Abstract
Objective. To develop, implement, and evaluate “Test2Learn” a program to enhance pharmacogenomics education through the use of personal genomic testing (PGT) and real genetic data.
Design. One hundred twenty-two second-year doctor of pharmacy (PharmD) students in a required course were offered PGT as part of a larger program approach to teach pharmacogenomics within a robust ethical framework. The program added novel learning objectives, lecture materials, analysis tools, and exercises using individual-level and population-level genetic data. Outcomes were assessed with objective measures and pre/post survey instruments.
Assessment. One hundred students (82%) underwent PGT. Knowledge significantly improved on multiple assessments. Genotyped students reported a greater increase in confidence in understanding test results by the end of the course. Similarly, undergoing PGT improved student’s self-perceived ability to empathize with patients compared to those not genotyped. Most students (71%) reported feeling PGT was an important part of the course, and 60% reported they had a better understanding of pharmacogenomics specifically because of the opportunity.
Conclusion. Implementation of PGT in the core pharmacy curriculum was feasible, well-received, and enhanced student learning of pharmacogenomics.
Keywords: pharmacogenomics, curriculum, active learning, personal genomic testing, genetics
INTRODUCTION
Pharmacogenomics, the study of how genetic variation impacts drug response, has been implemented in clinical practice based on the premise that it improves medication outcomes.1 As medication experts, pharmacists are well-positioned to ensure genetic data are used safely and effectively to tailor medication use to achieve “precision medicine,” defined by the National Institutes of Health (NIH) as: “tailoring of medical treatment to the individual characteristics of each patient.”2,3 Professional position statements advocate for pharmacists to play a leadership role in pharmacogenomics-based patient care.2,3 However, the majority of pharmacists are not confident with pharmacogenomics data.2,4-7
Because the 2016 Accreditation Council for Pharmacy Education (ACPE) Standards include pharmacogenomics, it is the responsibility of pharmacy schools to prepare pharmacists entering clinical practice to effectively use genetic data in the delivery of precision medicine.8 Furthermore, the American Association of Colleges of Pharmacy (AACP),4 the NIH-funded Genetics/Genomics Competency Center (G2C2),9 and the National Coalition for Health Professional Education in Genetics (NCHPEG)10 have created genetics competencies applicable to pharmacists. However, the state of pharmacogenomics instruction at most pharmacy schools was reported as “poor” or “not at all adequate” in a 2010 survey.11 New methods of instruction may be needed to drive better learning and retention.
A participatory education model in which students undergo personal genomic testing (PGT) enhanced classroom learning in limited trials but remains controversial.12-15 To explore pharmacy student interest at our institution, we polled over 200 students at the University of Pittsburgh School of Pharmacy in 2013 and found the majority (77%) were interested in undergoing PGT to help learn pharmacogenomics. We therefore sought to scale this innovative educational approach to the core doctor of pharmacy (PharmD) curriculum to attain its educational benefits within an appropriate ethical framework. In this report, we describe our development of “Test2Learn” (www.test2learn.org, trademark pending), an educational program focused on using PGT as a pedagogical tool, its implementation in a required course in the second-year of the PharmD curriculum at the University of Pittsburgh, and outcomes derived from objective measures of learning and student surveys. We hypothesized that the integration of PGT would engage students and enable them to achieve high-level pharmacogenomics competencies through active-learning experiences.
DESIGN
At the University of Pittsburgh, PharmD students are introduced to pharmacogenomics in the second year of the curriculum in the course Drug Development II. This required course teaches pharmacokinetics and pharmacodynamics concepts, which are critical to student understanding of the scientific basis of variable patient drug response. It bridges the foundational content in Biochemistry, Principles of Drug Action (including pharmacology), and Drug Development I (clinical trials and the diversity of patient populations) courses taught in year one with therapeutics courses in the third year. Drug Development II is team-taught by 10 clinical and research faculty members and takes place in the school’s new state-of-the-art classroom designed to facilitate group activities using computer/mobile technology.
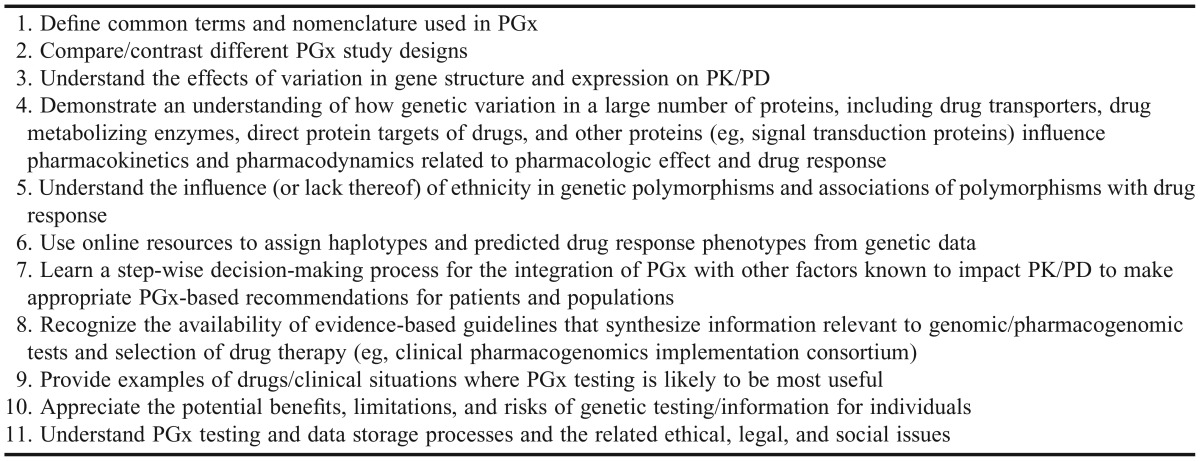
In summer 2014, course faculty members were offered PGT through the direct-to-consumer genetic testing company, 23andMe (Mountain View, CA). The objective of this “teach the teacher” approach was to expose faculty members to PGT as a pedagogical tool and to demonstrate how to incorporate pharmacogenomics and genetic data into their lessons, if desired. Drug Development II course objectives (Table 1) were designed through a formal process of review and integration of pharmacogenomics competencies and learning outcomes from the following sources: AACP,4 NCHPEG,10 the G2C2 Pharmacist Competencies,9 ACPE 2016 accreditation standards,8 a recent American College of Clinical Pharmacy commentary,16 and the existing University of Pittsburgh PharmD program curricular and course outcomes.
Table 1.
Pharmacogenomics-focused Drug Development II Learning Objectives
With these defined instructional goals, we developed a strategy to enrich course content using PGT data as a core component. The structure of Drug Development II, with initial lectures focusing on the science, followed by lectures involving application, was not changed. Rather, it was supplemented with the Test2Learn program: (1) new ethics content provided by faculty members from the Department of Human Genetics; (2) recorded videos used as an out-of-class genetics review; (3) revised lectures and practica; (4) a guest lecturer from 23andMe; and (5) new exercises leveraging the students’ experience with PGT and available genetic data. The intent of the revisions was to teach an effective process for pharmacogenomics-based clinical decision making rather than only specific drug-gene variant-phenotype associations.
In didactic lectures, students were taught gene variant nomenclature; pharmacogenomics information retrieval from national databases, such as dbSNP and the Pharmacogenomics Knowledgebase (PharmGKB);17 and genetic testing procedures, which were later applied in hands-on practica sessions. Information retrieval exercises asked students to answer questions about specific genes, variants, or phenotypes not previously covered in the course to test students’ ability to find accurate information. Students also identified how variants were assembled to determine haplotypes, assigned “patient” diplotypes, and predicted likely phenotypes in case-based activities. Students gained experience using PharmGKB pharmacogenomics translation tables and Clinical Pharmacogenomics Implementation Consortium (CPIC) guidelines routinely employed in research and in clinical practice.
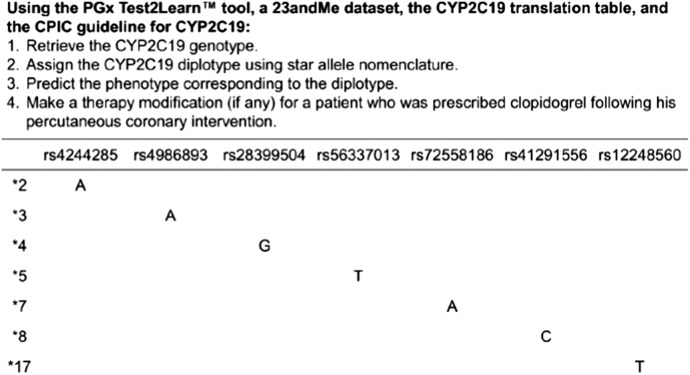
In one example exercise, students interpreted their own data as the basis for recommending appropriate anti-platelet therapy following percutaneous coronary intervention (Figure 1). Following the lecture teaching the pharmacogenomics of cardiovascular drugs (eg, warfarin and CYP2C9/VKORC1; simvastatin and SLCO1B1; clopidogrel and CYP2C19), the genetic data were also used in patient cases to teach students to integrate pharmacogenomics data with other clinical information (eg, laboratory data, medication history, drug interactions) for more advanced decision making. Students further learned to communicate pharmacogenomics information through small group activities during which students played the role of a pharmacist, patient, or another health care practitioner. The “pharmacist” conveyed recommendations based on the pharmacogenomics-based decision-making process, and performance was evaluated individually and through group assignments and reports to the entire class.
Figure 1.
Excerpt from the exercise in which students used custom software, individual genetic data (their own or an external dataset), simplified translation table adapted from PharmGKB,17 and CPIC guidelines to identify their diplotype and phenotype to make a clinical decision.
To help convey the concepts of genotyping and phenotyping, a phenotyping experiment was performed to focus students’ thinking on variability in populations and whether it could be predicted. The ability to taste bitterness of phenylthiocarbamide (PTC) impregnated paper strips (Frey Scientific, Nashua, NH) is a well-established experiment used to demonstrate simple genetic concepts.18 On the first day of class, every student was given a strip of PTC paper and asked to report anonymously via an audience response system if they could detect the distinct bitter taste. After students donated their raw data to the class pool, the distribution of “tasters” and “nontasters” was predicted from the genotype at rs713598 in the TAS2R38 gene for each student in the population dataset. To conclude this activity, distributions of phenotype tested tasters and genotype predicted tasters were presented to students.
The Test2Learn program underwent ethical, legal, and administrative reviews prior to launch. The project was initially evaluated at the university level by representatives of the Provost’s Office and received strong support by the School of Pharmacy Curriculum Committee, student representatives, and school faculty members in spring 2014. Since the planned surveys and genetic data used in class were anonymous, the program and its use of PGT did not meet the definition of research, according to the University of Pittsburgh Institutional Review Board (IRB). A bioethicist external to the school was also consulted to provide an additional independent review, and revisions were made to the project plan.
Personal genomic testing was explicitly presented to students as an optional exercise, and this understanding was documented through a written acknowledgement. To mitigate pressure to participate or coercion, decisions were confidential and blinding procedures were incorporated so that faculty members did not know who underwent testing. All students attended a mandatory ethics presentation delivered by a qualified bioethicist that discussed the risks of genetic testing.19 These lectures, supplemented by optional time set aside for faculty consultation, allowed students to make an informed decision over a 4-week time period regarding whether or not to undergo PGT. A genetic counselor was also made available as an additional safety net. To provide students opting out of PGT equal opportunity to work with real genetic data, anonymous datasets were downloaded from the Harvard Personal Genome Project (HPGP).20 This project allows individuals who wish to provide researchers their PGT data, such as 23andMe raw datasets, to upload their data to a publicly accessible repository. Each student was given a USB drive on which to store data for use in class exercises to maintain blinding.
Saliva-based genotyping kits were obtained from the direct-to-consumer genetic testing company, 23andMe, at a cost of approximately $80 per test. The company uses a custom genotyping array based on the HumanOmniExpress-24 panel (Illumina, San Diego, CA) that queries over 600 000 single nucleotide polymorphisms (SNPs) in addition to about 30 000 custom SNPs selected by 23andMe. The kits were ordered in bulk and distributed to students by a staff member unaffiliated with the program who tracked student names with the kit number received until they were successfully processed. Genotyped students signed 23andMe’s standard web-based consent to undergo PGT and created personal accounts on their website in order to receive ancestry reports and their raw genetic data directly. All costs were covered through grant support.
To complete a class population analysis, students were asked to submit their PGT data, but were under no obligation to do so. Students watched a short video tutorial on how to properly de-identify their personal data. To donate, students exchanged their USB drive containing their de-identified PGT data file for a new USB drive using designated bins, Genetic files were then extracted from the USB drives and added to a single database. Population data were compiled using a Python script and PLINK, v.1.07 (Shaun Purcell, Boston, MA).21 Population level diplotype frequencies for several pharmacogenes (CYP2C9, CYP2C19, SLCO1B1,VKORC1) were determined to show the prevalence of clinically relevant variation among the class. These data were also integrated into the cases and used to stimulate student thinking regarding the potential value of pre-emptive genotyping. Students were asked to identify drug-gene pairs to target based on the variant frequency data and integrate additional factors such a guidelines, medication use, test availability, and clinical outcomes into theoretical policy decisions.
We developed custom software, the Personal Genome Browser tool, for students to use in class to parse their PGT data for pharmacogenomics-related SNPs and genes. With it, students were able to efficiently access sequence-level genetic data for individual genes by Reference SNP Identification (RSID) number or to receive a list of all variant calls for a selected gene symbol to assign diplotypes and obtain interpretations. The tool was written using Python, v3.4.1 (Python Software Foundation, Beaverton, OR), custom libraries, and the Ensembl REST application program interface (European Bioinformatics Institute, Hinxton, UK)22 to pull gene IDs from RSID lookup. Importantly, the libraries could be modified to control what genes the tool could query, and the specific features, such as interpretations, could be remotely locked and/or unlocked.
Knowledge and attitudinal data were collected with anonymous pre/postsurveys completed by students on the first and last days of class, respectively. Surveys were adapted from previous publications and covered the following domains: knowledge of pharmacogenomics, genetics, and PGT; attitudes and beliefs regarding pharmacogenomics and PGT; and expectations of the Test2Learn program for achieving learning objectives.5,13,15,23,24 The postsurvey also asked students about whether or not they underwent PGT as part of the class, which allowed for comparisons based on the act of undergoing PGT vs using external anonymous data. The surveys were linked using an anonymous code generated by respondents for pre/post comparisons. Data were entered in a Microsoft Access database in duplicate and compared to ensure data integrity. To compare student performance relative to peers in the same course in prior years, a standardized quiz that assessed general knowledge of genetics and pharmacogenomics was administered as it has been every year since 2011.
For knowledge questions, students were scored based on the percent answered correctly and matched pre/postsurveys were compared using a paired t test. Likert scale questions (5=strongly agree, 4=agree, 3=neutral, 2=disagree, 1=strongly disagree) were compared using the Wilcoxon signed-rank test for paired data analysis and the Mann-Whitney U test for unpaired comparisons. Mean and standard deviation were provided for easy interpretation. Dichotomous responses (ie, yes/no) and proportions were evaluated using the chi-square test for unpaired comparisons or McNemar’s test for paired comparisons. Statistical analyses were performed using SPSS, v22 (IBM, New York, NY) and R, v3.1.2 (R Development Core Team, Vienna, Austria).
EVALUATION AND ASSESSMENT
Of the 122 students and 10 faculty members to whom genotyping was offered, 100 (82%) and 10 (100%), respectively, elected to undergo PGT. Turnaround time for results was approximately 3 weeks, and in only 2 instances did samples need to be recollected (one kit was lost in the mail, and one kit had insufficient DNA yield). No students had undergone PGT previously. Genotyped students cited “ancestry information,” “general curiosity,” and “it was a free test” for primary reasons driving their decision to undergo PGT. Those who chose not to be tested indicated “no interest,” “privacy concerns,” and “risk for incidental findings” as primary reasons. Of the students who elected to undergo testing and to complete the survey, 92 (100%) remained pleased with their decision, vs 11 (61.1%) who did not undergo PGT (p<0.001). Among those genotyped, 30 (32.6%) reported that they made further efforts outside of class to interpret their data.
Together, the planned redesign by the program team and the content revisions driven by individual faculty members within the “teach the teacher” model resulted in an additional 6 hours of pharmacogenomics or ethics content, as well as the modification of 4 hours of existing lectures/practica to achieve the new learning objectives. Time for the new material was made by eliminating some review content in the course or making it an out-of-class activity, replacing the existing pharmacogenomics material, and integrating new content with related areas of instruction (eg, pharmacokinetics of cardiovascular drugs).
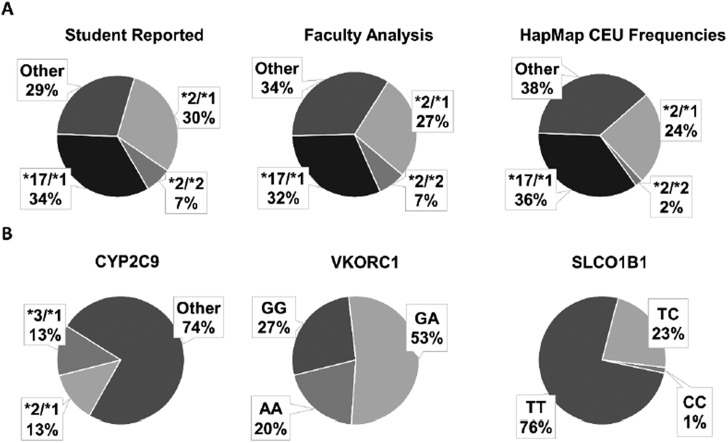
Students performed well on in-class exercises, and objective assessments demonstrated student understanding of pharmacogenomic concepts. For the PTC exercise, the proportion of students genotype-predicted to be a “taster” (74% of the 70 students carried at least one G allele at rs713598 in the TAS2R38 gene) closely resembled the self-reported phenotypic taster statuses (67% of the 117 students in the class reported they could taste bitterness). Figure 2A shows that the CYP2C19 diplotype assignments made by students were similar to results from faculty evaluators. The percentage of students with an “actionable” genotype (at leased one *2 allele) was 37% according to the students and 34% per the faculty analysis of student data. These data are consistent with diplotype frequencies from the HapMap population of Utah residents with ancestry from northern and western Europe (CEU).17,25 Similar exercises were completed for CYP2C9, VKORC1, and SLCO1B1 (Figure 2B). Overall, 86% of students had an actionable genotype in at least one of these 4 genes according to current CPIC guidelines. Evidence of each student’s ability to predict phenotype and to make recommendations in cases involving well-known drug-gene pairs was excellent with 74%, 80%, and 93% of students successfully completing case-based exercises involving CYP2C19-clopidogrel, VKORC1-warfarin, and SLCO1B1-simvastatin, respectively.
Figure 2.
A and B. CYP2C19 diplotype frequencies reported from individual data. (A) The concordance between student-faculty diplotype assignments and population similarity to an external established frequencies in Caucasian population as reported by HapMap-CEU phase 325 suggests that students were overall successful in completing the in-class exercise. (B) Class population-based frequencies of the pharmacogenes CYP2C9, VKORC1 rs9923231, and SLCO1B1 rs4149056 from students who submitted data (n=70). Findings showed that among these common genes, 86% of students had at least one actionable genotype.
The entire class (n=122) completed the presurvey and 90% (n=110) completed the postsurvey. Of the completed postsurveys, 89% (n=98) were successfully linked with the student’s presurvey allowing for robust pre/post comparisons. Reasons for unmatched surveys included students not filling out the linkage code or providing a linkage code that was illegible.
Student attitudes and beliefs regarding pharmacogenomics, PGT, and the role of a pharmacist, were evaluated. At the end of the course, more students said that they would recommend pharmacogenomic testing to a patient (58% pre vs 80% post, p<0.001), but more disagreed or strongly disagreed with the statement that patients would be able to accurately interpret genetic data on their own (73.5% pre vs 90.8% post; p<0.001). Positive opinions regarding the future use of pharmacogenomic testing and pharmacists’ roles did not change (Table 2). At baseline, respondents believed that testing would be routinely used to guide drug therapy (90.8% agree or strongly agree), that an ability to apply pharmacogenomics concepts on the topic would be important to their future career (89.3% agree or strongly agree), and that all pharmacists should have this knowledge (97% agree or strongly agree). A significant shift toward agreement that a pharmacist’s role should include counseling on genetic data was observed (66.3% pre vs 74.5% post; p=0.048).
Table 2.
Student Attitudes and Beliefs About Personal Genomic Testing and Pharmacogenomics (n=98)
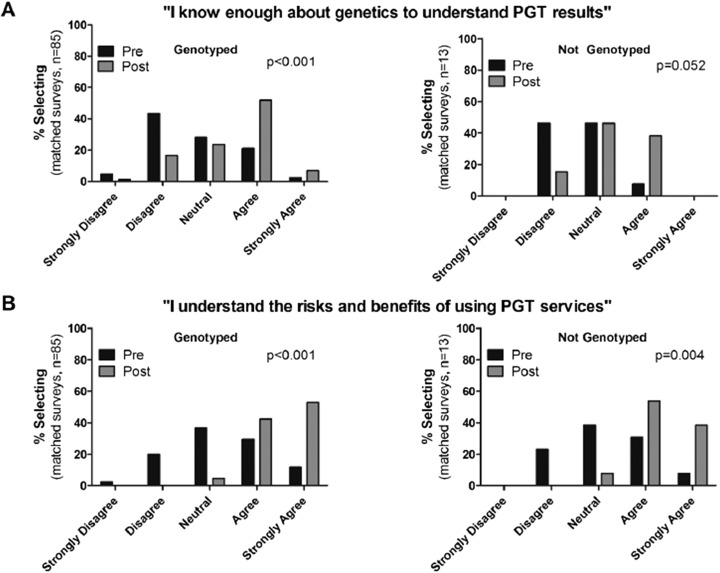
Genetics and pharmacogenomics knowledge as assessed by objective questions on the survey were significantly improved by the end of the course [82.9% (14.1) vs 90.5% (9.0)] correct on the presurvey vs postsurvey, respectively; p<0.001). Similarly, Table 3 shows improvement in quiz scores compared to previous years when students’ pharmacogenomics education was limited to traditional lecture material. Figure 3 shows student perceptions of their pharmacogenomics knowledge stratified by whether they personally underwent PGT. Among students who were genotyped, there was a significant shift in agreement that they knew enough about genetics to understand PGT results by the end of the course [2.7 (0.9) pre vs 3.5 (0.9) post; p<0.001]. The group that did not undergo PGT showed a similar trend, but it was not significant [2.6 (0.7) pre vs 3.2 (0.7) post; p=0.05]. Self-reported understanding of the risks and benefits of PGT services was more pronounced in students who underwent PGT, but significantly improved in both groups of students as a result of the program [genotyped students: 3.3 (1.0) pre vs 4.5 (0.6) post; p<0.001; nongenotyped students, 3.2 (0.9) pre vs 4.3 (0.6) post; p=0.004].
Table 3.
Excerpt From Quiz Evaluating General Knowledge of Genetics and Pharmacogenomics
Figure 3.
A and B. Student perceptions of their pharmacogenomics knowledge stratified by whether they underwent personal genomic testing (PGT). (A): Student-reported agreement with the statement that they know enough about genetics to understand PGT results stratified by whether students’ underwent PGT. Genotyped students showed a significant shift toward agreement with the statement on the postsurvey vs presurvey, while students who did not undergo genotyping showed only a trend toward shift in agreement on the postsurvey (B): Student reported agreement with the statement that they understand the risks and benefits of using PGT services in pre/postsurveys, stratified by the students' decision whether to undergo genotyping or not. Both groups of students showed a significant shift toward agreement with the statement in the postsurvey vs the presurvey.
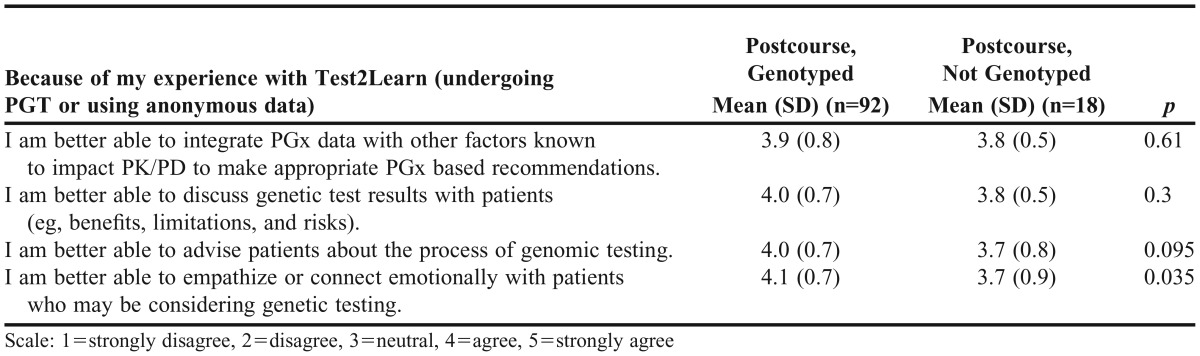
We also surveyed students regarding their self-efficacy at the end of the course (Table 4). Independent of whether they underwent PGT, students agreed they were able to integrate genetic data into practice, advise patients on genetic data, and discuss genetic test results. Student self-perceived ability to empathize with patients undergoing genetic testing was significantly higher in students who underwent the PGT process vs those who opted out (p=0.04).
Table 4.
Comparison of Student Perceived Ability to Work With and Integrate Personal Genomic Testing (PGT) into Patient Care (Postsurvey)
Finally, student opinions regarding the importance of program components and the success of its implementation were captured. Students thought observing the genetic diversity of their own class through the population-based exercises was an important part of the course (58% agreed and 23% strongly agreed). Similarly, 71% of the students who underwent genotyping felt that it was an important part of the course, and 60% felt that they had a better understanding of pharmacogenomics than those who did not undergo genetic testing. Finally, 81% of students felt that the pharmacogenomics material was well integrated and connected between lectures and practica. Most students found the ethics training to be valuable and, although no students sought genetics counseling, 84% agreed or agreed strongly that having such a professional available is an important part of a program that integrates PGT.
DISCUSSION
In this report we describe the development of Test2Learn, a PGT-based program designed to enhance pharmacogenomics education, and its successful implementation into a required course in the Doctor of Pharmacy curriculum at the University of Pittsburgh. We created an ethical framework for genetic testing of students, novel learning objectives, lecture materials, analysis tools, and exercises using individual- and population-level genetic data to achieve the high-level pharmacogenomics competencies. Integration into a required course in the core curriculum ensured that all students received specialized education in pharmacogenomics to help meet 2016 ACPE standards.8
At baseline, 90% of the second-year PharmD students we surveyed agreed or strongly agreed that PGT would be routinely used to guide drug therapy in the future, 88% agreed or strongly agreed that being able to apply pharmacogenomics concepts would be important in their future career as a pharmacist, and 97% agreed or strongly agreed that all pharmacists should have this knowledge. These beliefs did not significantly change at the end of the course. Our findings confirm forward-thinking attitudinal data regarding pharmacogenomics from other surveys of pharmacy students. Approximately 95% of pharmacy students at the University of Minnesota agreed or strongly agreed that “pharmacogenomics will be relevant in future medical practice” and 88.1% agreed or strongly agreed that “pharmacists should be required to have some comprehension of pharmacogenomics.”26 Similarly, 80% of those surveyed by Lee et al believed that pharmacists should be involved in educating patients and health care professionals.27
Several different educational models have been used to teach pharmacogenomics. Primer courses,28 introductory genetic testing experiences,14 elective courses involving small numbers of students,29 and shared curricula27 are used in PharmD programs. However, the topic is complex, and lessons from a continuing education program where only marginal improvements in knowledge retention were achieved suggest it requires a comprehensive educational effort to be successful.30 A participatory educational model involving PGT provided students with the opportunity to experience genetic testing and work with real data first-hand, but its educational value is debated.12,24 Stanford University was among the first to successfully implement comprehensive PGT in an elective genomics course through safeguards to address the complex ethical, legal, and social issues associated with students working with their own genomes.12 Salari and colleagues reported genotyped students gained greater improvements in pre/post course knowledge and believed that PGT was an important part of their learning.13 Positive experiences, albeit on a smaller scale, have been reported in pharmacy schools. Krynetskiy and Calligro added a laboratory exercise in a Temple University pharmaceutics class in which students determined their own genotype for a single drug metabolism gene. The majority of students strongly agreed that PGT “helped them to understand why pharmacogenomics is so important.”14 Knoell and colleagues also completed a genotyping exercise with 10 volunteers from a class of 115 in a clinical pharmacogenomics course at The Ohio State University College of Pharmacy. In their experience, 85% of the 115 students in the course agreed or agreed strongly that the PGT was beneficial in helping them connect to course content. Student feedback recommended that future genotyping exercises be expanded to all students.15
Through our program, students performed well on information retrieval and genotype interpretation assignments that were designed to meet learning objectives. Genetics and pharmacogenomics knowledge improved by the end of the course and in comparison to previous years. Student self-efficacy for tasks necessary to integrate pharmacogenomics in clinical practice was rated highly and their understanding of the risks and benefits of PGT significantly improved at the end of the course. Students were highly engaged as demonstrated by the 82% PGT participation rate. The overall program significantly increased student confidence (whether students thought they could understand test results) and, not surprisingly, their self-perceived ability to empathize with future patients who may be tested vs students who were not genotyped. Overall, results are consistent with, and extend, outcome data involving PGT in the medical school elective course at Stanford and early testing experiences in pharmacy curricula.13-15 Our pharmacy students specified PGT was an important part of the course, and those who were genotyped thought they had a better understanding of pharmacogenomics concepts because of this opportunity.
Aggregating student data and conducting population analyses was a useful exercise for several reasons. First, the PTC activity provided a simple and accessible example of phenotype prediction from genetic data. Second, the high frequency of actionable diplotypes (86%) showed potential clinical relevance. Consistent with findings from Knoell et al, students thought that identifying the heterogeneity in classmates was a powerful finding to help them appreciate population diversity.15
The risks to implementing PGT in an educational setting involve privacy and confidentiality, coercion vs a right to know (or not know), maintaining equal learning opportunities, psychosocial issues, and incidental findings.12,19 We believe that the Test2Learn implementation model created a clear path forward to mitigate these risks. Problems associated with privacy, confidentiality, and coercion in a classroom setting were mitigated through the use of blinding, USB drives, the Personal Genome Browser Tool, careful design so students understood the activity was optional, and ensuring that no identifiable data was collected. In particular, the Personal Genome Browser Tool provided a needed blinding mechanism, which using the 23andMe website didn’t provide, and a way to deliver alternative nonpersonal datasets to students not genotyped. In addition, since the program was implemented in a core curriculum, the large number of students who participated made population exercises feasible while still maintaining blinding and procedures designed to eliminate any pressure to participate.
Because 23andMe no longer provides health reports, the incidental finding risk is significantly lower. Importantly, the Food and Drug Administration did not restrict 23andMe or any other company from returning raw data as it is not trivial for nonexperts to get from sequence to a disease risk prediction (for many genes/diseases this is a challenge even for experts). However, to remain conservative, the Personal Genome Browser Tool gene lookup list was also limited to genes relevant to course activities, and course faculty members were careful to not direct students to third-party interpretative services or example genetic test reports that students might try to use to extend their data beyond the course’s pharmacogenomics focus. The postsurvey data indicated that only a small subset of students sought such interpretive services on their own, and no students used the genetics counselor we had made available. However, 84% of students agreed or strongly agreed that having a professional available is an important component of a program that integrates PGT. This apparent contradiction between perceived need and lack of use of counseling services is consistent with other educational trials of PGT.13,31 We believe the program’s focus on ethics training (expert lecturer, active discussions, the 4-week PGT decision-making period, and genetics counselor availability) was important in supporting students regarding potential psychosocial issues. Other investigators further advocate for student access to a multidisciplinary approach to consultative services (not just genetic counselors).31
The use of commercial genetic testing eliminated the need for faculty members to manage student personal genetic data and to create a local informed consent process. It made blinding easier, and, in general, is more realistically implemented in institutions where onsite genetic testing is not available, or there are concerns with storing student genetic data. Barriers to entry included test costs and availability of faculty members with the knowledge to create material based around PGT. We did not assess willingness of students to pay for testing, but these costs are comparable to or less than some required textbooks. Faculty training programs such as the Pharmacogenomics Education Program (PharmGenEd) show a shared curriculum and the “train-the-trainer” approach to be effective in educating faculty members who have little to no experience using or teaching pharmacogenomics.27,32 We similarly found that introducing PGT via a teach-the-teacher approach effectively engaged faculty members and helped them develop course material that allowed the students to use PGT.
The implementation approach and its evaluation had several limitations. The use of nonpersonal datasets alongside PGT provided an opportunity to measure the impact of PGT directly, but, since the design was nonrandomized, results were subject to a selection bias. It was also challenging to distinguish the act of being tested from being in the program that discussed being tested and using genetic data. Additionally, the survey was not validated, so it is possible that some results may not be generalizable. With the exception of survey questions designed to stratify those who underwent PGT, improvements in student knowledge and perceptions were assumed to be driven by the program as a whole. We did not have a sufficient number of students opting to use anonymous genetic data to provide a robust comparison of all measures. Finally, all outcomes were short-term. Future analyses will evaluate student learning and retention longitudinally. In spring 2015, we expanded the Test2Learn program into our second cohort of students in the first-year class, Drug Development I. Our vision is that pharmacogenomics education and use of PGT as a pedagogical tool should not be in a single course with a single focus, but rather be a curricular thread that extends through the entire PharmD program.
SUMMARY
The Test2Learn program was designed to enhance pharmacogenomics education in the core PharmD curriculum. This was accomplished through careful integration of PGT within an ethical framework, development of new educational materials, and collection of population level genetic data. Objective assessments and survey data show its implementation was highly successful and engaged students in achieving established pharmacogenomics competencies. Students who underwent genetic testing improved significantly in several areas when compared to students who used nonpersonal data. The PGT implementation was well-received, and we believe it is both feasible and transferable to other educational settings based on the testing availability, risks, costs, and outcomes produced vs course time requirements.
ACKNOWLEDGMENTS
The authors would like to thank the University of Pittsburgh Provost’s Office and the Advisory Council on Instructional Excellence and the National Institutes of Health (KL2 TR000146) for grant support. We would also like to thank Sherri Peterson for organizing student kit distribution and collection and Dr. Esther Kim at 23andMe for guest lecturing in the course and working closely with us to accomplish testing efficiently within a tight academic schedule. This work was presented at the 2015 AACP Annual meeting in Washington, DC as one of the winners of the 2015 AACP Innovations in Teaching Competition.
REFERENCES
- 1.Dunnenberger HM, Crews KR, Hoffman JM, et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol. 2015;55:89–106. doi: 10.1146/annurev-pharmtox-010814-124835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ASHP statement on the pharmacist's role in clinical pharmacogenomics. Am J Health-Syst Pharm. 2015;72(7):579–581. doi: 10.2146/sp150003. [DOI] [PubMed] [Google Scholar]
- 3. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, DC: National Academies Press; 2011. [PubMed]
- 4.Johnson JA, Bootman JL, Evans ME, et al. Pharmacogenomics: a scientific revolution in pharmaceutical sciences and pharmacy practice. Report of the 2001-2002 Academic Affairs Committee. Am J Pharm. Educ. 2002;66(4) supplement. [Google Scholar]
- 5.Tuteja S, Haynes K, Zayac C, Sprague JE, Bernhardt B, Pyeritz R. Community pharmacists' attitudes towards clinical utility and ethical implications of pharmacogenetic testing. Per Med. 2013;10(8) doi: 10.2217/pme.13.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roederer MW, Van Riper M, Valgus J, Knafl G, McLeod H. Knowledge, attitudes and education of pharmacists regarding pharmacogenetic testing. Personalized Medicine. 2011;9(1):19–27. doi: 10.2217/pme.11.87. [DOI] [PubMed] [Google Scholar]
- 7.de Denus S, Letarte N, Hurlimann T, et al. An evaluation of pharmacists' expectations towards pharmacogenomics. Pharmacogenomics. 2013;14(2):165–175. doi: 10.2217/pgs.12.197. [DOI] [PubMed] [Google Scholar]
- 8. Accreditation Standards and Key Elements for the Professional Program in Pharmacy Leading to the Doctor of Pharmacy Degree. Accreditation Council for Pharmacy Education. Standards 2016.
- 9.Calzone KA, Jerome-D'Emilia B, Jenkins J, et al. Establishment of the genetic/genomic competency center for education. J Nurs Scholarsh. 2011;43(4):351–358. doi: 10.1111/j.1547-5069.2011.01412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. National Coalition for Health Professional Education in Genetics. Core competencies in genetics essential for all health-care professionals. Lutherville, MD; 2007.
- 11.Murphy JE, Green JS, Adams LA, Squire RB, Kuo GM, McKay A. Pharmacogenomics in the curricula of colleges and schools of pharmacy in the United States. Am J Pharm Educ. 2010;74(1):Article 7. doi: 10.5688/aj740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salari K, Pizzo PA, Prober CG. Commentary: to genotype or not to genotype? Addressing the debate through the development of a genomics and personalized medicine curriculum. Acad Med. 2011;86(8):925–927. doi: 10.1097/ACM.0b013e3182223acf. [DOI] [PubMed] [Google Scholar]
- 13.Salari K, Karczewski KJ, Hudgins L, Ormond KE. Evidence that personal genome testing enhances student learning in a course on genomics and personalized medicine. PLoS One. 2013;8(7):e68853. doi: 10.1371/journal.pone.0068853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krynetskiy E, Lee Calligaro I. Introducing pharmacy students to pharmacogenomic analysis. Am J Pharm Educ. 2009;73(4):Article 71. doi: 10.5688/aj730471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knoell DL, Johnston JS, Bao S, Kelley KA. A genotyping exercise for pharmacogenetics in pharmacy practice. Am J Pharm Educ. 2009;73(3):Article 43. doi: 10.5688/aj730343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavallari LH, Overholser BR, Anderson D, et al. Recommended basic science foundation necessary to prepare pharmacists to manage personalized pharmacotherapy. Pharmacotherapy. 2010;30(6):228e–235e. [Google Scholar]
- 17.Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92(4):414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299(5610):1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- 19.Parker LS, Grubs R. Ethical considerations regarding classroom use of personal genomic information. J Microbiology and Biology Educ. 2014;15(2):191–196. doi: 10.1128/jmbe.v15i2.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Church GM. The personal genome project. Molecular Systems Biology. 2005;1(1) doi: 10.1038/msb4100040. 2005.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. Sep 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yates A, Beal K, Keenan S, et al. The Ensembl REST API: ensembl data for any language. Bioinformatics. 2015;31(1):143–145. doi: 10.1093/bioinformatics/btu613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roederer MW, Van Riper M, Valgus J, Knafl G, McLeod H. Knowledge, attitudes and education of pharmacists regarding pharmacogenetic testing. Personalized Medicine. 2012;9(1):19–27. doi: 10.2217/pme.11.87. [DOI] [PubMed] [Google Scholar]
- 24.Ormond KE, Hudgins L, Ladd JM, Magnus DM, Greely HT, Cho MK. Medical and graduate students' attitudes toward personal genomics. Genet Med. 2011;13(5):400–408. doi: 10.1097/GIM.0b013e31820562f6. [DOI] [PubMed] [Google Scholar]
- 25.International HapMap C, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations NatureSep 2 2010467731152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moen M, Lamba J. Assessment of healthcare students' views on pharmacogenomics at the University of Minnesota. Pharmacogenomics. 2012;13(13):1537–1545. doi: 10.2217/pgs.12.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee KC, Hudmon KS, Ma JD, Kuo GM. Evaluation of a shared pharmacogenomics curriculum for pharmacy students. Pharmacogenomics. 2015;16(4):315–322. doi: 10.2217/pgs.14.181. [DOI] [PubMed] [Google Scholar]
- 28.Nickola TJ, Munson AM. Pharmacogenomics primer course for first professional year pharmacy students. Pharmacogenomics. 2014;15(1):39–48. doi: 10.2217/pgs.13.197. [DOI] [PubMed] [Google Scholar]
- 29.Springer JA, Iannotti NV, Kane MD, Haynes K, Sprague JE. Pharmacogenomics training using an instructional software system. Am J Pharm Educ. 2011;75(2):Article 32. doi: 10.5688/ajpe75232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Formea CM, Nicholson WT, McCullough KB, et al. Development and evaluation of a pharmacogenomics educational program for pharmacists. Am J Pharm Educ. Feb 12 2013;77(1):Article 10. doi: 10.5688/ajpe77110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vernez S, Salari K, Ormond KE, Lee S. Personal genome testing in medical education: student experiences with genotyping in the classroom. Genome Med. 2013;5(3):24. doi: 10.1186/gm428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee KC, Ma JD, Hudmon KS, Kuo GM. A train-the-trainer approach to a shared pharmacogenomics curriculum for US colleges and schools of pharmacy. Am J Pharm Educ. 2012;76(10):Article 193. doi: 10.5688/ajpe7610193. [DOI] [PMC free article] [PubMed] [Google Scholar]