Abstract
Genome-wide association studies (GWAS) have identified >100 independent susceptibility loci for prostate cancer, including the hot spot at 8q24. However, how genetic variants at this locus confer disease risk hasn’t been fully characterized. Using circularized chromosome conformation capture (4C) coupled with next-generation sequencing and an enhancer at 8q24 as “bait”, we identified genome-wide partners interacting with this enhancer in cell lines LNCaP and C4-2B. These 4C-identified regions are distributed in open nuclear compartments, featuring active histone marks (H3K4me1, H3K4me2 and H3K27Ac). Transcription factors NKX3-1, FOXA1 and AR (androgen receptor) tend to occupy these 4C regions. We identified genes located at the interacting regions, and found them linked to positive regulation of mesenchymal cell proliferation in LNCaP and C4-2B, and several pathways (TGF beta signaling pathway in LNCaP and p53 pathway in C4-2B). Common genes (e.g. MYC and POU5F1B) were identified in both prostate cancer cell lines. However, each cell line also had exclusive genes (e.g. ELAC2 and PTEN in LNCaP and BRCA2 and ZFHX3 in C4-2B). In addition, BCL-2 identified in C4-2B might contribute to the progression of androgen-refractory prostate cancer. Overall, our work reveals key genes and pathways involved in prostate cancer onset and progression.
Prostate cancer (PCa) is one of the most common types of cancer affecting men worldwide. According to American Cancer Society: Cancer Facts and Figures 2015, around 220,800 new cases and 27,540 deaths are expected in the United States in 2015. Genetic predisposition to prostate cancer has been established by epidemiological studies. One of the most important goals of research in the field is to uncover how genetic variations affect phenotype. Genome-wide association studies (GWAS) have been successfully applied to identify genetic variants associated with a specific trait1,2. Many GWAS explored the association between single nucleotide polymorphisms (SNPs) and diseases, especially ones of a complex genetic nature. GWAS in diverse populations (European ancestry, African ancestry, Japanese ancestry, and Latino ancestry, reviewed in Al Olama et al.3 and Han et al.4) have identified >100 independent prostate cancer susceptibility loci and hundreds of potentially functional SNPs3,4,5,6,7,8,9,10,11,12,13. Of note, the vast majority of these SNPs or their surrogates in linkage disequilibrium are within putative enhancers12 in non-coding regions. It was shown that variants in risk enhancers could significantly affect androgen sensitivity in prostate cancer cells12. That leads to the question of how these “non-coding” loci alter cell property and confer disease risk. However, limited work has been done to tackle this problem and little is known about the functional mechanism of variants in these non-coding loci.
In this study, we focused on 8q24, an extensively studied locus that has been confirmed as a hotspot increasing prostate cancer risk14. Al Olama et al.15 confirmed three regions at 8q24 as risk regions for prostate cancer — region 1: 128.54–128.62 Mb, region 2: 128.14–128.28 Mb, and region 3: 128.47–128.54 Mb, and also characterized additional linkage disequilibrium blocks. Of note, regions 1 and 3 displayed minimal transcriptional activity16, and region 2 showed no association between risk SNPs and the gene expression of nearby transcripts17. In addition, cancer risk variants at 8q24 are mostly distributed in a block depleted of genes, with the closest gene being MYC, a proto-oncogene that is located ~200 kb telomeric16. Thus, we hypothesize that the genetic variation at the 8q24 locus may influence cancer risk by dysregulating the expression of distally located genes. Previous studies suggest that this long-range connection might be possible, often with loci in far-cis and even in trans. For example, in colorectal and prostate cancer a region encompassing risk variant rs6983267, also located in 8q24, interacts with MYC which resides approximately 335 kb telomeric17,18.
Jia et al.16 has fully characterized the functional enhancers at the gene-poor 8q24 locus by combining RNA expression, histone modifications and transcription factor binding profiles, and verified their activity using reporter assays. Among the 15 tested enhancer regions called AcP1 through AcP15, AcP10 has the most significant enhancer activities in prostate cancer risk region. Thus we set out to map the genome-wide contacts of the 8q24 risk enhancer AcP10 region in prostate cancer cells, hoping to understand (1) what target genes are influenced by this risk enhancer at a genome-wide scale, and (2) how these interactions contribute to prostate cancer risk. Utilizing circularized chromosome conformation capture (4C) coupled with next-generation sequencing, we can identify all the interacting partners of a given locus of interest at a genome-wide scale19,20. Previously, our group applied 4C-seq to explore the contact landscapes around the Oct4 distal enhancer in mouse and human pluripotent stem cells21,22,23. We employed a similar 4C-seq strategy to discover all genomic loci interacting with the enhancer at the 8q24 prostate cancer risk locus.
We selected two representative prostate cancer cell lines: lymph node cancer of the prostate (LNCaP) cells, which were isolated from lymph node metastasis and are androgen sensitive24, and C4-2B cells, which were derived from bone metastasis of the LNCaP parental line25 and can grow in an androgen independent way, but are still responsive to androgen in certain aspects26. We reasoned that the contact landscapes around the 8q24 risk locus revealed by 4C-seq in these two cell lines may shed light on the network(s) regulating prostate cancer initiation and/or progression.
Results
We identified 8q24 enhancer interactomes in LNCaP and C4-2B cells
To identify partners interacting with the 8q24 risk enhancer genome-wide, we applied the 4C technique (Fig. 1A), using enzymatic digestion by BglII as the primary fragmentation method as applied in many other nuclear organization studies19,20,21,22. Cells were fixed with paraformaldehyde, and chromosome segments in physical proximity were crosslinked. Then chromatin was digested by BglII, and the ends of interacting chromatin regions were ligated with T4 ligase. We designed primers (Fig. 1B, see Methods) to specifically amplify interacting partners with the bait for library construction. Two replicate libraries for LNCaP and C4-2B were subjected to next-generation sequencing on the Illumina platform. The sequencing reads were analyzed with our custom analysis pipeline27. Circos plots depicting the interaction profiles centered on the bait are shown in Supplementary Fig. S1.
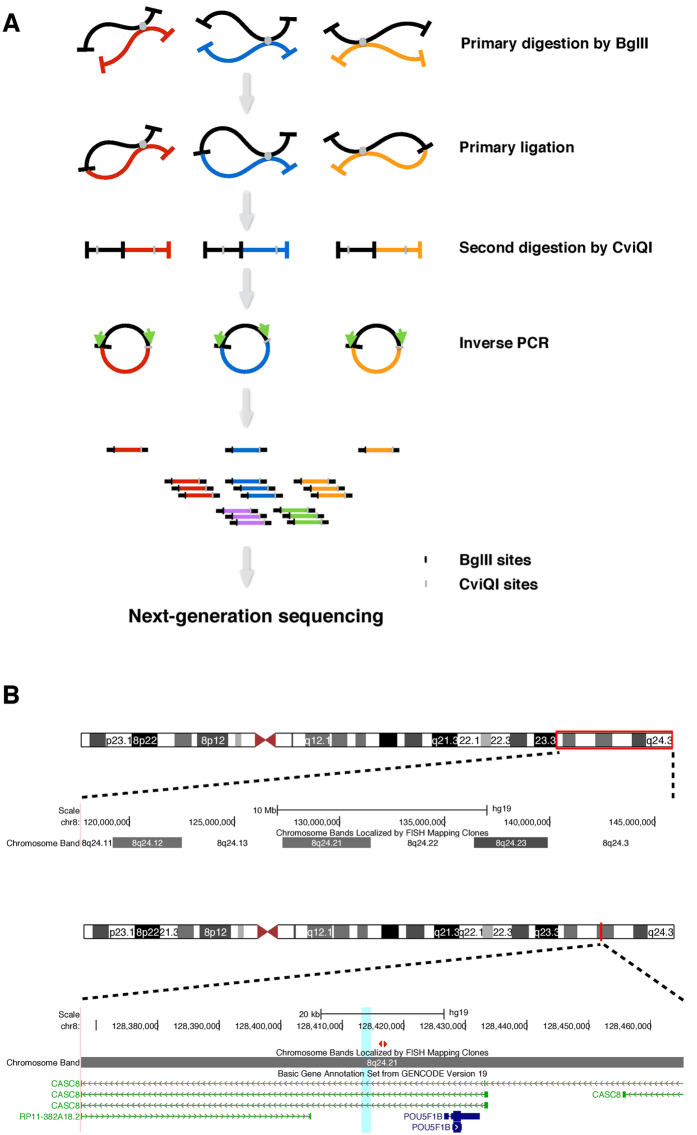
Figure 1. Overview of 4C-seq process and bait region at 8q24 locus.
(A) “Bait” segment is shown in black while “capture” segments are shown in red, blue and orange. Two rounds of digestion are included, and inverse PCR is performed to amplify unknown captured sequence. (B) Upper panel shows a broad view of human 8q24 locus, and lower panel shows a detailed view of AcP10 region at 8q24 locus. AcP10 region is highlighted in cyan. The pair of primers for inverse PCR is shown in red triangles. In gene annotation track, coding gene (POU5F1B) is shown in blue, while non-coding genes (CASC8 and RP11-382A18.2) are shown in green.
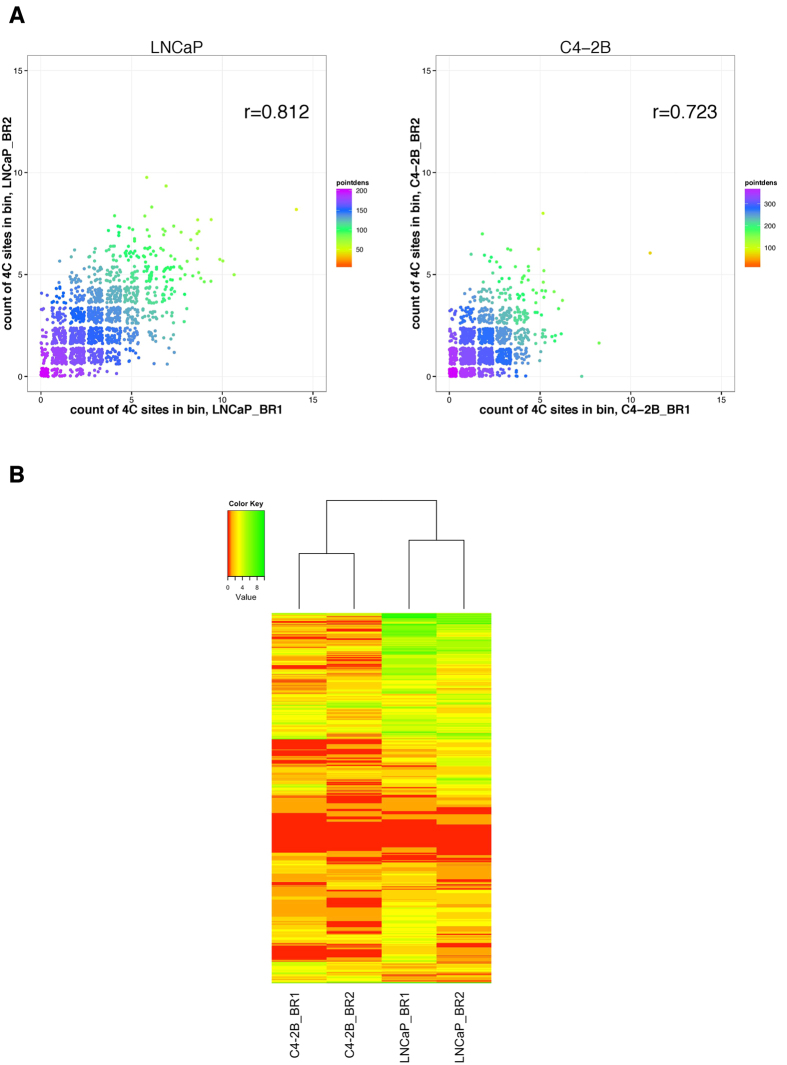
We evaluated reproducibility of interactions between biological replicates by counting trans interactions in every 2 Mb genomic bin and cis interactions in every 1 Mb bin. The Pearson’s correlation coefficient was 0.812 between replicates of LNCaP and 0.723 between replicates of C4-2B (Fig. 2A), indicating good consistency between replicates. Furthermore, clustering results showed that replicates of LNCaP and replicates of C4-2B separated well into two clusters (Fig. 2B). This suggests that replicates of the same cell line have consistent 4C results and the risk enhancer interaction profiles of LNCaP and C4-2B differ to some extent.
Figure 2. Consistency between replicates.
(A) Scatter plot showing interactions density. The numbers of interaction sites in each genomic bin (2Mb trans and 1Mb cis) in two replicates are plotted. Color scale indicates interaction density and Pearson correlation coefficient is shown in the panel. (B) Heatmap and dendrogram showing the interactions density and clustering of samples. Complete linkage method with Euclidean distance measure is used. Color scale indicates interaction density. BR: biological replicate.
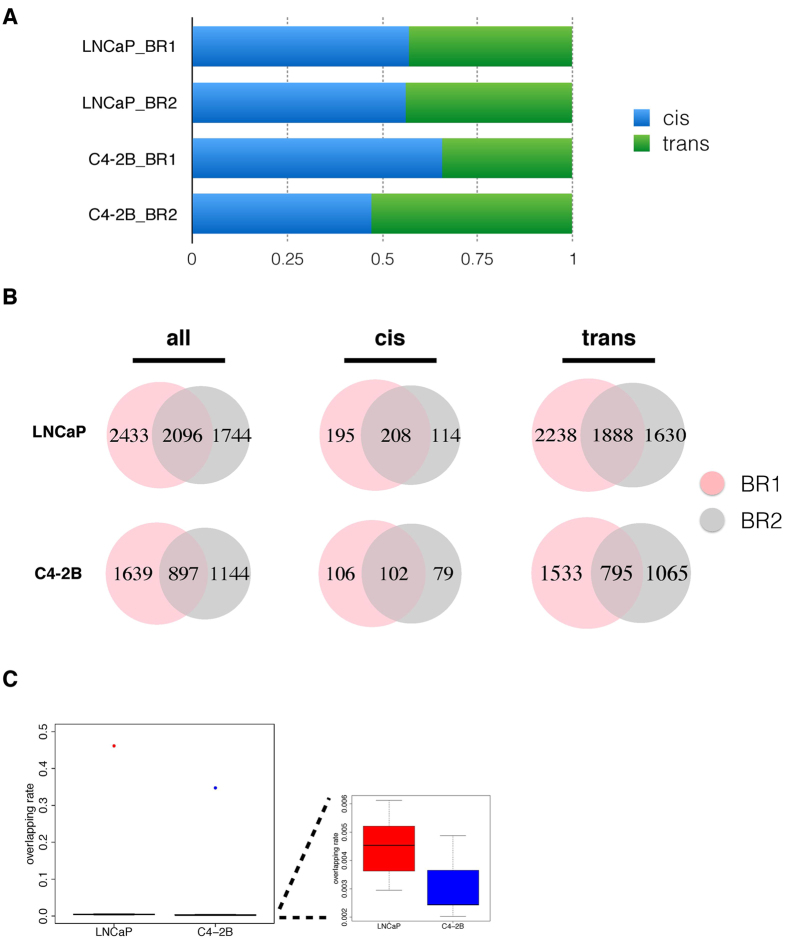
The analysis showed that around 47%~66% of the total reads in four datasets are distributed on the same chromosome where the bait is located (Fig. 3A). This conforms to the “cis/overall ratio of >40%” criteria proposed by Van De Werken et al.28, indicating good quality experiments. After merging reads aligned to the same site, our analysis revealed more than 3,000 interacting sites in LNCaP and more than 2,000 sites in C4-2B (Table 1). Of note, 2,096 sites and 897 sites were reproducibly identified in the two biological replicates of LNCaP and C4-2B, respectively (Table 1, Fig. 3B). Besides, these interactions consist of both cis and trans sites, with trans interactions accounting for the majority (~90%) (Table 1, Fig. 3B). To show that the overlapping between replicates is not by chance, we simulated 10 random sets of equally sized and number of sites for LNCaP and C4-2B respectively, and compared the overlapping rates in true versus random datasets. The median of overlapping rate was 0.45% for LNCaP random sets and 0.24% for C4-2B random sets (Fig. 3C). In comparison, the overlapping rate between biological replicates was much higher, 46.3% and 35.4% for LNCaP and C4-2B respectively, indicating that the overlap observed was not due to chance. In addition, the overlapping rates are comparable to similar studies investigating genome organizations. A previous ChIA-PET study of CTCF-mediated chromatin interactome in pluripotent cells reported the overlapping rate between biological replicates as 38%29. We reason that 4C technique takes a snapshot of the chromatin interaction patterns across hundreds of thousands of cells where chromatin-chromatin interactions occur in a highly dynamic fashion. These reproducible sites were thus considered to be high-fidelity interacting sites and would be used to conduct integrative association analysis.
Figure 3. Distribution of 4C reads and sites.
(A) Stacked bar plot showing the percentage of mapped reads in cis and in trans in two replicates of LNCaP and C4-2B. (B) A Venn diagram showing the number of overlapping sites between two biological replicates in LNCaP and C4-2B. (C) Boxplot showing the distributions of overlapping between biological replicates (red dot for LNCaP and blue dot for C4-2B) and between randomly simulated data.
Table 1. Summary of metrics in 4C-seq analysis in LNCaP and C4-2B.
| cell line | BR | # read pairs | # non-random sites | # overlapping non-random sites | # cis-sites | # overlapping cis-sites | # trans-sites | # overlapping trans-sites |
|---|---|---|---|---|---|---|---|---|
| LNCaP | 1 | 27,518,509 | 4,529 | 2,096 | 403 (8.9%) | 208 (9.9%) | 4,126 (91.1%) | 1,888 (90.1%) |
| 2 | 38,637,799 | 3,840 | 322 (8.4%) | 3,518 (91.6%) | ||||
| C4-2B | 1 | 25,060,263 | 2,536 | 897 | 208 (8.2%) | 102 (11.4%) | 2,328 (91.8%) | 795 (88.6%) |
| 2 | 39,250,858 | 2,041 | 181 (8.9%) | 1,860 (91.1%) | ||||
| Percentages are shown in brackets. Two biological replicates (BRs) were performed for each cell line. | ||||||||
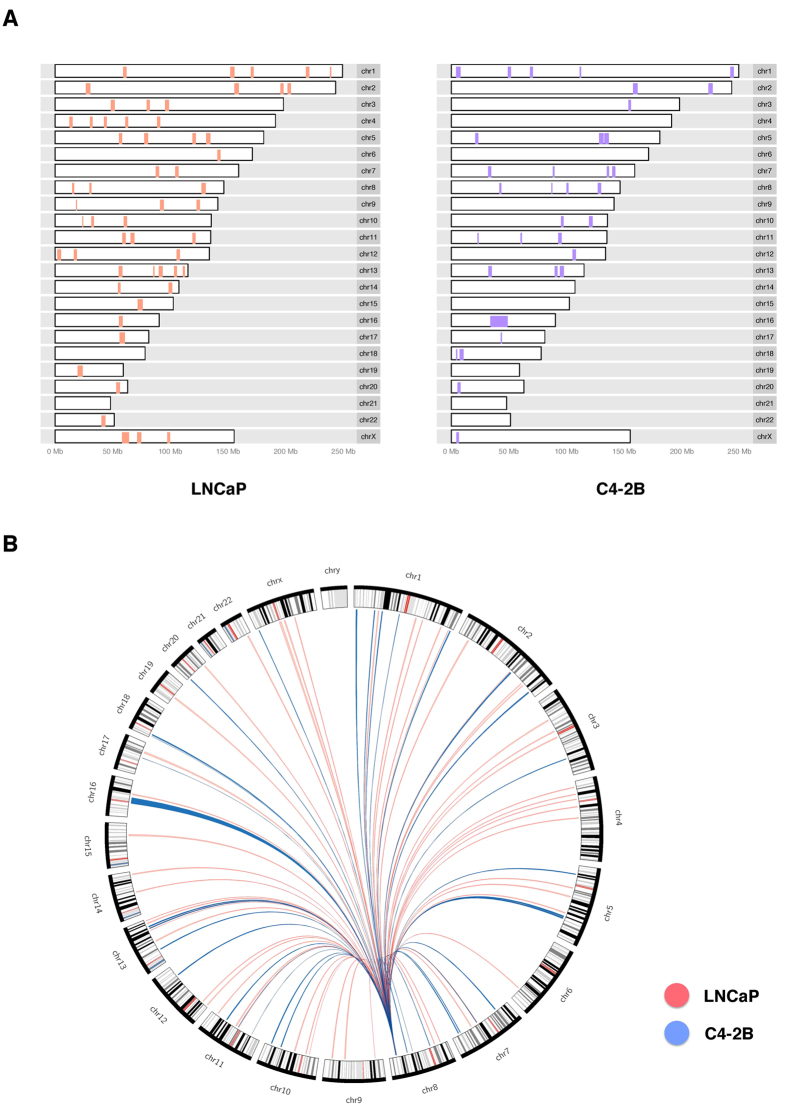
We further applied a binomial model based screen to call out significant domains (Fig. 4), and noticed that the distributions of sites interacting with the 8q24 enhancer were very different. This revealed that the chromosome organizations around the enhancer at 8q24 were very different in LNCaP and C4-2B cells. This in turn implies that the enhancer exerts influence over different sets of genes in LNCaP and C4-2B, leading to distinct expression profiles and cancer properties between LNCaP and C4-2B cells.
Figure 4. Significant 4C domains.
Genome-wide distribution of significant domains in LNCaP (red) and C4-2B (blue) is shown in tabular fashion (A) and circos plot (B).
The 8q24 enhancer interactomes occupy open chromosomal compartments
Following identification of 8q24 interacting regions, we speculated that the bait-interacting regions could share similar properties. We examined publicly available datasets related to prostate cancer (see Methods) and conducted association analysis following the two-compartment model proposed by Hi-C studies30,31, where chromosomes are distributed into open compartments enriched with active marks and closed compartments depleted of such marks.
Methylation of lysine 4 on Histone H3 — including H3K4me1, H3K4me2 and H3K4me3 — is a hallmark of open chromatin, often featuring promoters and enhancers of actively transcribed genes32,33. Moreover, these histone modifications facilitate the opening of chromatin by recruiting further chromatin-remodeling factors34,35,36. H3K36me3 is also a mark of active transcription37. In addition, H3K27Ac is a reliable and cell-type specific mark of active enhancers and promoters12. We examined the enrichment of H3K4 mono-, di- or tri-methylation, H3K36 tri-methylation and H3K27Ac marks around 4C interactome, and found that H3K4me1, H3K4me2, and H3K27Ac were enriched around identified 4C sites (+/− 500 kb) in both LNCaP (H3K4me1: p = 1.088e-6, H3K4me2: p = 7.357e-5, H3K27Ac: p = 5.882e-6; Wilcoxon rank sum test, Fig. 5A) and C4-2B cells (H3K4me1: p = 4.648e-7, H3K4me2: p = 5.435e-5, H3K27Ac: p = 1.903e-6; Wilcoxon rank sum test, Fig. 5A). However, we only observed marginally significant enrichment of H3K4me3 around 4C sites in LNCaP (p = 5.7e-2; Wilcoxon rank sum test, Fig. 5A) and C4-2B (p = 4.24e-2; Wilcoxon rank sum test, Fig. 5A) cells. Besides, H3K36me3 was marginally enriched in LNCaP (p = 2.036e-2) but significantly enriched in C4-2B (p = 6.359e-4). This result suggests that targeted regions of the 8q24 enhancer tend to be transcriptionally active regions marked by H3K4me1, H3K4me2 and H3K27Ac in prostate cancer cells — which could be one of the reasons for their cancerous property.
Figure 5. Enrichment of histone marks and transcription factor bindings.
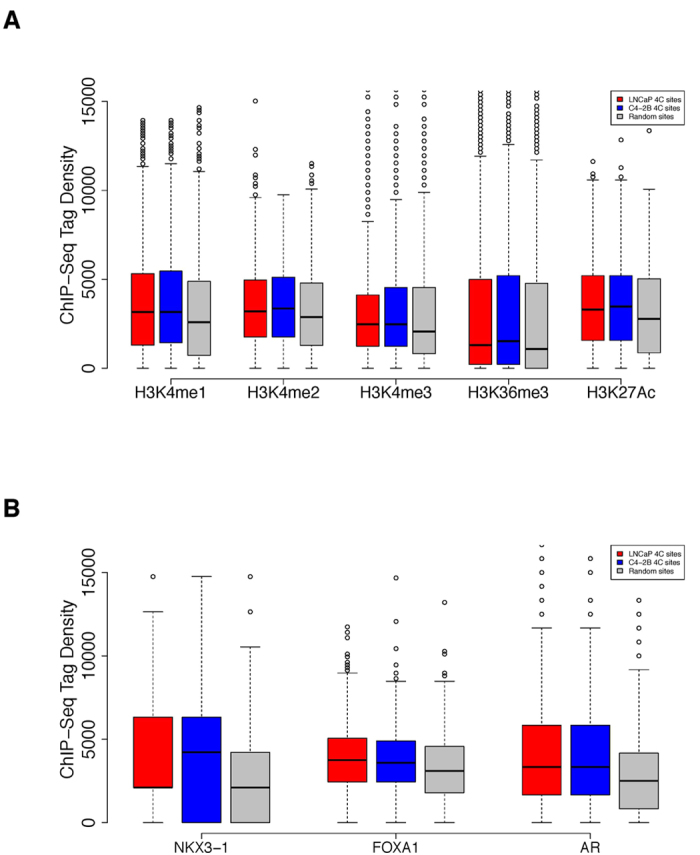
(A) Boxplot of different open chromatin marks distribution around the interacting sites in LNCaP (red), C4-2B (blue), and random sites (grey). ChIP-seq tags within +/− 500 kb around interacting sites were counted and normalized to 10 million. (B) Boxplot of transcription factors binding profile around the interacting sites in LNCaP (red), C4-2B (blue), and random sites (grey). ChIP-seq tags within +/− 500 kb around interacting sites were counted and normalized to 10 million.
The enhancer interactomes are enriched with key transcription factor bindings
Accessible chromatin marked by active histone features is closely related to the occupancy of regulatory factors38,39. NKX3-1, FOXA1 and AR (androgen receptor)40,41,42 play critical roles in the progression of prostate cancer. We thus compared the ChIP-seq profiles of the three factors and examined their peaks enrichment around our 4C sites (+/−500 kb), revealing that they were significantly enriched around the 4C loci compared to random sites in both LNCaP (NKX3.1: p = 7.214e-7, FOXA1: p = 7.871e-15, AR: p = 5.707e-12; Wilcoxon rank sum test, Fig. 5B) and C4-2B cells (NKX3.1: p = 4.593e-6, FOXA1: p = 6.989e-10, AR: p = 1.786e-11; Wilcoxon rank sum test, Fig. 5B). Therefore, we think these transcription factors can bind to accessible 4C regions and play a critical role in prostate cancer cell function.
The 4C sites are in close proximity to prostate cancer risk loci
We asked whether 4C sites identified in LNCaP and C4-2B are associated with annotated SNPs in the 100 independently identified prostate cancer risk loci3,5,6,7,8,9,10,11,12. Among the total 112 index-SNPs summarized so far (Supplementary Table S1), 70 and 49 were around +/−1 Mb of 4C sites in LNCaP and C4-2B, respectively. Compared to random sites, 4C sites in both LNCaP and C4-2B are closer to these index-SNPs (p = 6.474e-4 and 7.542e-3 for LNCaP and C4-2B respectively). In particular, rs8014671(14q24: TTC9), rs4242382 (8q24.21: POU5F1B, MYC), rs4242384 (8q24.21: POU5F1B, MYC), rs7000448 (8q24.21: LOC727677, MYC), rs1447295 (8q24.21: POU5F1B, MYC), rs6983267 (8q24.21: PCAT1, MYC), rs7837688 (8q24.21: POU5F1B, MYC) and rs188140481 (8q24.21: PCAT1, MYC) are in close proximity to 4C sites within 10kb in LNCaP. Similarly, rs1447295 (8q24.21: POU5F1B, MYC), rs4242382 (8q24.21: POU5F1B, MYC), rs6983267 (8q24.21: PCAT1, MYC), rs4242384 (8q24.21: POU5F1B, MYC) and rs7837688 (8q24.21: POU5F1B, MYC) are within 10kb around 4C sites of C4-2B. We reason that a considerable fraction (>44%) of risk SNPs are around our 4C sites. We think that our 4C pool can facilitate the identification of novel risk loci and risk SNPs, considering that some of the 4C sites are not correlated with those risk loci or SNPs that have been identified by GWAS. Moreover, since we observed the interaction between risk locus 8q24 and other risk loci, we raised the possibility that multiple risk loci could interact with each other forming foci, implicated in cancer onset and development.
The 4C regions are enriched with pathways and genes involved in prostate cancer activity
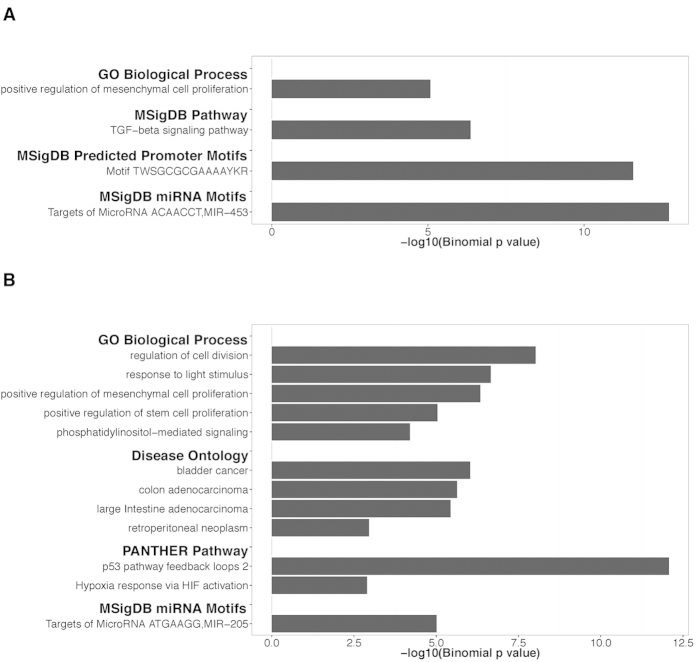
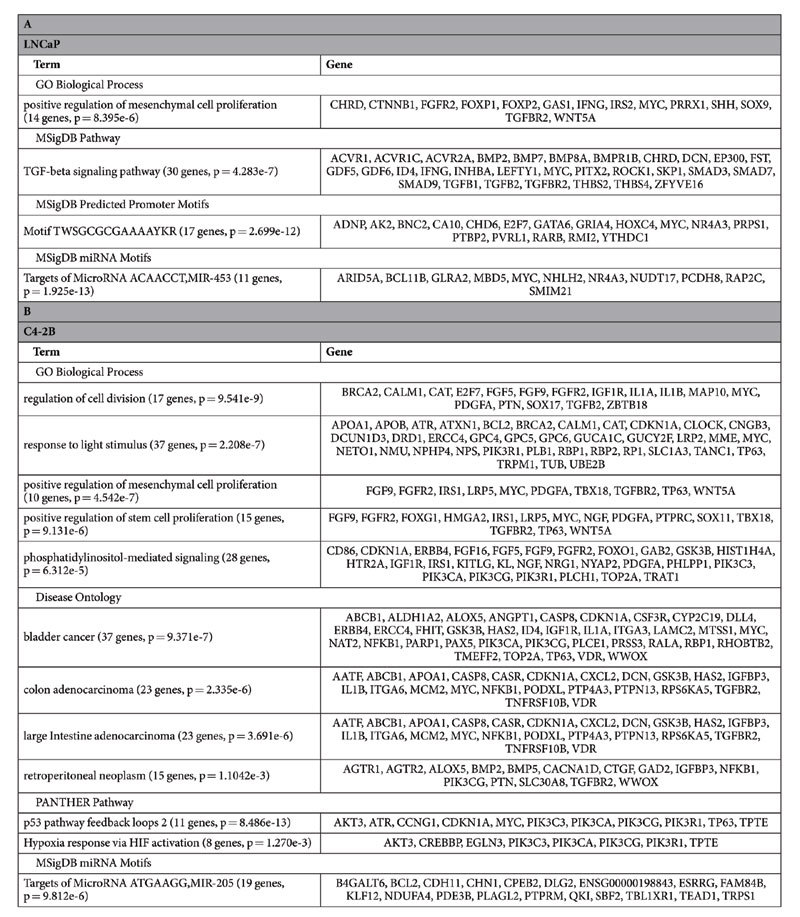
We input reproducible 4C sites in LNCaP and C4-2B to GREAT43 (Genomic Regions Enrichment of Annotations Tool) to analyze functional significance of these sites (Fig. 6, Table 2). We observed significant enrichment of positive regulation of mesenchymal cell proliferation in both LNCaP (p = 8.395e-6) and C4-2B (p = 4.542e-7). A recent study44 applying 3C-MTS (chromosome conformation capture based multiple target sequencing) technology to investigate 8q24’s physical interactions with multiple loci across the genome also showed the enrichment of this term. Other terms, including regulation of cell division (p = 9.541e-9), positive regulation of stem cell proliferation (p = 9.131e-6), phosphatidylinositol-mediated signaling (p = 6.312e-5), were found enriched in C4-2B as well. Genes in these terms might be closely involved in prostate cancer activities. In addition, we observed that response to light stimulus was shown as an enriched term in C4-2B, which is interesting considering previous studies proposing melatonin might reduce prostate cancer cell growth45 and lower prostate cancer risk46. As for pathway analysis, TFG-beta signaling pathway was enriched in LNCaP (p = 4.283e-7), while p53 pathway (p = 8.486e-13) and hypoxia response via HIF activation (p = 1.270e-3) were enriched in C4-2B. These pathways have been demonstrated or proposed as critical in the development of prostate cancer and the regulation of DNA replication, transcription and cell cycle in prostate cancer cells47,48,49,50,51. The 4C sites were also enriched for some miRNA motifs. For example, MIR-453 (ACAACCT) and MIR-205 (ATGAAGG) were significantly enriched in LNCaP (p = 1.925e-13) and C4-2B (p = 9.812e-6) respectively. Besides, we observed the enrichment of the transcription factor with binding motif TWSGCGCGAAAAYKR in LNCaP (p = 2.699e-12). We found this is an E2F motif in the inverted orientation (TTTSSCGC), implying the role of E2F in prostate cancer development. All the four disease ontology terms (bladder cancer: p = 9.371e-7, colon adenocarcinoma: p = 2.335e-6, large intestine adenocarcinoma: p = 3.691e-6, retroperitoneal neoplasm: p = 1.104e-3) in C4-2B were related to cancer, but no counterparts were found significantly enriched in LNCaP. Of note, under MSigDB Perturbation, the top 4 hits in LNCaP and top 2 hits in C4-2B are all related to cancer (Supplementary Table S2).
Figure 6. Gene enrichment analysis.
Results of gene enrichment analysis in LNCaP (A) and C4-2B (B).
Table 2. Significant enrichment of ontology annotations of genes associated with 4C sites in LNCaP (A) and C4-2B (B).
Next, we retrieved a set of seed genes associated with prostate cancer from Phenolyzer52 (Phenotype Based Gene Analyzer), a tool to discover genes associated with a disease/phenotype term. By comparing GREAT identified genes with the reference genes from Phenolyzer, we found 159 genes in LNCaP and 84 genes in C4-2B related to prostate cancer development (Table 3). 25 prostate cancer related seed genes commonly exist in LNCaP and C4-2B. Among them, MYC and POU5F1B are the highest ranked genes. Previous studies have shown that irregular chromatin looping and abnormal expression at the MYC locus could be a critical, oncogenic driving factor in prostate cancer53,54. Meanwhile, we noted some seed genes exclusively exist in LNCaP or C4-2B. The highest ranked genes in LNCaP are ELAC2 and PTEN; in comparison, the counterparts in C4-2B are BRCA2 and ZFHX3. As a comparison, the result of Du et al.44 strengthens our observation of the cis-interaction with MYC and trans-interaction with FAM84B (Table 3). In addition to MYC and FAM84B, Du et al.44 also showed CD96, PVT1, GSDMC, CXorf36, RRP12, USP14, SMIN3 as interaction genes. However, except for GSDMC in C4-2B, none of these genes were found in our 4C results for LNCaP or C4-2B. Based on Phenolyzer’s analysis, none of these 7 genes is annotated as seed genes associated with prostate cancer, thus it is possible that these interactions with 8q24 locus are not as important as others.
Table 3. Prostate cancer associated seed genes (annotated by Phenolyzer) identified by 4C-seq in LNCaP and C4-2B.
| Cell line | prostate cancer seed genes from Phenolyzer |
|---|---|
| LNCaP | (159 genes) ELAC2(7), PTEN(10), MSR1(15), EHBP1(16), MAD1L1(22), MYC(30), TNRC6B(38), CTBP2(47), FGF10(49), RAD23B(54), POU5F1B(60), PDLIM5(61), VGLL3(64), PKHD1(68), HAPLN1(69), ANK2(88), CWC22(89), CD180(90), INSC(95), ACTBL2(99), CNTN6(101), TMTC2(103), PTPRU(105), PRPF6(125), ZNF512B(127), KLF4(162), SOX9(163), SIDT1(164), KIAA2018(166), SPICE1(167), BOC(169), SP8(178), ABCB5(179), RAD51B(211), ZFP36L1(212), ATP9B(230), SALL3(234), KRT8(272), DCST2(304), ZBTB7B(308), BOD1(331), STC2(333), PKIA(337), PTGFRN(338), SYK(344), TSN(345), AMIGO2(349), NR2F2(351), INHBA(355), CHL1(358), ZNF608(365), BAI3(366), HEPH(371), GGTLC1(375), FOLH1(381), ERG(385), ESR2(391), PLAU(406), TNFSF10(411), VIM(416), TUSC3(428), CCND1(430), PTGS2(434), TGFB1(437), HIF1A(440), CYP19A1(441), NAT2(446), ESR1(447), CTNNB1(448), MME(450), FHIT(459), TLR4(466), NAT1(467), DAB2IP(479), ADAM9(482), TGFBR2(502), APC(503), GCNT1(507), VAV3(508), MDM2(511), FOXA1(514), CCND2(517), ITPR1(519), BMP7(520), CDH13(526), GJA1(527), GADD45A(528), GHR(531), IL16(539), TPT1(541), BMPR1B(545), PRKACB(550), EIF3H(554), PIK3CD(559), SEPP1(567), PRNP(582), PTPRK(584), FASLG(587), ANXA3(603), PPP3CA(612), APPL2(619), FOXC1(624), GNG5(630), HAO1(631), PCDH8(649), BCAS1(650), CST1(653), TPBG(659), RUNX1(660), DNAJC3(666), ZFP36L2(669), NR3C1(673), PRRX1(674), EMP1(675), PAWR(692), NPR3(708), USO1(710), TLR5(712), SP5(729), TPD52L1(733), NAGK(742), TMEFF2(743), ATAD3A(753), RFK(763), S100P(773), CDYL2(782), SCGB1D1(783), CEP290(786), ID4(788), CHGA(812), PDZD2(813), PKIB(814), SSTR1(823), SV2B(825), LEF1(827), STEAP4(828), TGIF1(829), SLC45A3(836), FST(838), GRP(850), PDC(857), SYT7(861), RCN1(867), ACPP(870), TPCN2(888), FAM84B(891), CBLN1(901), PEX2(902), IRX4(907), PIP4K2A(912), CADM2(915), SLC38A4(921), FSCB(922), GJD2(923), LIG4(948), SGCZ(949), SETBP1(951), SLC10A2(958), KRT78(960) |
| C4-2B | (84 genes) BRCA2(8), ZFHX3(18), HSD17B3(21), MYC(30), JAZF1(46), FOXP4(51), ANO7(57), POU5F1B(60), ITGA6(65), CWC22(89), PRPF6(125), ZNF512B(127), MMP8(190), TMEM123(194), BIRC2(196), MMP10(197), TANC1(202), CEP57L1(286), ADAR(296), KCNN3(307), KLF11(311), GRHL1(317), FSHR(332), DNAH5(350), NR2F2(351), HEPH(371), APOB(374), GGTLC1(375), VDR(384), ERG(385), IGFBP3(388), UGT2B17(401), BCL2(402), EZR(417), GRPR(418), TUSC3(428), CDKN1A(445), NAT2(446), MME(450), FHIT(459), GSTA1(460), IRS1(481), GSK3B(484), TGFBR2(502), LPAR1(504), FOXA1(514), CYP7B1(518), PODXL(548), ADRB2(551), DAG1(553), SSBP2(556), PIK3CA(557), PARP1(563), ALDH1A2(574), CREBBP(577), ENPP5(617), LIFR(618), GRB7(622), HAO1(631), TOP2A(636), TOM1L1(641), TNFRSF21(645), SLC12A2(665), EMP1(675), MAP3K7(691), PDE4D(730), INADL(736), TMEFF2(743), SPOCK1(765), COX6C(778), ID4(788), TRPV6(792), PSAT1(795), SSTR1(823), FST(838), NOG(842), TCF7L2(843), FAM84B(891), IRX4(907), COL21A1(917), FSCB(922), USH2A(939), SGCZ(949), TES(954) |
Numbers enclosed with brackets () are ranks calculated by Phenolyzer. In the table, genes are listed with decreasing rank, and genes exist in both LNCaP and C4-2B results are underlined.
Discussion
Prostate cancer is the second leading cause of death for men in the United States. Many studies have identified cancer risk associated loci and pathways involved in prostate cancer onset and development. In this study, we employed an unbiased “one-versus-all” technique – 4C-seq to uncover genome-wide interacting partners of a risk enhancer at the 8q24 prostate cancer risk locus. Using LNCaP and C4-2B cell lines for our 4C-seq analysis, we were able to compare these representative models of androgen sensitive and insensitive prostate cancer. Our analysis provides insight into key regions and genes interacting with the risk loci, contributing to our understanding of prostate cancer progression.
As shown above, LNCaP and C4-2B share similar genes interacting with the risk enhancer at 8q24, suggesting a general signature expression landscape for prostate cancer cells. Meanwhile, we also see specific differences in each cell line, where different genes are affected in the same or different enriched pathways. This is understandable given the fact that C4-2B is an osteotrophic and androgen insensitive prostate cancer cell line, unlike its parental LNCaP cell line. The difference in function relies on different set of genes. For example, BCL-2 gene was an interacting candidate in C4-2B but not in LNCaP. BCL-2 is an oncogene encoding inner mitochondrial membrane protein that is anti-apoptotic55. It has been shown that the transition to a hormone unresponsive state is accompanied with elevated BCL-2 expression, increased proliferation and decreased apoptosis56,57,58. Besides, there is an inverse relation between androgen receptor and BCL-2 expression59, which supports the involvement of BCL-2 in androgen-insensitive cells. To characterize the different picture of gene expression levels in LNCaP and C4-2B, we think RNA-seq can be conducted, which combined with 4C-seq results, will add to our knowledge of how prostate cancer cells transition into hormone-refractory state.
It came to our attention that some pathways and genes related with pluripotent stem cells are enriched, for instance, pathway—positive regulation of stem cell proliferation, and genes—MYC, POU5F1B. This implies the involvement of cancer stem cells in prostate cancer. Normal pluripotent stem cells and cancer stem cells are similar in many aspects, and the most striking one might be the ability to perpetuate themselves by self-renewal. Normal stem cells can be the targets of transforming mutations and cancer stem cells can drive the cancerous cells proliferation60. In addition, Du et al.44 and our results showed that positive regulation of mesenchymal cell proliferation was an enriched pathway in both LNCaP and C4-2B. Prostate cancer is one type of carcinoma, developing from epithelial cells that line the surface of glands and ducts61. Thus it is compelling to see that positive regulation of mesenchymal cell proliferation is an enriched pathway, given that transformed cells of mesenchymal origin will usually lead to sarcoma made of cancerous bone, cartilage, muscle, vascular, hematopoietic tissues and so on. Despite that, it has been shown that during tumorigenesis concomitant changes occur in cells surrounding epithelial cells as well62. These cells, called as stromal cells and composed of endothelial, fibroblastic, smooth muscle and nerve cells, play a supportive role for epithelial layer61. Several investigations showed that normal molecular dialogue between embryonic epithelial and mesenchymal cells form the basis of tissue/organ function63,64,65, while the disruption of that homeostasis might confer tumorigenesis. That sets the concept framework for the involvement of mesenchymal cells in prostate cancer.
In addition, we realized that some other cancers (for example, bladder cancer, breast cancer and lung cancer, Table 2, Supplementary Table S2) are shown enriched in our ontology analysis of genes interacting with 8q24. 8q24 was found to harbor common alleles in prostate, breast and colon cancer14,66,67. Besides, breast, prostate, colon and lung cancers are common type of carcinomas derived from epithelial cells. Thus it is probable that the risk locus 8q24 interact with similar genes and have common mechanism underlying the association with cancer.
We observed some miRNA motifs enrichment in 4C sites of LNCaP and C4-2B, implying that miRNAs might be implicated in prostate cancer. Over the years, miRNAs have been shown to play a role in regulating gene expression at post-transcriptional level68,69,70. Moreover, evidence is mounting that miRNAs are associated with various diseases, especially cancer71,72,73. Of note, recently an increasing number of studies are dedicated to taking computational approaches to characterize the association between miRNA and disease74,75,76. Our findings might contribute to the knowledge of miRNA-cancer relationship.
In our analysis, we have obtained a list of potentially biologically significant regions and genes in LNCaP and C4-2B cells. We can confirm interactions between the risk enhancer and candidate genes by performing 3C and fluorescent in situ hybridization (FISH). To test the biological importance of the interaction with the risk enhancer, we can apply CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 genome editing to knock out the enhancer region and compare RNA-seq expression profiles between edited and unmodified prostate cancer cells. Recently, Tak et al.77 has successfully applied this strategy to examine the effect of deletion of a distal element on transcriptome in colon cancer. The genes proposed by computational analysis and experimental validation will be important targets for drug development and intervention therapy.
Methods
Cell culture
LNCaP and C4-2B cells were maintained in RPMI 1640 supplemented with 5% (v/v) FBS (fetal bovine serum). LNCaP was obtained from the American Type Culture Collection (ATCC; Manassas, VA), and C4-2B was obtained from ViroMed Laboratories (Minneapolis, MN).
4C library preparation
4C library preparation was performed following previously described protocol28,78 with modifications. 5 million cells were trypsinized and resuspended as single cells in 0.5 ml RPMI 1640/10% FBS. For crosslinking, 9.5 ml of 2% formaldehyde/10% FBS in PBS was added. After being incubated for 10 minutes, fixation was quenched with 0.6 ml of 2.5 M glycine and cells were lysed in 1 ml lysis buffer 1 (50 mM HEPES-KOH, pH7.5, 140 mM NaCl, 1 mM EDTA, 10% Glycerol, 0.5% NP-40, 0.25% Triton X-100, protease inhibitors) for 10 minutes at 4 °C with rotating. Nuclei were pelleted by centrifugation and washed with lysis buffer 2 (10 mM Tris-HCl, pH8.0, 200 mM NaCl, 1 mM EDTA, protease inhibitors). Pelleted nuclei were resuspended in 0.5 ml of 1.2 fold restriction enzyme buffer and incubated with 0.3% SDS for 1 hour at 37 °C with shaking and further incubated with 2% Triton X-100 for 1 hour. BglII restriction enzyme were added and incubated overnight at 37 °C with shaking. Digestion was stopped after adding 1.6% SDS and incubated at 65 °C for 25 minutes. Samples were then diluted in 6.125 ml 1.15 fold ligation buffer and 100 Weiss unit of T4 DNA ligase was added. Samples were kept at 16 °C for 4 hours and then at 25 °C for 30 minutes. The ligated chromatin was digested by proteinase K and then incubated with RNaseA. Samples were purified by phenol-chloroform extraction and ethanol precipitation. Samples were further digested by CviQI and circularized using T4 DNA ligase. After purification, 8 parallel PCR reactions, each containing 1 ug of circularized DNA, were performed (primers: Index primer for replicate 1 –caagcagaagacggcatacgagatTCAAGTgtgactggagttcagacgtgtgctcttccgatcGGAAGTAGAGTAGCAATTCTTG; Index primer for replicate 2 – caagcagaagacggcatacgagatCTGATCgtgactggagttcagacgtgtgctcttccgatcGGAAGTAGAGTAGCAATTCTTG; Universial primer – aatgatacggcgaccaccgagatctacactctttccctacacgacgctcttccgatctATGGAAATCAAGCAGCAGATCT). The amplicons were extracted by E-gel. The bar-coded DNA libraries were sequenced as 50 bp single-end reads using the Illumina HiSeq2000 platform.
4C-seq data analysis
Reads with 5’ end mapped to the forward inverse PCR primer sequence were selected. The rest of the selected reads (including the BglII sites) were mapped to hg19 assembly by Burrows-Wheeler Aligner79 (BWA). The ligation sites were determined in that process. To get rid of off-target aligned reads, we applied a second round alignment, where the mapped ligated BglII sites were further mapped to a library that includes the locations of genome-wide BglII sites. Sites with only one read were eliminated to filter out random noise. Reproducible sites were defined as exact match of 4C sites with coverage >1 in both biological replicates.
To call out statistically significant regions, we applied a binomial model19,28. Every interacting site i on chromosome W with length LW was examined within window w with length lw. The number of interacting sites was defined as 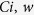 , and a z score was assigned to the window based on the following calculation:
, and a z score was assigned to the window based on the following calculation:
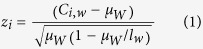 |
in which μW is the expected number of interacting sites in window w on chromosome W .
A window size of 500 was used in counting the number of ligated sites to define inter-chromosomal interactions, while a window size of 200 was set to define intra-chromosomal interactions. To define intra-chromosomal interactions, we set background window size to 5,000, so as to calculate the expected number of ligated sites in a window. We used type I error rate of 0.05 as the threshold to identify non-random chromosomal interactions.
Integrative association study
We integrated publicly available datasets that are associated with prostate cancer or LNCaP cells. From the ENCODE Project Portal, we retrieved ChIP-seq data for H3K4me3 (GSM945240). We also retrieved H3K27Ac ChIP-seq data (GSE51621), NKX3-1 ChIP-seq data (GSM699633), FoxA1 ChIP-seq data (GSM699634, GSM699635), and Androgen Receptor ChIP-seq data (GSE28219). The ChIP-seq results for other histone marks were downloaded from the Cistrome database.
GREAT and Phenolyzer
Genes associated with 4C sites were retrieved by GREAT43 (http://bejerano.stanford.edu/great/public/html/), a tool to assign functional annotation to genomic regions. The background regions were chosen as the default whole genome. We used default basal plus extension association rule (proximal: 5 kb upstream, 1 kb downstream; distal: up to 1000 kb). Curated regulatory domains were included.
Seed genes associated with prostate cancer were discovered by Phenolyzer52 (http://phenolyzer.usc.edu/), a tool to discover genes based on user inputted disease/phenotype terms. “prostate cancer” was used as input term and a total of 964 genes with ranks were returned.
Other
Genomic feature annotations were retrieved from the UCSC genome bioinformatics site. Genomic coordinates in other assemblies were converted to GRCh37/hg19 using the liftOver tool. Statistical tests and plotting were performed using R.
Additional Information
How to cite this article: Cai, M. et al. 4C-seq revealed long-range interactions of a functional enhancer at the 8q24 prostate cancer risk locus. Sci. Rep. 6, 22462; doi: 10.1038/srep22462 (2016).
Supplementary Material
Acknowledgments
We thank people from the Lu lab for their inputs. The work was supported by a grant from NIH(2R01CA13692406A1).
Footnotes
Author Contributions W.L. conceived the project. S.K. performed experiments. M.C. analyzed the data. K.W. advised M.C. in bioinformatic analyses. P.J.F. provided inputs for data analysis. G.A.C. provided cell lines and inputs for data analysis. M.C. and W.L. prepared the manuscript. All authors reviewed the manuscript.
References
- Klein R. J. et al. Complement factor H polymorphism in age-related macular degeneration. Science 308, 385–389, doi: 10.1126/science.1109557 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T. A. Genomewide Association Studies and Assessment of the Risk of Disease. New Engl J Med 363, 166–176 (2010). [DOI] [PubMed] [Google Scholar]
- Al Olama A. A. et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nature genetics 46, 1103–1109, doi: 10.1038/ng.3094 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. et al. Integration of multiethnic fine-mapping and genomic annotation to prioritize candidate functional SNPs at prostate cancer susceptibility regions. Human molecular genetics 24, 5603–5618, doi: 10.1093/hmg/ddv269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. et al. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nature genetics 44, 1231–1235, doi: 10.1038/ng.2424 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J. et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nature genetics 41, 1122–1126, doi: 10.1038/ng.448 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh C. L. et al. Genetic variants associated with predisposition to prostate cancer and potential clinical implications. Journal of internal medicine 271, 353–365, doi: 10.1111/j.1365-2796.2012.02511.x (2012). [DOI] [PubMed] [Google Scholar]
- Akamatsu S. et al. Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nature genetics 44, 426–429, S421, doi: 10.1038/ng.1104 (2012). [DOI] [PubMed] [Google Scholar]
- Amin Al Olama A. et al. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Human molecular genetics 22, 408–415, doi: 10.1093/hmg/dds425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J. et al. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nature genetics 44, 1326–1329, doi: 10.1038/ng.2437 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeles R. A. et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nature genetics 45, 385–391, 391e381–382, doi: 10.1038/ng.2560 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelett D. J. et al. Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS genetics 10, e1004102, doi: 10.1371/journal.pgen.1004102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenson K. et al. Common variants at the CHEK2 gene locus and risk of epithelial ovarian cancer. Carcinogenesis, doi: 10.1093/carcin/bgv138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman C. A. et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nature genetics 39, 638–644, doi: 10.1038/ng2015 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Olama A. A. et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nature genetics 41, 1058–1060, doi: 10.1038/ng.452 (2009). [DOI] [PubMed] [Google Scholar]
- Jia L. et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS genetics 5, e1000597, doi: 10.1371/journal.pgen.1000597 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadiyeh N. et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci USA 107, 9742–9746, doi: 10.1073/pnas.0910668107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz M. M. et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nature genetics 41, 882–884, doi: 10.1038/ng.403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E. et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes & development 25, 1371–1383, doi: 10.1101/gad.633311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z. et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nature genetics 38, 1341–1347, doi: 10.1038/ng1891 (2006). [DOI] [PubMed] [Google Scholar]
- Wei Z. et al. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell stem cell 13, 36–47, doi: 10.1016/j.stem.2013.05.010 (2013). [DOI] [PubMed] [Google Scholar]
- Gao F., Wei Z., An W., Wang K. & Lu W. The interactomes of POU5F1 and SOX2 enhancers in human embryonic stem cells. Scientific Reports 3, 1588, doi: 10.1038/srep01588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M. et al. Analysis of a transgenic Oct4 enhancer reveals high fidelity long-range chromosomal interactions. Scientific Reports 5, 14558, doi: 10.1038/srep14558 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horoszewicz J. S. et al. LNCaP model of human prostatic carcinoma. Cancer research 43, 1809–1818 (1983). [PubMed] [Google Scholar]
- Thalmann G. N. et al. Androgen-Independent Cancer Progression and Bone Metastasis in the Lncap Model of Human Prostate-Cancer. Cancer research 54, 2577–2581 (1994). [PubMed] [Google Scholar]
- Jia L. & Coetzee G. A. Androgen receptor-dependent PSA expression in androgen-independent prostate cancer cells does not involve androgen receptor occupancy of the PSA locus. Cancer research 65, 8003–8008, doi: 10.1158/0008-5472.CAN-04-3679 (2005). [DOI] [PubMed] [Google Scholar]
- Gao F., Wei Z., Lu W. & Wang K. Comparative analysis of 4C-Seq data generated from enzyme-based and sonication-based methods. BMC genomics 14, 345, doi: 10.1186/1471-2164-14-345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Werken H. J. et al. 4C technology: protocols and data analysis. Methods in enzymology 513, 89–112, doi: 10.1016/B978-0-12-391938-0.00004-5 (2012). [DOI] [PubMed] [Google Scholar]
- Handoko L. et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nature genetics 43, 630–638, doi: 10.1038/ng.857 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293, doi: 10.1126/science.1181369 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Marti-Renom M. A. & Mirny L. A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nature reviews. Genetics 14, 390–403, doi: 10.1038/nrg3454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature genetics 39, 311–318, doi: 10.1038/ng1966 (2007). [DOI] [PubMed] [Google Scholar]
- Ozsolak F., Song J. S., Liu X. S. & Fisher D. E. High-throughput mapping of the chromatin structure of human promoters. Nature biotechnology 25, 244–248, doi: 10.1038/nbt1279 (2007). [DOI] [PubMed] [Google Scholar]
- Flanagan J. F. et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438, 1181–1185, doi: 10.1038/nature04290 (2005). [DOI] [PubMed] [Google Scholar]
- Li H. et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95, doi: 10.1038/nature04802 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C. M. et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome research 17, 691–707, doi: 10.1101/gr.5704207 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R. J. & Reinberg D. Processing the H3K36me3 signature. Nature genetics 41, 270–271, doi: 10.1038/ng0309-270 (2009). [DOI] [PubMed] [Google Scholar]
- Gross D. S. & Garrard W. T. Nuclease Hypersensitive Sites in Chromatin. Annual review of biochemistry 57, 159–197, doi: 10.1146/Annurev.Bi.57.070188.001111 (1988). [DOI] [PubMed] [Google Scholar]
- Felsenfeld G., Boyes J., Chung J., Clark D. & Studitsky V. Chromatin structure and gene expression. P Natl Acad Sci USA 93, 9384–9388, doi: 10.1073/Pnas.93.18.9384 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurel B. et al. NKX3.1 as a Marker of Prostatic Origin in Metastatic Tumors. Am J Surg Pathol 34, 1097–1105, doi: 10.1097/PAS.0b013e3181e6cbf3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H. J., Zhao J. C., Ogden I., Bergan R. C. & Yu J. D. Androgen Receptor-Independent Function of FoxA1 in Prostate Cancer Metastasis. Cancer research 73, 3725–3736, doi: 10.1158/0008-5472.CAN-12-3468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein C. A. & Chang C. S. Androgen receptor in prostate cancer. Endocr Rev 25, 276–308, doi: 10.1210/er.2002-0032 (2004). [DOI] [PubMed] [Google Scholar]
- McLean C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nature biotechnology 28, 495–501, doi: 10.1038/nbt.1630 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M. et al. Prostate cancer risk locus at 8q24 as a regulatory hub by physical interactions with multiple genomic loci across the genome. Human molecular genetics 24, 154–166, doi: 10.1093/hmg/ddu426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz R. M. et al. Melatonin reduces prostate cancer cell growth leading to neuroendocrine differentiation via a receptor and PKA independent mechanism. Prostate 63, 29–43, doi: 10.1002/pros.20155 (2005). [DOI] [PubMed] [Google Scholar]
- Carlson L. E., Speca M., Patel K. D. & Goodey E. Mindfulness-based stress reduction in relation to quality of life, mood, symptoms of stress and levels of cortisol, dehydroepiandrosterone sulfate (DHEAS) and melatonin in breast and prostate cancer outpatients. Psychoneuroendocrinology 29, 448–474 (2004). [DOI] [PubMed] [Google Scholar]
- Tu W. H. et al. The loss of TGF-beta signaling promotes prostate cancer metastasis. Neoplasia 5, 267–277, doi: NO_DOI (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo B. T. et al. TGF-beta effects on prostate cancer cell migration and invasion are mediated by PGE2 through activation of PI3K/AKT/mTOR pathway. Endocrinology 154, 1768–1779, doi: 10.1210/en.2012-2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca P. et al. BRCA1 and p53 regulate critical prostate cancer pathways. Prostate cancer and prostatic diseases 16, 233–238, doi: 10.1038/pcan.2013.12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman I. et al. Inactivation of the p53 pathway in prostate cancer: impact on tumor progression. Clinical cancer research: an official journal of the American Association for Cancer Research 5, 2082–2088 (1999). [PubMed] [Google Scholar]
- Anastasiadis A. G. et al. Tumor cell hypoxia and the hypoxia-response signaling system as a target for prostate cancer therapy. Curr Drug Targets 4, 191–196, doi: 10.2174/1389450033491136 (2003). [DOI] [PubMed] [Google Scholar]
- Yang H., Robinson P. N. & Wang K. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nature methods 12, 841–843, doi: 10.1038/nmeth.3484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming W. H. et al. Expression of the c-myc protooncogene in human prostatic carcinoma and benign prostatic hyperplasia. Cancer research 46, 1535–1538 (1986). [PubMed] [Google Scholar]
- Gurel B. et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Modern Pathol 21, 1156–1167, doi: 10.1038/modpathol.2008.111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockenbery D., Nunez G., Milliman C., Schreiber R. D. & Korsmeyer S. J. Bcl-2 Is an Inner Mitochondrial-Membrane Protein That Blocks Programmed Cell-Death. Nature 348, 334–336, doi: 10.1038/348334a0 (1990). [DOI] [PubMed] [Google Scholar]
- Dorkin T. J. & Neal D. E. Basic science aspects of prostate cancer. Seminars in cancer biology 8, 21–27, doi: 10.1006/Scbi.1997.0049 (1997). [DOI] [PubMed] [Google Scholar]
- Raffo A. J. et al. Overexpression of Bcl-2 Protects Prostate-Cancer Cells from Apoptosis in-Vitro and Confers Resistance to Androgen Depletion in-Vivo. Cancer research 55, 4438–4445 (1995). [PubMed] [Google Scholar]
- Mcdonnell T. J. et al. Expression of the Protooncogene Bcl-2 in the Prostate and Its Association with Emergence of Androgen-Independent Prostate-Cancer. Cancer research 52, 6940–6944 (1992). [PubMed] [Google Scholar]
- Kolar Z. et al. Relation of Bcl-2 expression to androgen receptor, p21WAF1/CIP1, and cyclin D1 status in prostate cancer. J Clin Pathol-Mol Pa 53, 15–18 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T., Morrison S. J., Clarke M. F. & Weissman I. L. Stem cells, cancer, and cancer stem cells. Nature 414, 105–111, doi: 10.1038/35102167 (2001). [DOI] [PubMed] [Google Scholar]
- Olumi A. F. et al. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer research 59, 5002–5011 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnov-Jessen L., Petersen O. W. & Bissell M. J. Cellular changes involved in conversion of normal to malignant breast: importance of the stromal reaction. Physiological reviews 76, 69–125 (1996). [DOI] [PubMed] [Google Scholar]
- Haffen K., Kedinger M., Simon-Assmann P. M. & Lacroix B. Mesenchyme-dependent differentiation of intestinal brush-border enzymes in the gizzard endoderm of the chick embryo. Progress in clinical and biological research 85(Pt B), 261–270 (1982). [PubMed] [Google Scholar]
- Cunha G. R. et al. Normal and abnormal development of the male urogenital tract. Role of androgens, mesenchymal-epithelial interactions, and growth factors. Journal of andrology 13, 465–475 (1992). [PubMed] [Google Scholar]
- Kratochwil K. Tissue combination and organ culture studies in the development of the embryonic mammary gland. Developmental biology 4, 315–333 (1986). [DOI] [PubMed] [Google Scholar]
- Haiman C. A. et al. A common genetic risk factor for colorectal and prostate cancer. Nature genetics 39, 954–956, doi: 10.1038/ng2098 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton D. F. et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447, 1087–1093, doi: 10.1038/nature05887 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature 431, 350–355, doi: 10.1038/nature02871 (2004). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Meister G. & Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343–349, doi: 10.1038/nature02873 (2004). [DOI] [PubMed] [Google Scholar]
- Calin G. A. & Croce C. M. MicroRNA signatures in human cancers. Nature reviews. Cancer 6, 857–866, doi: 10.1038/nrc1997 (2006). [DOI] [PubMed] [Google Scholar]
- Iorio M. V. et al. MicroRNA gene expression deregulation in human breast cancer. Cancer research 65, 7065–7070, doi: 10.1158/0008-5472.CAN-05-1783 (2005). [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A. et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 7, 759–764 (2008). [DOI] [PubMed] [Google Scholar]
- Zeng X., Zhang X. & Zou Q. Integrative approaches for predicting microRNA function and prioritizing disease-related microRNA using biological interaction networks. Briefings in bioinformatics, doi: 10.1093/bib/bbv033 (2015). [DOI] [PubMed] [Google Scholar]
- Zou Q., Li J., Song L., Zeng X. & Wang G. Similarity computation strategies in the microRNA-disease network: a survey. Briefings in functional genomics, doi: 10.1093/bfgp/elv024 (2015). [DOI] [PubMed] [Google Scholar]
- Zou Q. et al. Prediction of MicroRNA-Disease Associations Based on Social Network Analysis Methods. BioMed research international 2015, 810514, doi: 10.1155/2015/810514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak Y. G. et al. Effects on the transcriptome upon deletion of a distal element cannot be predicted by the size of the H3K27Ac peak in human cells. Nucleic acids research, doi: 10.1093/nar/gkv1530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M., Kooren J. & de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nature methods 4, 895–901, doi: 10.1038/nmeth1114 (2007). [DOI] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760, doi: 10.1093/bioinformatics/btp324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.