Abstract
To explore the transcriptomic global response to osmotic stress in roots, 18 mRNA-seq libraries were generated from three triploid banana genotypes grown under mild osmotic stress (5% PEG) and control conditions. Illumina sequencing produced 568 million high quality reads, of which 70–84% were mapped to the banana diploid reference genome. Using different uni- and multivariate statistics, 92 genes were commonly identified as differentially expressed in the three genotypes. Using our in house workflow to analyze GO enriched and underlying biochemical pathways, we present the general processes affected by mild osmotic stress in the root and focus subsequently on the most significantly overrepresented classes associated with: respiration, glycolysis and fermentation. We hypothesize that in fast growing and oxygen demanding tissues, mild osmotic stress leads to a lower energy level, which induces a metabolic shift towards (i) a higher oxidative respiration, (ii) alternative respiration and (iii) fermentation. To confirm the mRNA-seq results, a subset of twenty up-regulated transcripts were further analysed by RT-qPCR in an independent experiment at three different time points. The identification and annotation of this set of genes provides a valuable resource to understand the importance of energy sensing during mild osmotic stress.
Functional genomics studies in plants are mostly performed on model species or species characterized to a great extent. However, numerous non-model plants are important food, feed or energy sources. In addition, they may exhibit some features and processes that are unique and cannot be approached via model plants. Banana (Musa spp.), including the sweet and starchy types, is a typical non-model crop which ranks among the top ten staple foods, with a total production that exceeded 145 million tons in 2013 (FAOstat). Modern cultivars are hybrids from one or both major diploid ancestors, M. acuminata and M. balbisiana, which contributed the A- and B- genomes, respectively1. Most of these cultivars are seedless triploids (2n = 3x = 33) with an AAA, AAB or ABB genome constitution. Being highly sterile, the commercial dessert bananas are produced based on clonal propagation of only a few genotypes (Cavendish, AAA genome group). This narrow, inflexible genotypic background makes the crop more susceptible to diseases, pests and environmental issues. Therefore, the large genetic diversity in Musa must be exploited to move away from the few restricted commercially exploited cultivars, while still meeting consumer’s expectations.
Drought stress is one of the major abiotic factors limiting banana production. Even though the crop is grown in the humid tropics and subtropics, in many locations rainfall is not sufficient or evenly distributed throughout the year. Thus, when there is no access to irrigation, mild drought conditions are responsible for considerable yield losses. For instance, East African highland bananas (AAAh genome group) generally receive 1200–1300 mm year−1 and every 100 mm shortage of water induces losses of 8–10% bunch weight2. On the other hand, black leaf streak (better known as black Sigatoka), economically the most important fungal disease that threatens commercial banana production, thrives in humid climates whereas drier areas are natural borders for the disease. In this context, cultivating more drought tolerant bananas in drier areas with lower infection rate would become an option3. Hence, increasing the understanding of drought tolerance in banana at the molecular and physiological level remains a critical objective for successful, knowledge-based crop improvement and varietal selection4,5. However, the identification of drought tolerant banana varieties in natural environments remains difficult due to complications in field management, variation in phenotype and unexpected rainfall events. To facilitate the process, initial screening protocols under controlled conditions have been developed5 and polyethylene glycol (PEG) treatment has demonstrated to simulate the occurrence of drought stress in drying soil6. Genome-wide gene expression analyses in banana under abiotic stress have been sparse with only a proof a concept for drought stress with microarrays7 or more recent transcriptomic studies on salt or cold stress8,9, but so far no large-scale transcriptomic analysis has been reported on the response to osmotic stress.
Transcriptome research conducted in various plant species has revealed that drought stress tolerance is a multigenic trait. During the response, a large number of genes are modified in their expression involving a precise regulation of extensive gene interacting networks, which further cause a series of physiological and biochemical alterations. Initial plant response mechanisms prevent or alleviate cellular damage caused by the stress, re-establish homeostatic conditions and allow continuation of growth10. Therefore, equilibrium recovery of the energetic and redox imbalances imposed by the stressor are the first targets of a plant’s immediate response. Despite significant progress over the past decade aiming to understand the metabolic pathways affected by drought stress, little is known about their dynamics in non-model crops. Recent advances in next-generation sequencing (NGS) technologies and associated bioinformatic tools have revolutionized plant transcriptomics research. mRNA-seq offers a precise way to measure transcript levels while simultaneously providing sequence information11. This efficient, cost-effective sequencing technology has been widely used to characterize the transcriptomes of plants for gene discovery, marker development and understanding gene regulatory networks of important biological processes. However, the use of mRNA-seq to evaluate global gene expression patterns is complicated in non-model species, particularly when they are polyploid, like banana. Short reads matching multiple loci can be allocated to a single transcript or be removed from the analysis, affecting accurate quantification of expression levels. Also gene duplication and genome reorganization events contribute to such complexity12. The availability of a reference genome helps the alignment of reads and dealing with paralogs or allelic variants. Moreover, mRNA-seq can provide additional information to identify previously unknown or wrongly annotated coding sequences. Recently, an A- and a draft B- Musa genome have been released13,14, providing the first complete catalogue of all predicted genes and largely facilitating genomic/transcriptomic analyses in the genus as well as comparative studies with other plant genomes15.
In the present study, banana plants were exposed to 5% PEG-8000. Thus, water availability was in the mid-range of naturally occurring soil water potentials, representing mild water deficit conditions16. The objective of our study was to characterize the general osmotic stress reactions in banana roots. We performed large-scale transcriptome sequencing using Illumina technology on three banana genotypes representing three important subgroups of cultivated bananas with diverse genomic constitutions and different origin/geographical distribution. This work contributes to a better understanding of the molecular mechanisms and provides a workflow to study responses to water deficit in a non-model crop. We put forward that genes commonly altered in the three genotypes are more likely to play a general role in the reaction to mild osmotic stress in all banana genotypes and possibly in many crops. By selecting a subset of these genes and validating them by RT-qPCR in an independent experiment, we confirm the success of RNA-seq for transcriptome evaluation of a non-model crop.
Results
General landscape of the banana root transcriptome under mild osmotic stress
Using the root tip as source of mRNA, a total of 18 cDNA libraries were generated from three biological replicates of the three genotypes and the two conditions, control (0% PEG) and mild osmotic stress (5% PEG treatment). This resulted in 600 million single raw reads (100 bp) of which 94.6% passed Illumina quality filtering (Table 1). 79.6% of the high quality reads mapped to the M. acuminata reference genome, with about 85% of them aligning to a single location. Reads with multiple locations, ambiguous or with no match (69 million, 7%) were discarded. It reduced the number of reads that uniquely matched exons and, thus, were used for the differential expression analyses, to 383 million. On average, at least 5 reads spanned 29,931 genes, which represent 80.6% of the total number of genes in the M. acuminata genome (Table 1). Statistics were very similar among samples under stress and control conditions in each genotype (Supplementary Table S1).
Table 1. Summary of the banana transcriptome sequencing dataset.
| Genotype | aCachaco (ITC0643) | aGrande Naine (ITC0180) | aMbwazirume (ITC0084) | bTotal/cAverage |
|---|---|---|---|---|
| No. raw reads (106) | 194 | 214 | 192 | 600b |
| No. high-quality reads (106) (and %) | 185 (95) | 201 (94) | 182 (95) | 568 (94.6)b |
| No. aligned reads (106) (and %) | 130 (70) | 169 (84) | 153 (84) | 452 (79.6)b |
| No. unique reads on exons (106) | 108 | 143 | 132 | 383b |
| Genes covered (%) | 79 | 81.8 | 81 | 80.6c |
| Average No. reads by transcript | 486 | 640 | 593 | 573c |
| Median No. reads by transcript | 106 | 140 | 128 | 125c |
aAverage values of the 6 libraries per genotype.
bTotal or.
caverage values of the three genotypes.
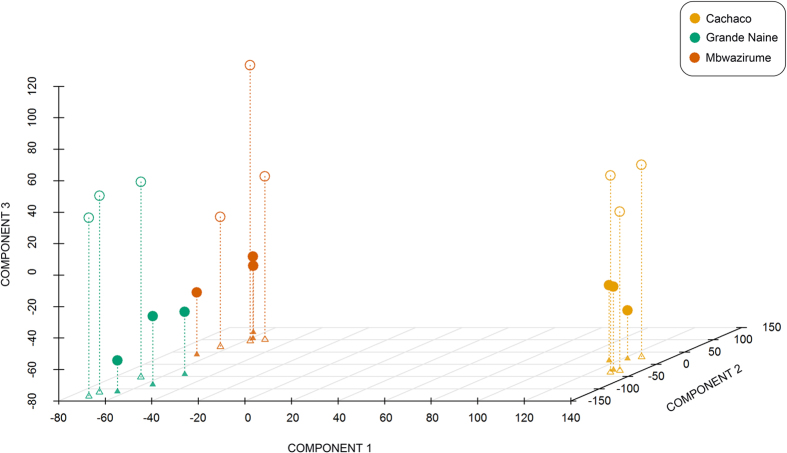
Results from Partial Least Square (PLS) analysis (Fig. 1) indicated that the banana root transcriptome is considerably different for A and B genomes, as component 1 noticeably separated genotype ABB (Cachaco) from both AAA genotypes (Grande Naine and Mbwazirume). Component 2 was able to distinguish between Grande Naine and Mbwazirume, which belong to different subgroups of cultivated varieties (Cavendish and East African highland bananas, respectively), as previously reported17. Besides, component 3 clearly separated the samples according to the treatment in all three genotypes.
Figure 1. Partial Least Squares analysis of the banana root transcriptome under mild osmotic stress.
Empty circles: stress conditions (5% PEG); filled circles: control conditions (0% PEG). Projections over components 1 and 2 are indicated by triangles in the 2D-space.
Identification, functional annotation and characterization of differentially expressed genes (DEGs)
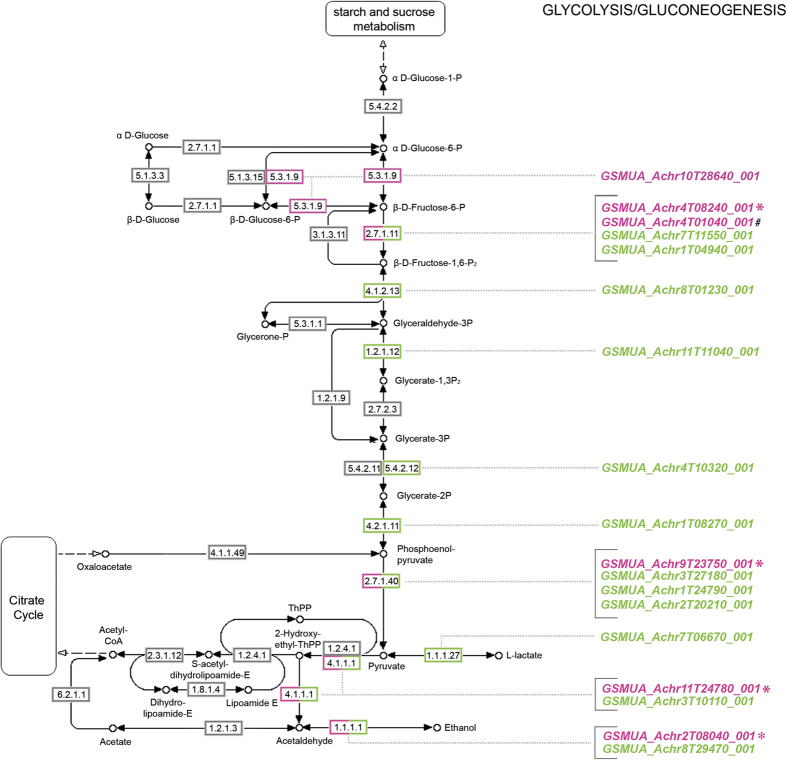
As shown in Table 2, nearly double number of DEGs (670) were detected in Grande Naine as compared to Cachaco (337 DEGs) or Mbwazirume (302 DEGs). Likewise, a higher proportion of DEGs specific to Grande Naine was observed, as 563 (~84%) out of the 670 DEGs were not shared with the other two genotypes. By contrast, Cachaco and Mbwazirume showed very similar proportions (~68%) of specific DEGs, since 229 out of the 337 DEGs in Cachaco and 206 out of the 302 DEGs in Mbwazirume were distinctive of each genotype (Supplementary Figure S1). To classify the differentially expressed transcripts to putative homologs of known genes, we performed a sequence similarity search against known protein sequence datasets (NCBI nr) by using the BLASTp option from BLAST2GO18. In total, 319 (94.7%) of the differentially expressed transcripts in Cachaco, 636 (94.9%) in Grande Naine and 292 (96.7%) in Mbwazirume showed significant (e-value < 10−3) sequence similarity to entries of NCBI nr (Supplementary Table S2). Our aim is to characterize the differential transcriptome for banana in general so we focus further on the DEGs commonly detected in all three genotypes. Using the union of the results provided by two different statistical approaches (see Methods section), 92 DEGs were detected in all three genotypes (Supplementary Figure S2). BLASTp results from BLAST2GO identified homologous proteins for 89 (96.7%) of them and gene ontology (GO) terms were assigned to 81 (91%) out of the 89 genes. Additionally, enzyme codes were assigned to 26 (32.1%) out of the 81 genes with associated GO terms (Supplementary Table S3). GO enrichment analysis was performed to discover significantly over-represented functional categories by comparing the annotated set of DEGs to all banana genes (GO terms were available for 26,097 of the 36,542 sequenced genes). In total, 24 GO terms were significantly enriched using Fisher’s exact test at p < 0.01 (Table 3). Overrepresented GO terms in the mRNA-seq data provides new insights into mild osmotic stress-induced processes and functions. Most significant enriched GO terms related to biological processes were grouped into “response to (low) oxygen levels” (GO:0001666, p-value: 5.0 × 10−5; GO:0036293, p-value: 5.3 × 10−5 and GO:0070482, p-value: 5.9 × 10−5), “oxidation-reduction process” (GO:0055114, p-value: 2.5 × 10−4), “protein hydroxylation” (GO:0018126 and GO:0019511, p-value: 5.2 × 10−4) and “metabolic processes” (GO:0044710, p-value: 5.2 × 10−4 and GO:0006091, p-value: 1.3 × 10−3). This enrichment analysis points towards an important function for classes associated with respiration, glycolysis and fermentation. KEGG pathway analyses using the DEGs revealed that glycolysis coupled to fermentation was significantly induced after three days of 5% PEG treatment (Fig. 2; Supplementary Table S4). To gain a broader overview of the changes in the pathway, all Musa genes coding for glycolytic and fermentative enzymes were checked in each genotype. Six genes corresponding to five different enzymes were induced in the three genotypes. Additionally, twelve genes corresponding to nine enzymes showed up-regulation in one or two genotypes. In total, eighteen genes corresponding to ten important enzymatic steps in the glycolysis-fermentation pathway were up-regulated in at least one genotype (Fig. 2). In the steps catalysed by 6-phosphofructokinase (EC 2.7.1.11), pyruvate kinase (EC 2.7.1.40), pyruvate decarboxylase (EC 4.1.1.1) and alcohol dehydrogenase (EC 1.1.1.1), more than one gene encoding for the same enzyme showed increased expression.
Table 2. Number of differentially expressed, up- and down-regulated genes detected by edgeR-RLE in the root of each genotype during mild osmotic stress.
| Genotype | No. DEGs | No. Up-regulated genes | No. Down-regulated genes |
|---|---|---|---|
| Cachaco (ABB) | 337 | 260 | 77 |
| Grande Naine (AAA) | 670 | 310 | 360 |
| Mbwazirume (AAA) | 302 | 251 | 51 |
Number of biological replicates (stress/control): n = 3/3; False discovery rate (FDR) ≤ 0.05; DEGs: differentially expressed genes.
Table 3. Enriched GO terms (over representation) for biological processes detected by Fisher’s Exact Test at p < 0.01 on TopGO package78.
| GO ID | Term | Annotateda | Significantb | Expectedc | Significance |
|---|---|---|---|---|---|
| GO:0001666 | response to hypoxia | 71 | 4 | 0.2 | 5.00 × 10−5 |
| GO:0036293 | response to decreased oxygen levels | 72 | 4 | 0.21 | 5.30 × 10−5 |
| GO:0070482 | response to oxygen levels | 74 | 4 | 0.21 | 5.90 × 10−5 |
| GO:0055114 | oxidation-reduction process | 1922 | 15 | 5.47 | 2.50 × 10−4 |
| GO:0018126 | protein hydroxylation | 12 | 2 | 0.03 | 5.20 × 10−4 |
| GO:0019511 | peptidyl-proline hydroxylation | 12 | 2 | 0.03 | 5.20 × 10−4 |
| GO:0044710 | single-organism metabolic process | 6929 | 32 | 19.73 | 9.40 × 10−4 |
| GO:0006091 | generation of precursor metabolites and energy | 759 | 8 | 2.16 | 1.35 × 10−3 |
| GO:0044699 | single-organism process | 11993 | 46 | 34.15 | 1.50 × 10−3 |
| GO:0006067 | ethanol metabolic process | 1 | 1 | 0 | 2.85 × 10−3 |
| GO:0006069 | ethanol oxidation | 1 | 1 | 0 | 2.85 × 10−3 |
| GO:0009233 | menaquinone metabolic process | 1 | 1 | 0 | 2.85 × 10−3 |
| GO:0009234 | menaquinone biosynthetic process | 1 | 1 | 0 | 2.85 × 10−3 |
| GO:0009817 | defense response to fungus, incompatible interaction | 31 | 2 | 0.09 | 3.52 × 10−3 |
| GO:0009814 | defense response, incompatible interaction | 398 | 5 | 1.13 | 5.48 × 10−3 |
| GO:0006522 | alanine metabolic process | 2 | 1 | 0.01 | 5.69 × 10−3 |
| GO:0009078 | pyruvate family amino acid metabolic process | 2 | 1 | 0.01 | 5.69 × 10−3 |
| GO:0018345 | protein palmitoylation | 2 | 1 | 0.01 | 5.69 × 10−3 |
| GO:0006096 | glycolytic process | 262 | 4 | 0.75 | 6.62 × 10−3 |
| GO:0010310 | regulation of hydrogen peroxide metabolic process | 143 | 3 | 0.41 | 7.91 × 10−3 |
| GO:0006103 | 2-oxoglutarate metabolic process | 3 | 1 | 0.01 | 8.52 × 10−3 |
| GO:0006531 | aspartate metabolic process | 3 | 1 | 0.01 | 8.52 × 10−3 |
| GO:0090470 | shoot organ boundary specification | 3 | 1 | 0.01 | 8.52 × 10−3 |
| GO:0048856 | anatomical structure development | 3042 | 16 | 8.66 | 9.66 × 10−3 |
aNumber of genes mapped to the GO term from all annotated Musa genes.
bNumber of genes mapped to the GO term in the DEGs common to the three genotypes, and
cexpected number these genes mapped to the GO term if they were randomly distributed over all GO terms.
Figure 2. Pathway visualization of glycolysis-fermentation associated enzymes and corresponding transcripts up-regulated in banana root under mild osmotic stress (modified from KEGG pathway in plants).
Enzyme codes: 5.3.1.9: Glucose-6-phosphate isomerase; 2.7.1.11: 6-phosphofructokinase; 4.1.2.13: fructose-bisphosphate aldolase; 1.2.1.12: glyceraldehyde-3-phosphate dehydrogenase; 5.4.2.12: phosphoglycerate mutase; 4.2.1.11: phosphopyruvate hydratase; 2.7.1.40: pyruvate kinase; 4.1.1.1: pyruvate decarboxylase, 1.1.1.27: L-lactate dehydrogenase; 1.1.1.1: alcohol dehydrogenase. Enzymes and transcripts coded pink are induced in all three genotypes; those coded green are induced in one or two genotypes. *genes validated by RT-qPCR. #not detected at FDR ≤ 0.05.
Validation of up-regulated genes by quantitative real time RT-PCR (RT-qPCR)
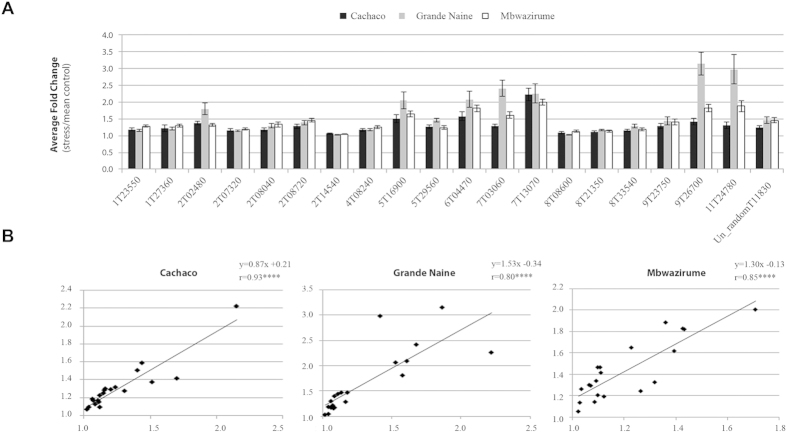
To confirm the accuracy and reproducibility of the mRNA-seq results and the robustness of the statistics, a subset of twenty genes commonly up-regulated in the three genotypes (Table 4) was selected as described in Methods section for validation by RT-qPCR. Therefore, an independent experiment was set up with 6 biological replicates per genotype and three different time points: 6 hours, 3 days and 7 days after 0 and 5% PEG treatment. ANOVA test on the RT-qPCR data indicated a significant genotype-independent treatment effect (p < 0.05) for 18 genes at the earliest time point (6 h) and for all 20 genes at day 3 and day 7 (Table 5, Supplementary Table S5 and Supplementary Figure S3). At day 3 (same time point as the mRNA-seq results), treatment effects were significant (p < 0.05) in all three genotypes for 18 out of the 20 candidate genes (Supplementary Figure S3). In general, fold changes of RT-qPCR expression values were very similar among the three genotypes (Fig. 3A). Besides, highly significant correlation coefficients were found (r = 0.80–0.93; p < 0.0001) when comparing RNA-seq and RT-qPCR results in each genotype, indicating a good consistency between the two analysis techniques and the two independent experiments (Fig. 3B). Similar correlation coefficients between mRNA-seq and RT-qPCR have been obtained in recent transcriptomic studies conducted in other plant crops19,20.
Table 4. List of the 20 candidate genes up-regulated in root during mild osmotic stress and selected for RT-qPCR validation.
| Gene IDa | FC (p-value)b |
Arabidopsis thalianaortholog(s) | Functionc | Musafamily IDd(No. paralogs) | ||
|---|---|---|---|---|---|---|
| Cach | GN | Mbw | ||||
| GSMUA_Achr1T23550_001 | 2.0 × 101(1 × 10−5) | 4.0 × 100(2 × 10−2)# | 6.3 × 100(4 × 10−3)# | AT2G19350 AT4G29850 | Unknown function (DUF872) | CF158574 (4) |
| GSMUA_Achr1T27360_001 | 2.9 × 101(2 × 10−9) | 6.3 × 100(2 × 10−3)# | 5.0 × 100(6 × 10−3)# | AT1G53580 | Mononuclear Fe(II)-containing member of the b-lactamase fold superfamily (ETHE1-like) | CF158578 (2) |
| GSMUA_Achr2T02480_001 | 3.4 × 103(2 × 10−12) | 3.7 × 103(9 × 10−17) | 2.5 × 102(3 × 10−7) | AT1G03475 | Coproporphyrinogen III oxidase | CF104144 (3) |
| GSMUA_Achr2T07320_001 | 3.1 × 101(9 × 10−9) | 1.2 × 101(2 × 10−5) | 2.0 × 101(2 × 10−7) | AT1G17290 AT1G72330 | Alanine aminotransferase (AlaAT) | CF104167 (3) |
| GSMUA_Achr2T08040_001* | 1.0 × 101(1 × 10−4) | 5.6 × 100(2 × 10−2)# | 2.1 × 101(8 × 10−7) | AT1G77120 | Alcohol dehydrogenase (ADH) | CF104189 (5) |
| GSMUA_Achr2T08720_001 | 1.8 × 102(2 × 10−15) | 1.5 × 101(3 × 10−5) | 3.0 × 101(1 × 10−6) | AT2G16060 AT3G10520 | Class I nonsymbiotic haemoglobin (HB) | CF158581 (3) |
| GSMUA_Achr2T14540_001 | 2.5 × 100(6 × 10−2)# | 2.0 × 100(2 × 10−1)# | 2.5 × 100(6 × 10−2) | AT3G18170 AT3G18180 | Glycosyltransferase family 61 protein | CF158591 (5) |
| GSMUA_Achr4T08240_001* | 1.2 × 101(4 × 10−6) | 4.0 × 100(2 × 10−2)# | 4.0 × 100(1 × 10−2)# | AT4G26270 AT4G29220 AT5G56630 AT4G32840 | 6-phosphofructokinase (6PFK) | CF158617 (5) |
| GSMUA_Achr5T16900_001 | 9.1 × 102(4 × 10−8) | 3.0 × 103(1 × 10−9) | 6.8 × 101(1 × 10−5) | AT4G10040 AT1G22840 | Cytochrome c (CYTC) | CF158619 (5) |
| GSMUA_Achr5T29560_001 | 6.5 × 102(9 × 10−9) | 1.2 × 102(6 × 10−10) | 4.3 × 102(8 × 10−8) | AT5G27760 AT3G05550 | Hypoxia responsive family protein | CF158650 (6) |
| GSMUA_Achr6T04470_001 | 1.0 × 103(7 × 10−10) | 2.1 × 103(6 × 10−9) | 9.9 × 102(8 × 10−6) | AT3G28490 AT3G28480 | Prolyl 4-hydroxylase, alpha subunit (P4H) | CF158570 (3) |
| GSMUA_Achr7T03060_001 | 8.9 × 101(4 × 10−7) | 2.9 × 103(9 × 10−10) | 6.0 × 102(2 × 10−5) | AT1G43800 | Stearoyl-acyl-carrier-protein desaturase family protein (S-ACP-DES) | CF158628 (2) |
| GSMUA_Achr7T13070_001 | 2.5 × 105(1 × 10−26) | 3.9 × 105(3 × 10−10) | 8.1 × 103(8 × 10−10) | AT1G21890 AT1G44800 AT4G08290 AT2G37460 | Nodulin MtN21-like transporter family protein (UMAMIT) | CF104204 (5) |
| GSMUA_Achr8T08600_001 | 5.0 × 100(3 × 10−3)# | 6.3 × 100(7 × 10−3)# | 4.0 × 100(9 × 10−3)# | AT5G19140 | Unknown function (DUF3700) | CF158620 (4) |
| GSMUA_Achr8T21350_001 | 1.8 × 101(7 × 10−6) | 1.6 × 101(9 × 10−7) | 1.6 × 101(4 × 10−5) | AT3G10920 AT3G56350 | Manganese superoxide dismutase (MSD) | CF158635 (4) |
| GSMUA_Achr8T33540_001 | 2.7 × 101(4 × 10−7) | 5.0 × 101(3 × 10−9) | 2.3 × 101(2 × 10−5) | AT5G44730 | Haloacid dehalogenase-like hydrolase (HAD) superfamily protein | CF158624 (2) |
| GSMUA_Achr9T23750_001* | 6.7 × 101(7 × 10−8) | 1.4 × 101(1 × 10−4) | 1.8 × 101(7 × 10−5) | AT5G08570 AT5G63680 | Pyruvate kinase family protein (PK) | CF158643 (5) |
| GSMUA_Achr9T26700_001 | 1.0 × 105(1 × 10−8) | 4.2 × 104(1 × 10−10) | 7.2 × 103(10 × 10−9) | AT3G29970 | B12D protein | CF158632 (2) |
| GSMUA_Achr11T24780_001* | 1.5 × 102(4 × 10−8) | 4.7 × 102(1 × 10−7) | 5.2 × 102(1 × 10−8) | AT5G01330 AT5G01320 AT4G33070 AT5G54960 | Thiamine pyrophosphate dependent pyruvate decarboxylase family protein (PDC) | CF158639 (6) |
| GSMUA_AchrUn_randomT11830_001 | 7.9 × 101(1 × 10−14) | 4.1 × 101(3 × 10−9) | 2.3 × 101(4 × 10−8) | AT2G32720 | Member of Cytochromes b5 (CB5-B) | CF158596 (4) |
aMusa candidate genes (abbreviation in bold) selected from RNA-seq data.
bFold Change (stress vs. control) calculated by edgeR-RLE, and associated p-value.
cFunction of the A. thaliana ortholog(s) with the highest sequence similarity to the Musa candidate gene (underlined).
dFamily ID for Custom families created in GreenPhyl database (http://www.greenphyl.org/) and number of Musa paralogous genes since divergence with A. thaliana. Cach: Cachaco, GN: Grande Naine, Mbw: Mbwazirume.
*Candidate genes involved in the glycolysis-fermentation pathway.
#not detected at FDR≤0.05, but detected by the non-parametric test combined with PLS.
Table 5. ANOVA results showing the significance level of the genotype, treatment and genotype x treatment effects for the 20 candidate genes analysed by RT-qPCR.
| Gene ID | Genotype effect | Treatment effect | Genotype × treatment effect |
|---|---|---|---|
| p-value | |||
| GSMUA_Achr1T23550_001 | *** | **** | ns |
| GSMUA_Achr1T27360_001 | *** | **** | ns |
| GSMUA_Achr2T02480_001 | *** | **** | ns |
| GSMUA_Achr2T07320_001 | * | **** | ns |
| GSMUA_Achr2T08040_001 | ns | **** | ns |
| GSMUA_Achr2T08720_001 | ns | **** | ns |
| GSMUA_Achr2T14540_001 | **** | * | ns |
| GSMUA_Achr4T08240_001 | * | **** | ns |
| GSMUA_Achr5T16900_001 | ** | **** | ns |
| GSMUA_Achr5T29560_001 | * | **** | ns |
| GSMUA_Achr6T04470_001 | * | **** | ns |
| GSMUA_Achr7T03060_001 | * | **** | * |
| GSMUA_Achr7T13070_001 | ns | **** | ns |
| GSMUA_Achr8T08600_001 | ns | ** | ns |
| GSMUA_Achr8T21350_001 | ** | **** | ns |
| GSMUA_Achr8T33540_001 | *** | **** | ns |
| GSMUA_Achr9T23750_001 | ns | **** | ns |
| GSMUA_Achr9T26700_001 | *** | **** | ns |
| GSMUA_Achr11T24780_001 | ** | **** | ns |
| GSMUA_AchrUn_randomT11830 | * | **** | ns |
Genotypes used: Cachaco, Grande Naine and Mbwazirume. Number of biological replicates (stress/control): n = 6/6. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns: not significant. Candidate genes involved in the glycolysis-fermentation pathway are underlined.
Figure 3. Confirmation of expression profiles by RT-qPCR for the 20 selected candidate genes and comparison with RNA-seq data.
(A) Relative expression levels (fold changes of log transformed data) at day 3; gene ID abbreviations according to Table 4. Musa genes EF-1, L2 and ACT-1 were used as internal controls to normalize the expression data. (B) Correlations between RNA-seq and RT-qPCR results at day 3 in each genotype. X-axis: average fold change (stress vs. mean control) in RNA-seq; Y-axis: average fold change (stress vs. mean control) in RT-qPCR; error bars: standard error of the means in each genotype; r: Pearson correlation coefficient; ****p < 0.0001.
Identification of Musa paralogs and corresponding gene expression patterns
To infer as accurately as possible the functions of the candidate genes, all genes with similar sequences were identified in the Musa genome and their orthology relationships established with genes from Arabidopsis thaliana. For each candidate, one or more paralogs (i.e. genes derived by duplication in the Musa specific lineage) were identified (Table 4). As shown for the genes related to the glycolysis-fermentation pathway, paralogs can exhibit an expression pattern significantly correlated (p < 0.01) to that of the candidate gene or can show different expression patterns (Supplementary Figure S4). In the first case, the genes appear as redundant copies (at least in the tissues and conditions analysed) whereas sub-functionalization after duplication events can be postulated for the genes whose expression pattern diverged21.
Discussion
Transcriptome analysis in Musa under of mild osmotic stress
In this study, mRNA-seq was used to analyse transcriptomic changes in the roos of three triploid banana genotypes subjected to mild osmotic stress. The biggest challenge to perform mRNA-seq on a non-model crop, such as banana, is its ploidy level. A few studies of the banana transcriptomic response to abiotic/biotic stresses have been reported for triploid genotypes9,22,23 and different approaches were applied depending on the availability or not of the diploid reference genome. Here we opted for a mapping-first approach of short reads12. As expected, the percentage of reads that mapped to the reference genome was higher for both AAA genotypes (84%) than for ABB Cachaco (70%), since the latter contains two copies of the B genome and only one copy of the A genome (Table 1). However, the number of high quality reads and mapped genes was comparable among the three genotypes and ensured a good coverage of the Musa genome (Table 1 and Supplementary Table S1).
The varying number of DEGs in the three genotypes and the Partial Least Square analysis (Fig. 1) point towards genotype specific reactions. Provided similar percentage of mapped reads in Grande Naine and Mbwazirume (both AAA, Table 1), Grande Naine seems to be the most reactive genotype, since nearly double number of DEGs were detected when comparing to Cachaco or Mbwazirume (Table 2).
Physiological impact of mild osmotic stress
Enhanced oxidative respiration and reactive oxygen species (ROS) production
Stress responses in plants occur at various organ levels, among which root specific processes are particularly relevant24. Roots are big sinks of energy and the main consumers of carbon fixed in photosynthesis during the vegetative stage. During stress, a higher proportion of dry matter is allocated to the root in order to satisfy its increased energy demand25. As non-green tissues, roots entirely depend on glycolysis and mitochondrial respiration for their energy production. Stress causes a higher energy consumption and, thus, enhances respiration, one of the major cellular pathways dependent on oxygen26. Three Musa genes identified in our study (GSMUA_Achr9T26700_001, GSMUA_Achr5T16900_001 and GSMUA_Achr5T29560_001) are involved in mitochondrial respiration (Table 4).
Gene GSMUA_Achr9T26700_001, (Fig. 4), shows a strong induction in all three genotypes (Table 4 and Supplementary Figure S3). It has been annotated as a subunit of the respiratory complex I, also known as NADH:ubiquinone oxidoreductase (Table 4 and Supplementary Table S3), a major component of the mitochondrial electron transport chain. This complex couples the oxidation of NADH to the reduction of ubiquinone with the generation of a proton gradient used for ATP synthesis27. We found one paralogous gene, GSMUA_Achr6T27380_001, that is also strongly up-regulated in the three genotypes and shows an expression pattern significantly correlated (p < 0.01) to that of GSMUA_Achr9T26700_001 (Supplementary Figure S5).
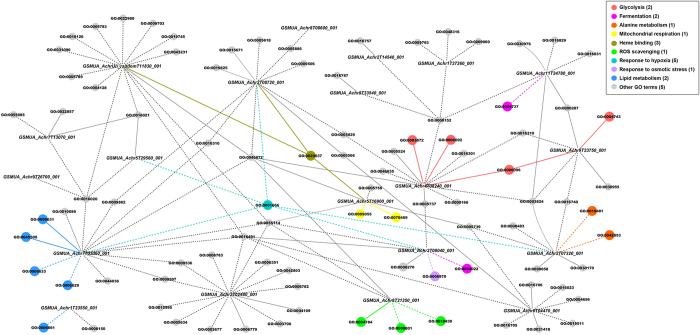
Figure 4. Interactive network of candidate genes and associated GO terms depicting key root processes affected under mild osmotic stress.
Solid lines: GO terms assigned via Uniprot (http://www.uniprot.org/). Dashed lines: GO terms assigned via cross-species annotation. Relevant GO terms are highlighted in different colors and the number of genes associated to them is indicated between brackets.
Gene GSMUA_Achr5T16900_001, associated to electron carrier activity and the respiratory chain (GO:0009055 and GO:0070469; Fig. 4), codes for a cytochrome c (Table 4 and Supplementary Table S3), which is involved in electron transfer between the respiratory complex III (ubiquinone-cytochrome c oxidoreductase) and complex IV (cytochrome c oxidase (COX))28.
Gene GSMUA_Achr5T29560_001, related to integral component of membrane (GO:0016021; Fig. 4), has been annotated as an hypoxia responsive family gene (Table 4) and as a respiratory super complex factor 2 (Rcf2) homolog (Supplementary Table S3). Rcf2 is a cytochrome c oxidase (COX) subunit required for optimal enzyme activity and the correct assembly of the cytochrome bc1-COX super complex, which belongs to the conserved hypoxia-induced gene 1 (Hig1) protein family29. We have found other three Musa paralogs with a significantly correlated (p < 0.01) expression pattern to that of GSMUA_Achr5T29560_001 (Supplementary Figure S6).
The identification of GSMUA_Achr9T26700_001, GSMUA_Achr5T16900_001 and GSMUA_Achr5T29560_001 and of some paralogs with correlated expression patterns supports the assumption of an enhanced respiration rate under mild osmotic stress. We hypothesize that the enhanced respiration is driven by the increased energy demand and serves to cope with the adverse conditions25.
In stressed plants, a direct link between the mitochondrial electron transport respiratory chain and ROS production has been demonstrated. ROS can act as important signalling molecules involved in the stress signal transduction pathway, while excessive ROS may induce oxidative damage to cellular components and structures30. Plants have developed an antioxidant system to remove the excess of superoxide (O2−) radicals, a type of ROS, which includes superoxide dismutases. These enzymes convert toxic O2− to hydrogen peroxide and water31. Gene GSMUA_Achr8T21350_001 was annotated to superoxide dismutase activity (GO:0004784; Fig. 4) and, thus, to ROS scavenging, and described as a superoxide dismutase. The gene shows a significantly induced expression in each genotype after 3 days of PEG treatment and in two genotypes after 7 days of PEG treatment, while no significant induction was detected in the earliest time point (Supplementary Figure S3). The product of AT3G10920, ortholog in Arabidopsis (Table 4), is a manganese superoxide dismutase (MSD) located in the mitochondria which also accumulates under osmotic stress31. We suggest that banana roots trigger the complex antioxidant network to regulate ROS production and to facilitate appropriate signalling during mild osmotic stress.
Fermentation and carbon allocation
Metabolically active cells, such as those in the root tip, have a high oxygen demand and are particularly prone to suffer from hypoxia, i.e. low oxygen levels32. According to Aguilar et al. (2003), respiratory oxygen consumption in banana roots decreases substantially with distance from the apex and the stele33. We hypothesize that higher respiration rates in both, the root apex and the stele, lead to a shift from more aerobic to more anaerobic metabolism for ATP production. The options are alternative respiration and NAD+ regeneration via fermentation34. Fermentative ATP production is much less efficient and increases the demand for carbohydrates. Consistently, we observed a generalized induction of transcripts for enzymes involved in glycolysis and fermentation (Fig. 2 and Supplementary Table S4). Protein accumulation of enzymes belonging to this pathway has also been reported in banana plants and meristems under osmotic stress5,35 and, recently, an increase of glycolysis-related proteins has been found in soybean roots submitted to drought stress36. In our study, there were multiple steps in the glycolysis-fermentation pathway where genes were significantly induced (Fig. 2). The increased expression of the genes GSMUA_Achr4T08240_001 (6-phosphofructokinase; 6PFK), GSMUA_Achr9T23750_001 (pyruvate kinase; PK), GSMUA_Achr11T24780_001 (pyruvate decarboxylase; PDC) and GSMUA_Achr2T08040_001 (alcohol dehydrogenase; ADH) has been verified by RT-qPCR (Supplementary Figure S3). All 4 genes were annotated to the glycolytic process (GO:0006096) and GSMUA_Achr11T24780_001 (pyruvate decarboxylase; PDC) and GSMUA_Achr2T08040_001 (alcohol dehydrogenase; ADH) are involved in the ethanolic fermentation process (Fig. 4). Since 6-phosphofructokinase, pyruvate kinase and pyruvate decarboxylase catalyse irreversible reactions, they represent important control points. The up-regulation of the genes under stress ensures a continuous flow of metabolites throughout the pathway. This is supported by the fact that all four candidate genes have at least another paralog with a significantly correlated (p < 0.01) expression pattern (Supplementary Figure S4). Pyruvate, the final product of glycolysis, can either be converted into lactate by lactate dehydrogenase or to ethanol by pyruvate decarboxylase and alcohol dehydrogenase. As an initial reaction to oxygen deprivation, lactic acid fermentation is activated causing a decrease in cytosolic pH. This reduces the activity of the responsible enzyme and lactic acid fermentation is followed by alcoholic fermentation37. The observed up-regulation of genes encoding pyruvate decarboxylase and alcohol dehydrogenase in Musa is in agreement with previous studies where the corresponding Arabidopsis orthologs also showed induction under low oxygen conditions38,39. Interestingly, ethanol production and alcohol dehydrogenase induction was also found in plants under other abiotic stresses, including dehydration40,41, which confirms the hypothesis that fermentation plays a role under environmental stress. In our study, the majority of up-regulated genes involved in glycolysis were predicted to be in the cytosol (Supplementary Table S4), an indication that most of the carbon source is channeled via cytosolic glycolysis to feed fermentative pathways. Plants regulate the balance between respiration and fermentation to be able to control the internal oxygen level42. The induction of the fermentative enzymes pyruvate decarboxylase and alcohol dehydrogenase is not exclusively dependent on the oxygen concentration, but is also linked to changes in the energy status (ratio of ATP to ADP). Consequently, sensing the energy status would be an important component for optimizing plant metabolism.
Pyruvate can also serve as precursor for the synthesis of alanine by the enzyme alanine amino transferase (AlaAT). This enables to conserve carbon skeletons that otherwise would be lost by fermentation and also prevents cytoplasmic acidification by avoiding lactic acid production and proton consumption43,44. Other possible benefits of alanine accumulation during hypoxia have been reviewed by Menegus et al. (1993) and include prevention of ammonium toxicity, provision of a reduced nitrogen store and generation of osmotic pressure45. Hypoxia induced transcripts of genes encoding alanine amino transferases has been reported in other plant species as Arabidopsis, wheat or soybean39,46,47. Gene GSMUA_Achr2T07320_001 has been annotated as alanine amino transferase (AlaAT; Fig. 4, Table 4 and Supplementary Table S3). The Arabidopsis ortholog AT1G17290 also shows an induction under low oxygen conditions38,39 and, consistently, we observed up-regulation of GSMUA_Achr2T07320_001 already at 6 hours of PEG treatment (Supplementary Figure S3). This suggests that the banana root tip starts rebalancing carbon allocation as soon as 6 hours.
Alternative respiration and haemoglobin (Hb)/nitric oxide (NO) cycle
As an alternative to oxygen-based respiration and classic fermentation, a process involving stress-induced class I haemoglobins (Hbs) has been described in plants under low oxygen48. Under such conditions, root mitochondria use nitrite as final electron acceptor instead of oxygen producing NO49, which is toxic to cells and scavenged by haemoglobin proteins. In Arabidopsis and barley plants under hypoxia, a rapid induction of haemoglobin expression has been detected50,51. However, this induction would rather respond to cell energy/redox status than to low oxygen levels52. Interestingly, overexpression of haemoglobin in hypoxic maize cell cultures resulted in lower ethanolic fermentation, since a greater turnover of NO in the Hb/NO cycle increased NADH oxidation, replacing to some extent the requirement for alcohol dehydrogenase activity53. Gene GSMUA_Achr2T08720_001, connected to oxygen binding/transport and heme binding (GO:0019825, GO:0015671 and GO:0020037; Fig. 4), has been annotated as class I nonsymbiotic haemoglobin (Table 4 and Supplementary Table S3) and its Arabidopsis ortholog AT2G16060 is also up-regulated under hypoxia50. Remarkably, cytochrome b5 reductases have been proposed to play a role in the NO/Hb cycle, particularly in the reduction of metahaemoglobin to haemoglobin48. According to this hypothesis, up-regulation of GSMUA_AchrUn_randomT11830_001, also connected to heme binding (GO:0020037; Fig. 4) and annotated as a member of the cytochrome b5 (Table 4 and Supplementary Table S3), could be linked to the induction of our haemoglobin gene (GSMUA_Achr2T08720_0010).
Detoxification
Apart from ROS and NO, other compounds can be toxic to cells when they accumulate in the mitochondria. An example is hydrogen sulfide, generated from cysteine degradation and considered a potent inhibitor of aerobic respiration. Its effects change from physiological to toxic within a narrow concentration range. Thus, regulatory mechanisms are needed to keep endogenous sulfide levels under control54. Gene GSMUA_Achr1T27360_001 has been annotated as ETHE1-like, a member of the b-lactamase fold superfamily (Table 4 and Supplementary Table S3). The ortholog in Arabidopsis is AT1G53580, which encodes a mitochondrial sulfur dioxygenase (AtETHE1) involved in detoxification of hydrogen sulfide55. AtETHE1 and OsETHE1 from rice have proved to show high root-specific and stress-inducible expression, suggesting a potential role of this gene in the stress response56,57. Due to its higher metabolism, the root tip undergoes quick protein turnover and sulfate acquired by roots constitute the primary sulfur source for growth, justifying the role of ETHE1 as part of the sulfur catabolism pathway in roots. Recently, a key function in the use of amino acids as alternative respiratory substrates during carbohydrate starvation has been attributed to AtETHE158.
Other root transcripts up-regulated during mild osmotic stress
One of our candidates, GSMUA_Achr6T04470_001, was linked to oxidoreductase activity (GO:0016706; Fig. 4) and has been annotated as a prolyl-4-hydroxylase alpha subunit (P4H; Table 4 and Supplementary Table S3). In a study carried out in chickpea roots after dehydration treatment, prolyl 4-hydroxylase alpha subunits belonged to the most up-regulated group of transcripts59. Interestingly, overexpression in Arabidopsis of different AtP4H genes has proven to increase root hair length/density, a response that would facilitate both nutrient and water assimilation by the plant60. Gene GSMUA_Achr2T14540_001, associated to the transfer of glycosyl groups (GO:0016757; Fig. 4), has been annotated as a glycosyl transferase 61 family protein (Table 4 and Supplementary Table S3). Differential expression of glycosyl transferases was also found in Arabidopsis root cultures under hypoxia treatment and in cotton roots subjected to drought stress, where they were linked to cell wall processes38,61. Gene GSMUA_Achr7T03060_001, related to fatty acid metabolism and desaturase activity (GO:0006631 and GO:0045300; Fig. 4), has been annotated as a stearoyl-[acyl-carrier-protein] 9-desaturase (Table 4 and Supplementary Table S3), a gene involved in the biosynthesis of polyunsaturated fatty acids62. Its paralog, GSMUA_Achr8T32640_001, is also up-regulated under the applied stress and exhibits an expression pattern significantly correlated (p < 0.01) to that of GSMUA_Achr7T03060_001 (Supplementary Figure S7). Recently, the ortholog in Arabidopsis, AT1G43800, has been shown to increase the levels of unsaturated fatty acids in crown galls under hypoxia and drought stress conditions63. Remarkably, candidate GSMUA_Achr1T23550_001 (gene of unknown function, Table 4) has also been associated to lipid metabolism (GO:0006661; Fig. 4). Gene GSMUA_Achr7T13070_00 is linked to transmembrane transporter activity and integral component of membrane (GO:0022857 and GO:0016021; Fig. 4), and has been annotated as a nodulin MtN21-like transporter family protein with a strong induction in the three analysed genotypes (Table 4). Interestingly, AT1G75500, another nodulin MtN21-like protein, has been found up-regulated in Arabidopsis root cultures under hypoxia, but its specific involvement in the stress response has not yet been determined38.
Given the advances in genomic technology platforms, the unique ability to compare transcriptomes across several species can be exploited to cross-reference information concerning genes and gene functions. However, it is still a challenge to infer gene functions in a non-model crop and to rely in cross-species annotation, as exemplified for candidates GSMUA_Achr1T23550_001 (GO:0006661 and GO:0008150; Fig. 4) and GSMUA_Achr8T08600_001 (GO: 0005886; Fig. 4), since their gene functions could not be inferred even when comparing with Arabidopsis or other plant species (Table 4 and Supplementary Table S3).
Conclusion
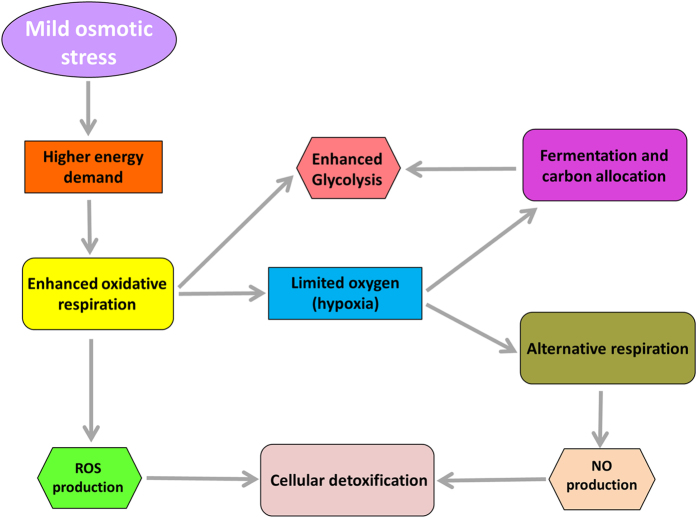
Transcriptome profiling in the polyploid non-model crop Musa indicated that the roots change the broad spectrum of energy metabolism after applying mild osmotic stress. We hypothesize that osmotic stress leads to a drop in energy level, which induces a metabolic shift towards (i) a higher oxidative respiration, (ii) alternative respiration and (iii) fermentation (Fig. 5). By validating a subset of genes by RT-qPCR, we confirm the success of RNA-seq for evaluation of a non-model crop. This work contributes to a better understanding of the molecular mechanisms and provides a workflow to study responses to osmotic stress.
Figure 5. Summary of the main processes induced during osmotic stress in banana roots.
Mild osmotic stress causes a higher energy demand which enhances aerobic respiration and leads to local hypoxia in the root tip. In this situation, alternative respiration and fermentation/carbon allocation take place. Due to the induction of fermentation and the increase in respiration, the glycolytic pathway is also enhanced. Respiratory activity generates toxic compounds, as reactive oxygen species (ROS) and nitric oxide (NO), which are detoxified by cells.
Methods
Plant material, growth conditions and osmotic stress treatment
In vitro banana plants of the genotypes ‘Cachaco’ (Bluggoe ABB, ITC0643), ‘Grande Naine’ (Cavendish AAA, ITC0180) and ‘Mbwazirume’ (East African highland banana AAAh, ITC0084) were supplied by the Bioversity International Transit Centre hosted at KU Leuven, Belgium. In vitro plants were grown for 35 days in autotrophic conditions prior to the start of the experiment and roots were covered to protect from light. Each plant (3 biological replicates per genotype) was grown in a 500 mL PP container with 305 mL medium: 3.61 g/L KNO3, 1.21 g/L K2SO4, 1.61 g/L MgSO4. 7H2O, 1.81 g/L MgCl2.6H2O, 0.6 g/L Sequestrene, 0.0114 g/L H3BO3, 0.027 g/L MnSO4.H2O, 0.0023 g/L ZnSO4.7H2O, 0.0016 g/L CuSO4.5H2O, 0.0007 g/L NaMo4.2H2O, pH = 6 (modified from Swennen et al.64). Plants were grown in a phytotron (Aralab Fitoclima Bio 600) with a 12 h/12 h light/dark period (average light intensity of 183 ± 29 μmol photons m−2 s−1). Humidity and temperature were kept constant at 75% and 25 °C, respectively. At the start of the treatment, the group of the stressed plants received fresh medium containing five percent (W/W) of PEG-8000 (Sigma, USA), while control plants received the same fresh medium without PEG-8000. Subsequently, in both groups (stress/control), the medium was refreshed when it reached 55% of the initial volume in at least one plant. Three days after 5% PEG treatment the plants were sacrificed and root tips (segments of on average 4 cm from the apex) were collected for RNA-sequencing. An independent experiment was set up for RT-qPCR validation, including six biological replicates per genotype and three different time points: 6 hours, 3 days and 7 days after 5% PEG or control treatment.
RNA extraction and cDNA library preparation
Root material from the two independent experiments was snap frozen in liquid nitrogen and stored at −80 °C. Total RNA was extracted as described in65. Samples were treated with TurboTM DNase I (Ambion, Austin, TX, USA) for 45 min at 37 °C followed by a phenol-chloroform/ethanol purification step to eliminate gDNA traces. Real-time PCR was performed using DNase-treated RNA as template and primers for the Elongation factor 1α (EF1) genomic sequence to verify absence of gDNA. Only samples with undetected amplification after 40 cycles were used. RNA quantity and quality (A260/230, A260/280) were determined using Nanodrop ND-1000TM spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
For cDNA library construction, RNA integrity was checked by ExperionTM (BIO-RAD Laboratories, Inc. USA; RQI > 9.4) and BioAnalyzer (Agilent;RIN > 7.8). The TruSeq RNA Sample Seq kit (Illumina Inc.) was used according to the manufacturer’s protocol to generate the libraries. In brief, poly-A containing RNA molecules were purified from 1 μg total RNA using oligo-dT magnetic beads and fragmented by adding the fragmentation buffer and heating at 94 °C for 8 min in a thermocycler. First strand cDNA was synthesized using random primers. Second strand cDNA synthesis, end repair, A-tailing and adapter ligation was done in accordance with the manufacturer’s instructions. Purified cDNA templates were enriched by 15 cycles of PCR for 10 s at 98 °C, 30 s at 65 °C and 30 s at 72 °C using Illumina’s proprietary primers and Phusion DNA polymerase. Each indexed cDNA library was verified using a DNA 1000 Chip on a Bioanalyzer 2100, quantified by RT-PCR with the KAPA Library Quantification Kit for Illumina Sequencing Platforms (Kapa Biosystems Ltd, SA) and diluted to 10 nM using Illumina’s resuspension buffer.
Illumina sequencing and mapping
Multiplex sequencing on an Illumina HiSeq2000 was performed as 100 bp, single reads at GenomiX, Montpellier, France. For each lane of sequencing, 5 libraries were equimolarly pooled, denatured using 0.1N NaOH and diluted to a final concentration of 7 pM using Illumina’s HT1 buffer. 120 μL of the dilution was then transferred into a 200 μL strip tube and placed on ice before loading onto the cBot. The flow cell was clustered using Single Read Cluster Generation Kit (Illumina Inc.), according to the Illumina SR_amplification_Linearization_Blocking _PrimerHyb recipe. The flow cell was loaded onto the Illumina HiSeq 2000 instrument following the manufacturer’s instructions and sequencing was performed with the 100 cycles, single read, indexed protocol. Image analyses and base calling were performed using the HiSeq Control Software (HCS) and Real-Time Analysis component (RTA). Demultiplexing was carried out using CASAVA. Data quality was assessed using fastQC (Babraham Institute, USA) and the Illumina software SAV. RNA-seq reads were quality filtered using Illumina purity filter and aligned to the M. acuminata assembly v113 from the Banana Genome Hub66 with gene model annotations, using the splice junction mapper TopHat 1.4.167 and Bowtie 0.12.8 (default parameters). Gene counting was calculated using HTSeq v0.5.3p9 (http://www-huber.embl.de/users/anders/HTSeq/) in union mode. Reads not aligned on exons or with multiple hits were disregarded.
Differential gene expression analyses and selection of candidate genes
Differential gene expression between stress and control conditions was evaluated using edge R v2.6.268 in the R statistical environment v2.15.069, and data were normalized using RLE70. p-value was adjusted for multiple testing by controlling the false discovery rate (FDR) at ≤5%. A second method was applied to detect DEGs. For the normalization, raw read counts were divided by the total number of reads in each library and results were log-transformed to assess normality by Shapiro-Wilk test71 using STATISTICA 7.0 (StatSoft, Inc. USA). As normality was not always achieved and due to the limited number of biological replicates, the non-parametric Kolmogorov-Smirnov test72 was applied (p < 0.1) using STATISTICA 7.0 (StatSoft, Inc. USA). Partial Least Square Analysis (PLS) was carried out to reveal the most important variables (genes) and to provide a rank to all DEGs73. A total of 20 candidate genes up-regulated in the three genotypes were selected for subsequent analyses. Of them, 4 were detected by edgeR-RLE, 6 by the non-parametric test and Partial Least Square Analysis, and 10 by both methods simultaneously. Differential expression analyses and Partial Least Square Analysis were performed excluding genes with less than 15 reads in at least one genotype when combining stress and control libraries. In total 7,680 transcripts (20.4% of all sequenced transcripts) were excluded.
Identification of orthologous and paralogous genes
For each candidate gene, a genome wide analysis was performed to identify putative Musa paralogs, according to first clustering level in GreenPhyl DB74. Annotations were manually inspected and, when necessary, corrections were made based on mapping data of the RNA-Seq libraries and comparisons with similar genes annotated in Vitis vinifera and A. thaliana. Following an approach based on definition of multi-specific orthologous groups15, for each candidate the orthologous gene(s) was identified in A. thaliana. All Musa genes included in the same orthologous group were considered real paralogs. To identify significant correlations between expression levels of the candidate genes and their corresponding paralog(s), Spearman rank correlation analysis (p < 0.01) on the normalized read counts were performed in R package.
Annotation, GO enrichment and pathway analysis
For the DEGs, protein homologues were searched using NCBI Blast option from BLAST2GO18, which examined sequences against public non-redundant databases using BLASTp algorithms (e-value < 10−3). Gene ontology (GO) terms were assigned and clustered based on biological process, cellular component or molecular function. Mapping and annotation were performed using default parameters (e-value hit filter 10−6, annotation cutoff 55, GO weight 5, HSP-hit-coverage cut-off 0). Alternatively, GO terms for the selected candidate genes were retrieved from Uniprot75. Specific gene products were associated to biological pathways as determined by the KEGG pathway mapping function76 from BLAST2GO. For the candidate genes, associated GO terms obtained via Uniprot and/or BLAST2GO were imported into Cytoscape to generate the corresponding interaction network77. GO term enrichment analysis was conducted using the R package TopGO78 and significance was calculated based on Fisher’s exact test with a cut-off threshold of p < 0.01.
Design and optimization of RT-qPCR primers
For each candidate gene and the Musa reference genes ribosomal protein L2 (L2), Actin-1 (ACT-1), Tubulin β-1 chain (TUB1) and Elongation factor 1α (EF1), copy-specific primers were designed to amplify part of the 3′ or 5′ untranslated regions using Primer3 (version 0.4.0). Chosen parameters were: product size range 100–150 bp, primer size 20–22 bp, primer Tm 57–60 °C (with maximum Tm difference = 2 °C) and GC content 45–60%. Primer combinations were custom-ordered from a commercial supplier (Integrated DNA Technology, USA) and tested at two concentrations (100 and 150 nM) and with two cDNA dilutions (x3 and x48). Amplicon sizes were checked by 2% agarose gel electrophoresis and ethidium bromide staining. Primer specificities were confirmed with the melting-curve after amplification by RT-qPCR. A standard curve of five serial four-fold dilutions of pooled cDNA, a no template control, was made to calculate gene-specific amplification efficiencies (E) and correlation coefficients (R2) (Supplementary Table S6).
RT-qPCR analysis and determination of gene expression levels
For RT-qPCR validation of RNA-seq data, root tip RNA was isolated from the independent experiment. For each DNA-free RNA sample, 1 μg was reversed-transcribed to cDNA by using an oligo(dT)18 primer and the RevertAid H Minus First Strand cDNA Synthesis kit (Fermentas, St-Leon Rot, Germany) according to the manufacturer’s instructions. RT-qPCR design, calculations and statistics used followed the MIQE guidelines79. RT-qPCR was carried out in 96-well plates and in duplicated volumes of 15 μL using the SYBR Green I technology. All reactions were analysed with the StepOnePlusTM Real-Time PCR System (Applied Biosystems, USA). The master mix containing 1 × ABsoluteTM QPCR SYBR® Green Mix (Thermo Scientific, Epsom, UK), 100 or 150 nM of each forward and reverse primer (Supplementary Table S6) and 125 ng λ-DNA (Roche Diagnostics, Vilvoorde, Belgium) was mixed with 2 μL of a 50x diluted template cDNA, control gDNA or water. λ-DNA was added as carrier DNA to minimise absorption and Poisson effects. The following amplification program was used: polymerase activation at 95 °C for 15 min., 40 cycles of 95 °C for 15 s, 60–62 °C for 20 s and 72 °C for 20 s. A standard curve as the above mentioned and the cDNA samples were run concomitantly in each assay. Cq values were imported into qBase+ software (Biogazelle) and relative expression values were determined using the most stable reference genes (EF-1, L2 and ACT-1). Data were log-transformed to assess normality by Shapiro-Wilk test using STATISTICA 7.0 (StatSoft, Inc. USA). As normality was achieved, a two-way analysis of variance (ANOVA; p < 0.05) was applied to calculate the effects of the genotype, treatment and genotype × treatment using STATISTICA 7.0 (StatSoft, Inc. USA). Fisher’s least significant difference (LSD) mean comparison was used as the post-hoc test.
Additional Information
How to cite this article: Zorrilla-Fontanesi, Y. et al. Differential root transcriptomics in a polyploid non-model crop: the importance of respiration during osmotic stress. Sci. Rep. 6, 22583; doi: 10.1038/srep22583 (2016).
Supplementary Material
Acknowledgments
The authors would like to thank Edwige André and Saskia Windelinckx for the in vitro experiments, and Els Thiry for isolating total RNA for RNA sequencing. This work was supported by the Bioversity International project ‘Adding value to the ITC collection through molecular and phenotypic characterization’, financed by the Belgian Development Cooperation and the CGIAR Research Program on Roots, Tubers and Bananas (RTB).
Footnotes
Author Contributions M.R., R.S., N.R. and S.C. conceived and designed the experiments. Y.Z.-F., H.D., E.D. and S.N. performed the experiments. Y.Z.-F., M.R., A.C., E.K., H.D., E.D., S.N. and S.C. analysed the data. Y.Z.-F., M.R., A.C. and S.C. wrote the manuscript. R.S., N.R. and S.C. supervised the study. All authors reviewed and approved the final manuscript.
References
- Simmonds N. W. & Shepherd K. The taxonomy and origins of the cultivated bananas. J. Linn. Soc., Bot. 55, 302–312 (1955). [Google Scholar]
- Van Asten P. J. A., Fermont A. M. & Taulya G. Drought is a major yield loss factor for rainfed East African highland banana. Agric. Water Manage. 98, 541–552 (2011). [Google Scholar]
- de Lapeyre de Bellaire L., Fouré E., Abadie C. & Carlier J. Black Leaf Streak Disease is challenging the banana industry. Fruits 65, 327–342 (2010). [Google Scholar]
- Kissel E., van Asten P., Swennen R., Lorenzen L. & Carpentier S. C. Transpiration efficiency versus growth: exploring the banana biodiversity for drought tolerance. Sci. Hortic. 185, 175–182 (2015). [Google Scholar]
- Vanhove A.-C., Vermaelen W., Panis B., Swennen R. & Carpentier S. C. Screening the banana biodiversity for drought tolerance: can an in vitro growth model and proteomics be used as a tool to discover tolerant varieties and understand homeostasis. Front. Plant Sci. 2, 176 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J. et al. Isolation and analysis of water stress-induced genes in maize seedlings by subtractive PCR and cDNA macroarray. Plant Mol. Biol. 55, 807–823 (2004). [DOI] [PubMed] [Google Scholar]
- Davey M. W. et al. Heterologous oligonucleotide microarrays for transcriptomics in a non-model species; a proof-of-concept study of drought stress in Musa. BMC Genomics 10, 436 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. S. et al. Transcripts and microRNAs responding to salt stress in Musa acuminata Colla (AAA Group) cv. Berangan roots. PLoS One 10, e0127526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.-S. et al. Comparative transcriptomics analysis reveals difference of key gene expression between banana and plantain in response to cold stress. BMC Genomics 16, 446 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Z., Apse M. P. & Blumwald E. Engineering salinity and water-stress tolerance in crop plants: getting closer to the field. Adv. Bot. Res. 57, 405–443 (2011). [Google Scholar]
- Wang Z., Gerstein M. & Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. A., Ponnala L. & Weber C. A. Strategies for transcriptome analysis in nonmodel plants. Am. J. of Bot. 99, 267–276 (2012). [DOI] [PubMed] [Google Scholar]
- D’Hont A. et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488, 213–217 (2012). [DOI] [PubMed] [Google Scholar]
- Davey M. W. et al. A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter- and intra-specific Musa hybrids. BMC Genomics 14, 683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci A., Guignon V., Roux N. & Rouard M. Genomic analysis of NAC transcription factors in banana (Musa acuminata) and definition of NAC orthologous groups for monocots and dicots. Plant Mol. Biol. 85, 63–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Geen A. Soil water dynamics. Nature Educ. Knowledge 3(6), 12 (2012). [Google Scholar]
- Perrier X. et al. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proc. Natl. Acad. Sci. USA 108, 11311–11318 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S. et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L. et al. Transcriptomic analysis reveals the roles of microtubule-related genes and transcription factors in fruit length regulation in cucumber (Cucumis sativus L.). Sci. Rep. 5, 8031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Transcriptional response to petiole heat girdling in cassava. Sci. Rep. 5, 8414 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. & Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics 154, 459–473 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. De novo characterization of the banana root transcriptome and analysis of gene expression under Fusarium oxysporum f. sp. Cubense tropical race 4 infection. BMC Genomics 13, 650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. et al. Transcriptome profiling of resistant and susceptible Cavendish banana roots following inoculation with Fusarium oxysporum f. sp. cubense tropical race 4. BMC Genomics 13, 374 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewin V. An underground revolution. Nature 466, 552–553 (2010). [DOI] [PubMed] [Google Scholar]
- Setter T. L. Transport/harvest index: Photosynthate partitioning in stressed plants. In Stress response in plants: Adaption and acclimation mechanisms (eds. Alscher R. G. & Cumming J. R. ) 17–36 (New York: Wiley-Liss, 1990). [Google Scholar]
- Rizhsky L., Liang H. & Mittler R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 130, 1143–1151 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt U. Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75, 69–92 (2006). [DOI] [PubMed] [Google Scholar]
- Millar A. H., Whelan J., Soole K. L. & Day D. A. Organization and regulation of mitochondrial respiration in plants. Annu. Rev. Plant Biol. 62, 79–194 (2011). [DOI] [PubMed] [Google Scholar]
- Strogolova V., Furness A., Robb-McGrath M., Garlich J. & Stuart R. A. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol.Cell. Biol. 32, 1363–1373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Shulaev V. & Mittler R. Reactive oxygen signaling and abiotic stress. Physiol. Plant. 133, 481–489 (2008). [DOI] [PubMed] [Google Scholar]
- Ndimba B. K., Chivasa S., Simon W. J. & Slabas A. R. Identification of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 5, 4185–4196 (2005). [DOI] [PubMed] [Google Scholar]
- Drew M. C. Oxygen deficiency and root metabolism: injury and acclimation during hypoxia and anoxia. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 223–250 (1997). [DOI] [PubMed] [Google Scholar]
- Aguilar E. A., Turner D. W., Gibbs D. J., Armstrong W. & Sivasithamparam K. Oxygen distribution and movement, respiration and nutrient loading in banana roots (Musa spp. L.) subjected to aerated and oxygen-depleted environments. Plant Soil. 253, 91–102 (2003). [Google Scholar]
- Dennis E. S. et al. Molecular strategies for improving waterlogging tolerance in plants. J. Exp. Bot. 51, 89–97 (2000). [PubMed] [Google Scholar]
- Carpentier S. C. et al. Sugar-mediated acclimation: the importance of sucrose metabolism in meristems. J. Proteome Res. 9, 5038–5046 (2010). [DOI] [PubMed] [Google Scholar]
- Oh M. & Komatsu S. Characterization of proteins in soybean roots under flooding and drought stresses. J. Proteomics. 114, 161–181 (2015). [DOI] [PubMed] [Google Scholar]
- Roberts J. K., Callis J., Wemmer D., Walbot V. & Jardetzky O. Mechanisms of cytoplasmic pH regulation in hypoxic maize root tips and its role in survival under hypoxia. Proc. Natl. Acad. Sci. USA 81, 3379–3383 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok E. J. et al. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell. 14, 2481–2494 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E., Poggi A., Novi G., Alpi A. & Perata P. A genomewide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol. 137, 1130–1138 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolferus R., Jacobs M., Peacock W. J. & Dennis E. S. Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol. 105, 1075–1087 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmerer T. W. & Kozlowski T. T. Ethylene, ethane, acetaldehyde, and ethanol production by plants under stress. Plant Physiol. 69, 840–847 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabalza A. et al. Regulation of respiration and fermentation to control the plant internal oxygen concentration. Plant Physiol. 149, 1087–1098 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good A. G. & Muench D. G. Long-term anaerobic metabolism in root tissue (metabolic products of pyruvate metabolism). Plant Physiol. 101, 1163–1161 1168 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiani R., Nebuloni M., Mattana M. & Brambilla I. Anaerobic accumulation of amino acids in rice roots: role of the glutamine synthetase/glutamate synthase cycle. Amino Acids 18, 207–217 (2000). [DOI] [PubMed] [Google Scholar]
- Menegus F., Cattaruzza L., Chersi A. & Fronza G. Rice and wheat seedlings as plant models of high and low tolerance to anoxia. In “Surviving hypoxia: mechanisms of adaptation and control”. 53–64 (CRC Press: Boca Raton, FL, 1993).
- Kendziorek M., Paszkowski A. & Zagdańska B. Differential regulation of alanine aminotransferase homologues by abiotic stresses in wheat (Triticum aestivum L.) seedlings. Plant Cell Rep. 31, 1105–1117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa C. A. F. & Sodek L. Alanine metabolism and alanine aminotransferase activity in soybean (Glycine max) during hypoxia of the root system and subsequent return to normoxia. Environ. Exp. Bot. 50, 1–8 (2003). [Google Scholar]
- Igamberdiev A. U. & Hill R. D. Nitrate, NO and haemoglobin in plant adaptation to hypoxia: an alternative to classic fermentation pathways. J. Exp. Bot. 55, 2473–2482 (2004). [DOI] [PubMed] [Google Scholar]
- Stoimenova M., Igamberdiev A. U., Gupta K. J. & Hill R. D. Nitrite-driven anaerobic ATP synthesis in barley and rice root mitochondria. Planta 226, 465–474 (2007). [DOI] [PubMed] [Google Scholar]
- Liu F. et al. Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol. 137, 1115–1129 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E. R., Nie X. Z., MacGregor A. W. & Hill R. D. A cereal haemoglobin gene is expressed in seed and root tissues under anaerobic conditions. Plant Mol. Biol. 24, 853–862 (1994). [DOI] [PubMed] [Google Scholar]
- Nie X. & Hill R. D. Mitochondrial respiration and hemoglobin gene expression in barley aleurone tissue. Plant Physiol. 114, 835–840 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa A. W., Duff S. M. G., Guy P. A. & Hill R. D. Altering hemoglobin levels changes energy status in maize cells under hypoxia. Proc. Natl. Acad. Sci. USA 95, 10317–10321 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillaud F. & Blachier F. Mitochondria and sulfide: a very old story of poisoning, feeding, and signaling? Antioxid. Redox Signal. 15, 379–391 (2011). [DOI] [PubMed] [Google Scholar]
- Holdorf M. M. et al. Arabidopsis ETHE1 encodes a sulfur dioxygenase that is essential for embryo and endosperm development. Plant Physiol. 160, 226–236 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafiz A., Singh A. K., Pareek A., Sopory S. K. & Singla-Pareek S. L. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genomics. 11, 293–305 (2011). [DOI] [PubMed] [Google Scholar]
- Kaur C. et al. Expression of multiple stress inducible ETHE1-like protein from rice is higher in roots and is regulated by calcium. Physiol. Plant. 152, 1–16 (2014). [DOI] [PubMed] [Google Scholar]
- Krüßel L. et al. The mitochondrial sulfur dioxygenase ETHYLMALONIC ENCEPHALOPATHY PROTEIN1 is required for amino acid catabolism during carbohydrate starvation and embryo development in Arabidopsis. Plant Physiol. 165, 92–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina C. et al. SuperSAGE: the drought stress-responsive transcriptome of chickpea roots. BMC Genomics. 9, 10.1186/1471-2164-9-553 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez S. M. et al. O-glycosylated cell wall proteins are essential in root hair growth. Science 332, 1401–1403 (2011). [DOI] [PubMed] [Google Scholar]
- Ranjan A. et al. Comparative transcriptomic analysis of roots of contrasting Gossypium herbaceum genotypes revealing adaptation to drought. BMC Genomics 13, 680 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson G. A. et al. Primary structures of the precursor and mature forms of stearoyl-acyl carrier protein desaturase from safflower embryos and requirement of ferredoxin for enzyme activity. Proc. Natl. Acad. Sci. USA 88, 2578–2582 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkenberg J. et al. Two fatty acid desaturases, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 and FATTY ACID DESATURASE3, are involved in drought and hypoxia stress signaling in Arabidopsis crown galls. Plant Physiol. 164, 570–583 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swennen R., De Langhe E., Janssen J. & Decoene D. Study of the root development of some Musa cultivars in hydroponics. Fruits 41, 515–524 (1986). [Google Scholar]
- Podevin N., Krauss A., Henry I., Swennen R. & Remy S. Selection and validation of reference genes for quantitative RT-PCR expression studies of the non-model crop Musa. Mol. Breed. 30, 1237–1252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droc G. et al. The banana genome hub. Database 2013, 1–14, 10.1093/database/bat035 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core Team. R. R: A language and environment for statistical computing. R Foundation for Statistical Computing, 2013. http://www.R-project.org. [Google Scholar]
- Anders S. & Huber W. Differential expression analysis for sequence count data. Genome Biol. 11, 1–12 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro S. S. & Wilk M. B. An analysis of variance test for normality (complete samples). Biometrika 52, 591–611 (1965). [Google Scholar]
- Massey F. J. The Kolmogorov-Smirnov test for goodness of fit. JASA. 46, 68–78 (1951). [Google Scholar]
- Boulesteix A. L. & Strimmer, K. Partial least squares: a versatile tool for the analysis of high-dimensional genomic data. Brief. Bioinform. 8, 32–44 (2007). [DOI] [PubMed] [Google Scholar]
- Rouard M. et al. GreenPhyl DB v2.0: comparative and functional genomics in plants. Nucleic Acids Res. 39, 1095–1102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. & Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A., Rahnenfuhrer J. & Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607 (2006). [DOI] [PubMed] [Google Scholar]
- Bustin S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemist. 55, 611–622 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.