitions P1 to P4 (“fecal community types”) in the investigated stool samples. Each vertical line of the bubble plot represents one individual stool sample, each horizontal line represents a bacterial genus identified at the right ordinate. The size of the squares indicates the percentage of 16S rRNA gene sequences represented by this genus in the given stool sample (code at the lower left corner).
B. The relative percentage of partitions P1 to P4 is given for all 56 investigated diarrhea patients for the indicated day of hospitalization (day D01 to 05) and the return visit (day D21) in comparison with age-matched local healthy controls (H).
4. Discussion
In the current PT trial, T4-like phages were chosen for safety concerns (Sarker et al., 2012). T4-like coliphages are obligate lytic viruses, their genomes are well characterized and devoid of bacterial virulence genes and they were propagated on a safe production strain (E. coli K-12, itself devoid of virulence genes and inducible prophages) (Denou et al., 2009). However, the in vivo replication potential of T4 was previously only explored in mice where it was found to be inferior to T7 phage (Weiss et al., 2009). Whether the prolonged T7 phage excretion is an advantage for PT over the shorter elimination time of T4 phage (Bruttin and Brüssow, 2005) is currently unclear. The commercial Russian phage cocktail consisted of at least 17 different phage types including T7 phage (McCallin et al., 2013). The Microgen recipients showed indeed a higher fecal phage titer than T4 recipients, but they did not show a higher prevalence of T7 over T4 in the stool. The higher fecal output in the M recipients is partly explained by a higher oral phage input.
In acute cases of E. coli diarrhea, the barrier function of the gut mucosa in the small intestine is compromised. When the pathogen is lysed by phage, in vivo released endotoxins and enterotoxins can thus potentially cross into the circulation. However, no systemic reaction to endotoxin was observed (Yang et al., 2010) which is not surprising since no significant in vivo replication of the phage was seen, hence no significant lysis of cells with concomitant toxin release had occurred. While oral coliphages as virions are safe in diarrhea patients, it is still unclear whether oral phages are safe when acting as infectious virus lysing the E. coli target cell in the gut of diarrhea patients.
Neither the T4-like nor the Russian coliphage cocktail showed a clinical benefit over standard care in terms of stool output, stool frequency or rehydration. This observation should be interpreted with caution since it could still represent a false-negative outcome because at interim analysis only half of the planned patients were enrolled. The reported trial was thus not powered to detect a treatment effect, but only to assess safety issues and to gain a mechanistic insight into PT. Notably, we did not observe a substantial in vivo replication of the orally applied phage which is a basic requirement for PT. Despite a careful screening, only 60% of the 120 enrolled patients harbored an E. coli pathogen in the stool. E. coli is thus a less frequent cause of diarrhea (18% of all screened patients) than we had anticipated for Bangladeshi children (36% of all screened patients) (Albert et al., 1999). Furthermore, even in microbiologically defined E. coli diarrhea less than 5% of the total fecal bacteria were E. coli. When only ETEC pathogens were counted, a median titer of 105 cfu ETEC per g stool was detected. With an estimated in vivo burst size of 10 (Brüssow, 2013), no phage amplification beyond 106 pfu/g stool was therefore expected, which is close to the observed peak fecal phage titers. That a microbiologically defined E. coli diarrhea was not characterized by high intestinal pathogen titers can also be deduced from the observation that ETEC and EAEC infections in the present study showed only 10-fold higher fecal E. coli titers than diarrhea patients lacking an E. coli pathogen. Likewise, careful quantitative PCR analysis in stools from Peruvian children with EPEC diarrhea showed titers of 3 × 105 EPEC/g stool which were just 10-fold higher than EPEC titers in healthy controls (Barletta et al., 2011). Since the in vitro replication threshold for T4 phage on a laboratory indicator E. coli strain was determined to 103 cfu/ml (Wiggins and Alexander, 1985), the pathogenic E. coli titer in confirmed cases of E. coli diarrhea might simply not be high enough to sustain phage replication in vivo.
From our data it is impossible to predict what percentage of the diarrheal episodes is caused by E. coli. Only a minority of the screened children had an E. coli diarrhea by standard tests. Even less showed a high fecal abundance of E. coli by molecular tests and stool titrations. We cannot deduce from our data whether a low abundance bacterial group could be responsible for diarrhea in children. However, to overcome limitations for future phage therapy trials, we strongly recommend to control first and foremost whether the targeted pathogen occurs with sufficient titers to support in vivo phage replication.
There are still other limitations which have to be considered when interpreting the negative clinical outcome of the present study. Only half of the patients contained phage-sensitive E. coli colonies in the stool. The Russian phage cocktail was obviously not customized to the regional patients and we had to adapt our T4 phage cocktail to the ecological situation in Bangladesh to achieve a reasonable coverage (Bourdin et al., 2014b). Since the phages were isolated several years before the start of the trial, a constant adaption of the phage cocktail to the prevailing ecological situation is needed. However, it is not clear whether phage coverage was indeed the limiting factor in the current trial since the phage susceptibility of the stool isolates did not correlate with fecal phage titers. Both phage preparations were given without antacid. Buffering of gastric acidity by a slightly alkaline mineral water was perhaps too weak and an additional dose of bicarbonate as recommended by the commercial supplier might have increased phage transit through the stomach. Since it would also increase the risk of nosocomial cross-infection in a diarrhea hospital, such a treatment was excluded for ethical reasons. However, we do not believe that pharmaceutic formulation of the phage was a limiting factor in the current trial since children have a higher (more alkaline) stomach pH than adults. Indeed, in previous trials sensitive biologicals were given to children with diarrhea (antibodies, probiotics), which were recovered intact from the feces (Hilpert et al., 1987) or achieved clinical effects in Bangladesh (Sarker et al., 1998, Sarker et al., 2005). However, a higher oral phage dose might be desirable since children in Bangladesh showed a less good fecal bioavailability of phage than adults from Switzerland (Bruttin and Brüssow, 2005) suggesting some loss during gastrointestinal passage.
Regarding the lack of in vivo susceptibility of E. coli to coliphages, several factors need consideration. Prominent are physical barriers for phages to reach E. coli (increased peristalsis in diarrhea, reduced phage diffusion rate in mucus layer, pathogen protection in biofilms) or slow growth rate of bacterial cells in the stationary phase (Brüssow, 2013). It will be important to study E. coli-phage interaction in vivo with animal experiments. However, E. coli does not induce diarrhea in small laboratory animals and infections with Citrobacter rodentium in mice, commonly used as a surrogate, are not a suitable model system since it induces a colitis (Collins et al., 2014). Experiments in animals like piglets or calves which suffer naturally from E. coli infection in the small intestine and diarrhea (Fairbrother et al., 2005) are clearly needed.
Another important limitation of our study is the question whether our observations in the fecal samples represent what is happening within the gut. From an analytical viewpoint it would be desirable to analyze bacteria and phages in samples from the upper small intestine. ETEC and EPEC infections occur in the upper parts of the gut and the bacteria excreted with stool might not any longer be in the same growth phase as in the small intestine (stationary vs. logarithmic growth). However, such sampling is invasive and ethically difficult to justify in a normally self-limiting disease in children. Again, dissection experiments in experimentally infected and phage-treated pigs might provide the needed insight. In fact, data from pig have supported the “kill-the-winner” model of phage–bacterium population dynamics also for the gut ecosystem (Allen et al., 2011) suggesting that high titers of target bacteria are needed for PT.
ETEC showed only a short-lived stool peak titer which corresponds to recent reports describing in adult cholera patients from Bangladesh an even shorter peak titer of V. cholerae maintained for just 6 h (Hsiao et al., 2014). Curiously, the peak titer of ETEC occurred on the second day of hospitalization as if ETEC was not the cause for hospital admission. In fact, the role of E. coli as a pathogen has recently been challenged for its low pathogenicity index. Only certain E. coli pathotypes in selected age groups of children and geographical areas were by this criterion significantly linked with diarrhea (Taniuchi et al., 2013, Liu et al., 2014, Kotloff et al., 2013). In fact, E. coli diarrhea in children from developing countries is increasingly considered as a polymicrobial infection (Taniuchi et al., 2013, Kotloff et al., 2013) as was also seen in the present study.
Microbiota analysis by 16S rRNA gene sequencing confirmed that only a small fraction of the stool microbiota from children hospitalized with a microbiologically diagnosed E. coli diarrhea were represented by Escherichia. At admission, the dominant stool bacteria were fecal streptococci independently whether ETEC, EAEC or non-E. coli diarrhea cases were considered. Comparison with healthy local controls suggests a marked streptococcal dysbiosis in the acute phase of diarrhea. Notably, the relative abundance of streptococci was positively correlated with stool output, it predicted the length of diarrhea duration and in EAEC or non-E. coli diarrhea it decreased with the resolution of diarrhea. Is a fecal streptococcal dysbiosis thus a potential biomarker for diarrhea? Since all these observations apply to the average of the studied population and not to individuals, who showed great variation, this marker is of limited use for the clinical microbiologist. However, fecal streptococci diagnosed as S. salivarius also represented a prominent part of the acute diarrhea microbiota in adult cholera patients from Bangladesh (Hsiao et al., 2014). E. coli diarrhea in adults and children from Bangladesh showed an ordered gut microbiota succession and fecal streptococci belonged to this sequence (David et al., 2015). In the GEMS survey children from Bangladesh displayed a significant increase in fecal streptococci during diarrhea. This was, although at lower level, also observed in African children (Pop et al., 2014). Are fecal streptococci therefore candidate new enteropathogens as suggested by Chinese researchers (Jin et al., 2013)? This is unlikely for four reasons: first, using effluents from ileostoma patients, fecal streptococci of the S. bovis complex have been identified as typical small intestine commensal in humans (Booijink et al., 2010). Fecal streptococci are fast-growing, efficient fermenters of simple carbohydrates (van den Bogert et al., 2013) possibly allowing a transient outgrowth when competing gut bacteria are purged from the gut by increased peristalsis in diarrhea. Second, no diarrhea-relevant virulence genes were detected in the three sequenced genomes of fecal streptococci isolated from diarrhea patients. Notably, a biologically proven virulence (pilus) gene identified in a S. gallolyticus isolated from an endocarditis patient (Danne et al., 2011) was not detected in the stool isolates. Third, an increased abundance of S. gallolyticus was also seen in colon cancer patients (Boleij and Tjalsma, 2013). Bacteria of the S. gallolyticus species complex might adhere to the chronically (colon cancer) or transiently (diarrhea) injured gut mucosa. Interestingly, the replacement of the streptococci by fecal bifidobacteria occurs with the same kinetics as the repair of the gut mucosa with newly synthesized enterocytes. Finally, the analysis of the fecal microbial community types showed that elevated fecal Streptococcus abundance had no pathogenic potential as long as they are counter-balanced by an elevated Bifidobacterium abundance. This could mean that our understanding of diarrhea etiology will have to change in the future: not only need quantitative pathogen titers to be determined, pathogen constellations and pathogens interactions with commensal bacteria need to be considered for the understanding of diarrhea. In our dataset, loss of bifidobacteria was in fact the best biomarker for diarrhea.
Based on these considerations and the presented microbiological and clinical observations, we decided to stop the PT trial at interim analysis. In summary, while phage therapy is still one very important alternative tool against antibiotic resistance, we need to understand in detail the in vivo interactions between phage and its host bacterium to judge the reliability and efficacy of such approaches. At the moment it is risky to conclude whether phage therapy is or is not a viable option against antibiotic-resistant bacterial pathogens.
Competing Interests
The authors listed under institutions b and c are paid employees of the two specified research centers of Nestec Ltd., but have otherwise no financial interest in phage therapy.
The sponsor (Nestlé Nutrition and Nestlé Health Science) was not involved in data evaluation.
Author Contribution
SAS is the PI of the clinical study responsible for study design, protocol writing, for defense of the protocol to institutional review boards, data collection tool development, data analysis, interpretation and manuscript writing; SS is the Co-PI of the clinical study responsible for defending the protocol at IRB, data collection tool development and data collection; GR did stool phage counting and streptococcal isolation; DM did library preparation, the sequencing and the assembly of the Strep genomes; PD did design the de novo genome assembly and data interpretation; FC did the phage susceptibility testing of fecal E. coli isolates; GB did the T4-like coliphages cocktail development, GMP production and release; SMC did the PCR detection of E. coli virulence genes in stool DNA; CN-B contributed to the bioinformatic analysis of the microbiota and the streptococcal genomes; TN did the statistical analysis; MA did the viable E. coli counts in fresh stools; SH collected the clinical data; FQ contributed to study design, data interpretation and supervised the etiology analysis in the clinical microbiology lab; KT did clinical microbiology data collection; MK did streptococcal genome data submission and annotation; MD did the qPCR analysis of elt and est genes; CL did the qPCR of total stool bacteria; YD searched the streptococcal genomes for virulence genes; SEA did the Dirichlet Multinomial Mixtures analysis; BB did acquisition, analysis and interpretation of microbiota data, and contributed to the writing of manuscript; HB designed with the PI the clinical protocol, coordinated and supervised the project and wrote the manuscript.
Acknowledgments
We thank our colleagues Lionel Philippe (Clinical Development Unit); Thierry von der Weid and Nelly Conus (Nestlé Nutrition) and Martinas Kuslys (Nestlé Health Science) for their help in the execution of the clinical trial. The Nestlé Research Centre, Lausanne, Switzerland, provides support to icddr,b for its operations and research. icddr,b gratefully acknowledges their support and commitment to icddr,b's research efforts. Our heartfelt thanks also go to the nurses and research assistants from icddr,b for their support and help.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.12.023.
Appendix A. Supplementary data
Supplementary Fig. 1 Fecal coliphage counts in diarrhea patients differentiated according to etiology and fecal E. coli phage susceptibility. A. The distribution of fecal phage titers in pfu/g stool (ordinate) is given for individual stool samples from patients without E. coli diagnosis (left vertical column), ETEC diagnosis (middle column) or EPEC and EAEC diagnosis (right column). B. The distribution of fecal phage titers is given as a scattergram with in pfu/g stool on the ordinate and individual stool samples on the abscissa (stool samples 1–60 did not contain fecal E. coli colonies susceptible to the oral coliphages while stool samples 61 to 120 contained phage-susceptible E. coli colonies).
Supplementary Fig. 2 Differentiation of T4- and T7-like phages in the stool samples by plaque assay. Large and small plaques from Microgen phage cocktail displayed on E. coli indicator cell WG-5 (A). Phages from a large plaque were amplified and identified as T7-like phages by negative stain electron microscopy (B). Phages from small plaques were likewise amplified and identified as T4-like phages by electron microscopy (C). Small and large plaques were counted in stool samples from Microgen cocktail recipients and the ratio of small/large plaques was plotted for 18 individual stool samples aligned along the abscissa. Ratios close to unity indicate that the about 1:1 proportion of T4 over T7 phages found in the oral cocktail was not distorted after gastrointestinal passage.
Supplementary Fig. 3 Genome comparisons for the isolated and sequenced fecal streptococci. A: Alignment of the genome from fecal S. gallolyticus isolate S1 from a diarrhea patient in Bangladesh with the genome of a S. gallolyticus isolate UCN_34 from an endocarditis patient. The arrow marks the virulence genes in UNC_34 (Danne et al., 2011) missing in strain S1. B: Alignment of the Streptococcus isolates S3, S1, S6 and S5 (from top to bottom) with the S. lutetiensis strain 0033 isolated from a Chinese child with diarrhea without identified enteropathogen (Jin et al., 2013).
Supplementary Figure 4 Association of bacterial genera with diarrhea resolution. Relative abundance of bacteria belonging to the genera Bifidobacterium, Escherichia, and Streptococcus as percent of total (ordinate, bars not reaching 100% contain still other bacterial genera, color code in box at bottom right) in individual stool samples from healthy controls (H) and diarrhea patients separated according to etiology (ON: other bacteria, no pathogen; PA: EPEC or EAEC infection; T: ETEC infection). The patients are further separated on the abscissa according to the days of hospitalization as specified in the color codes at the bottom left.
Supplementary tables
References
- Ahmed T., Bhuiyan T.R., Zaman K., Sinclair D., Qadri F. Vaccines for preventing enterotoxigenic Escherichia coli (ETEC) diarrhoea. Cochrane Database Syst. Rev. 2013;7 doi: 10.1002/14651858.CD009029.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M.J., Faruque A.S., Faruque S.M., Sack R.B., Mahalanabis D. Case–control study of enteropathogens associated with childhood diarrhea in Dhaka, Bangladesh. J. Clin. Microbiol. 1999;37:3458–3464. doi: 10.1128/jcm.37.11.3458-3464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen H.K., Looft T., Bayles D.O. Antibiotics in feed induce prophages in swine fecal microbiomes. MBio. 2011;2 doi: 10.1128/mBio.00260-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barletta F., Ochoa T.J., Mercado E. Quantitative real-time polymerase chain reaction for enteropathogenic Escherichia coli: a tool for investigation of asymptomatic versus symptomatic infections. Clin. Infect. Dis. 2011;53:1223–1229. doi: 10.1093/cid/cir730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boleij A., Tjalsma H. The itinerary of Streptococcus gallolyticus infection in patients with colonic malignant disease. Lancet Infect. Dis. 2013;13(8):719–724. doi: 10.1016/S1473-3099(13)70107-5. [DOI] [PubMed] [Google Scholar]
- Booijink C.C. High temporal and inter-individual variation detected in the human ileal microbiota. Environ. Microbiol. 2010;12(12):3213–3227. doi: 10.1111/j.1462-2920.2010.02294.x. [DOI] [PubMed] [Google Scholar]
- Bourdin G., Navarro A., Sarker S.A. Coverage of diarrhoea-associated Escherichia coli isolates from different origins with two types of phage cocktails. Microb. Biotechnol. 2014;7:165–176. doi: 10.1111/1751-7915.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdin G., Schmitt B., Marvin Guy L. Amplification and purification of T4-like Escherichia coli phages for phage therapy: from laboratory to pilot scale. Appl. Environ. Microbiol. 2014;80:1469–1476. doi: 10.1128/AEM.03357-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüssow H. Phage therapy: the Escherichia coli experience. Microbiology. 2005;151:2133–2140. doi: 10.1099/mic.0.27849-0. [DOI] [PubMed] [Google Scholar]
- Brüssow H. What is needed for phage therapy to become a reality in Western medicine? Virology. 2012;434:138–142. doi: 10.1016/j.virol.2012.09.015. [DOI] [PubMed] [Google Scholar]
- Brüssow H. Bacteriophage–host interaction: from splendid isolation into a messy reality. Curr. Opin. Microbiol. 2013;16:500–506. doi: 10.1016/j.mib.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Bruttin A., Brüssow H. Human volunteers receiving Escherichia coli phage T4 orally: a safety test of phage therapy. Antimicrob. Agents Chemother. 2005;49:2874–2878. doi: 10.1128/AAC.49.7.2874-2878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casswall T.H., Sarker S.A., Faruque S.M. Treatment of enterotoxigenic and enteropathogenic Escherichia coli-induced diarrhoea in children with bovine immunoglobulin milk concentrate from hyperimmunized cows: a double-blind, placebo-controlled, clinical trial. Scand. J. Gastroenterol. 2000;35:711–718. doi: 10.1080/003655200750023372. [DOI] [PubMed] [Google Scholar]
- Chen L., Xiong Z., Sun L., Yang J., Jin Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 2012;40(Database issue):D641–D645. doi: 10.1093/nar/gkr989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibani-Chennoufi S., Sidoti J., Bruttin A. Isolation of Escherichia coli bacteriophages from the stool of pediatric diarrhea patients in Bangladesh. J. Bacteriol. 2004;186:8287–8294. doi: 10.1128/JB.186.24.8287-8294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C.S. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10(6):563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- Collins J.W., Keeney K.M., Crepin V.F. Citrobacter rodentium: infection, inflammation and the microbiota. Nat. Rev. Microbiol. 2014;12:612–623. doi: 10.1038/nrmicro3315. [DOI] [PubMed] [Google Scholar]
- Danne C. Molecular characterization of a Streptococcus gallolyticus genomic island encoding a pilus involved in endocarditis. J. Infect. Dis. 2011;204:1960–1970. doi: 10.1093/infdis/jir666. [DOI] [PubMed] [Google Scholar]
- David L.A. Gut microbial succession follows acute secretory diarrhea in humans. MBio. 2015;6(3) doi: 10.1128/mBio.00381-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denou E., Bruttin A., Barretto C., Ngom-Bru C., Brüssow H., Zuber S. T4 phages against Escherichia coli diarrhea: potential and problems. Virology. 2009;388:21–30. doi: 10.1016/j.virol.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Ding T., Schloss P.D. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother J.M., Nadeau E., Gyles C.L. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev. 2005;6:17–39. doi: 10.1079/ahr2005105. [DOI] [PubMed] [Google Scholar]
- Gardner S.N., Hall B.G. When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One. 2013;8(12):e81760. doi: 10.1371/journal.pone.0081760. (2013 Dec 9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith C.S., Miller S.E. Modern use of electron microscopy for detection of viruses. Clin. Microbiol. Rev. 2009;22:552–563. doi: 10.1128/CMR.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guion C.E., Ochoa T.J., Walker C.M., Barletta F., Cleary T.G. Detection of diarrheagenic Escherichia coli by use of melting-curve analysis and real-time multiplex PCR. J. Clin. Microbiol. 2008;46:1752–1757. doi: 10.1128/JCM.02341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán C., Mocé-Llivina L., Lucena F., Jofre J. Evaluation of Escherichia coli host strain CB390 for simultaneous detection of somatic and F-specific coliphages. Appl. Environ. Microbiol. 2008;74:531–534. doi: 10.1128/AEM.01710-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A.M., Chowdhury F., Begum Y.A. Shifting prevalence of major diarrheal pathogens in patients seeking hospital care during floods in 1998, 2004, and 2007 in Dhaka, Bangladesh. Am.J.Trop. Med. Hyg. 2008;79:708–714. [PMC free article] [PubMed] [Google Scholar]
- Hilpert H., Brüssow H., Mietens C., Sidoti J., Lerner L., Werchau H. Use of bovine milk concentrate containing antibody to rotavirus to treat rotavirus gastroenteritis in infants. J. Infect. Dis. 1987;156:158–166. doi: 10.1093/infdis/156.1.158. [DOI] [PubMed] [Google Scholar]
- Holmes I., Harris K., Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao A., Ahmed A.M., Subramanian S. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z.D., Lowe B., Verenkar M.P. Prevalence of enteric pathogens among international travelers with diarrhea acquired in Kenya (Mombasa), India (Goa), or Jamaica (Montego Bay) J. Infect. Dis. 2002;185:497–502. doi: 10.1086/338834. [DOI] [PubMed] [Google Scholar]
- Jin D. Dynamics of fecal microbial communities in children with diarrhea of unknown etiology and genomic analysis of associated Streptococcus lutetiensis. BMC Microbiol. 2013;13 doi: 10.1186/1471-2180-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junick J., Blaut M. Quantification of human fecal bifidobacterium species by use of quantitative real-time PCR analysis targeting the groEL gene. Appl. Environ. Microbiol. 2012;78:2613–2622. doi: 10.1128/AEM.07749-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff K.L., Nataro J.P., Blackwelder W.C. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case–control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- Liu B., Pop M. ARDB—antibiotic resistance genes database. Nucleic Acids Res. 2009;37(Database issue):D443–D447. doi: 10.1093/nar/gkn656. (2009 Jan) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Kabir F., Manneh J. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect. Dis. 2014;14:716–724. doi: 10.1016/S1473-3099(14)70808-4. [DOI] [PubMed] [Google Scholar]
- Martinez L. Options for Action. WHO; Geneva: 2012. The evolving threat of antimicrobial resistance. [Google Scholar]
- McCallin S., Sarker S.A., Barretto C. Safety analysis of a Russian phage cocktail: from MetaGenomic analysis to oral application in healthy human subjects. Virology. 2013;443:187–196. doi: 10.1016/j.virol.2013.05.022. [DOI] [PubMed] [Google Scholar]
- Międzybrodzki R., Borysowski J., Weber-Dąbrowska B. Clinical aspects of phage therapy. Adv. Virus Res. 2012;83:73–121. doi: 10.1016/B978-0-12-394438-2.00003-7. [DOI] [PubMed] [Google Scholar]
- Nadkarni M.A., Martin F.E., Jacques N.A., Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- Piddock L.J. The crisis of no new antibiotics—what is the way forward? Lancet Infect. Dis. 2012;12:249–253. doi: 10.1016/S1473-3099(11)70316-4. [DOI] [PubMed] [Google Scholar]
- Pop M. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol. 2014;15(6):R76. doi: 10.1186/gb-2014-15-6-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadri F., Svennerholm A.M., Faruque A.S., Sack R.B. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 2005;18:465–483. doi: 10.1128/CMR.18.3.465-483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Sanchez M., Darimont C., Drapeau V. Effect of Lactobacillus rhamnosus CGMCC1.3724 supplementation on weight loss and maintenance in obese men and women. Br. J. Nutr. 2014;111:1507–1519. doi: 10.1017/S0007114513003875. [DOI] [PubMed] [Google Scholar]
- Sarker S.A., Casswall T.H., Mahalanabis D. Successful treatment of rotavirus diarrhea in children with immunoglobulin from immunized bovine colostrum. Pediatr. Infect. Dis. J. 1998;17:1149–1154. doi: 10.1097/00006454-199812000-00010. [DOI] [PubMed] [Google Scholar]
- Sarker S.A., McCallin S., Barretto C. Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology. 2012;434:222–232. doi: 10.1016/j.virol.2012.09.002. [DOI] [PubMed] [Google Scholar]
- Sarker S.A., Sultana S., Fuchs G.J. Lactobacillus paracasei strain ST11 has no effect on rotavirus but ameliorates the outcome of nonrotavirus diarrhea in children from Bangladesh. Pediatrics. 2005;116:e221–e228. doi: 10.1542/peds.2004-2334. [DOI] [PubMed] [Google Scholar]
- Schlegel L., Grimont F., Ageron E., Grimont P.A., Bouvet A. Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. Int. J. Syst. Evol. Microbiol. 2003;53(Pt 3):631–645. doi: 10.1099/ijs.0.02361-0. [DOI] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 2011;77:3219–3226. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeoni U., Berger B., Junick J. Gut microbiota analysis reveals a marked shift to bifidobacteria by a starter infant formula containing a synbiotic of bovine milk-derived oligosaccharides and Bifidobacterium animalis subsp. lactis CNCM I-3446. Environ. Microbiol. 2015 doi: 10.1111/1462-2920.13144. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Stanton T.B. A call for antibiotic alternatives research. Trends Microbiol. 2013;21:111–113. doi: 10.1016/j.tim.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Sulakvelidze A., Alavidze Z., Morris J.G., Jr. Bacteriophage therapy. Antimicrob. Agents Chemother. 2001;45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenungsson B., Lagergren A., Ekwall E. Enteropathogens in adult patients with diarrhea and healthy control subjects: a 1-year prospective study in a Swedish clinic for infectious diseases. Clin. Infect. Dis. 2000;30:770–778. doi: 10.1086/313770. [DOI] [PubMed] [Google Scholar]
- Taniuchi M., Sobuz S.U., Begum S. Etiology of diarrhea in Bangladeshi infants in the first year of life analyzed using molecular methods. J. Infect. Dis. 2013;208:1794–1802. doi: 10.1093/infdis/jit507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bogert B., Boekhorst J., Herrmann R., Smid E.J., Zoetendal E.G., Kleerebezem M. Comparative genomics analysis of Streptococcus isolates from the human small intestine reveals their adaptation to a highly dynamic ecosystem. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0083418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M., Kruger E., Durán C. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. J. Clin. Microbiol. 2005;43:5362–5365. doi: 10.1128/JCM.43.10.5362-5365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M., Denou E., Bruttin A., Serra-Moreno R., Dillmann M.L., Brüssow H. In vivo replication of T4 and T7 bacteriophages in germ-free mice colonized with Escherichia coli. Virology. 2009;393:16–23. doi: 10.1016/j.virol.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Wiggins B.A., Alexander M. Minimum bacterial density for bacteriophage replication: implications for significance of bacteriophages in natural ecosystems. Appl. Environ. Microbiol. 1985;49:19–23. doi: 10.1128/aem.49.1.19-23.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A., Hawkins C.H., Anggård E.E., Harper D.R. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 2009;34:349–357. doi: 10.1111/j.1749-4486.2009.01973.x. [DOI] [PubMed] [Google Scholar]
- Yang C.J., Lee N.Y., Lin Y.H. Jarisch–Herxheimer reaction after penicillin therapy among patients with syphilis in the era of the HIV infection epidemic: incidence and risk factors. Clin. Infect. Dis. 2010;51:976–979. doi: 10.1086/656419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 Fecal coliphage counts in diarrhea patients differentiated according to etiology and fecal E. coli phage susceptibility. A. The distribution of fecal phage titers in pfu/g stool (ordinate) is given for individual stool samples from patients without E. coli diagnosis (left vertical column), ETEC diagnosis (middle column) or EPEC and EAEC diagnosis (right column). B. The distribution of fecal phage titers is given as a scattergram with in pfu/g stool on the ordinate and individual stool samples on the abscissa (stool samples 1–60 did not contain fecal E. coli colonies susceptible to the oral coliphages while stool samples 61 to 120 contained phage-susceptible E. coli colonies).
Supplementary Fig. 2 Differentiation of T4- and T7-like phages in the stool samples by plaque assay. Large and small plaques from Microgen phage cocktail displayed on E. coli indicator cell WG-5 (A). Phages from a large plaque were amplified and identified as T7-like phages by negative stain electron microscopy (B). Phages from small plaques were likewise amplified and identified as T4-like phages by electron microscopy (C). Small and large plaques were counted in stool samples from Microgen cocktail recipients and the ratio of small/large plaques was plotted for 18 individual stool samples aligned along the abscissa. Ratios close to unity indicate that the about 1:1 proportion of T4 over T7 phages found in the oral cocktail was not distorted after gastrointestinal passage.
Supplementary Fig. 3 Genome comparisons for the isolated and sequenced fecal streptococci. A: Alignment of the genome from fecal S. gallolyticus isolate S1 from a diarrhea patient in Bangladesh with the genome of a S. gallolyticus isolate UCN_34 from an endocarditis patient. The arrow marks the virulence genes in UNC_34 (Danne et al., 2011) missing in strain S1. B: Alignment of the Streptococcus isolates S3, S1, S6 and S5 (from top to bottom) with the S. lutetiensis strain 0033 isolated from a Chinese child with diarrhea without identified enteropathogen (Jin et al., 2013).
Supplementary Figure 4 Association of bacterial genera with diarrhea resolution. Relative abundance of bacteria belonging to the genera Bifidobacterium, Escherichia, and Streptococcus as percent of total (ordinate, bars not reaching 100% contain still other bacterial genera, color code in box at bottom right) in individual stool samples from healthy controls (H) and diarrhea patients separated according to etiology (ON: other bacteria, no pathogen; PA: EPEC or EAEC infection; T: ETEC infection). The patients are further separated on the abscissa according to the days of hospitalization as specified in the color codes at the bottom left.
Supplementary tables