Abstract
Neutrophil extracellular traps (NETs) were first reported in 2004, and since their discovery, there has been an increasing interest in NETs, how they are formed, their role in controlling infections, and their contribution to disease pathogenesis. Despite this rapid expansion of our understanding of NETs, many details remain unclear including the role of reactive oxygen species (ROS) in the formation of NETs. Further, to study NETs, investigators typically require a large number of cells purified via a lengthy purification regimen. Here, we report a microfluidic device used to quantify both ROS and NET production over time in response to various stimulants, including live bacteria. This device enables ROS and NET analysis using a process that purifies primary human neutrophils in less than 10 minutes and requires only a few microliters of whole blood. Using this device we demonstrate the ability to identify distinct capabilities of neutrophil subsets (including ROS production and NET formation), the ability to use different stimulants/inhibitors, and the ability to effectively use samples stored for up to 8 hours. This device permits the study of ROS and NETs in a user-friendly format and has potential for widespread applications in the study of human disease.
Introduction
Neutrophils are important players in innate immunity, and are considered the first responders of the immune system.1, 2 As part of their antimicrobial repertoire, neutrophils extrude their chromatin mixed with the cytotoxic peptides from their granules and form neutrophil extracellular traps (NETs) to immobilize and kill invading microorganisms.3 The antimicrobial role of these NETs has been established through in vitro killing assays and in vivo studies using mouse models of bacterial and fungal infection.4-6
There is still a debate about the signaling pathways that regulate NET formation (NETosis).7-9 Patients suffering from chronic granulomatous disease (CGD), a disease in which reactive oxygen species production is adversely affected, cannot form NETs in the presence of pathogen-associated molecular patterns (PAMPs), indicating the requirement of reactive oxygen species (ROS) for NETosis after stimulation by microbial products.10 However, other studies have demonstrated that immune complexes can induce NETosis, independent of ROS production.8 With an association between NETs and autoimmune diseases becoming more apparent, there is a need to elucidate the signaling cascades that drive NETosis in the context of disease.11, 12 Should ROS or other molecules be required for NETosis, they might serve as targets in the treatment of disease.
Single-cell analysis of NETosis can offer important insights into the process of NET formation. With multiple studies now suggesting the presence of different neutrophil subsets,13, 14 characterizing NETosis in single cells could offer insights into whether or not specific neutrophil phenotypes release more or fewer NETs and how they do so. For example, Yousefi et al. identified viable neutrophils that produced NETs composed of mitochondrial DNA instead of nuclear DNA.15 In support of this finding, McIlroy et al. revealed that NETs released in patients after major trauma were composed solely of mitochondrial DNA.16 Furthermore, Yousefi et al. used ROS inhibitors to show that NETs composed of mitochondrial DNA following stimulation with granulocyte/macrophage colony-stimulating factor (GM-CSF) and subsequent toll-like receptor 4 (TLR4) agonist or complement factor 5a (C5a) are ROS dependent.15 Therefore, studying NETosis in individual cells might lead to novel findings related to neutrophil type and NET DNA origin.
Currently, neutrophil functional assays separately quantify ROS and NETosis, and do not allow same-cell comparisons of ROS and NET production.7, 17, 18 Immunostaining fixed samples, a common approach for NET analysis, only provides information at a single time-point without capturing the dynamic process of NETosis. To study the active process of NETosis, researchers have used cell impermeable nucleic acid dyes to quantify extracellular DNA during time course experiments.17, 19, 20 However, these dyes are unable to capture early NETosis-related changes prior to DNA extrusion, including chromatin decondensation and the spilling of the nucleoplasm into the cytoplasm. With ROS production being one of the first steps of NETosis following PAMP-activation,17 a tool is required that allows the analysis of ROS as well as all stages of NETosis in a single cell to clarify the pathways involved with NETosis.
Within the past decade, microfluidic tools have been developed and used to study neutrophil adhesion and chemotaxis.21-23 Recently, these microfluidic devices were used in clinical studies to characterize neutrophils from burn patients and individuals with asthma, 24, 25 and in murine studies of arthritis.26 In addition to their well-characterized chemotactic gradients and novel neutrophil isolation techniques, these microfluidic devices require small volumes of sample. In comparison, non-microfluidic assays for studying chemotaxis or NETosis require a substantial population of purified neutrophils from a large volume of whole blood. The sample size can be prohibitive, especially in the case of small animal models, where the animals must be euthanized prior to neutrophil isolation in order to obtain large enough neutrophil numbers for use in assays. To address these limitations, we have developed a microfluidic device which enables purification of neutrophils from a drop of blood (< 10 μl) and simultaneously measures production of ROS and NET formation. This device utilizes Hoechst nucleic acid stain to identify in real-time, DNA within intact neutrophils as well as NETs. Concomitantly, the fluorescent dye dihydrorhodamine-123 (DHR) is used to quantify ROS production within the same cell. Using this device, we illustrate that production of extracellular traps by neutrophils in the presence of surface coated P-selectin requires additional stimuli such as Phorbyl 12-myristate 13-acetate (PMA) or Pseudomonas aeruginosa. Further, we illustrate localization of Pseudomonas aeruginosa to NETs and demonstrate formation of NETs in the presence of heparin from stored blood. The presence of NETs was supported by inhibiting the formation of NETs using the NADPH-oxidase inhibitor diphenyleinedium (DPI), which inhibits ROS and NETs in a dose dependent manner.
Material and Methods
Reagents
Dihydrorhodamine-123 (DHR-123), Hoechst 33342 and Sytox Green nuclear dyes were purchased from Molecular Probes (Eugene, OR) and stored at −20°C. Phorbyl 12-myristate 13-acetate (PMA) and Diphenyleneiodonium chloride (DPI) were purchased from Sigma Aldrich (Milwaukee, WI). PMA was dissolved in DMSO at a stock solution concentration of 1.6mM and stored at −20°C. DPI was dissolved in de-ionized water at a concentration of 10mM, and stored at room temperature. Recombinant human P-selectin (R&D Systems, Minneapolis, MN) was re-suspended in deionized water (100 μg/mL) and stored at 4°C. Phycoerythrin conjugated anti-human CD14 (Clone 61D3) was purchased from eBioScience (San Diego, CA). AlexaFluor 647 conjugated anti-mouse/human CD11b conjugated (Clone M1/7) and AlexaFluor 790 conjugated anti-human CD45 (Clone H130) were purchased from BioLegend (San Diego, CA).
Microfluidic Device
Microfluidic devices were fabricated using soft lithography techniques and PolyDimethylSiloxane (PDMS, Sylgaard 184, Dow Corning, Midland, MI) as previously described.27-29 A multilayered mold was created by coating SU-8 photoresist (Microchem Corp., Westborough, MA) on a silicon wafer. The channels were 100 μm in height, and the openings into the channels were 250 μm in height.
Device Use
Microfluidic channels were placed on a non-tissue culture treated 6 cm petri dish (Corning Inc., Corning, NY, USA). The channels were coated with 3 μl of recombinant human P-selectin (100 μg/mL) at 4°C for 30 minutes. After coating, the P-selectin containing solution was washed out of the channels with 3 μl of PBS. Peripheral blood was isolated from a healthy donor with prior approval from the University of Wisconsin Institutional Review Board using a disposable safety lancet (Fisher Scientific, Pittsburgh, PA). 4 μl of blood was pipetted into a reservoir containing 24 μl of PBS and mixed gently, for a final 1:6 dilution of blood. 1 μl of dilute whole blood was passed through each microchannel two times, with each pass 30 seconds apart. After allowing neutrophil capture for 3 minutes, a staining mix containing 10 μM DHR-123 (Life Technologies, Grand Island, NY) and 10 μg/ml Hoechst stain (Life Technologies , Grand Island, NY) in PBS was added to the channels. For neutrophil activation, the staining solution contained approximately 1 μM of PMA. For ROS inhibition, the staining solution contained various concentrations of DPI. Each condition was replicated in duplicate or in triplicate on a single device.
Microscopy
Phase contrast and fluorescent images were taken using an Olympus IX-81 optical microscope with a 10X objective with a numerical aperture (NA) of 0.30; the images were captured using the computer program Slidebook (3i, Denver, CO). Neutrophils were imaged every 3 minutes for 2 hours.
Staining of Captured Neutrophils
To verify that captured cells are neutrophils, microfluidic devices were coated with P-selectin or bovine serum albumin (BSA) at 100 μg/ml as described. Diluted blood was placed within these channels, and captured cells were stained for 30 minutes with Hoechst stain (1:250), anti-CD14-PE (1:30), anti-CD45-790 (1:10), and anti-CD11b-Alexa 647 (1:10). Cells were imaged on a Nikon TI-eclipse, at 10X magnification. Images of the channels were rolling ball background subtracted using Image J (NIH, Bethesda, MD), and stained cells were manually quantified using Image J.
Bacteria
The Pseudomonas aeruginosa strain PAK, which carries a constitutively expressed red fluorescent protein mCherry (pMKB1::mCherry) was kindly provided by S. Moskowitz, University of Washington, Seattle, WA, USA.30 After streaking for single colony isolation on Luria-Bertani (LB) agar supplemented with 200 μg/mL carbenicillin (LB carbenicillin-200), a single clone was cultured overnight in 5 mL of LB carbenicillin-200 broth at 37°C with shaking (200rpm). 1 mL of overnight culture was pelleted by centrifugation at 8000 rpm for 5 min. The bacteria were then washed with 1 mL of 1X phosphate-buffered saline (PBS) before final resuspension in 1 mL of 1X PBS. Bacteria used in the microfluidic device were diluted from this final inoculum.
CYBA Knockdown in PLB-985 Cells
CYBA (gene encoding p22phox) knockdown was performed by lentiviral transduction of shRNA as previously described.31, 32 Briefly, lentiviral shRNA constructs in the pLKO.1 Puro vector (Open Biosystems, Thermo Fisher Scientific) were ordered. Targets (sense) used for these studies were scrambled control (Ctrl) 5’- GTCTCCGAACGTGT CACGTT-3’ and CYBA (TRCN0000064581) 5’-CGCTTCACCCAGTGGTACTTT-3’. 3.5×106 HEK293T cells, which were used to generate viral particles, were plated on a tissue culture treated 10cm dish the day before transfection. One 10cm plate was transfected per shRNA construct by the calcium phosphate method using the following constructs: 6 μg control or CYBA shRNA construct, 0.6 μg vesicular stomatitis virus (VSV-G), and 5.4 μg cytomegalovirus (CMV 8.9.1). Supernatant was collected after 72 hr. and centrifuged to remove debris. 1×106 PLB-985 were mixed with the viral supernatant in the presence of 4 μg/ml polybrene in a 6 well plate and centrifuged at 1000× g in a swinging bucket centrifuge for 30 minutes at 32°C. Stable lines were created by selection of transduced cells with 1 μg/ml puromycin.
Western Blotting
PLB-985 cells differentiated for 6 days in the presence of 1.3% DMSO were resuspended in ice cold lysis buffer (20 mM Tris pH 7.6, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1 μg/ml Pepstatin A, 200 μM PMSF, 1 μg/ml Leupeptin, and 2 μg/ml Aprotinin) and placed on ice for 10 min. Lysates were sonicated for 20 seconds at 20% power using a QSONICA Q125 sonicator and centrifuged at 14,000 × g. A BCA assay kit (Thermo Fisher Scientific) was used to determine protein concentration according to the manufacturers instructions. 50 μg of protein was loaded for each sample, and PageRuler Plus Prestained Protein Ladder (Thermo Fisher Scientific) was used to approximate molecular weight. Following transfer to nitrocellulose, the membrane was blocked in 5% milk in TBS-T (15mM Tris pH 7.4, 150mM NaCl, 0.1% Tween 20) and probed with rabbit anti-p22phox (1:50) and mouse anti-α-tubulin DM1a (1:2000) in TBS-T overnight. Infrared-labeled secondary antibodies, goat anti-mouse 800 (Rockland Immunochemicals Inc.) and goat anti-rabbit 680 (Invitrogen) (1:10,000) were used for detection on a Li-Cor Odyssey infrared scanner (Li-Cor Biosciences, Lincold, NE).
Blood Collection
Blood was obtained from healthy donors with prior IRB approval. Blood was collected and stored in Vacutainer® tubes containing sodium heparin or sodium citrate for 8 hours at room temperature or 4° Celsius for experiments analyzing neutrophil function from stored anti-coagulated blood. For all other experiments, blood was collected from a healthy donor using a Lancer puncture as described earlier.
Image Analysis
Captured fluorescent images were analyzed using Image J (NIH, Bethesda, MD). For quantification of ROS production, images from the Hoechst stain were used to identify all cells present within the viewing window by the following procedure; (1) Hoechst images were background subtracted using a rolling ball function (radius = 50 pixels); (2) Nuclei were identified by a pre-selected threshold; (3) The threshold Hoechst images were dilated using a 3×3 matrix; (4) The Image J “analyze particles” command was used to select the regions defining each cell. Hoechst-defined cells were further analyzed for Rhodamine-123 fluorescent staining. Mean Rhodamine-123 fluorescent values within a cell structure were calculated and recorded for analysis. A single image frame captured during time-lapse imaging was used for quantification. For NETosis, single image frames were captured at the same time, after 3 hours of stimulation. Collected images were analyzed manually using Image J “multipoint selection” function.
Human Subjects
Peripheral blood was obtained with informed consent from healthy donors at University of Wisconsin-Madison. All experiments were performed according to the national and institutional guidelines and approved by the University of Wisconsin Institutional Review Board (The University of Wisconsin Institutional Review Board; Protocol #2011-0463).
Statistics
Log transformed mean fluorescence values of DHR representing ROS production and percent NETs present within a microfluidic device were graphed using the Open Source R statistics package. Student's T-test was used to compare the formation of NETs identified by Hoechst stain to Sytox Green, and one-way analysis of variance (ANOVA) in the Open Source R statistics package was used to compare NETs produced within multi condition experiments.
Results
To enable analysis of ROS and NETs from a few microliters of blood, we optimized the device shown in Figure 1. P-selectin is coated onto the bottom of a channel to capture and purify neutrophils. Fluid handling of the microfluidic device is based on passive pumping principles;33 therefore its operation requires only a pipette (Figure 1). Functional readouts are performed using two different dyes, Hoechst DNA stain and DHR, which enable real-time measurement of NETosis and production of ROS, respectively. Captured cells were confirmed to be neutrophils using immunocytochemistry staining. Neutrophils were identified as CD11b positive, CD14 negative and CD45 negative cells (Suppl. Figure 1).
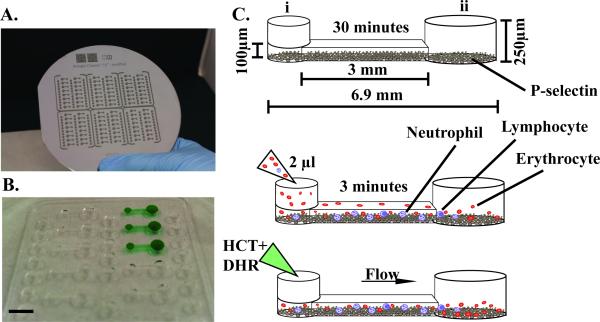
Figure 1. Photograph of microfluidic device and steps of operation.
A, Photograph of a silicone wafer mold of the device. B, Photograph of an arrayed polydimethylsiloxane (PDMS) device with 16 microfluidic channels. Black bar indicates 5 mm. C-E, Device operation. C, Add coating of P-selectin within the microfluidic device for 30 minutes. Dimensions of device channel are noted. Port i denotes the input port where fluids are added, port ii denotes the output port. D, Dilute blood within a reservoir and pipette ~2μl of blood within the channel, and allow neutrophils to adhere for 3 minutes. E, Add fluorescent dye mixture containing Hoechst (HCT) and dihydrorhodamine-123 (DHR). Fluid will flow from port I to port ii due to the difference in surface tension.
Real-time Identification of NETosis By Hoechst Staining
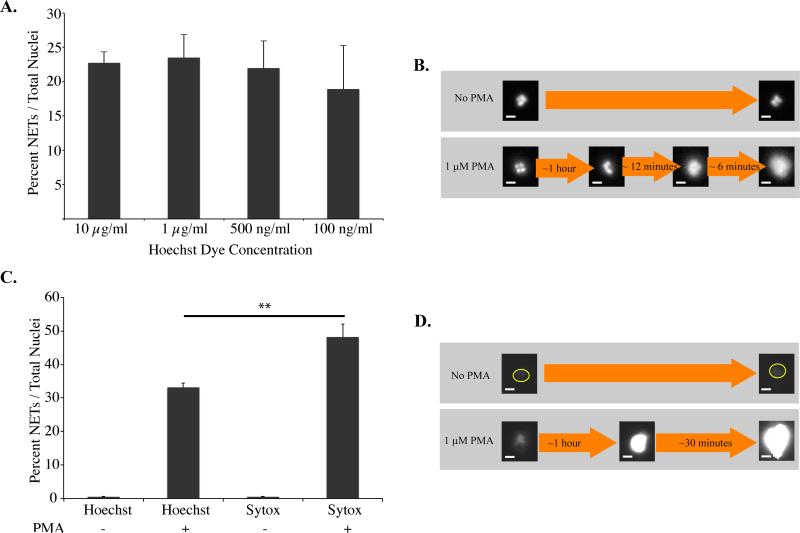
Pre-staining with Hoechst nucleic acid dye enabled visualization of NETs and chromatin of intact neutrophils (Figure 2). This dye fluoresces after binding to minor grooves of DNA using a mechanism that could theoretically inhibit chromatin decondensation and NETosis. Therefore, for purposes of the current microfluidic device, it was important to characterize the effects of Hoechst staining on NETosis. In controlled experiments, varying concentrations of Hoechst stain did not affect the percent NETs formed in response to PMA (Figure 2, A). An important advantage of Hoechst staining compared to other DNA dyes, including 4',6-diamidino-2-phenylindole (DAPI) and Sytox® green, is its ability to stain and monitor chromatin changes within live intact cells. Hoechst stain enabled visualization of NETosis associated changes, including chromatin decondensation, spilling of the nucleoplasm into the cytoplasm, and cytoplasmic perforation (Figure 2, B). The observed timeline of these steps were similar to previously reported studies.7, 17 For example, after one hour there was loss of multi-lobed neutrophil chromatin. Within 9-12 minutes of this time-point, there was loss of chromatin features and the presence of more diffuse fluorescence, matching previously reported timeline for nucleoplasm spilling into the cytoplasm. Within 3-6 minutes of this stage, there was an additional increase in diffuse Hoechst staining, suggesting perforation of cytoplasm and spilling of DNA. There were no additional changes observed after this step, further supporting this as the terminal step of NET formation. To further support the formation of NETs within our device, NETs identified by Hoechst staining were compared to NETs identified by Sytox Green (Figure 2, C-D). NETs were formed on the same timeline in the presence of either stain after stimulation by PMA. Sytox Green staining identified on average 10 percent more NETs in comparison to Hoechst staining (Figure 2, C).
Figure 2. Hoechst stained NETs.
A, Percent NETs formed after 2 hours in Phorbyl 12-myristate 13-acetate (PMA) activated neutrophils stained with varying concentrations of Hoechst stain relative to total number of visualized nuclei. Differences between varying concentrations of Hoechst stain were statistically not significant as determined by one-way ANOVA (p-value < 0.2). B, Control (no PMA) and PMA activated neutrophils indicating changes in chromatin decondensation within the neutrophils over time. C, Percent NETs formed after 2 hours in PMA, stained with Hoechst stain (10 μg/ml) or Sytox Green (5 μM) stain. Statistically significant d D, Control (no PMA) and PMA activated neutrophils indicating increased Sytox Green staining after 90 minutes with PMA. Sytox Green staining identified 10 percent more NETs in comparison to Hoechst stain as determined by Student's T-test (**: p-value < 0.01). All data was repeated in duplicate or in triplicate at least three times. Neutrophils activated with PMA at 1 μM. White bar indicates 5 μm.
Real-time Quantification of Reactive Oxygen Species
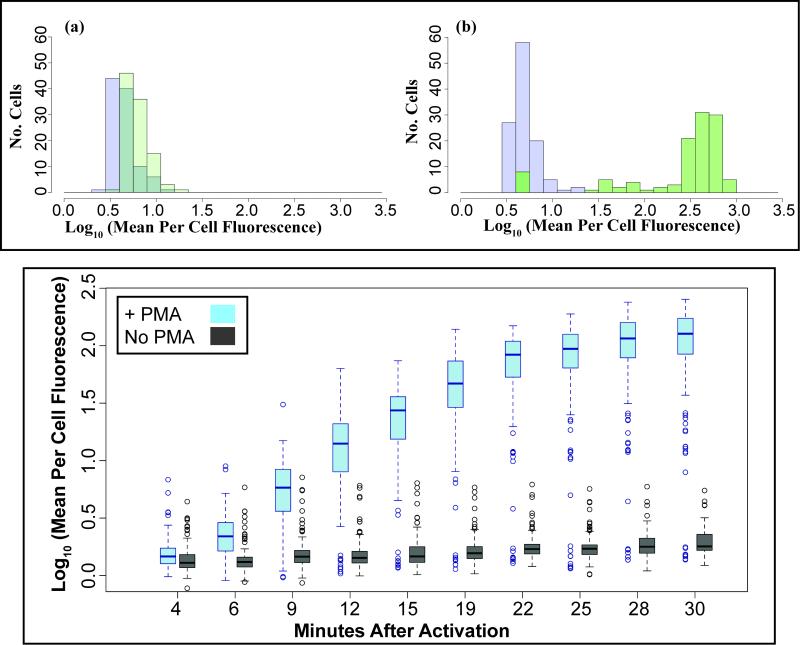
The microfluidic device utilizes the superoxide activated fluorescent dye, DHR to quantify ROS production. The optimal DHR concentration was experimentally determined to be 10 μM for distinguishing PMA-activated neutrophils from non-activated neutrophils (data not shown). Additional experiments were completed to analyze PMA activated neutrophils stained with 10 μM DHR. The microfluidic device enabled the analysis of ROS production by each cell over time (Figure 3). There is an increase in the mean log fluorescence per cell after the addition of PMA (Figure 3, B) compared to non-activated control cells (Figure 3, A). The dynamic analysis indicated plateauing of the fluorescence read-out corresponding to ROS production approximately twenty minutes post PMA activation (Figure 3, C).
Figure 3. Dynamic measurement of neutrophil ROS production detected by DHR.
A-B, Histogram of log transformed dihydrorhodamine-123 (DHR) fluorescence of (A) non-activated neutrophils and (B) PMA-activated neutrophils at four minutes (purple) and 30 minutes (green) after experimental setup. C, Box-plot figure of log transformed fluorescence of non-activated and PMA-activated neutrophils over-time indicating plateauing of fluorescence at 24 minutes post PMA activation. All data was repeated in duplicate or in triplicate at least three times.
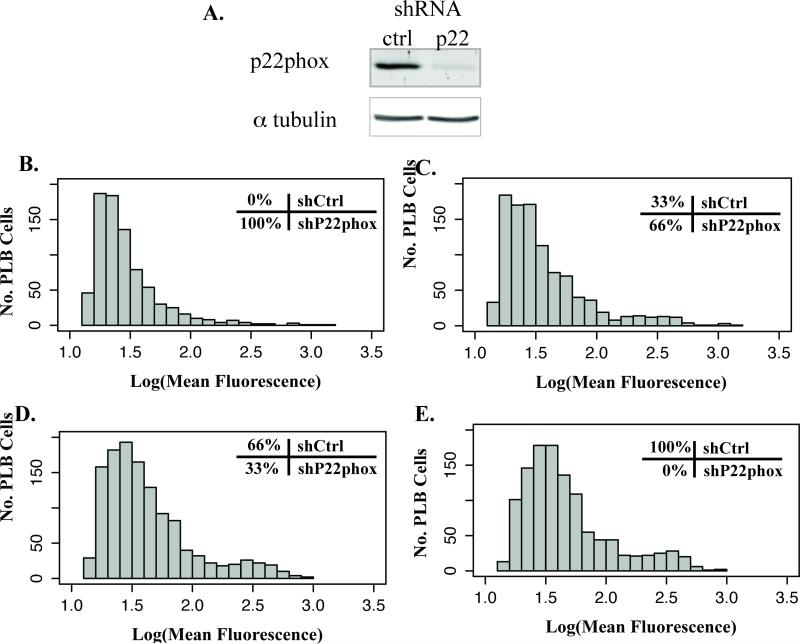
Furthermore, the microfluidic device enabled identification of sub-populations of ROS-producing activated neutrophils, which can be important to the pathophysiology of disease. For example, individuals who are carriers of chronic granulomatous disease (CGD) possess a subpopulation of neutrophils that have limited NADPH-oxidase activity.34 To better characterize the ability of the device to distinguish sub-populations of activated neutrophils, the immature myeloid cell line PLB-985 was used as a model to study deficient ROS production. In these cells, P22phox, a component of the NADPH oxidase complex, was knocked down using short hairpin RNA. These P22phox-deficient cells were then mixed in varying ratios with control PLB cells (Figure 4). Comparisons of these mixtures indicated the presence of a sub-population of control cells.
Figure 4. Comparison of ROS in heterogeneous cell line population.
The enzyme NADPH oxidase was knockdown in the myeloid cell line PLB-985 (shP22phox) and mixed with wild-type PLB-985 (shCtrl) in varying ratios. (A) Western blot analysis of PLB-985 lysate indicating successful p22phox knockdown. Histogram of log-transformed fluorescence of these population mixtures activated for 30 minutes with PMA 1μM indicates an increase in median fluorescence with increasing ratios of wild-type PLB-985. (B) shP22phox only, (C) 66% shP22phox and 33% shCtrl, (D) 33% shP22phox and 66% shCtrl, (E) shCtrl only.
PMA Dose Dependent Production of ROS and NETosis
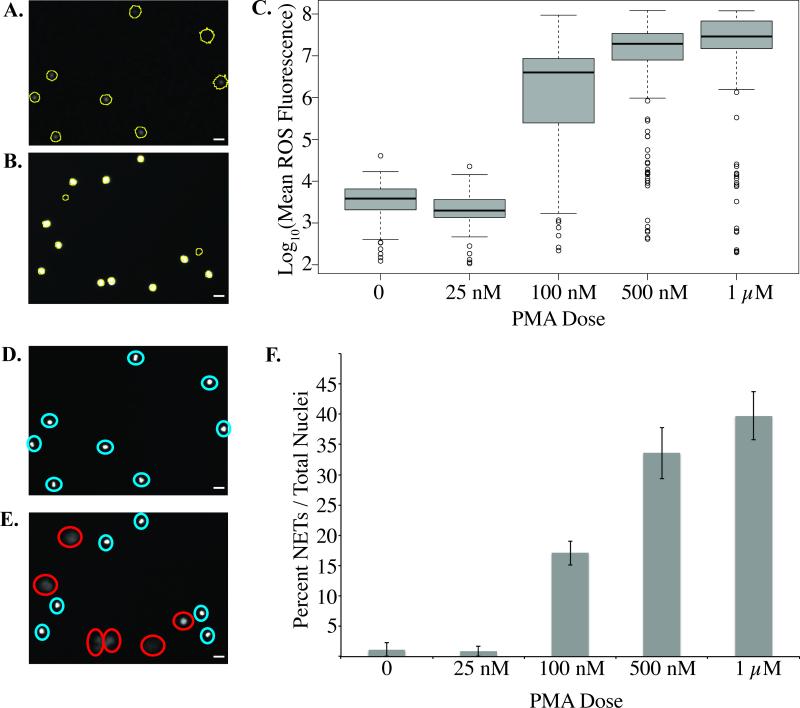
To determine the optimal PMA dose for activating ROS production and NETosis in neutrophils, we tested different PMA concentrations ranging from 25 nM up to 1 μM (Figure 5 and Suppl. Figure 2). A significant increase in the production of ROS and the formation of NETs was noted with increasing concentrations of PMA (Figure 5). There are obvious differences in neutrophils in channels without PMA compared to cells in channels with PMA, both in terms of ROS production as measured by DHR (Figure 5, A and B), and in terms of NET release as measured by the diffusion of Hoechst stain (Figure 5, D and E). Both the PBS and the 25 nM PMA samples showed little to no ROS or NET production, suggesting a threshold value for inducing ROS and NET formation within the devices (Figure 5, C and F). Optimal ROS production was achieved with a PMA concentration of 1 μM (Figure 5, C), and optimal NETosis was reached with a PMA concentration of 1 μm (Figure 5, F). Therefore, a PMA concentration of 1 μM was used in subsequent experiments in order to analyze ROS and NET production.
Figure 5. Dose-dependent PMA activation of neutrophils.
A-B, Representative images of dihydrorhodamine-123 (DHR) fluorescent image of (A) non-activated and (B) PMA-activated neutrophils at 30 minutes. Cells are circled in yellow. C, Box-plot of neutrophil reactive oxygen species (ROS) production at 30 minutes with varying PMA dosing. D-E, Representative images of Hoechst nuclear stain fluorescent image of (D) non-activated and (E) PMA-activated neutrophils. Intact nuclei are labeled by a blue circle and NETs are labeled by a red circle. Please note the fuzzy NET structures in comparison to the sharply stained intact neutrophils. F, Percent NETs formed in 3 hours with varying doses of PMA relative to total number of visualized nuclei. There is a significant increase in NETs with increasing concentrations of PMA as determined by one-way ANOVA (p-value < 0.05). All data was repeated in duplicate or in triplicate at least three times. White bars indicates 10 μm.
Diphenyleneidonium (DPI) Inhibits ROS and NET Formation of Captured Neutrophils
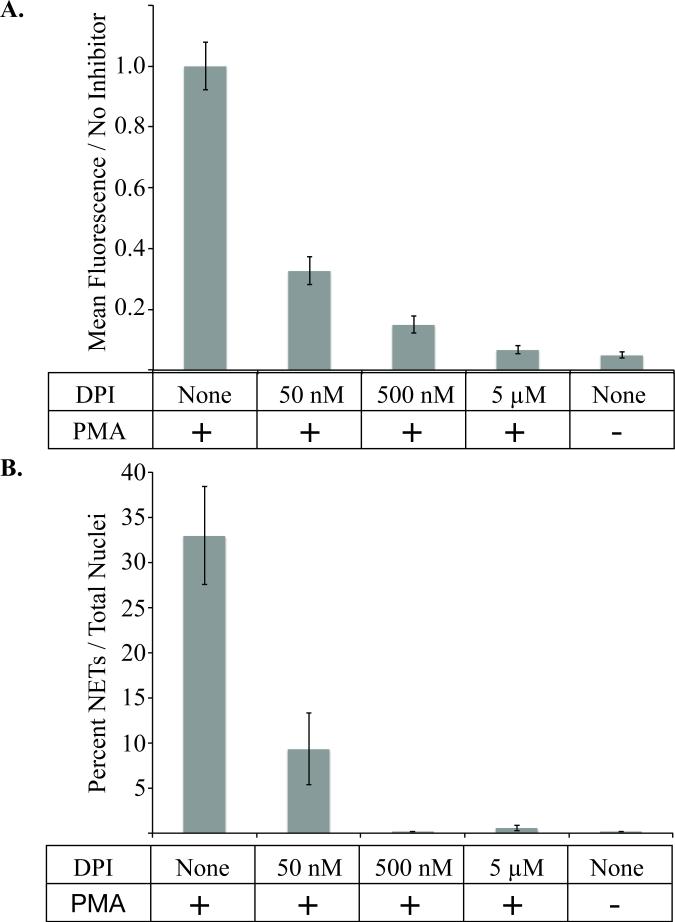
The microfluidic NET device was further validated using diphenyleneidonium chloride (DPI) to inhibit the reactive oxygen species-producing enzyme NADPH-oxidase (Figure 6). There was an observed dose-dependent DPI inhibition of ROS production (Figure 6, A) and NET formation (Figure 6, B) in neutrophils activated with 1 μM PMA. The correlation between ROS and NET inhibition with DPI suggests that ROS is essential for NET production following PMA stimulation in our microfluidic device.
Figure 6. Diphenyleineideom (DPI) inhibits PMA induced neutrophil activation.
A, The mean fluorescence intensity of cells treated with 1μM PMA for 30 minutes with or without the addition of varying concentrations of DPI inhibitor. Fluorescence is measured by dihydrorhodamine-123 (DHR) signal relative to positive control cells activated with 1μM of PMA without inhibitor. There is a statistically significant decrease in formation of reactive oxygen species measured by DHR signal as determined by oneway ANOVA (p-value < 0.001). B, The percent of NETs produced in 3 hours after activation with 1μM PMA in the presence or absence of the indicated concentration of DPI. All data was repeated in duplicate or in triplicate at least three times.
Pseudomonas aeruginosa Induces Neutrophil Extracellular Traps within the Microfluidic Device
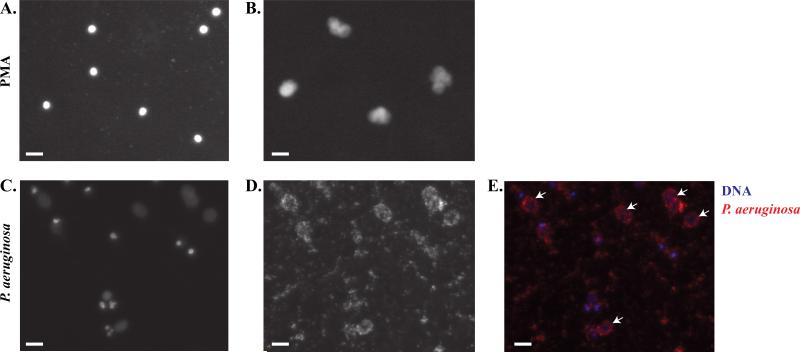
Various pathogens, including P. aeruginosa, are known to induce NET formation.35, 36 To mimic biologic conditions, neutrophils isolated using the microfluidic device were incubated with P. aeruginosa expressing the mCherry fluorescent peptide (Figure 7, C-E). Channels with neutrophils stimulated with or without PMA on the same device acted as controls to measure NETosis (Figure 7, A and B). In comparison to cells exposed to PMA that induced NET formation after 2 hours, the labeled bacteria were observed to induce NETs after 8 hours of incubation (Figure 7, C). Furthermore, P. aeruginosa localized to the NETs (Figure 7, E) and neutrophils were observed to migrate in the presence of P. aeruginosa (Suppl. Video 1).
Figure 7. Pseudomonas aeruginosa induced NETosis.
A-B, Representative images of NETosis with Hoechst staining of (A) untreated neutrophils and (B) 1μM PMA treated neutrophils after 3 hour incubations. C-E, Representative images of NETs produced following stimulation with overnight cultures of P. aeruginosa for 8 hours. (C) Hoechst staining of neutrophils cultured with P. aeruginosa. (D) mCherry labeled P. aeruginosa added to neutrophils. (E) Overlay of blue Hoechst staining with red mCherry labeled P. aeruginosa. Arrows indicate localization of bacteria to NETs. All data was repeated in duplicate or in triplicate at least three times. White bar indicates 10 μm.
NETosis Measurements From Collected Blood
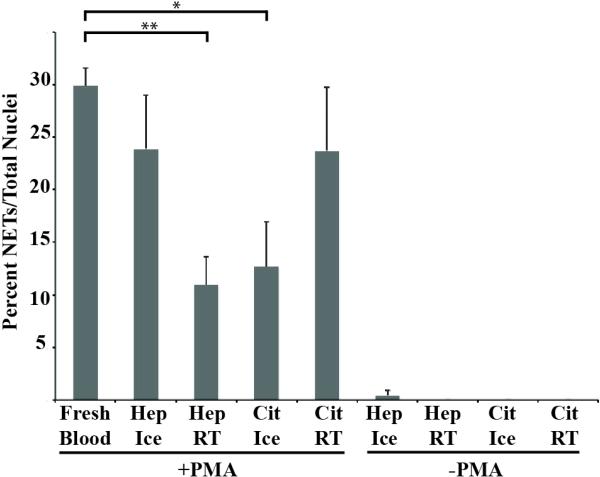
The collaborative nature of research requires analysis of samples that are transported from distant locations. This is especially important while studying rare human diseases, where blood is collected at satellite sites and transported to central laboratories. To test and optimize our methods for stored blood, we quantified NETs produced in response to PMA by neutrophils after storing blood for 8 hours at room temperature or 4° Celsius within Vacutainer® tubes containing sodium heparin or sodium citrate. Percent NETs formed from anti-coagulated stored blood and fresh blood were compared using one-away ANOVA and Tukey's post hoc test. There was a decrease in the percent NETs formed in citrate anti-coagulated blood stored at 4° Celsius in comparison to citrate anticoagulated blood stored at room temperature, but this difference was not statistically significant as determined by ANOVA and Tukey's post hoc test. Similar quantities of NETs were formed by neutrophils from heparinized blood stored on ice and citrate anticoagulated blood stored at room temperature. Heparinized blood stored on ice had higher NETs formed in comparison to heparinized blood stored at room temperature and citrate anti-coagulated blood stored on ice, but this difference was not statistically significant. There was a statistically significant difference in percent NETs formed between fresh blood and heparinized blood stored at room temperature and citrated blood stored on ice (Figure 8). We also counted the number of captured neutrophils and found that there were significantly fewer neutrophils captured when the samples were stored at room temperature compared to on ice in the presence of either anticoagulant (data not shown). Accounting for both percent NETs and the number of captured neutrophils, we determined heparinized blood maintained at 4°C to be the ideal condition.
Figure 8. NETosis from stored blood.
Percent NET formation from blood stored for 8 hours at room temperate (RT) or 4°C Celsius (Ice) within Vacutainer® tubes containing sodium heparin (Hep) or sodium citrate (Cit) (*: p-value < 0.05, **: p-value < 0.01 by one-way ANOVA and Tukey's post hoc test).
Discussion
In this study we report a simple microfluidic device for quantification and analysis of NET formation. With this device, we can isolate neutrophils from a drop of whole blood, simultaneously identify NETs and quantify neutrophil production of ROS in a single cell using Hoechst and DHR fluorescent dyes, and can potentially track neutrophil migration. Immunocytochemistry confirmed the capture of neutrophils in the microfluidic device with P-selectin coating, and the absence of neutrophils in channels coated with BSA alone further supporting the requirement of P-selectin for neutrophil enrichment. This microfluidic device represents a powerful tool to analyze NETosis within a single cell.
We induced NETosis in the microfluidic device using PMA, a protein kinase C (PKC) activating molecule.37 After testing different concentrations of PMA, we determined 1 μM of PMA to be an optimal concentration to stimulate NETosis in our device. We also induced ROS production with this concentration of PMA, with maximal ROS production reached in approximately 24 minutes after PMA addition. This PMA dose is significantly higher than previously reported PMA concentrations used for inducing NET formation.3 Possible explanations for this high dose could include absorption of PMA by the polydimethylsiloxane (PDMS) microfluidic channel. Our laboratory has previously reported absorption of hydrophobic molecules, including estrogen, by PDMS.38 Therefore it is possible that PMA, a small hydrophobic molecule could be absorbed by the PDMS. Nonetheless, our results support previous findings that PMA stimulates both ROS and NET production in cells,7, 18, 39 demonstrating the usefulness of the device in analyzing ROS production and NETosis.
Our results demonstrate that PMA stimulation was required to induce NETosis within our microfluidic channels. Activated endothelial cells and platelets are reported to induce NET formation. Gupta et al. reported that neutrophils form NETs when co-cultured with endothelial cells activated by tumor necrosis factor alpha (TNF-α) or PMA.40 In addition, Caudrillier et al. showed that platelets can induce NETosis when activated by thrombin receptor-activating peptide.41 When activated, both endothelial cells and platelets express P-selectin on the cells surface. However, P-selectin coating alone in our device did not induce NETosis. Rather, PMA activation was required to induce NETs within these channels.
An additional advantage of the reported approach for quantification and identification of NETs is the ability to visualize changes in chromatin structure over-time. Analyzing these changes will be important for understanding the dynamic process of NETosis. By using a single nucleic acid dye, we were able to visualize chromatin decondensation and the loss of the multi-lobed chromatin. Additionally, by observing the diffusion of Hoechst staining over-time, we were able to identify the loss of nuclear membrane integrity and of cytoplasmic membrane perforation. In concurrence with other papers, we found that initial changes in chromatin structure were observed approximately 1 hour after induction and NETs were released into the extracellular space approximately after 75 to 90 minutes of incubation.7, 17 In addition to the ROS measurements, visualizing the nuclear decondensation process and loss of membrane integrity will enable better understanding of the timeline and steps of NETosis.
After we demonstrated that PMA could induce NET production, we tested a biologically relevant stimulant, P. aeruginosa, in our device. While we were able to generate NETs in the channels, it took 8 hours before we saw NET release from the cells. Although the timing of NET production after PMA activation is similar in our channels compared to previous reports,7 the timeline of NET release after P. aeruginosa is much longer in our devices than in other studies (8 hours compared to 2-3 hours).36, 42 This discrepancy might be due to the fact that in previous reports, P. aeruginosa was centrifuged onto neutrophils so that the two were in contact. In our channels, however, the bacteria are not spun down. Therefore, it might take longer for the neutrophils in our microfluidic devices to come into contact with the bacteria and to become activated, delaying NET release. Regardless, not only can soluble molecules activate NETosis in our device, but live, whole microbes can also activate NET production in our device. Additional experiments in which NETs are formed and P. aeruginosa are placed within the channel would serve to assess the interaction between formed NETs and bacteria. However, the fragility of formed NETs within the microfluidic device limits any additional fluid manipulation including the placement of suspended bacterial culture.
During initial diagnosis of a patient, reliable and rapid tests that use minimal amounts of patient tissue are ideal, and our device fulfills these requirements. Our device can quantify the number of cells able to create ROS and the relative amount of ROS produced in each cell in just 30 minutes. Additionally, the microfluidic device allows for live imaging of the cells, providing visualization for any unusual characteristics or delays during ROS production. Furthermore, depending on the stimulant, if the cells are allowed to incubate for an additional 2-3 hours, NET production can be monitored and quantified. With multiple autoimmune diseases linked to deregulation or NETosis,11, 12 this device could be useful in screening for a number of autoimmune diseases. Already, we have tested the efficacy of common storage methods of patient whole blood in producing NETs in our device, finding that samples can be reliably stored for 8 hours on ice.
This paper reports a technique for the quantification of the formation of neutrophil extracellular traps and the production of reactive oxygen species from neutrophils captured out of a drop of blood. The technique enables the analysis of the different stages of NETosis based on the changing features of the neutrophil chromatin. Furthermore, the technique was used to determine the timeline between these different steps. We have shown that this device is an effective tool that will allow investigators to better understand the signaling cascades involved in NETosis, which stimulants induce specific pathways over others, if certain neutrophil subtypes have propensities for NETosis, and how mutations affect NETosis, ultimately resulting in the development of disease.
Supplementary Material
Insight, innovation, integration.
The biological insight is, formation of neutrophil extracellular traps in the presence of surface coated P-selectin requires additional stimulants including live bacteria or activators of protein kinase C. And extracellular traps can be formed in the presence of heparin from blood stored for up to 8 hours. The innovation is the application of a microfluidic device to capture neutrophils, analyze per cell changes in chromatin structure and quantify per cell production of reactive oxygen species over time. The integration of this device will streamline neutrophil function tests, and will help elucidate the role of neutrophil extracellular traps in autoimmune disorders and infection through time-dependent cellular level analysis.
Acknowledgments
This work was supported by University of Wisconsin Carbone Cancer Center Support Grant P30 CA014520 and NIH R01EB010039 BRG-Understanding cell migration through microscale in vitro models. KMM was supported by NIH training grant # T32GM007215. FMH was supported by the Clinical and Translational Science Award (CTSA) program (NIH grant UL1TR000427) and University of Wisconsin Medical Scientist Training Program training grant (NIH grant GM008692). TWS received predoctoral funding support from an NIH, National Cancer Institute F30 fellowship (NIH grant HL114143), the University of Wisconsin Hematology training grant (NIH grant HL007899), and the University of Wisconsin Medical Scientist Training Program training grant (NIH grant GM008692). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. D.J.B. and D.J.G. hold equity in Salus Discovery LLC which has licensed technology described in this article. D.J.B. and D.J.G. hold equity in Tasso, Inc. D.J.B. holds equity in Bellbrook Labs, LLC. The authors would like to thank Dr. Erwin Berthier who provided specific guidance during the project. The authors would like to thank Ms. Alice Puchalski for her assistance with manuscript edits.
References
- 1.Kolaczkowska E, Kubes P. Nature reviews. Immunology. 2013;13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 2.Mayadas TN, Cullere X, Lowell CA. Annual review of pathology. 2014;9:181–218. doi: 10.1146/annurev-pathol-020712-164023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 4.Douda DN, Jackson R, Grasemann H, Palaniyar N. J Immunol. 2011;187:1856–1865. doi: 10.4049/jimmunol.1004201. [DOI] [PubMed] [Google Scholar]
- 5.Urban CF, Reichard U, Brinkmann V, Zychlinsky A. Cell Microbiol. 2006;8:668–676. doi: 10.1111/j.1462-5822.2005.00659.x. [DOI] [PubMed] [Google Scholar]
- 6.Ramos-Kichik V, Mondragon-Flores R, Mondragon-Castelan M, Gonzalez- Pozos S, Muniz-Hernandez S, Rojas-Espinosa O, Chacon-Salinas R, Estrada- Parra S, Estrada-Garcia I. Tuberculosis (Edinb) 2009;89:29–37. doi: 10.1016/j.tube.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, Weinrauch Y, Brinkmann V, Zychlinsky A. J Cell Biol. 2007;176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen K, Nishi H, Travers R, Tsuboi N, Martinod K, Wagner DD, Stan R, Croce K, Mayadas TN. Blood. 2012;120:4421–4431. doi: 10.1182/blood-2011-12-401133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrd AS, O'Brien XM, Johnson CM, Lavigne LM, Reichner JS. J Immunol. 2013;190:4136–4148. doi: 10.4049/jimmunol.1202671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianchi M, Niemiec MJ, Siler U, Urban CF, Reichenbach J. J Allergy Clin Immunol. 2011;127:1243–1252. e1247. doi: 10.1016/j.jaci.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 11.Kaplan MJ, Radic M. J Immunol. 2012;189:2689–2695. doi: 10.4049/jimmunol.1201719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper PR, Palmer LJ, Chapple IL. Periodontology. 2000;2013:63, 165–197. doi: 10.1111/prd.12025. [DOI] [PubMed] [Google Scholar]
- 13.Tsuda Y, Takahashi H, Kobayashi M, Hanafusa T, Herndon DN, Suzuki F. Immunity. 2004;21:215–226. doi: 10.1016/j.immuni.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Pillay J, Ramakers BP, Kamp VM, Loi AL, Lam SW, Hietbrink F, Leenen LP, Tool AT, Pickkers P, Koenderman L. J Leukoc Biol. 2010;88:211–220. doi: 10.1189/jlb.1209793. [DOI] [PubMed] [Google Scholar]
- 15.Yousefi S, Mihalache C, Kozlowski E, Schmid I, Simon HU. Cell Death Differ. 2009;16:1438–1444. doi: 10.1038/cdd.2009.96. [DOI] [PubMed] [Google Scholar]
- 16.McIlroy DJ, Jarnicki AG, Au GG, Lott N, Smith DW, Hansbro PM, Balogh ZJ. Journal of critical care. 2014;29:1133, e1131–1135. doi: 10.1016/j.jcrc.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 17.Remijsen Q, Vanden Berghe T, Wirawan E, Asselbergh B, Parthoens E, De Rycke R, Noppen S, Delforge M, Willems J, Vandenabeele P. Cell research. 2011;21:290–304. doi: 10.1038/cr.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keshari RS, Verma A, Barthwal MK, Dikshit M. J Cell Biochem. 2013;114:532–540. doi: 10.1002/jcb.24391. [DOI] [PubMed] [Google Scholar]
- 19.Brinkmann V, Laube B, Abu Abed U, Goosmann C, Zychlinsky A. J Vis Exp. 2010 doi: 10.3791/1724. DOI: 10.3791/1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta AK, Giaglis S, Hasler P, Hahn S. PLoS One. 2014;9:e97088. doi: 10.1371/journal.pone.0097088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sackmann EK, Berthier E, Young EW, Shelef MA, Wernimont SA, Huttenlocher A, Beebe DJ. Blood. 2012;120:e45–53. doi: 10.1182/blood-2012-03-416453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D, Haynes CL. Anal Chem. 2013;85:10787–10796. doi: 10.1021/ac4020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, Newbold MA, Haynes CL. Analyst. 2015;140:5055–5064. doi: 10.1039/c5an00967g. [DOI] [PubMed] [Google Scholar]
- 24.Butler KL, Ambravaneswaran V, Agrawal N, Bilodeau M, Toner M, Tompkins RG, Fagan S, Irimia D. PLoS One. 2010;5:e11921. doi: 10.1371/journal.pone.0011921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sackmann EK, Berthier E, Schwantes EA, Fichtinger PS, Evans MD, Dziadzio LL, Huttenlocher A, Mathur SK, Beebe DJ. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1324043111. DOI: 10.1073/pnas.1324043111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shelef MA, Sokolove J, Lahey LJ, Wagner CA, Sackmann EK, Warner TF, Wang Y, Beebe DJ, Robinson WH, Huttenlocher A. Arthritis & rheumatology. 2014;66:1482–1491. doi: 10.1002/art.38393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jo BH, Van Lerberghe LM, Motsegood KM, Beebe DJ. J Microelectromech S. 2000;9:76–81. [Google Scholar]
- 28.Sung KE, Su G, Pehlke C, Trier SM, Eliceiri KW, Keely PJ, Friedl A, Beebe DJ. Biomaterials. 2009;30:4833–4841. doi: 10.1016/j.biomaterials.2009.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berthier E, Surfus J, Verbsky J, Huttenlocher A, Beebe D. Integr Biol (Camb) 2010;2:630–638. doi: 10.1039/c0ib00030b. [DOI] [PubMed] [Google Scholar]
- 30.Brannon MK, Davis JM, Mathias JR, Hall CJ, Emerson JC, Crosier PS, Huttenlocher A, Ramakrishnan L, Moskowitz SM. Cell Microbiol. 2009;11:755–768. doi: 10.1111/j.1462-5822.2009.01288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavnar PJ, Berthier E, Beebe DJ, Huttenlocher A. J Cell Biol. 2011;193:465–473. doi: 10.1083/jcb.201010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starnes TW, Cortesio CL, Huttenlocher A. Methods Mol Biol. 2011;769:111–136. doi: 10.1007/978-1-61779-207-6_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker GM, Beebe DJ. Lab Chip. 2002;2:131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- 34.Segal BH, Leto TL, Gallin JI, Malech HL, Holland SM. Medicine (Baltimore) 2000;79:170–200. doi: 10.1097/00005792-200005000-00004. [DOI] [PubMed] [Google Scholar]
- 35.Guimaraes-Costa AB, Nascimento MT, Wardini AB, Pinto-da-Silva LH, Saraiva EM. Journal of parasitology research. 2012;2012:929743. doi: 10.1155/2012/929743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young RL, Malcolm KC, Kret JE, Caceres SM, Poch KR, Nichols DP, Taylor-Cousar JL, Saavedra MT, Randell SH, Vasil ML, Burns JL, Moskowitz SM, Nick JA. PLoS One. 2011;6:e23637. doi: 10.1371/journal.pone.0023637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, Koebis M, Suo S, Ohno S, Ishiura S. Biochem Biophys Res Commun. 2012;423:212–217. doi: 10.1016/j.bbrc.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 38.Regehr KJ, Domenech M, Koepsel JT, Carver KC, Ellison-Zelski SJ, Murphy WL, Schuler LA, Alarid ET, Beebe DJ. Lab Chip. 2009;9:2132–2139. doi: 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirchner T, Moller S, Klinger M, Solbach W, Laskay T, Behnen M. Mediators of inflammation. 2012;2012:849136. doi: 10.1155/2012/849136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gupta AK, Joshi MB, Philippova M, Erne P, Hasler P, Hahn S, Resink TJ. FEBS Lett. 2010;584:3193–3197. doi: 10.1016/j.febslet.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Caudrillier A, Kessenbrock K, Gilliss BM, Nguyen JX, Marques MB, Monestier M, Toy P, Werb Z, Looney MR. The Journal of clinical investigation. 2012;122:2661–2671. doi: 10.1172/JCI61303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halverson TW, Wilton M, Poon KK, Petri B, Lewenza S. PLoS pathogens. 2015;11:e1004593. doi: 10.1371/journal.ppat.1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.