Three-dimensional (3D) cell culture is becoming more widely used and several studies have shown that cells cultured in 3D behave differently compared to cells cultured in 2D, largely due to interactions of cells with the surrounding extracellular matrix (ECM).[1-3] Organotypic in vitro culture systems that provide an additional level of spatial organization are gaining popularity in an effort to more faithfully recapitulate in vivo structure and function. The spatial organization of cells cultured in 3D has been shown to affect cellular behavior such as proliferation and migration.[4-8] This may be due to concentration gradients of soluble factors, cell adhesion and mechanical forces.[9,10] Similarly, we have observed a considerable change in cytokine secretion by both endothelial cells and epithelial cells when cultured in an in vivo-like structure (i.e., a lumen) compared to 2D and 3D cultures.[11,12] Lumens (i.e. tubular structures) are ubiquitous in vivo being present in blood vessels, mammary ducts, and the lymphatic system. Lumen structures of varying size and geometry are involved in key normal and disease processes including angiogenesis, cancer development and drug delivery.[13,14] Therefore, there is a need for practical methods to create various lumen structures to advance organotypic culture platforms for increased physiological relevance.
A number of approaches for creating lumen structures in vitro exist. A stamping approach was developed to fabricate lumen structures in vitro by stamping channels on ECM gels followed by cellular addition.[15-19] Microfluidic channels have been coated with ECM proteins followed by lining with cells to mimic lumen structures.[20] 3D-printers have been used to print ECM gels with luminal channels within the gels or to print sacrificial structures like carbohydrate-glass networks and agarose structures that are encapsulated in hydrogels and the degraded later.[21-23] The degree of lumen structure complexity that can be achieved by the 3D-printers is an advantage over other methods; however, the 3D-printing based methods often require an extra step to degrade the sacrificial structures. The size and shape of the sacrificial structures are limited by the nozzle type and size if a traditional 3D-printer (e.g. single nozzle having a circular cross-section) is used.[21,22] Lumen structures have been achieved by patterning ECM gel using fluid flow (e.g. viscous fingering in which a less viscous fluid flows through a more viscous fluid, subsequently creating a circular hollow ECM structure) or needles to pattern a gel.[24-29] Although viscous fingering and needles have simplified the process of creating lumens, viscous fingering and needles are limited to circular straight lumens, limiting the capabilities of these methods. While previous methods have greatly contributed to advancing our ability to create a lumen structure in vitro, none have the combination of attributes that we desired. These attributes include 1) the ability to create fully enclosed lumen structures having different cross-sectional shapes; 2) the ability to form a lumen network (i.e., mimicking a branched network of vessels) across a range of ECM materials (e.g., various natural ECM materials and synthetic materials); and 3) the adaptability for use with existing high-throughput infrastructure such as liquid handlers.
Here, we introduce a new method, which allows for fabrication of 3D embedded lumens where size, structure, distance and configuration can be controlled using standard poly-dimethylsiloxane (PDMS) micromolding methods. The method enables multiple 3D lumen structures to be created within a natural or synthetic ECM gel placed in a microfluidic chamber, facilitating biophysical and biochemical signaling between cells in different lumen compartments (e.g. blood vessels adjacent to organ ducts) and the surrounding ECM. With this new method, multiple lumens can be linked together to create a complex lumen network (e.g., branching from one primary lumen to multiple subsequent secondary and tertiary lumens). In addition, the method is compatible with existing high throughput infrastructure, allowing efficient investigation of cell-cell and cell-ECM interactions in more in vivo-like environments.
In our method, which we call LumeNEXT, lumen structures are created by utilizing a removable PDMS rod. The PDMS rod is initially placed in a microfluidic chamber. The chamber is filled with an unpolymerized ECM solution that is subsequently polymerized. Once the ECM is completely polymerized, the PDMS rod is removed without disrupting the integrity of the surrounding ECM gel, creating a lumen structure that mimics the geometry of the PDMS rod. By controlling the properties of the PDMS rod, the size, shape, and orientation of a lumen within a natural or synthetic ECM gel can be controlled.
LumeNEXT uses two components - a microfluidic chamber and a removable PDMS rod. PDMS rods can be created using 1) hypodermic needles or 2) fabricated PS molds. First, if only simple straight lumens are desired, PDMS rods were simply prepared by using needles loaded with PDMS (Figure S1, Supporting Information). After loading, the uncured PDMS solution was heated at 100°C for two hours. Due to the elastomeric properties of PDMS, the rods can be removed from the needles without difficulties. With this technique we were able to make rods of different cross-sectional diameter by using needles of different gauge sizes. Secondly, fabricated polystyrene (PS) molds were used to facilitate the fabrication of PDMS rods with many other shapes beside the cylindrical PDMS rod that is obtained from using needles. Sandwiched PS molds (Figure 1a) are fabricated by using a milling machine.[30] CNC milling of the PS molds enabled rapid fabrication of molds with different shapes and dimensions. To obtain a circular cross-section of the PDMS rod a ball-end end mill was used to create a smooth half-circle on a PS sheet. The two mirror imaged PS molds were clamped together to form a circular cross-section lumen. The molds were then filled with uncured PDMS solution, which was heated at 100°C for two hours (Figure 1a). After heating, the two mold pieces were separated to reveal the PDMS rods resembling the shape of the sandwiched PS molds as seen in Figure 1a. By altering the milled PS molds, rods with different geometries and cross-sectional shape and size can be obtained. For example, instead of a ball-end end mill, a flat-end end mill was used to create a square shape on a PS sheet. This technique was used to make all the different configurations in Figure S2 (Supporting Information). Particularly for the half-circle cross-section of the lumen (Figure S2a, Supporting Information), a PS mold containing the half circle pattern was sandwiched with a flat PS sheet.
Figure 1.
Fabrication process of rods and assembly of the device. (a) Two mirror-image PS milled molds are connected to make a complete lumen mold, which is filled with PDMS to create the rods. (b) PDMS rod is placed in the chamber on two placeholders (red dashed frames just for illustration) followed by the addition of hydrogel and subsequent removal of the rod.
The microfluidic chamber can be fabricated using your material and method of choice. We used traditional soft-lithography to create PDMS chambers (Figure 1b) and a milling machine to fabricate PS chambers. The chamber that houses the PDMS rods, and the subsequent lumen structures, consists of two separate layers. The bottom layer features an open-topped chamber where the PDMS rod is placed and an unpolymerized ECM solution is loaded to create a lumen (Figure 1b). The hexagonal shape provides more stability to the gel by providing larger surface to volume ratio than rectangular shape, preventing the gel from being disturbed when removing the rod. In addition, there are two placeholders near ports (shown in Figure 1b) for the rods to rest on. These placeholders determine the position of the rod (i.e., the location of a lumen within a ECM gel), and the PDMS rod is self-supported, thus will be suspended inside the chamber. This allows the gel to completely surround the rod in all dimensions including the top and bottom. This capability is particularly beneficial when aligning multiple lumens precisely in a chamber by pre-determining the location of each lumen within the same ECM gel. The second layer consists of the same hexagonal chamber shape but has a thin layer of PDMS that encloses the chamber and provides a support to the collagen. We found that when the chamber was open to the air, the collagen collapsed on the lumen after removing the rod, which resulted in difficult cell loading and affected the cross-sectional shape of the patterned lumen. Finally, the top layer has four loading ports; two for gel loading and two that are connected to the lumen to facilitate cell and media loading. The ports interconnected to the lumen are used to flow fluid into the lumens using passive pumping.[31] In short, the passive pumping phenomenon occurs due to the larger drop that has a lower internal pressure than the smaller drop. Although we have mainly focused on a static condition via passive pumping, our system can be connected to external pumps and tubing without any structural modification if flow condition is desired. Figure 1b illustrates the chamber assembly and the process for lumen formation. Figure S3 (Supporting Information) shows an example of an assembled device. The PDMS rod was stained in blue (Figure S3a, Supporting Information) and yellow colored solution was added in the chamber as an example of where the ECM gel is located (Figure S3b, Supporting Information). This fabrication method is also compatible with other chamber materials such as PS, which may be a more suitable material for certain applications, particularly for hormone signaling studies (Figure S2c, Supporting Information).[32] The device can be modified to hold more rods and different configurations. Figure S2d (Supporting Information) is an example of a double lumen device containing two PDMS rods (red and blue).
After the chamber is assembled, the chamber was sterilized under UV light for 10 minutes. To achieve maximum adhesion between collagen type-1 and PDMS to reduce undesirable contraction of collagen, we used a two-step coating of 2% Poly(ethyleneimine) followed by 0.1% Gluteraldehyde solution.[33] The chamber was washed twice with sterile water to remove any residual coating solution, and collagen type-1 was added into the chamber. After incubation at 37°C for 10 minutes, the rods were removed from the chamber using sterile tweezers as seen in Figure 1b. The rods came out cleanly without any residual collagen on them, making the rods reusable. The removal of the rod left behind a lumen representing the shape of the rod. The ease of the removal of the rod is attributed to the physical properties of PDMS. The PDMS rods are very smooth and flexible, exerting little stress in the collagen when pulled out. Also, the flexibility of the PDMS rod allowed us to pull the rods approximately at a 45° angle (Figure 1b). This allowed us to incorporate the inner and outer ports that are oriented orthogonally to the main lumen enabling the use of passive pumping and eliminating the need for tubing.[31]
The ideal sacrificial structure to create lumens by LumeNEXT should be flexible, biocompatible, high mechanical integrity (self-supported, high elastic modulus), low protein absorption, low ECM adhesion, easy to cast, an off-the-shelf inexpensive material, and reusable for practicability. PDMS fits all this requirements. In addition, PDMS rods do not require any special coating to prevent ECM gel from adhering to it. When other materials like a needle was used to create lumen structures, the needle surface had to be coated to prevent collagen adhesion to the surface of the needle and make the removal without disruption of the gel.[27] Other materials with similar physical properties can be employed. For example, in preliminary experiments we found that materials such as nylon monofilaments and PEG rods can be used to make straight lumens. Nylon monofilaments are commercially available, but casting it to make more complex sacrificial structures can be challenging. PEG rods have lower mechanical integrity than PDMS rods, and thus break easily during the lumen manufacturing process. In addition, PEG rods swell considerable upon contact with liquid, and can not be reused because it can absorb chemical substances that can be released when reused.
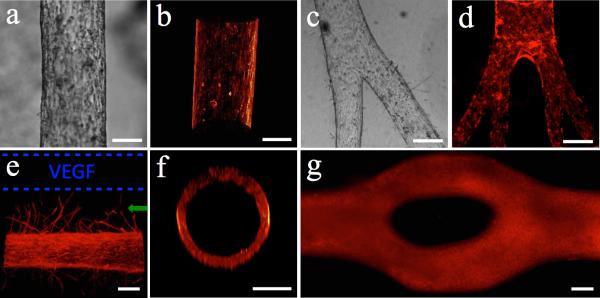
As an example application of the method, we lined a lumen with endothelial cells to create a biomimetic in vitro blood vessel. Figure 2a-d shows the confluent monolayer of endothelial cells that lined the lumen. 3D volume reconstruction was used to show the 3D structures (Figure 2b) as well as the circular cross-section (Figure 2f) of a cell-lined lumen. We have created lumens with cross-sections as large as 1.15mm and as small as 50μm (Figure S4, Supporting Information). To validate the perfusability of the lumens, generation of confluent monolayer and presence of tight junctions, Texas Red Dextran 70kDa was added into a cell-free lumen and an endothelial cell-lined lumen. The endothelial cell-lined lumens showed significantly reduced diffusion of the Dextran molecule into the surrounding gel (Figure S5, Supporting Information), confirming the formation of tight junctions of endothelial cells in the lumen (Figure S6c, Supporting Information). Since the fluorescence signal of the Dextran dims with the distance from the source as the diffusion of the Dextran occurs (Figure S5a, Supporting Information), a heat map approach was used to illustrate the diffusion of the dye out of the lumens (Figure S5b, c, e and f, Supporting Information). Using the double lumen device shown in Figure S3d (Supporting Information), we demonstrated how this device could be employed in angiogenesis assays. In a device with two lumens separated by 400μm, we lined one lumen with endothelial cells and loaded the other with 100ng/ml vascular endothelial growth factor (VEGF) solution. As shown in Figure 2e, there was directed generation of new microvessels (green arrow) with diameters of 20-30μm towards the growth factor source (blue dashed lines) after 3 days of culture. The endothelial cells in the lumen are responding to the VEGF gradient and when such gradient is not provided no sprouts are observed (Figure 2a). To show cell viability, a LIVE/DEAD® assay was performed. Figure S6f (Supporting Information) shows that after three days of culture the endothelial cells cultured in a lumen were viable. A device containing multiple lumens can be prepared using LumeNEXT to conduct compartmentalized co-culture. Figure S7 (Supporting Information) shows an endothelial lumen (HUVECs) co-cultured with kidney cancer cells (786-O). After 3 day of culture, the cancer cells were invading the ECM and migrated towards the endothelial lumen.
Figure 2.
Endothelial cell-lined lumens and possible morphologies. (a) Image of endothelial cell-lined lumen and (b) 3D volume reconstruction of the biomimetic blood vessel using F-actin staining (phalloidin). (c) Bright field image of bi-branched lumen and (d) multi-branched (tertiary branches) with phalloidin staining. (e) Angiogenesis via vascular endothelial growth factor (VEGF) gradient from source (dashed blue lines). (f) Cross-section of circular lumen lined with endothelial cells (phalloidin staining). (g) Bifurcated lumen lined with endothelial cells. Bar=150μm.
Another advantage of our system is that it facilitates temporal flexibility, where PDMS-rods can be removed at different time points. The temporal flexibility of rod removal is an advantage for co-culture, particularly when a first cell line needs to be differentiated for several days prior the addition of a second cell line. The lumen diameter increases with time after rod removal if the cells are not added (Figure S8, Supporting Information). This is partly due to collagen contraction. Keeping the rod inside provides more stability to the gel because the rod acts as a backbone to the gel. After lining the lumen with cells, the cells subsequently provide a similar support to the collagen. We have not observed cell-lined lumens to increase size after several days of culture (Figure S8e, Supporting Information). If the lumen is not lined with cells after the removal of rods, the lumen shape deforms, and in some cases in an asymmetric way (Figure S8f, Supporting Information), affecting the structural reproducibility. By keeping the rod inside we can ensure that the lumen dimensions will not change over time, hence we can still provide reproducible spatial control between the two cell types. Methods that require sacrificial structure degradation are incapable of this temporal flexibility. Other methods using needles can provide it but only for straight lumens.
The method also allows creation of branching lumen structures. We built a branched PS mold in order to create branched PDMS rods. For this method the rod is pulled out from the unbranched side. Figure 2c shows a bi-branched lumen and Figure 2d is an example of a tertiary branched lumen. In order to minimize mechanical stress applied on the gel while removing the branched PDMS rods that could fracture a surrounding ECM gel, the sum of the diameter and volume of the branched rods is smaller than the diameter and volume of the unbranched side of the rod. This way, when pulling out the rod, the branched rods fit in the volume of the unbranched side, lowering the stress exerted on the gel. The capability of creating branched lumens is important because we can generate structures that mimic blood vessel networks in vivo. Another advantage of our method is the capability of controlling the cross-sectional area of the lumens to create lumens with different cross-sectional shapes. It has been shown that vessels in human body do not always have a rounded cross-sectional shape. For example, Goel et al. have shown that tumor vessels have very irregular cross sections, including triangular cross-sections with sharp corners, where angiogenic sprouts are generated.[34] Similarly, Padera et al. describes that the abnormalities in vessels’ cross sections are due to cancer cells compressing tumor vessels, changing their cross-sectional shape and presenting an obstacle for the transport of therapeutics drugs into tumors.[35] Using our method we can create lumens with different cross-sectional shapes and study the tumor vessels in vitro. For example we have created lumens with various cross-sectional shapes (Figure S2c-d, Supporting Information) by using rods with the respective shape. In addition, it allows the study of the effect of corners and bifurcations on cellular behavior. In preliminary experiments (Figure S2a-b, Supporting Information) we have seen that where a lumen splits into two separated lumens there are new endothelial sprouts (green arrow) in between the lumens. This is consistent with Nelson's studies where sharp corners of epithelial tube-like structures produced more sprouts due to a concentration gradient of soluble factors.[4,9] A bifurcated lumen (i.e., lumen that converges into two lumens and diverges back to one) as shown in Figure 2g, was generated by using a PDMS rod with the same shape but with a very small partial trans-sectional incision created in the middle of the rod (Figure S9a, Supporting Information). The incision was made half way through the rod, which is controlled by removing the top layer of the PS rod mold to expose only half of the rod cross-sectional area for cutting. The other half of the PDMS rod was protected by the bottom layer of the PS rod mold. A razor blade was used to create a partial incision in the middle of the bifurcated rods (Figure S9a, Supporting Information). After assembling the device and polymerizing the ECM, the rod was removed by using two tweezers to pull out from both sides at the same time (Figure S9b, Supporting Information). The incision creates a weak point, where the rod preferentially break making possible the extraction of more complex structures. Previously described similar methods, such as the use of needles, lack the ability to create such complex lumen structures that branch and lack control over the cross-sectional shapes.[29]
In addition to enabling enhanced dimensional control and multiple geometries of lumens, our method is advantageous because 1) the method does not cause significant alteration of collagen concentration before and after polymerization and 2) the method does not solely rely on thermal polymerization method of ECM. Previous methods such as viscous fingering employ two different liquids of different viscosities, which often causes dilution of the collagen solution, thereby changing final concentration of collagen after polymerization (Figure S10, Supporting Information). Also, the viscous fingering method relies on the viscous and thermal properties of collagen, inhibiting the use of novel synthetic hydrogels. Our new system is highly reproducible and compatible with various natural and synthetic hydrogels that are rigid enough to allow the pulling of the PDMS rod without fracturing the ECM gel. For instance, for collagen type-1, a concentration as low as 2.5mg/ml can generate reproducible lumen structures and, for PEG gels, a concentration as low as 15kDa MW can be used. With the method shown in this paper, the synthetic hydrogel can be loaded, polymerized and the PDMS rod can easily be removed to reveal a lumen. Synthetic hydrogels are gaining popularity in organotypic research since they are cheaper, easier to manipulate and have lower batch-to-batch variation as compared to natural ECMs like collagen. Figure S11 (Supporting Information) shows endothelial cell-lined lumens in different Poly(ethylene glycol) (PEG) hydrogels. We were able to show how the morphology of the cells varies in different PEG gel stiffness. In softer PEG gels (20kDa) the endothelial cells arrange in a network-like structure like they would do on softer materials like Matrigel (Figure S11a-b, Supporting Information).[36] On the other hand, in rigid PEG gels (700kDa) the cells made a more confluent monolayer and had a rounded morphology (Figure S11c-d, Supporting Information). The system we present in this paper can be very useful to elucidate the behavior of different luminal cells on the presence of synthetic biomaterials.
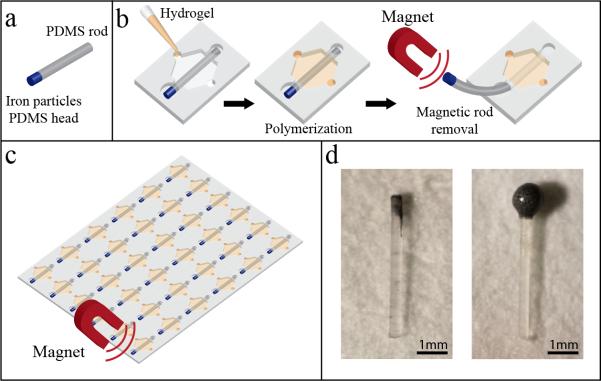
Our method can be extended even further to be compatible with high-throughput screening platform by embedding iron particles into the PDMS rod used to make flexible metallic or magnetic rods. An arrayed high-throughput platform that is compatible with automation and high-throughput screening system will serve as a very useful tool to efficiently investigate the interactions of other microenvironmental components, the response of various primary cells isolated from different patients to certain microenvironmental conditions, and drug screening. In fact, by using similar platforms, our lab has shown that the microchannel array can be utilized to look at the effects of different ECM proteins, different patient samples, and different drugs.[37-39] To achieve higher-throughput using our method, multiple magnetic rods can be removed simultaneously using a magnet to rapidly generate multiple lumens. The use of rods with an iron-PDMS head is shown in Figure 3a. By creating a rod with one end presenting iron particles, magnets can be used to remove multiple rods at once (Figure 3b-c) (Figure S12 and Video S1, Supporting Information). This method is faster and minimizes the human error that can happen by removing one rod at a time with tweezers. In addition, a magnet can be incorporated into automated instruments such as a liquid handler, allowing ECM gel loading into the chamber, automated rod removal and subsequent cell and reagents loading by the liquid handler.
Figure 3.
Magnetic rods and applications. (a) Illustration of an iron-PDMS rod. (b) Method for extraction of iron-PDMS rods. (c) Illustration of proposed high-throughput system for removal of rods using magnetic rod. (d) Pictures of magnetic rods.
The magnetic rods can be achieved by two methods. The first is to partially fill a PDMS rod mold with PDMS followed by the addition of iron-PDMS solution. After baking, the rod will have a PDMS body with an iron-PDMS head like shown in Figure 3d on the left picture. Alternatively, previously formed PDMS rods can be dipped in iron-PDMS solution and baked. This will form a bigger iron-PDMS head that is very magnetic even to weak magnets.
We have presented a method that can advance the fabrication of lumens in natural and synthetic gels. It is an accessible, low-cost, user-friendly approach that can easily be incorporated in most laboratories without the need of additional specialized equipment like microfluidic pumps, 3D printers, etc. Another important advantage of our system is that the entire process can be achieved in less than 6 hours for a total of 48 devices with double rods (96 lumens total). The simplicity and scalability of this method makes it an attractive system that can be used in many research areas, including drug screening, organotypic modeling, studying cell interactions, and tissue engineering.
Experimental Section
SU-8 mold and PDMSpreparation
The top and bottom layers of the microfluidic channels were fabricated using soft lithography. The molds were designed using Adobe Illustrator and printed on a transparency. The layers were spun with SU-8 100 (Y13273 1000L 1GL, MICRO CHEM, Newton, MA) according to the manufacturer's specifications on a silicon wafer (CC-1385, WRS, San Jose, CA). After the photoresist was soft-baked on a hot plate, a UV light source was used to transfer the device pattern to the photoresist. A post-exposure hard-baking step was executed. This process was repeated for additional layers. Upon completing all the layers, the mold was developed for 45 minutes in SU-8 developer (PGMEA, 537543, Sigma, St. Louis, MO) and washed with acetone and iso-propyl alcohol. Poly-dimethylsiloxane (Sylgard 184 silicon elastomer base, 3097366-1004, Dow Corning, Salzburg, MI) (PDMS) was prepared at a ratio of 1:10 curing agent (Sylgar 184 silicone elastomer curing agent, 3097358-1004, Dow Corning, Salzburg, MI) and degassed in a vacuum for 30 minutes. The PDMS was then poured over the SU-8 silicon mold on a hot plate and baked at 80 °C for 4 hours.
Polystyrene channel fabrication
The top and bottoms layers of the (PS) channels were fabricated by CNC milling (PCNC 770, Tormach, Waunakee, WI) of 1.2 mm thick PS (ST313120, Goodfellow, Coraopolis, PA). The channels were designed using SOLIDWORKS. The two halves were assembled by acetonitrile bonding.[40] Briefly, acetonitrile (#271004, Sigma Aldrich, St. Louis, MO) was applied drop-wise to one layer, which was quickly placed onto the matching half layer. The excess acetonitrile was aspirated, and the two-layer device was heated on a hot plate at 70°C. Gentle pressure was applied to the device during heating for 30 seconds. The bonded PS pieces were glued to a glass cover slide using Norland optical adhesive (6801 Ultraviolet curing, Norland products INC., Cranbury, NJ) and cured under a UV light source for 20 seconds.
PDMS rod fabrication
To achieve complex PDMS rod shapes, PDMS rod molds were fabricated by CNC milling of 1.2 mm thick PS. The molds were designed using SOLIDWORKS. Once the molds were milled, they were clamped together and PDMS solution was allowed to fill the channels by capillary flow. After the channels were filled, the clamped PS mold was placed in an oven at 100 °C for 2 hours. The mold layers were carefully split apart and the PDMS rods were collected. Alternatively, to achieve circular cross-sectional PDMS straight rods, hypodermic needles (25G, 14-840-84, Fisher Scientific, Pittsburgh, PA) were filled with PDMS and baked a hot plate at 100°C for one hour (Figure S1, Supporting Information). After baking, the PDMS rods were carefully removed from the needle with tweezers. Magnetic rods were achieved by dipping the PDMS rods in a solution of 50%(w/v) iron powder (93-2601, Strem Chemicals, Newburyport, MA) and PDMS. The rods where baked at 100°C for 10 minutes after dipping in iron-PDMS solution.
LumeNEXT setup
First, the top and bottom layers of the chamber were combined resulting in the desired chamber unit. The PDMS rods were inserted into the chamber unit. Next, in order to bond the devices to a glass surface and to create a hydrophilic environment, the devices were oxygen-plasma treated (Femto, Thierry Corp., Royal Oak, MI). To sterilize, the devices were placed in a biosafety cabinet and exposed to UV light for 10 minutes. The devices were pretreated with 2% Poly(ethyleneimine) (03880, Sigma-Aldrich, St. Louis, MO) diluted in sterile deionized water for 10 minutes, and 0.1% Gluteraldehyde (G6257, Sigma-Aldrich, St. Louis, MO) diluted in sterile deionized water for 30 minutes, followed by three washes with sterile deionized water.
Collagen gel preparation
The following steps were carried out on ice to halt the polymerization of collagen. For a collagen solution with a final concentration of 6mg/ml, 80μl of rat tail collagen type-1 10mg/ml (354249, BD Biosciences, San Jose, CA) was combined with 20μl of 5X PBS and 3μl of 0.5 N Sodium Hydroxide (S318, Fisher Scientific, Pittsburgh PA). The mixture was incubated on ice for 20 minutes. Finally, 34μl of PBS was added for a final collagen concentration of 6mg/ml and a pH of 7.4. For 8mg/ml, 6.5mg/ml, 5mg/ml, 4mg/ml and 3.5mg/ml collagen concentrations, the amount of PBS in the previous step was adjusted to obtain the desired final concentration.
Creating lumens
After the devices were prepared, the collagen gel mixture was loaded and placed at ambient temperature for 10 minutes, followed by 10 minutes in a 37°C incubator. The PDMS rods were pulled out of the polymerized collagen gel from the output port resulting in a lumen structure in the collagen gel.
Cell culture and cell seeding
For the endothelial cell-lined lumen experiments, Human Umbilical Vein Endothelial Cells (HUVEC) were used (C2517A, Lonza, Allandale, NJ). HUVEC's were cultured in Endothelial Cell Growth Medium (EGM-2 BulletKit, CC-3162, Lonza, Allandale, NJ). Each lumen was loaded with 2μl of a cell solution at 50,000cells/μl. The devices were then rotated at 2RPM for 45 minutes at 37°C to allow cell attachment uniformly throughout the lumen. Cell-lined lumens were fed with 6μl of EGM-2 medium for 24 hours, and then the medium was replaced by EGM-2 without serum. For angiogenesis experiment, 100ng/ml of active human VEGF full length protein (ab9571, abcam, Cambridge, MA). Co-culture experiments were performed with renal cell carcinoma cells, 786-O (CRL-1932, ATCC, Manassas, VA). 786-O were cultured in RPMI 1640 Medium with L-glutamine (10-040-CV, Corning-cellgro, Manassas, VA) with 10% Fetal Bovine Serum (FBS, 35-010-CV, Corning-cellgro, Manassas, VA). The same protocol as HUVECs was used with the distinction of a final cell concentration of 250,000cells/ul in 6mg/ml collagen and they where not used to line the lumen but to fill it with a 3D suspension of the cells to recreate a tumor. All the cells were fixed and stained after 3 days of culture.
Immunofluorescent staining
Prior to imaging, the cell-lined lumens were first fixed and stained. A 4% paraformaldehyde (15700, EM Science, Hatfield, PA) solution was adde in the lumens at ambient temperature for 30 minutes. The paraformaldehyde was removed and washed with 0.1% Tween PBS three times. A 0.2% Triton® X-100 (807426, MP Biomedicals, Santa Ana, CA) solution was added in the lumens for 30 minutes at ambient temperature. The lumens were washed three times with 0.1% Tween PBS, and 3% Bovine Serum Albumin (BSA) (A9056-100G, Sigma-Aldrich, St. Louis, MO) blocking buffer solution was added and left in overnight. The next day the lumens were washed three times with 0.1% Tween PBS and 6μl of Texas Red®-X Phalloidin (T7471, Life Technologies, Grand Island, NY) 1:50, DAPI (D1306, Life Thechnologies, Eugene, OR) 1:100 and/or anti-ZO1 tight junction protein antibody (ab59720, abcam, Cambridge, MA) 1:25, all diluted in blocking buffer, were added and left in overnight. Prior to imaging, the lumens were washed three times with PBS to remove the dyes and minimize background.
Imaging and analysis
Brightfield images were acquired on an inverted microscope (IX81, Omlympus) using Slidebook 5.0 imaging system (Intelligent Imaging Innovations (3i), Inc.). F-actin and collagen fibers were imaged by using multiphoton laser scanning microscopy (with second harmonic filter for collagen). All multiphoton laser scanning microscopy (MPLSM) and Second Harmonic Generation (SHG) imaging was done on an optical workstation that was constructed around a Nikon Eclipse TE300. A MaiTai Deepsee Ti: sapphire laser (Spectra Physics, Mountain View, CA) excitation source tuned to 890 nm was utilized to generate both multiphoton excitation and SHG. The beam was focused onto the sample with a Nikon (Mehlville, NY) 20X Super Fluor air-immersion lens (numerical aperture (NA) = 1.2). All SHG imaging was detected from the back-scattered SHG signal with a H7422 GaAsP photomultiplier detector (Hamamatsu, Bridgewater, NJ), and the presence of collagen was confirmed by filtering the emission signal with a 445 nm (narrow-band pass) filter (TFI Technologies, Greenfield, MA) to isolate the SHG signal. Acquisition was performed with WiscScan (http://www.loci.wisc.edu/software/wiscscan), a laser scanning software acquisition package developed at LOCI (Laboratory for Optical and Computational Instrumentation, University of Wisconsin, Madison, WI).
Supplementary Material
Acknowledgements
We would like to thanks Joseph Ulbrich for his assistance in fabrication and data collection. David J. Beebe holds equity in Bellbrook Labs, LLC, Tasso, Inc., and Salus Discovery, LLC. This work was supported by NIH R01 EB010039, University of Wisconsin Carbone Cancer Center Support Grant NIH P30 CA014520 and the University of Wisconsin Graduate Engineering Research Scholars.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Lee GY, Kenny PA, Lee EH, Bissell MJ. Nat. Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, Gray JW, Bissell MJ. Mol. Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung KE, Su X, Bethier E, Pehlke C, Fried A, Beebe DJ. PLoS One. 2013;8(10):e76373. doi: 10.1371/journal.pone.0076373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori H, Gjorevski N, Inman JL, Bissell MJ, Nelson CM. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14890–14895. doi: 10.1073/pnas.0901269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bissell MJ, Radisky D. Nat. Rev. Cancer. 2001;1(1):46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cukierman E, Pankov R, Stevens DR, Yamada KM. Science. 2001;294(5547):1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 7.Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian U, van Rheenen J, Deryugina E, Friedl P. Semin. Cell Dev. Biol. 2009;20(8):931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blobel CP. Blood. 2010;115(25):5128–5130. doi: 10.1182/blood-2010-03-275271. [DOI] [PubMed] [Google Scholar]
- 9.Gjorevski N, Nelson CM. Integr. Biol. 2010;2:424–434. doi: 10.1039/c0ib00040j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffith LG, Swartz MA. Nat. Rev. Mol. Cell Biol. 2006;7(3):211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 11.Bischel LL, Sung KE, Jiménez-Torres JA, Mader B, Keely PJ, Beebe DJ. FASEB J. 2014;28(11):4583–4590. doi: 10.1096/fj.13-243733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant DM, Mostov KE. Nat. Rev. Mol. Cell Biol. 2008;9:887–901. doi: 10.1038/nrm2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson CM, van Duijn MM, Inman JL, Fletcher DA, Bissell MJ. Science. 2006;314(5797):298–300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan J, Shah S, Thomas A, Ou-Yang HD, Liu Y. Microfluid. Nanofluid. 2013;14(1-2):77–87. doi: 10.1007/s10404-012-1024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HY, Nelson CM. Bio-inspired Mater. Biomed. Eng. 2014;1:161–171. [Google Scholar]
- 16.Wray LS, Tsioris K, Gil ES, Omenetto FG, Kaplan DL. Adv. Funct. Mater. 2013;23:3404–3412. doi: 10.1002/adfm.201202926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi JS, Piao Y, Seo TS. Bioprocess Biosyst. Eng. 2013;35(12):1871–1878. doi: 10.1007/s00449-013-0961-z. [DOI] [PubMed] [Google Scholar]
- 18.Baker BM, Trappmann B, Stapleton SC, Toro E, Chen CS. Lab Chip. 2013;13:32–46. doi: 10.1039/c3lc50493j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, López JA, Stroock AD. Proc. Natl. Acad. Sci. U. S. A. 2012;109(24):9342–9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB, Kamm RD. Proc. Natl. Acad. Sci. U. S. A. 2012;108(34):13515–13520. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DHT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Nat. Mater. 2012;11:768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, Barabaschi G, Demarchi D, Dokmeci MR, Yange Y, Khademhosseini A. Lab Chip. 2014;14:2202–2211. doi: 10.1039/c4lc00030g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis Jennifer A. Adv. Mater. 2014;26:3124–3130. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 24.Bischel LL, Lee S, Beebe DJ. J. Lab. Autom. 2012;17(2):96–103. doi: 10.1177/2211068211426694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bischel LL, Young EWK, Mader B, Beebe DJ. Biomaterials. 2013;34:1471–1477. doi: 10.1016/j.biomaterials.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bischel LL, Sung KE, Beebe DJ. BMC Cancer. 2015;15:12. doi: 10.1186/s12885-015-1007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park JH, Chung BG, Lee WG, Kim J, Brigham MD, Shim J, Lee S, Hwang C, Durmus NG, Demirci U, Khademhosseini A. Biotechnol. Bioeng. 2010;106(1):138–148. doi: 10.1002/bit.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chroback KM, Potter DR, Tien J. Microvasc Res. 2006;71:185–196. doi: 10.1016/j.mvr.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen DHT, Stapleton SC, Yang MT, Cha SS, Choi CK, Galie PA, Chen CS. Proc. Natl. Acad. Sci. U. S. A. 2013;110(17):6712–6717. doi: 10.1073/pnas.1221526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guckenberger DJ, de Groot TE, Wan AMD, Beebe DJ, Young EWK. Lab Chip. 2015 doi: 10.1039/c5lc00234f. DOI: 10.1039/c5lc00234f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker GM, Beebe DJ. Lab Chip. 2002;2:131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- 32.Regehr KJ, Domenech M, Koepsel JT, Carver KC, Ellison-Zelski SJ, Murphy WL, Schuler LA, Alarid ET, Beebe DJ. Lab Chip. 2009;9(15):2132–2139. doi: 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran CNB, Walt DR. J. Coll. Int. Sci. 1989;132:373–381. [Google Scholar]
- 34.Goel S, Duda DG, Xu L, Munn LL, Boucher Y, Fukumura D, Jain RK. Physiol. Rev. 2011;91(3):1071–1121. doi: 10.1152/physrev.00038.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Padera TP, Stoll BR, Tooredman JB, Capen D, Tomaso E, Jain RK. [DOI] [PubMed]
- 36.Donovan D, Brown NJ, Bishop ET, Lewis CE. Angiogenesis. 2001;4(2):113–21. doi: 10.1023/a:1012218401036. [DOI] [PubMed] [Google Scholar]; Nature. 2004;427:695. doi: 10.1038/427695a. [DOI] [PubMed] [Google Scholar]
- 37.Montanez-Sauri SI, Sung KE, Berthier E, Beebe DJ. Integr. Biol. 2013;5:631–640. doi: 10.1039/c3ib20225a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su G, Sung KE, Beebe DJ, Friedl A. PLoS ONE. 2012;7(10):e46685. doi: 10.1371/journal.pone.0046685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pak C, Callander NS, Young EWK, Titz B, Kim K, Saha S, Chng K, Asimakopoulos F, Beebe DJ, Miyamoto S. Integr. Biol. 2015;7:643–654. doi: 10.1039/c5ib00071h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou P, Young L, Chen Z. Biomed. Microdevices. 2010;12(5):821–832. doi: 10.1007/s10544-010-9436-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.