Significance
RNase L, an antiviral enzyme activated during infection, degrades viral and cellular RNAs, inhibits protein synthesis, and restricts the replication and spread of diverse viruses. RNase L activation depends on 2′,5′-oligoadenylates synthesized by different oligoadenylate synthetases (OASs), i.e., OAS1, OAS2, and OAS3. OASs are induced by interferon and are activated by viral dsRNA. It has been unclear which of these OAS proteins is necessary and/or sufficient to activate RNase L during viral infections. We show that OAS3, but not OAS1 or OAS2, is required to activate RNase L and to restrict the replication of four different human viruses. These findings suggest that OAS3 may provide a target for antiviral therapies and that OAS1 and OAS2 may have alternative roles.
Keywords: ribonuclease L, oligoadenylate synthetase 3, antiviral response, type I interferon, 2-5A
Abstract
The 2′,5′-oligoadenylate (2-5A) synthetase (OAS)–RNase L system is an IFN-induced antiviral pathway. RNase L activity depends on 2-5A, synthesized by OAS. Although all three enzymatically active OAS proteins in humans—OAS1, OAS2, and OAS3—synthesize 2-5A upon binding dsRNA, it is unclear which are responsible for RNase L activation during viral infection. We used clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated protein-9 nuclease (Cas9) technology to engineer human A549-derived cell lines in which each of the OAS genes or RNase L is knocked out. Upon transfection with poly(rI):poly(rC), a synthetic surrogate for viral dsRNA, or infection with each of four viruses from different groups (West Nile virus, Sindbis virus, influenza virus, or vaccinia virus), OAS1-KO and OAS2-KO cells synthesized amounts of 2-5A similar to those synthesized in parental wild-type cells, causing RNase L activation as assessed by rRNA degradation. In contrast, OAS3-KO cells synthesized minimal 2-5A, and rRNA remained intact, similar to infected RNase L-KO cells. All four viruses replicated to higher titers in OAS3-KO or RNase L-KO A549 cells than in parental, OAS1-KO, or OAS2-KO cells, demonstrating the antiviral effects of OAS3. OAS3 displayed a higher affinity for dsRNA in intact cells than either OAS1 or OAS2, consistent with its dominant role in RNase L activation. Finally, the requirement for OAS3 as the major OAS isoform responsible for RNase L activation was not restricted to A549 cells, because OAS3-KO cells derived from two other human cell lines also were deficient in RNase L activation.
Critically important to understanding antiviral innate immunity is determining which host proteins are responsible for inhibiting different types of viruses. However, there are significant gaps in our knowledge about the specificity of many host antiviral proteins. The 2′,5′-oligoadenylate (2-5A) synthetase (OAS)–RNase L system (reviewed in ref. 1) is a case in point. OASs are pattern-recognition receptors for viral dsRNA, a common pathogen-associated molecular pattern for many types of RNA and DNA viruses. In humans, there are four OAS genes, all stimulated by IFN, but only three of these encode catalytically active proteins. OAS1, OAS2, and OAS3 contain one, two, and three core OAS units, respectively, but all three enzymes synthesize 2-5A from ATP upon binding dsRNA (2). OASL, containing one basic unit plus two ubiquitin-like domains, does not synthesize 2-5A but instead activates RIG-I signaling in response to dsRNA (3). In addition, OASs are structurally homologous to cGAS, a sensor of cytoplasmic DNA, often of microbial origin, that produces 2′,5′-cGMP-AMP activators of STING leading to type I IFN production (4).
The only well-established function of 2-5A is to activate RNase L, causing endonucleolytic cleavage of viral and cellular ssRNAs, thereby blocking viral replication. Many viruses encode antagonists of the OAS–RNase L pathway, providing evidence that RNase L is a potent antiviral protein (1, 5, 6). However, far less clear is which of the catalytically active OAS species are responsible for suppressing different types of viral infections in human cells. The main obstacle has been the absence of OAS-KO models, other than for murine Oasl1 (7) and Oasl2 (8). Also, although some genetics studies conclude that polymorphisms in OAS1 are associated with susceptibility to West Nile virus (WNV) (9), prostate cancer (10), diabetes (11), multiple sclerosis (12), and other pathological conditions, there is little, if any, evidence that this susceptibility is mediated through RNase L.
To study the impact of different OAS species on different viruses, we used clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated protein-9 nuclease (Cas9) gene-editing technology, which allows the convenient and efficient disruption of genes in mammalian cells (13, 14). Our results provide the surprising conclusion that, among the catalytically active forms of OAS proteins, OAS3 is mainly responsible for producing 2-5A activators of RNase L during infections by a wide range of different types of human viruses.
Results
Ablation of Different OAS Species Reveals a Role for OAS3 in the Cellular Response to dsRNA.
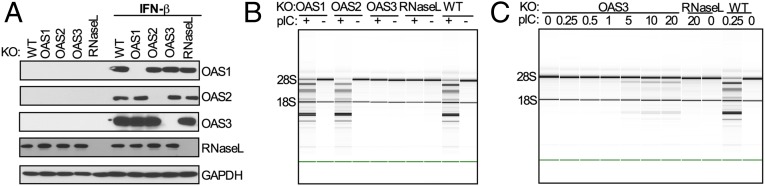
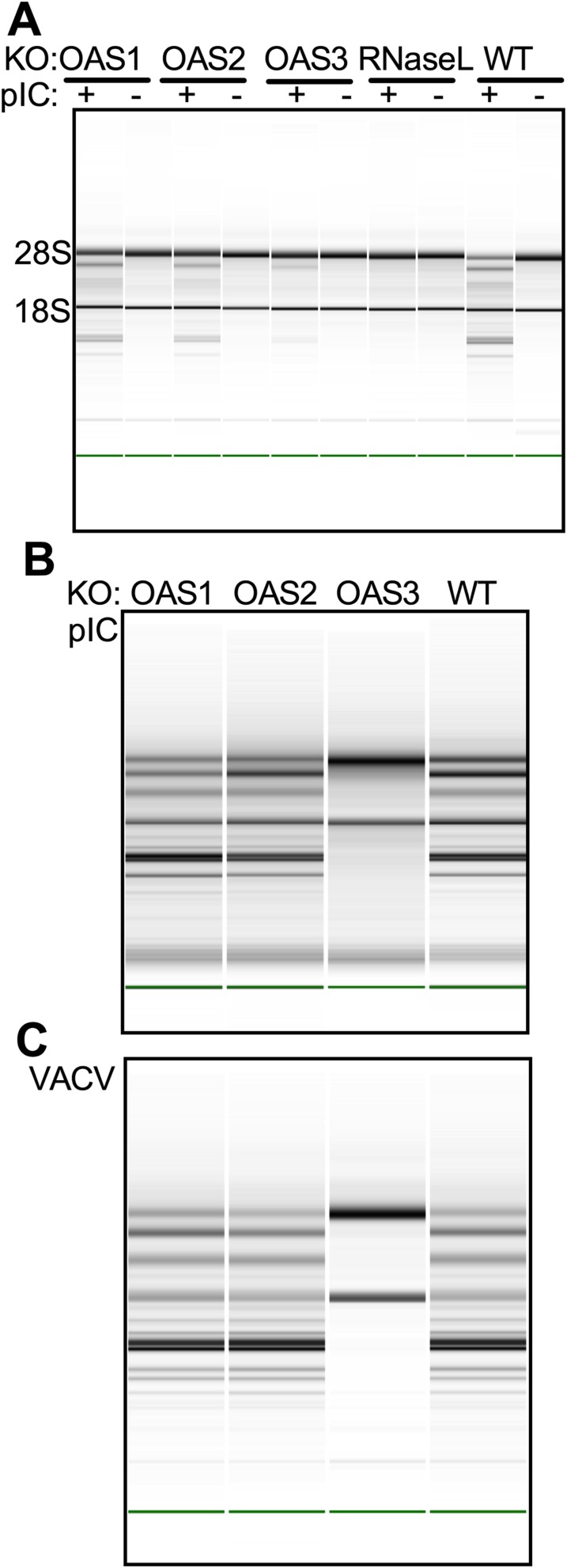
To investigate the relative antiviral activities of different OAS species, we used CRISPR-Cas9 technology to construct human lung carcinoma A549 cell lines individually lacking OAS1, OAS2, OAS3, or RNase L (13, 14). We selected two cell lines for each genotype, verified the interruption of each gene in each cell line by DNA sequencing (Tables S1–S3), and then verified the absence of protein expression by Western blot (Fig. 1A). Because each pair of cell lines had the same phenotypes, we show data for only one. We initially used poly(rI):poly(rC) (pIC) as a surrogate for viral dsRNA to induce activation of the OAS–RNase L pathway by monitoring rRNA degradation as an index of RNase L activation (15, 16). As expected, following the transfection of cells with 250 ng/mL of pIC, rRNA was degraded in parental A549 cells [RNA integrity number (RIN) = 5.8] but remained intact in cells lacking expression of RNase L (RIN = 10) (Fig. 1B). Surprisingly, the 18S and 28S rRNAs also remained intact in pIC-transfected cells lacking OAS3. In contrast rRNA degradation in pIC-transfected OAS1-KO and OAS2-KO cells was similar to that in pIC-transfected WT cells. As a control, cleavage of rRNA was restored by stable expression of Flag-tagged OAS3 in the OAS3-KO cells (Fig. S1 A and B). We next determined the minimal amount of pIC required to induce enough 2-5A for RNase L-mediated degradation of rRNA (Fig. 1C). At very high concentrations (5–20 μg/mL) of pIC, rRNA degradation was barely detectable in OAS3-KO cells (at 20 μg; RIN = 8.1). In contrast, WT cells showed much higher levels of rRNA degradation at pIC concentrations as low as 250 ng/mL (RIN = 5.8). These results indicate that OAS3 is largely responsible for the RNase L-mediated degradation of rRNA in response to pIC, whereas OAS1 and/or OAS2 have only a minimal effect.
Table S1.
Construction of the plasmids for knockout human OAS1, OAS2, OAS3, and RNase L by CRISPR/Cas9
| Gene | Primer | Nucleotide sequence (5′–3′)* | Targeting region |
| OAS1 | sgO1-2 Forward | CACCGGAAAAGTGGTGAGAGGACTG | Exon2 |
| Reverse | AAACCAGTCCTCTCACCACTTTTCC | ||
| sgO1-9 Forward | CACCGCAGGATCAGTTAAATCGCCG | Exon2 | |
| Reverse | AAACCGGCGATTTAACTGATCCTGC | ||
| OAS2 | sgO2-5 Forward | CACCGCAGCTTCTGAGCAGGCACCG | Exon1 |
| Reverse | AAACCGGTGCCTGCTCAGAAGCTGC | ||
| sgO2-9 Forward | CACCGGAAGCTGGGTTGGTTTATCC | Exon1 | |
| Reverse | AAACGGATAAACCAACCCAGCTTCC | ||
| OAS3 | sgO3-1 Forward | CACCGGCGATGCCCGCATCTCACTG | Exon2 |
| Reverse | AAACCAGTGAGATGCGGGCATCGCC | ||
| sgO3-9 Forward | CACCGGAAACGTGAGTCTCAGACCA | Exon2 | |
| Reverse | AAACTGGTCTGAGACTCACGTTTCC | ||
| RNASEL | sgRL-6 Forward | CACCGTTTGAGGCGAAAGACAAAGG | Exon1 |
| Reverse | AAACCCTTTGTCTTTCGCCTCAAAC |
Nucleotides sequences selected from the published database (13). The italicized bold type designates the sgRNA sequences.
Table S3.
OAS1-, OAS2-, OAS3-, and RNase L-KO cells generated by CRISPR/Cas9
| Parental cell line | Gene | sgRNA | Delivery method* | Knockout cell line† | Genotyping‡ |
| A549 | OAS1 | sgO1-2 | Electroporation | A549-O1-2 C1 | 1-nt insertion |
| sgO1-9 | Transfection | A549-O1-9 C1 | 1-nt insertion | ||
| OAS2 | sgO2-5 | Electroporation | A549-O2-5 C2 | ND | |
| sgO2-9 | Transfection | A549-O2-9 C1 | 1-nt insertion | ||
| OAS3 | sgO3-1 | Transfection | A549-O3-1 C1 | 1-nt insertion | |
| sgO3-9 | Transfection | A549-O3-9 C1 | 1-nt insertion | ||
| RNASEL | sgRL-6 | Electroporation | A549-RL-6 C1 | 1-nt insertion | |
| sgRL-6 | Transduction | A549-leRL-6 C1 | 1-nt insertion | ||
| HT1080 | OAS1 | sgO1-9 | Transfection | HT-O1-9 C1 | ND |
| OAS2 | sgO2-9 | Transfection | HT-O2-9 C2 | 7-nt insertion | |
| OAS3 | sgO3-1 | Transfection | HT-O3-1 C3 | 1-nt insertion | |
| RNASEL | sgRL-6 | Transfection | HT-RL-6 C1 | 1-nt deletion | |
| HME | OAS1 | sgO1-2 | Transduction | HME-O1-2 | ND |
| OAS2 | sgO2-9 | Transduction | HME-O2-9 | ND | |
| OAS3 | sgO3-1 | Transduction | HME-O3-1 | ND |
DNA with sgRNA constructs were delivered by transfection or electroporation or by Lenti-CRIPSR transduction.
All knockouts were confirmed by Western blotting.
The DNA fragments were amplified and sequenced and compared with the reference sequences of the genes; ND, not determined.
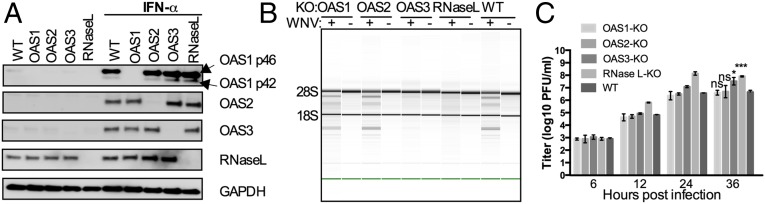
Fig. 1.
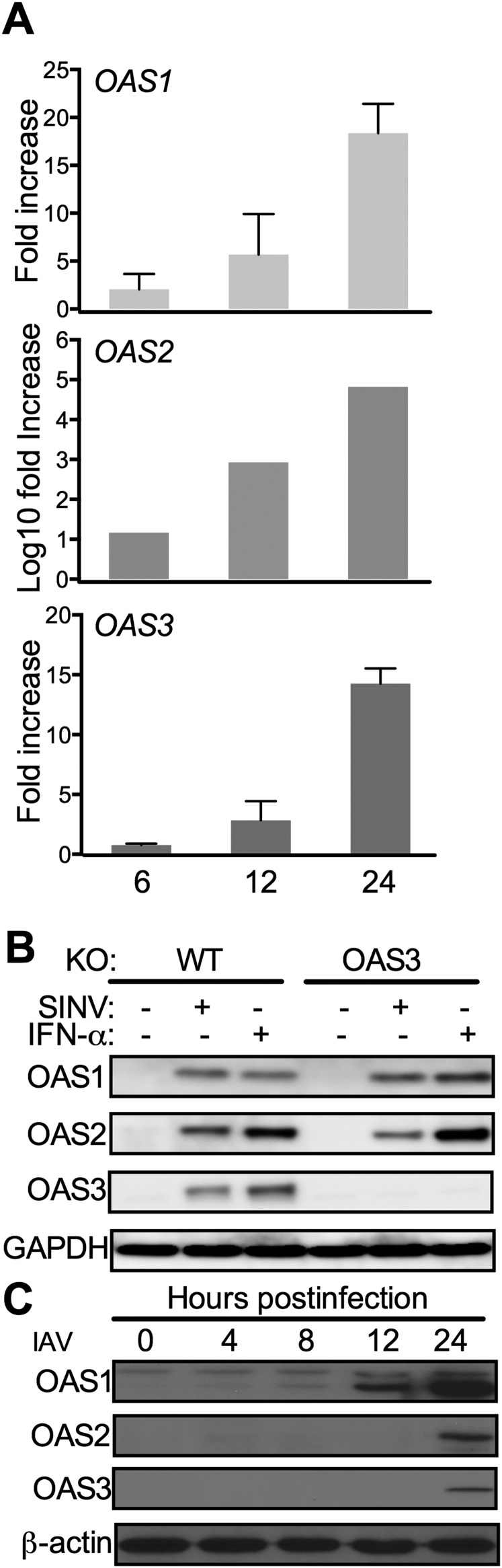
Activation of RNase L by pIC in A549 cells requires OAS3 expression. (A) OAS1-, OAS2-, OAS3-, and RNase L-KO A549 cells were mock-treated or treated with IFN-β (1,000 U/mL) overnight. Cells were lysed, and proteins were analyzed by immunoblotting with antibodies against OAS1, OAS2, OAS3, RNase L, and GAPDH. (B and C) WT and KO cells were transfected with 0.25 μg/mL of pIC (B) or OAS3-KO cells were transfected with increasing doses of pIC (0–20 μg/mL) (C), and RNase L-KO and WT A549 cells were transfected with 20 μg/mL and 0.25 μg/mL of pIC, respectively. At 4 hpi cells were lysed, and RNA integrity was assessed with a Bioanalyzer. The positions of 18S and 28S rRNA are indicated.
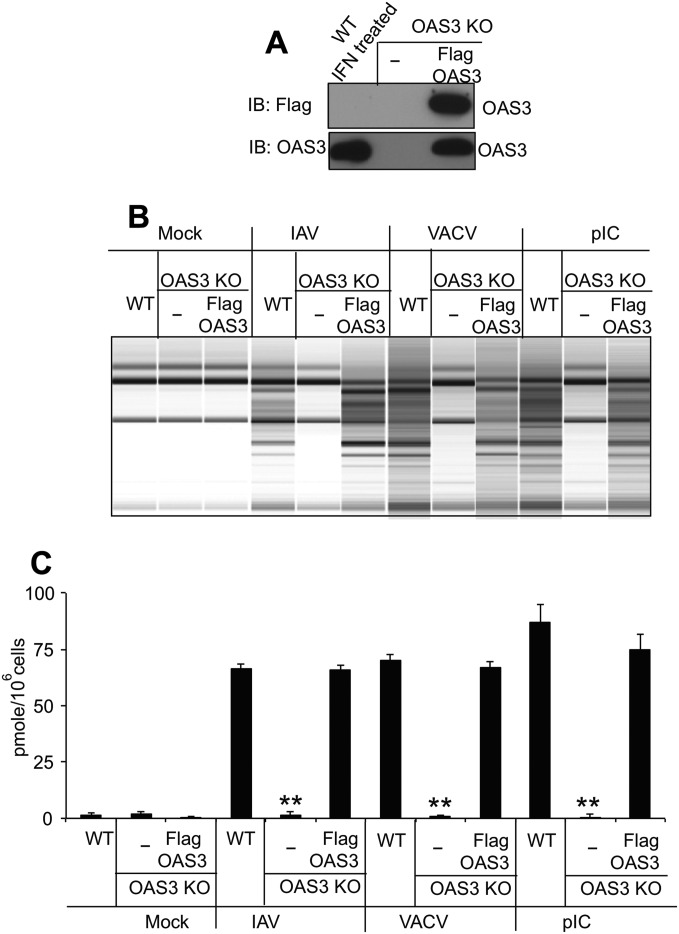
Fig. S1.
Expression of OAS3 protein in OAS3-KO cells restores 2-5A production and RNase L activation. (A) OAS3-KO cells were transfected with p3XFlag-tagged OAS3 cDNA, and a cell line was generated by neomycin selection. Cell lysates were analyzed by electrophoresis followed by immunoblotting with anti-Flag antibody (Upper) and monoclonal antibody against human OAS3 (Lower). (B) OAS3-KO cells and OAS3-KO cells in which OAS3 expression was restored (as indicated) were transfected with pIC (1 μg/mL) or were mock-infected or were infected with IAVΔNS1 (MOI = 10) or VACVΔE3L (MOI = 10). Cells were lysed at 4 (pIC) or 24 (virus) hpi, and total RNA was isolated and monitored for integrity on a Bioanalyzer. (C) OAS3-KO cells and OAS3-KO cells in which OAS3 expression was restored (as indicated) were transfected with pIC (1 μg/mL) or were mock-infected or infected with IAVΔNS1 (MOI = 1) or VACVΔE3L (MOI = 1). Cells were lysed at 4 (pIC) or 24 (virus) hpi, and intracellular levels of 2-5A were determined by FRET assay. **P < 0.0001.
Role for OAS3 in the Activation of RNase L During Infection by Diverse RNA and DNA Viruses.
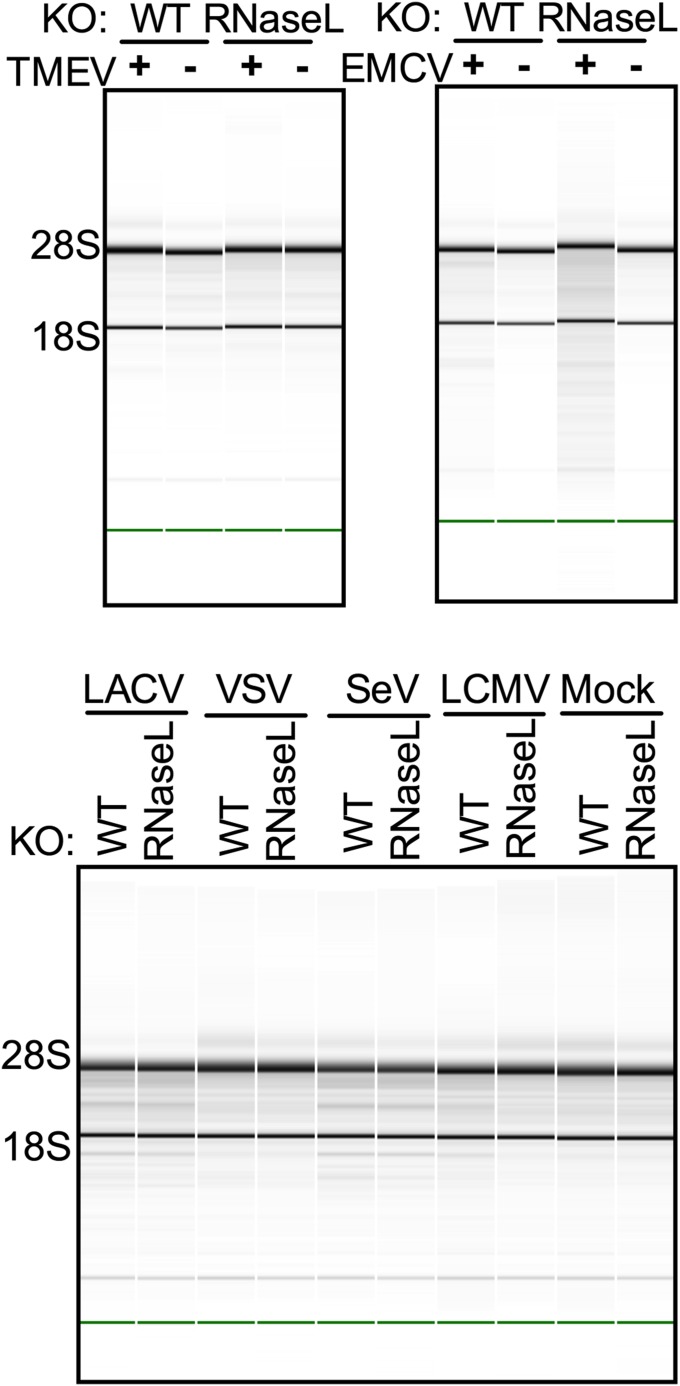
We next investigated which OAS genes are responsible for virus induction of RNase L activity. Initially, we infected both parental A549 and RNase L-KO cells with a variety of viruses representing diverse viral groups. Many viruses encode inhibitors of the OAS–RNase L pathway and do not activate RNase L, at least in some cell types. Among the viruses we tested were the picornaviruses Theiler murine encephalomyocarditis virus (TMEV) and encephalomyocarditis virus (EMCV), the bunyavirus La Crosse virus (LACV), the rhabdovirus vesicular stomatitis virus (VSV), the paramyxovirus Sendai virus (SeV), and the arenavirus lymphocytic choriomeningitis virus (LCMV). All failed to generate detectable levels of RNase L-mediated rRNA cleavage in A549 cells, indicating minimal or no activation of RNase L (Fig. S2). Thus, we were unable to use these viruses to probe the activation of RNase L. However, four other viruses from diverse groups, including three RNA viruses and one DNA virus, were able to activate RNase L in A549 cells and were used for further studies.
Fig. S2.
Viruses that cause minimal or no activation of RNase L as determined by monitoring rRNA integrity in A549 cells. Parental and RNase L-KO cells were infected at MOI = 20. Cells were lysed at 12 (LACV, VSV, SeV, EMCV), 21 (TMEV), or 60 (LCMV) hpi, and RNA integrity was analyzed on a Bioanalyzer. The positions of 18S and 28S rRNA are indicated.
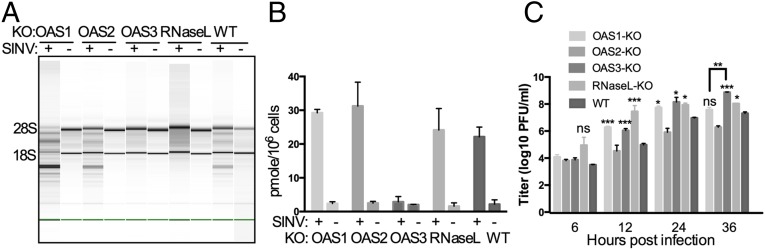
Parental A549 and OAS-KO cells were infected with Sindbis virus (SINV) a human alphavirus with a positive-stranded RNA genome, at a multiplicity of infection (MOI) of 1 pfu per cell, and at 24 h post infection (hpi) were assessed for rRNA degradation (Fig. 2A). Similar to the findings with pIC-induced RNase L activation, rRNA degradation was not observed in OAS3-KO cells or in RNase L-KO cells but was robust in infected cells lacking OAS1 or OAS2 expression and in WT A549 cells. The levels of 2-5A present in infected cells were measured by RNase L activation in an in vitro FRET-based assay (17). During SINV infections 2-5A accumulated to similar levels in OAS1-KO, OAS2-KO, RNase L-KO, and WT cells (Fig. 2B). However, consistent with the rRNA degradation results, 2-5A failed to accumulate in OAS3-KO cells, suggesting minimal OAS activity during infection in cells lacking OAS3 (Fig. 2 A and B). It is important to note that OAS1 and OAS2 mRNA and proteins were induced by SINV infection in OAS3-KO cells (Fig. S3 A and B) but were unable to compensate for loss of OAS3 and produce detectable 2-5A to promote RNA degradation (Fig. 2). Starting at 12 hpi, viral titers in cells lacking OAS1, OAS3, or RNase L were higher than in parental A549 or OAS2-KO cells. Interestingly, at 36 hpi viral titers were 30-fold higher in OAS3-KO cells than in parental A549 cells, and titers in OAS1-KO cells were significantly lower than in OAS3-KO cells (Fig. 2C).
Fig. 2.
Activation of RNase L during SINV infection of A549 cells requires OAS3 expression. (A) WT A549 and KO cells were infected with SINV (MOI = 1 pfu per cell), and at 24 hpi cells were lysed and RNA integrity was assessed. The positions of 18S and 28S rRNA are indicated. (B) Cells were infected with SINV (MOI = 1 pfu per cell), and at 24 hpi intracellular levels of 2-5A were quantified by FRET assay. (C) Cells were infected with SINV (MOI = 1 pfu per cell), and at the indicated time points infectious virus in the supernatant was titered by plaque assay on Vero cells. The data are from three biological replicates and are expressed as means ± SD; *P < 0.05, **P < 0.01, ***P < 0.001.
Fig. S3.
Infections with SINV or IAVΔNS1 induce up-regulation of OAS1, OAS2, and OAS3 gene expression in A549 cells. (A) A549 cells (number of experiments, n = 3) were infected with SINV (MOI = 5). Cells were lysed at 2, 6, 12, and 24 hpi, and RNA was isolated. OAS1, OAS2, and OAS3 mRNAs were quantified by qRT-PCR and expressed as fold-increase over levels at 2 hpi. Data are expressed as mean ± SD. (B) WT or OAS3-KO A549 cells were infected with SINV (MOI = 1) or treated with 1,000 U/mL of IFN-α. Cells were lysed 24 hpi or post IFN treatment, and proteins were analyzed by Western immunoblot with antibodies against OAS1, OAS2, OAS3, and GAPDH. (C) WT A549 cells were infected by IAVΔNS1 (MOI = 1). Cells were lysed at 0, 4, 8, 12, and 24 hpi, and proteins were analyzed by Western immunoblots with antibodies against OAS1, OAS2, OAS3, and β-actin.
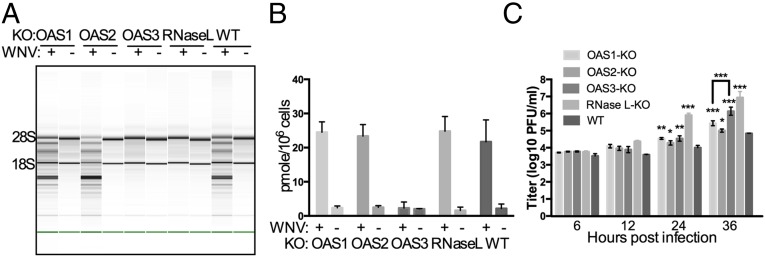
Infections of WT and OAS3-KO cells were carried out with another human positive-stranded RNA virus, a flavivirus, the Kunjin strain of WNV (MOI = 5 pfu per cell), and at 24 hpi cells were assessed for rRNA degradation (Fig. 3A). The results were similar to those obtained with SINV, in that rRNAs were intact in cells lacking OAS3 as well as in RNase L-KO cells at 24 hpi. Consistent with this observation, intracellular 2-5A levels increased in infected WT, OAS1-KO, OAS2-KO, and RNase L-KO cells but not in infected OAS3-KO cells (Fig. 3B). Replication of WNV was assessed at several times post infection in each cell type. At 24 and 36 hpi, viral titers in RNase L-KO cells were about 100-fold higher than in parental (WT) cells, and OAS3-KO cells had titers 20-fold higher than in parental cells at 36 hpi. Titers in OAS1- and OAS2-KO cells at 36 hpi also were higher than in parental cells but were significantly lower than in OAS3-KO cells (Discussion). Thus, a dependence on OAS3 expression for RNase L activation was observed for both WNV and SINV virus.
Fig. 3.
Activation of RNase L during WNV infection of A549 cells requires OAS3 expression. (A) WT A549 and KO cells were infected with WNV (MOI = 5 pfu per cell), and at 24 hpi RNA integrity was assessed. (B) WT A549 and KO cells were infected with WNV (MOI = 5 pfu per cell), and at 24 hpi 2-5A intracellular levels were quantified by FRET assay. (C) Cells were infected with WNV (MOI = 1 pfu per cell), and at the indicated time points infectious virus in the supernatant was titered by plaque assay. The data are from three biological replicates and are expressed as means ± SD; *P < 0.05, **P < 0.01, ***P < 0.001.
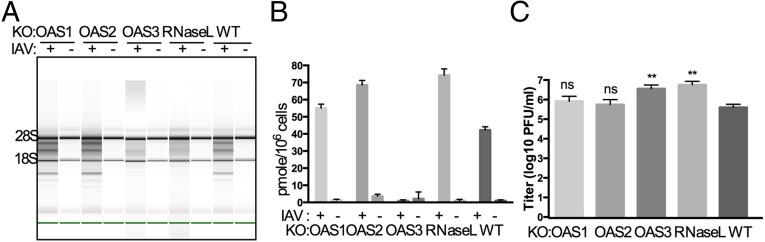
We carried out similar infections with two viruses from different groups, influenza A virus (IAV), a negative-stranded RNA virus with a segmented genome, and vaccinia virus (VACV), a poxvirus with a large DNA genome. WT IAV encodes the NS1 protein, an RNA-binding protein that inhibits the OAS–RNase L pathway (18); thus for these experiments we used an NS1 mutant of IAV (the mouse-adapted H1N1 strain A/PR/8/34), which activates the OAS–RNase L pathway in A549 cells (19, 20). Similar to our observations with WNV and SINV, cleavage of rRNA by RNase L occurred only in IAVΔNS1-infected parental A549, OAS1-KO, and OAS2-KO cells, whereas rRNA remained intact in IAV-infected OAS3-KO and RNase L-KO cells (Fig. 4A). Furthermore, there was no 2-5A accumulation in IAVΔNS1-infected cells lacking OAS3 (Fig. 4B). After 24 h of infection with IAVΔNS1, cells lacking OAS3 or RNase L had viral titers ∼10-fold higher than those in parental cells or in cells lacking OAS1 or OAS2 (Fig. 4C).
Fig. 4.
Activation of RNase L during IAVΔNS1 infection of A549 cells requires OAS3 expression. (A) WT A549 and KO cells were infected with IAVΔNS1 (MOI = 10), and at 24 hpi RNA integrity was assessed. (B) Cells were infected with IAVΔNS1 (MOI = 1 pfu per cell), and at 24 hpi 2-5A intracellular levels were quantified by FRET assay. (C) Cells were infected with IAVΔNS1 (MOI = 1 pfu per cell), and at 24 hpi infectious virus in the supernatants was titered by plaque assays. The data are from three biological replicates and are expressed as means ± SD; **P < 0.01.
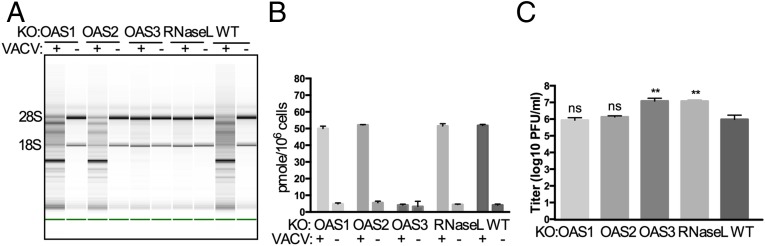
WT VACV inhibits OAS activation through the E3L RNA-binding proteins (21, 22). As with the other three viruses, following infection with VACVΔE3L the cleavage of rRNA by RNase L as well as the accumulation of intracellular 2-5A depended on the presence of OAS3 but not on OAS1 or OAS2 (Figs. 5 A and B). Viral titers were 12-fold higher in the absence of OAS3 or RNase L (Fig. 5C), indicating that OAS3-dependent activation of RNase L restricts VACV replication. The overexpression of OAS3 in OAS3-KO cells restores RNA degradation and 2-5A production (Fig. S1) during IAV and VACV infections as well as following pIC transfections, as is consistent with our findings that 2-5A production and rRNA degradation are dependent on OAS3 expression in A549 cells and that the OAS3-KO cells are competent to activate RNase L.
Fig. 5.
Activation of RNase L during VACVΔE3L infection of A549 cells requires OAS3 expression. (A) WT A549 cells and KO cells were infected with VACVΔE3L (MOI = 10 pfu per cell), and at 24 hpi RNA integrity was assessed. (B) Cells were infected with VACVΔE3L (MOI = 1 pfu per cell), and at 24 hpi 2-5A intracellular levels were quantified by FRET assay. (C) Cells were infected with VACVΔE3L (MOI = 1 pfu per cell), and at 24 hpi infectious virus in the supernatants was titered by plaque assays on BHK21 cells. The data are from three biological replicates and are expressed as means ± SD; **P < 0.01.
Activation of RNase L in HT1080 Cells That Express OAS1 p46 Is Also Dependent on OAS3.
OAS1 has a polymorphism causing the synthesis of splice variants to produce either p42 or p46 isoforms of OAS1 (23). A549 cells are homozygous for the gene that encodes the OAS1 p42 splice variant. We wanted to determine if the p46 isoform of OAS1 was similar to the OAS1 p42 in its relative lack of effect as compared with OAS3 during viral infections. Thus, we used the CRISPR-Cas9 techniques and the same guide RNAs to construct a similar set of cells with ablation of expression of OAS1, OAS2, OAS3, and RNase L in HT1080 cells, a human fibrosarcoma-derived cell line that is heterozygous for OAS1 genes that encode the splice variants p42 and p46 (23, 24). Sequencing of cellular DNA in the HT1080 cell lines indicated insertion or deletion mutations in the targeted genes (Table S3), and the OAS1, OAS2, OAS3 and RNase L knockouts were confirmed at the protein-expression level by Western blots. Expression of OAS1 p46 is greater than that of OAS1 p42 in HT1080 cells (Fig. 6A). We transfected the HT1080 cells with pIC (Fig. S4A) or infected the cells with WNV (Fig. 6B) and found that the levels of rRNA degradation in cells lacking OAS1 or OAS2 were similar to those in parental HT1080 cells, whereas rRNA remained intact in cells lacking OAS3. The viral titers from OAS3-KO HT1080 cells were ninefold higher than those from parental HT1080 cells (Fig. 6C). Thus, as with A549 cells, after infection with WNV, the loss of OAS1 or OAS2 had no detectable effects on the activation of RNase L, but OAS3 was required for the activation of RNase L. Similar results were obtained using pIC transfection in human mammary epithelial (HME) cells (p42 only) (Fig. S4B) and in VACVΔE3L infections (Fig. S4C). Thus the requirement for OAS3 expression was common to the three cell lines tested (A549, HT1080, and HME).
Fig. 6.
Activation of RNase L during WNV infection of HT1080 cells requires OAS3 expression. (A) OAS1-, OAS2-, OAS3-, and RNase L-KO HT1080 cells were mock-treated or were treated with IFN-α (2,000 U/mL) overnight. Proteins were analyzed by immunoblotting with antibodies against OAS1, OAS2, OAS3, and RNase L. (B) Cells were infected with WNV (MOI = 20 pfu per cell), and at 24 hpi RNA integrity was assessed. (C) Cells were infected WNV (MOI = 1 pfu per cell), and at the indicated time points infectious virus from the supernatant was titered by plaque assay. The data are from three biological replicates and are expressed as means ± SD; *P < 0.05, ***P < 0.001.
Fig. S4.
Activation of RNase L by pIC or VACVΔE3L in HT10810 or HME cells requires OAS3 expression. (A and B) HT1080 WT or KO cells were transfected with 500 ng/mL (A) or HME cells were transfected with1 μg/mL (B) of pIC, and at 4 h (HT1080 cells) or 3 h (HME cells) post transfection RNA was purified from cell lysates and analyzed on a Bioanalyzer. (C) WT or KO HME cells were infected with VACVΔE3L (MOI = 10). At 18 hpi RNA was purified from cell lysates and analyzed on a Bioanalyzer.
OAS3 Has a Higher Affinity for dsRNA than Do OAS1 and OAS2.
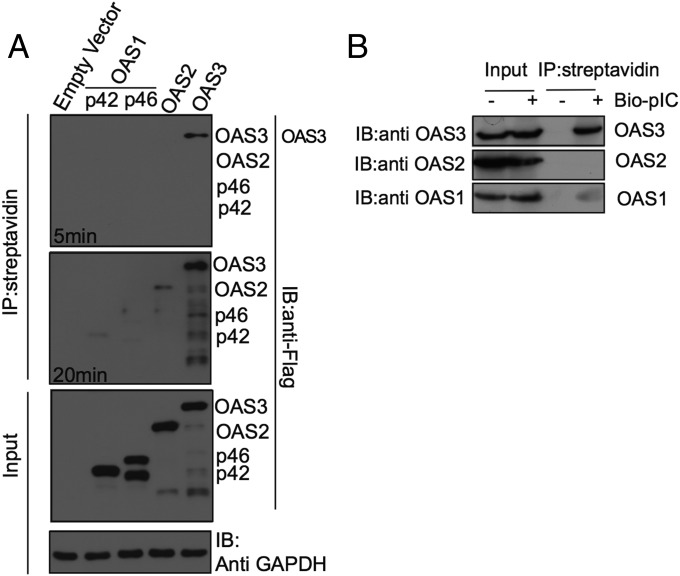
To investigate why OAS3 was required for activation of RNase L and why OAS1 and OAS2 were insufficient, we compared the interactions of each protein with pIC in intact cells. Thus, A549 cells were transfected with plasmids expressing Flag-tagged versions of OAS1 p42, OAS1 p46, OAS2, or OAS3 and 48 h were later were transfected with biotinylated pIC. Three hours after pIC transfection, cells were lysed, and complexes were purified on streptavidin-coupled beads. The bound proteins were separated by denaturing PAGE and were probed in Western blots with antibodies directed against the Flag epitope. Although OAS1 and OAS2 were barely detected after 20-min exposure, OAS3 was clearly present in pIC-bound complexes after only 5 min of exposure (Fig. 7A). These data suggest that OAS3 has a higher affinity for synthetic dsRNA than do OAS1 p42, OAS p46, or OAS2 and are consistent with a prior report (25). A similar experiment was carried out using IFN treatment to up-regulate endogenous OAS proteins. After transfection with biotinylated pIC and purification, proteins in complexes were separated by denaturing gel electrophoresis and were immunoblotted with antisera directed against each OAS. Although all species of OAS were detected in the unfractionated cell lysate (input), OAS3 was the predominant protein detected in the pIC-bound fraction; a trace amount of OAS1 also was evident. Therefore, similar to the overexpressed OASs, endogenous OAS3 displayed a higher affinity for pIC than did OAS1 or OAS2 (Fig. 7B).
Fig. 7.
OAS3 binds to dsRNA with higher affinity than do OAS1 or OAS2 in intact cells. (A) A549 cells were transfected with empty vector (mock) or plasmids encoding Flag-tagged OAS1 p42, OAS1 p46, OAS2, or OAS3 and then were transfected with pIC-biotin. Streptavidin bead pull downs or proteins were carried out and were analyzed by immunoblotting with anti-Flag antibody. The blot was exposed for 5 or 20 min. Immunoblots without pull down show input levels of OAS proteins and GAPDH loading and transfer control. The additional, faster-migrating band in the p46 lanes is most likely a breakdown product of p46. (B) A549 cells were treated with IFN-β (1,000 U/mL) and then were transfected with pIC-biotin. Streptavidin bead pull down of pIC-biotin bound proteins was carried out and probed by immunoblotting with antibodies against OAS1, OAS2, and OAS3. Input levels of proteins are also shown. Thirty-fold more protein was used for pull down compared with input protein on the immunoblot.
Discussion
We used CRISPR-Cas9 technology to construct human cell lines lacking expression of OAS1, -2, and -3 genes to determine which OAS proteins are required for RNase L-dependent antiviral activities. Using a diverse group of viruses as well as pIC, a synthetic surrogate for viral dsRNA, we found that OAS3 expression is necessary for activation of RNase L, as assessed by an rRNA degradation assay. Upon infection or pIC transfection, cells lacking OAS3 failed to synthesize detectable levels of 2-5A, whereas cells lacking OAS1 or OAS2 were able to produce amounts of 2-5A similar to those in the parental A549 cells. The FRET-based assay that we used for 2-5A quantification is an indirect assay based on the ability of 2-5A to activate RNase L (17). We conclude that OAS1 and OAS2 may be minimally activated, if at all, during these viral infections, although the expression levels of OAS1, OAS2, and OAS3 are up-regulated during IAV infection (Fig. S3C). Furthermore, all three OAS mRNAs are induced during SINV infection of A549 cells, and corresponding proteins are detectable by immunoblotting at 24 hpi in WT or OAS3-KO A549 cells (except that, as expected, OAS3 is not expressed in OAS3-KO cells) (Fig. S3 A and B). Alternatively, it is possible that OAS1 or OAS2 can synthesize forms of 2-5A that fail to activate RNase L. For instance, the FRET assay will detect the triadenylate 2′,5′-p3A3 but not the diadenylate 2′,5′-p3A2 because only the former of these two compounds activates RNase L (17). Thus, OAS3 was the dominant factor in the activation of RNase L during infection with all four viruses tested, i.e., two positive-stranded RNA viruses (WNV, a flavivirus, and SINV, an alphavirus), a negative-stranded RNA orthomyxovirus (IAV), and a DNA poxvirus (VACV). This finding was not restricted to A549 cells, because a similar dependence on OAS3 expression was observed in two other human cell types, HT1080 and HME cells (Fig. S4).
The antiviral effects of the OAS–RNase L pathway were clearly demonstrated by comparing WT to RNase L- and OAS3-KO cells for all four viruses (WNV, SINV, IAV, and VACV). There was at least a 10-fold increase in viral titer in cells lacking either RNase L or OAS3 as compared with parental A549 cells. However, WNV also replicated to higher titers in cells lacking OAS1 expression and to a lesser extent in OAS2-KO cells as compared with parental A549 cells. Similarly, SINV virus also replicated to higher titers in cells lacking OAS1 expression, suggesting that OAS1 has antiviral activities, although 2-5A or RNase L-mediated rRNA cleavage could not be detected in virus-infected OAS3-KO cells, which produce both OAS1 and OAS2. Thus, OAS1 and OAS2 may be less effective than OAS3 in producing 2-5A during viral infections, or viral countermeasures against OAS1 and OAS2 may be more effective than viral antagonism of OAS3. The latter explanation seems less likely, because as the same results were obtained using four diverse viruses that might be expected to have different antagonism patterns.
Interestingly, in overexpression studies OAS1 and OAS3, but not OAS2, were able to inhibit Dengue virus infection in A549 cells by an RNase L-dependent pathway (26). Similarly porcine OAS1 overexpression was more able to restrict Japanese encephalitis virus replication than porcine OAS2 (27). Overexpression of OAS3 restricted replication of Chikungunya virus (like SINV, an alphavirus) in HeLa cells (28), consistent with our finding that SINV replicated to a higher titer in OAS3-KO cells than in WT A549 cells (Fig. 3C). All these studies are consistent with our results and suggest that, although endogenous levels of OAS1 are not sufficient to activate RNase L, at least not as monitored by rRNA cleavage, overexpressed OAS1 may produce enough 2-5A to activate RNase L during virus infection.
A polymorphism in the OAS1 gene (rs10774671 SNP) results in the synthesis of splice variants, p42 or p46. Cells may be of the AA genotype, synthesizing p42, GG synthesizing p46, or AG, synthesizing both proteins (23). It has been reported that the AA genotype is associated with higher susceptibility to WNV infection and with progressive hepatitis C virus disease and that p46, when overexpressed, inhibits the replication of several flaviviruses in cell culture (9). Because A549 cells have the AA genotype and produced only OAS p42, we constructed similar knockout cells lines using parental HT1080 cells with the AG genotype. The phenotype of this series of cells was similar to that in corresponding A549-KO cells. Following transfection with pIC or viral infection, OAS3 expression was required for the activation of RNase L, but OAS1 and OAS2 were insufficient. Similar data were obtained with HME cells (encoding OAS1 p42). Thus, OAS3, a protein originally identified more than 30 y ago through protein purification efforts (29), was the dominant enzyme for the activation of RNase L in three cell types tested.
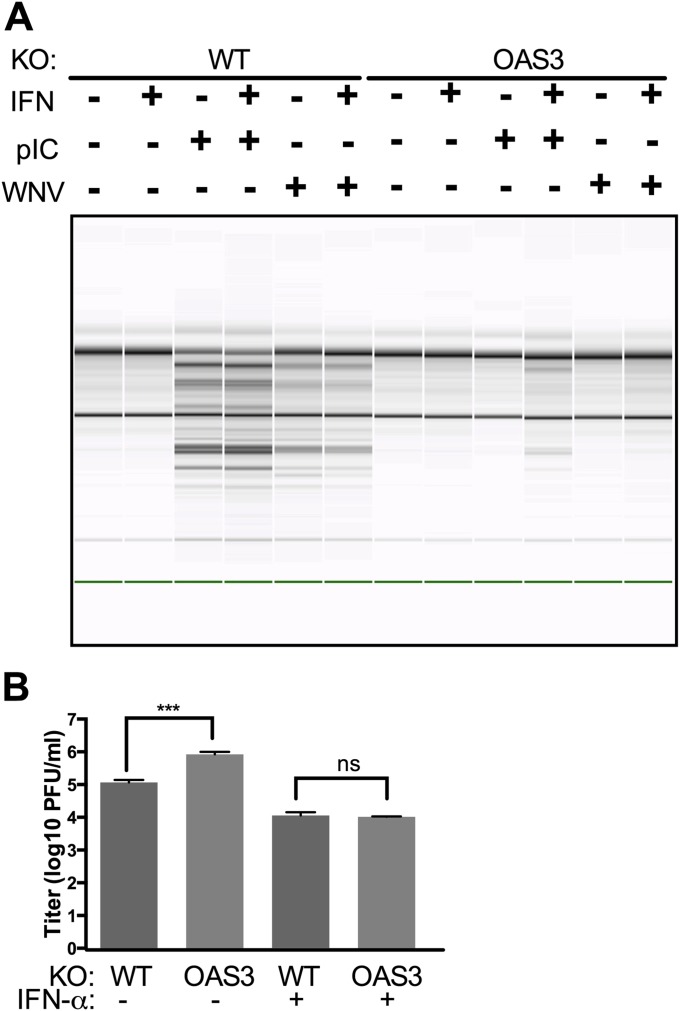
During infections in vivo, when some infected cells may produce large amounts of IFN, the level of expression of OAS genes, like other IFN-stimulated genes, is induced as shown in Western blots (Figs. 1A and 6A and Fig. S3). Thus, it is possible that later in infection the levels of OAS1 and OAS2 might be high enough to produce enough 2-5A to activate RNase L even in the absence of OAS3. However, when OAS3-KO cells were pretreated with IFN before infection with WNV, there was no detectable RNase L activation (Fig. S5A), despite the up-regulation of OAS1 and OAS2 during viral infection (Fig. S3). Although virus replicated to a higher titer in OAS3-KO cells than in WT cells, IFN treatment reduced titers to the same level in WT and OAS3-KO cells (Fig. S5B). Therefore, although RNase L was activated in IFN-treated WT cells, other IFN-induced antiviral proteins compensated for the loss of OAS3 in OAS3-KO cells.
Fig. S5.
IFN pretreatment reduces WNV titer in WT and OAS3-KO cells to the same level, independent of RNase L activation. (A) Cells were mock-treated or treated with 100 U of IFN-α overnight. Cells were transfected with 500 ng/mL of pIC or were infected with WNV (MOI = 20). Cells were lysed 4 h post transfection or 24 hpi, and RNA integrity was assessed on a Bioanalyzer. The position of 18S and 28S rRNA are indicated. (B) WT or OAS3-KO A549 cells were treated with 100 U of IFN-α overnight and then were infected with WNV (MOI = 1). At 36 hpi supernatants were harvested and titered for infectious virus. The data are pooled from three independent experiments carried out in duplicate. ***P < 0.001.
In previous studies comparing murine cell types, we found that RNase L is activated primarily in myeloid cells during infection with a murine coronavirus mutant. Similar to our current findings in human epithelial cells, pretreatment with IFN reduced the titer of infectious virus in all cell types regardless of whether RNase L was activated. In the murine system, IFN was not necessary to up-regulate OAS gene-expression levels to activate RNase L. Indeed, high basal levels of OAS expression were crucial to RNase activation L (30). Little is known about the basal levels of OAS gene expression in human cells and whether activation of RNase L is cell type-dependent, as it is in the murine system.
Two recent reports (31, 32) and an older one (33) have suggested that OAS3 is more active than OAS1 in synthesizing 2-5A. Recombinant OAS3 is activated at a lower concentration of dsRNA than is OAS1, and on average the 2-5As synthesized by OAS3 are longer than those synthesized by OAS1 and clearly are of sufficient length to activate RNase L (31). An early study of the enzymatic activity of purified OAS1 and OAS3 (then referred to as “33-kDa” and “110-kDa” OASs, respectively) also concluded that OAS3 is activated at lower RNA concentrations and that the two enzymes have different pH optima, suggesting that they may function under different conditions within cells (33). Differential subcellular localization has been reported among the OAS proteins (23, 26) and could influence their relative contributions to RNase L activation. In addition, a study that included the crystal structure of the N-terminal enzymatically inactive 2-5A synthetase domain of OAS3 (hOAS3.DI) in complex with 19-bp dsRNA indicated that this domain I (DI) subunit has high affinity for the binding of long (>50 bp) dsRNA, which then is presented to the enzymatically active C-terminal domain III (DIII) of OAS3 that produces 2-5A from ATP. In contrast, OAS1 can bind short or long dsRNA equally well but with lower affinity than OAS3 (25).
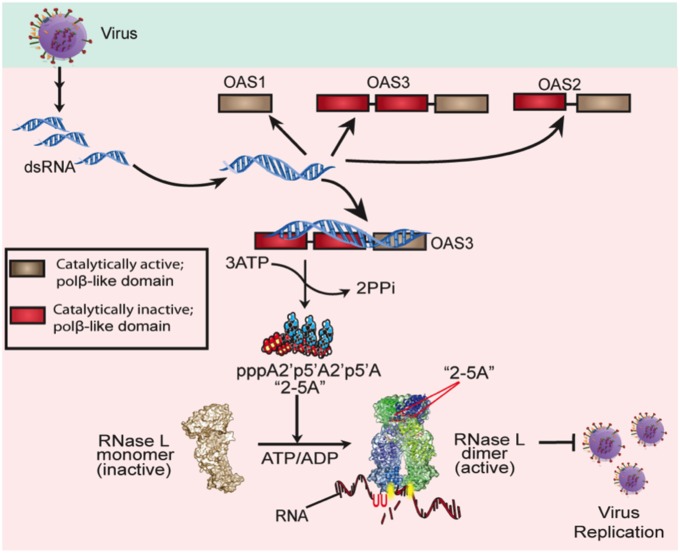
Our data demonstrate that OAS3 is the primary protein synthesizing 2-5A to activate RNase L during infections with four diverse viruses and in different cell types. Furthermore, our studies using viral infections in cells deficient in different OAS proteins validate a previous model for OAS activation (Fig. 8) (25). Viral dsRNA is sensed primarily by OAS3 as it binds initially to DI, spanning the subunits, and activates DIII to synthesize 2-5A, which in turn activates RNase L. RNase L restricts virus replication and spread directly by cleaving viral RNA and indirectly by its effects on host mRNA and subsequently on protein synthesis and its promotion of apoptosis (34, 35). The conservation of the catalytic domains in OAS1 and OAS2 during mammalian evolution suggests important biological functions that have yet to be defined fully, possibly including minor or supporting roles in restricting viral infections. Indeed WNV and SINV show enhanced replication in OAS1-KO and OAS2-KO cells at some time points, and there may be similar or additional effects in other cell types. Further investigation of the RNase L-dependent and possible RNase L-independent activities of OAS1 and OAS2 as well as differences among cell types in relative levels of OAS1, OAS2, and OAS3 gene expression and activity are important aims for future studies.
Fig. 8.
The OAS–RNase L pathway. Upon detection of viral dsRNA during infection with diverse human viruses (WNV, SINV, IAVΔNS1, VACVΔE3L), OAS3 produces 2-5A, which activates RNase L. RNase L degrades cellular and viral RNA, leading to the restriction of virus replication. [Modified with permission from ref. 25 and with permission from ref. 36; http://www.sciencedirect.com/science/journal/10972765.]
Materials and Methods
OAS and RNase L-KO cell lines were constructed with CRISPR-Cas9 gene editing using A549, HT1080, and HME cells. Parental and KO cells lines were assessed by Western immunoblots for OAS and RNase L protein expression with or without pIC transfection or infection. Parental and KO cells were transfected with pIC and with a group of diverse viruses and were assessed for activation of RNase L, intracellular levels of 2-5A, and virus replication. All these techniques are described in SI Materials and Methods. Primer sequences for Fig. S3A are listed in Table S4.
Table S4.
qRT-PCR primers for human actin and OAS genes
| Gene | Forward (5′–3′) | Reverse (5′–3′) |
| actin | ACTGGAACGGTGAAGGTGAC | GTGGACTTGGGAGAGGACTG |
| OAS1 | GAAGGCAGCTCACGAAACC | AGGCCTCAGCCTCTTGTG |
| OAS2 | TTCTGCCTGCACCACTCTTCAACGA | GCCAGTCTTCAGAGCTGTGCCTTTG |
| OAS3 | CCGAACTGTCCTGGGCCTGATCC | CCCATTCCCCAGGTCCCATGTGG |
SI Material and Methods
Cell Lines and Viruses.
Human A549 cells were cultured in Roswell Park Memorial Institute (RPMI) medium 1640 (Gibco) supplemented with 10% (vol/vol) FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Human HT1080 cells were cultured in DMEM (Gibco) supplemented with 10% (vol/vol) FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. hTERT-HME1 (HME) cells, a gift from George Stark, Cleveland Clinic, Cleveland, were cultured with mammary epithelial cell growth medium (CC-3150; Lonza). African green monkey kidney Vero cells were cultured in DMEM supplemented with 10% (vol/vol) FBS, 10 mM Hepes, 1 mM sodium pyruvate, 100 U/mL penicillin, 100 μg/mL streptomycin, and 50 μg/mL gentamicin. Baby hamster kidney (BHK-21) cells were cultured in DMEM supplemented with 10% (vol/vol) FBS, 5% (vol/vol) tryptose phosphate broth (Sigma-Aldrich), 100 U/mL penicillin, and 100 μg/mL streptomycin. Human HEK 293T cells were cultured in DMEM supplemented with 10% (vol/vol) FBS and 1 mM sodium pyruvate. MDCK-NS1-GFP cells [a gift from Adolfo Garcia-Sastre, Icahn School of Medicine at Mount Sinai, New York (37)] were grown in DMEM supplemented with 10% (vol/vol) FBS as described previously (19).
WNV Kunjin strain (38) was obtained from Paul Bates, University of Pennsylvania, Philadelphia and was propagated in Vero cells; SINV expressing GFP from a subgenomic promoter (39) was obtained from Sara Cherry, University of Pennsylvania, Philadelphia and was propagated in BHK-21 cells; TMEV (DA strain) (40) was obtained from Julian Leibowitz, Texas A & M University, College Station, TX and was propagated in BHK-21 cells; EMCV (41) was obtained from Ian M. Kerr, Cancer Research UK, London; SeV strain Cantell (42) was obtained from Carolina B. Lopez, University of Pennsylvania, Philadelphia; LACV (43) was obtained from Samantha Soldan, University of Pennsylvania, Philadelphia and was propagated in BHK-21 cells; LCMV was obtained from John Wherry, University of Pennsylvania, Philadelphia (44) and was propagated in BHK-21 cells. The mouse-adapted H1N1 strain A/PR/8/34, deleted for the NS1 gene (IAV∆NS1) (20), was obtained from Adolfo Garcia-Sastre and was grown in MDCK-NS1 cells as described previously (19). Mutant vaccinia virus (VACVΔE3L) was obtained from Bertram Jacobs, Arizona State University, Tempe, AZ and was grown as described previously (22).
Antibodies.
OAS p42 is also referred to as “p41.” Mouse anti-OAS1 p41 F3 antibody (1:1,000; Santa Cruz), rabbit anti-OAS1 p42/46 D1W3A monoclonal antibody (1:200; Cell Signaling), rabbit anti-OAS2 H-180 polyclonal antibody (1:500; Santa Cruz), goat anti-OAS3 N-18 (1:250; Santa Cruz), mouse anti-human RNase L (1:1,000) (45), mouse anti-GAPDH GA1R (1:1,000; Thermo Fisher), and mouse anti-Flag M2 antibody (Sigma-Aldrich) were used to detect OAS1 p41, OAS1 p42/p46, OAS2 p69, OAS3 p100, RNase L, and GAPDH, respectively. Secondary antibodies were anti-mouse antibody (1:5,000; Thermo Fisher), anti-rabbit antibody (1:1,500 for rabbit anti-OAS1 p41/46 or 1:5,000 for rabbit anti-OAS2 p69; Cell Signaling), and anti-goat antibody conjugated with HRP (1:5,000; Santa Cruz).
Construction of Plasmids and Pseudo Lentivirus.
The oligonucleotide sequences (Table S1) used for the generation of small guide RNAs (sgRNA) were selected from a published sgRNA database (46). Two sgRNA sequences were selected for each OAS1, OAS2, and OAS3 gene knockout, and one was selected for knockout of the RNase L gene (RNASEL). A pair of forward and reverse oligonucleotides for the generation of each sgRNA (synthesized by IDT) was annealed by published methods (47) and was inserted into plasmid vectors pSpCas9-BB (Addgene) and pLenti-CRISPR (Addgene) between the BbsI and BsmBI restriction sites. The resulting plasmids were pSpCas9-sgO1-2, pSpCas9-sgO1-9, pLenti-sgO1-2, and pLenti-sgO1-9 (targeting the OAS1 gene); pSpCas9-O2-5, pSpCas9-O2-9, pLenti-sgO2-5, and pLenti-sgO2-9 (targeting the OAS2 gene); pSpCas9-O3-1, pSpCas9-O3-9, pLenti-sgO3-1, and pLenti-sgO3-9 (targeting the OAS3 gene); and pSpCas9-sgRL-6, pLenti-sgRL-6 (targeting the RNASEL gene).
For packaging of pseudo lentiviruses, 1 × 106 HEK 293T cells were plated in one well of a six-well plate and on the next day were transfected with 5 μg pLenti-CRIPSR (with sgRNA), 3.5 μg psPAX2, and 1.25 μg of pCMV-VSV-G (obtained from Paul Bates, University of Pennsylvania, Philadelphia) using Lipofectamine 2000 (Invitrogen) (24 μL in 250 μL of DMEM). The supernatants were harvested at 24 and 48 h post transfection and were stored at −80 °C. The 48-h supernatants were used for further knockout experiments.
Construction of Cells with OAS1, OAS2, OAS3, and RNASEL Knockout.
To knock out the targeted genes using pSpCas9 plasmids, 5 × 105 A549 or HT1080 cells were plated into one well of a six-well plate and were transfected with 600 ng of plasmid in 250 μL of medium in 6 μL of Lipofectamine 2000 in 250 μL of medium. Alternatively, A549 cells were washed three times with ice-cold PBS and were suspended at 3 × 106 cells/mL in electroporation buffer (Bio-Bad). Cells (800 μL) were mixed with 20 μg of pSpCas9 plasmids, transferred to a 0.4-cm Gene Pulsar Cuvette (Bio-Rad), pulsed at 300 V, 500 μF, 1,000 Ω in a Gene Pulser II (Bio-Rad), and transferred to six-well plates containing 4 mL medium without antibiotics. Forty-eight hours post transfection or electroporation, fresh medium containing puromycin (1.5 μg/mL for A549 cells or 0.5 μg/mL for HT1080 cells) was added. After 3 d, medium containing puromycin was removed and replaced with fresh medium without puromycin. Surviving cells became confluent 1 or 2 wk later. Confluent A549 cells were treated with trypsin (Gibco) and were cloned by limited dilution. Briefly, cells were diluted to 2.5 cells/mL, and 200 μL of cells were added to one well of a 96-well plate. Single cells were selected for further amplification and genotyping. HT1080 cells were cloned similarly by limited dilution.
To construct knockout A549 or HME cells using Lenti-CRIPSR, 1 × 105 cells were plated into one well of a 24-well plate and were transduced with 100 μL of pseudo lentiviruses. Forty-eight hours post transduction, cells were cultured in medium containing 2 μg/mL of puromycin for 3 d. Puromycin-resistant cells were cloned by limited dilution. Single-cell clones were selected for further amplification and genotyping.
Genomic DNA was extracted using a DNeasy Blood and Tissue Kit (Qiagen). DNA fragments of each gene covering the region targeted by sgRNA were amplified by PCR using gene-specific primers (Table S2) mixed with GoTaq Master Mix (Promega), and the PCR products were sequenced. Cells with frame-shift mutations (deletion or insertion) in targeted genes were selected for further experiments. For A549 cells, two clones for each OAS1, OAS2, OAS3, and RNASEL knockout for each sgRNA were cultured for further experiments (Table S3). For HT1080 cells, one clone for each gene knockout by one sgRNA was cultured for further experiments (Table S3). Results from the two cell lines for each gene were identical; data are shown for one representative cell line of each gene ablation in the figures.
Table S2.
Primers for genotyping OAS1-, OAS2-, OAS3-, and RNase L-KO cells
| Gene | Primer | Nucleotide sequence (5′–3′) |
| OAS1 | Forward | TTATAGGAGTTTAAGACATGCAATG |
| Reverse | CAGGGCATCAAAGGCAGGCAGCACATCG | |
| OAS2 | Forward | CAAGAGTTGGTAAGCTCGCTGCAG |
| Reverse | AAGTGTGGGCCTCGCACATAATAG | |
| OAS3 | Forward | CTTGCCTCACTCAAGTTGCCTGCC |
| Reverse | CCAGGTCAAACAGCAAGTTGGTGG | |
| RNASEL | Forward | TGCCAGGTGGAATGTCAGAAGACTG |
| Reverse | CAGCAGATGCGTAATAGCCTCCAC |
Subcloning of Human OAS cDNAs.
Human OAS-1 cDNA was PCR amplified from Harvard Plasmid Repository clone HsCD00370585 (GenBank accession no. BC071981.1) and was subcloned into vector p3xFlag CMV-10 (Sigma) at restriction sites NotI and XbaI. Human OAS-2 cDNA was PCR amplified from Harvard Plasmid Repository clone HsCD00377272 (GenBank accession no. BC049215.1) and was subcloned into vector p3xFlag CMV-10 at restriction sites NotI and XbaI. Human OAS-3 cDNA was PCR amplified from Harvard Plasmid Repository clone HsCD00347055 (GenBank accession no. BC113746) and was subcloned into vector p3xFlag CMV-10 at restriction sites HindIII and XbaI.
The OAS1 p46 cDNA was cloned from HT1080 cells. Briefly, the cells were treated with 2,000 U/mL of IFN-α overnight, and the total cellular RNA was extracted using an RNeasy Mini Kit (Qiagen). The cDNA was synthesized by reverse transcription by using SuperScript III (Invitrogen) and a p46 mRNA-specific reverse primer: 5-cctctagactagaggatggtgcaggtccagtcctcttc-3. The DNA was amplified using PfuUltraII Fusion HS DNA polymerase (Agilent) with the forward primer 5-cttgcggccgctatgatggatctcagaaataccccag-3 and the reverse primer used for reverse transcription and was inserted into p3xFlag-CMV10 (Sigma-Aldrich) at the NotI and XbaI restriction sites to generate p3xFlag-OAS1-p46. All constructs were verified by nucleotide sequencing, and expression was confirmed by Western blot.
Stable Expression of Flag OAS3 in OAS3-KO A549 Cells.
A549 OAS3-KO cells (2.5 × 105) were transfected with Flag-OAS3–expressing plasmids (2 μg/mL) using Lipofectamine 2000. The cells then were selected with neomycin (500 μg/mL). Individual colonies were isolated, and OAS3 expression was monitored by Western blot using both OAS3 and anti-Flag antibody. A single clone with the highest level of OAS3 expression was selected for the experiment shown in Fig. S1.
Western Immunoblotting.
Confluent cells in six-well plates were mock-treated or treated with 1,000 U/mL of IFN-β (A549 cells) or 2,000 U/mL of IFN-α (HT1080 cells) (PBL Assay Science) overnight, harvested, washed in PBS, and lysed with Nonidet P-40 buffer with protease inhibitor mixture (Roche). Cell lysates were mixed with 4× Laemmli buffer, boiled at 95 °C for 5 min, and analyzed by electrophoresis on 4–12% or 4–15% gradient SDS gels. Proteins were transferred to PVDF membranes, which were treated with 5% nonfat milk in Tris⋅HCl buffer saline with 0.5% Tween-20 (TBST) blocking buffer for 1 h, followed by incubation overnight at 4 °C with antibodies diluted in TBST. Membranes then were washed three times with TBST, incubated with secondary antibodies for 1 h at room temperature, washed three times with TBST, and then incubated with SuperSignal West Dura Extended Duration substrate (Thermo Fisher). The signal was detected using an Amersham Imager 600 (GE Healthcare).
Virus Growth Kinetics.
To study the growth kinetics of WNV and SINV, cells were plated into six-well plates, 1 × 106 cells per well. The next day, cells were infected with the indicated viruses (three parallel wells per virus) at MOI = 1. At 6, 12, 24, and 36 h postinfection, 200 μL of supernatants were harvested and stored at −80 °C for titration. To study the growth kinetics of IAV and VACV, cells were washed two times with PBS and then were placed in a minimal amount of serum-free RPMI medium 1640. Cells were infected with mutant IAV (IAV/PR8/ΔNS1) at MOI 1–20, as indicated in the figures. Cells were incubated with virus for 1 h with gentle agitation every 10 min. The virus was removed, and cells were washed twice with PBS. The cells then were resuspended in complete medium until they were harvested. VACV (VACVΔE3L) infections were performed as described earlier (22).
Plaque Assays.
WNV and SINV were diluted in DMEM, and 200 μL were added to confluent Vero monolayers in six-well plates. The plates were incubated for 1 h at 37 °C and were rocked at 15-min intervals. Then the cells were overlaid with 3 mL warm DMEM containing 1% FBS and 0.7% Agar. Plaques were stained with neutral red at 36–48 hpi. IAV/PR8 (∆NS1) titers were determined in MDCK-NS1-GFP cells as described previously (19). VACV (VACVΔE3L) titers were determined in BHK-21 cells as described previously (22).
rRNA Cleavage Assay.
Cells were transfected with pIC (Merck-Millipore) or were infected with the indicated viruses. At 4–5 h post transfection or at the indicated hours post infection cells were harvested in RLT buffer (RNeasy Mini Kit, Qiagen) or TRIzol (Invitrogen). Total RNA was extracted and was resolved on RNA chips using an Agilent 2100 BioAnalyzer. The integrity of RNA was assessed by RIN (16).
Quantification of 2-5A.
Intracellular 2-5A was quantified by an indirect assay that measures the activation of RNase L activity (48). In brief, pIC-transfected or virus-infected cells were washed with PBS and lysed in preheated (95 °C) lysis buffer [50 mM Tris⋅HCl (pH 7.2), 0.15 M NaCl, 1% Nonidet P-40, 200 µM sodium orthovanadate, 2 mM EDTA, 5 mM MgCl2, 5 mM DTT] and heated to 95 °C for another 7 min. The cell extracts then were centrifuged for 10 min at 14,000 × g at room temperature, and cleared supernatants were collected. Levels of 2-5A were determined by RNase L-based FRET assays in comparison with a standard curve of authentic (2′-5′)p3A3, as described elsewhere (17).
Binding of OAS Proteins to pIC in Intact Cells.
For pIC pulldown of ectopically expressed OASs, plasmids expressing Flag-tagged OAS p42, OAS1 p46, OAS2, and OAS3 (10 µg) were transfected in A549 cells (2 × 106) using Lipofectamine 2000. After 48 h of transfection, cells were trypsinized, and 2 × 106 cells were seeded in fresh 10-cm dishes. After 12 h cells were transfected with 100 ng/mL biotinylated pIC (Invivogen) using Lipofectamine 2000 for 3 h. Cell were washed with ice-cold PBS, lysis buffer [50 mM Tris⋅HCl, 150 mM NaCl, 2.5 mM MgCl2, 0.5% Nonidet P-40, protease inhibitor (Roche), and RNase inhibitor (1 U/mL)]. The cell extracts were centrifuged for 12 min at 140,000 × g at 4 °C. The cleared supernatants were collected, and protein was quantified by Bio-Rad assays. Biotinylated pIC was precipitated from cell lysates (1.5 mg of protein) by streptavidin-agarose beads (Thermo Scientific) followed by immunoblotting with anti-Flag antibody (Sigma-Aldrich). For pIC pulldown of endogenous OASs, 2 × 106 A549 cells were treated with IFN-β (1,000 U/mL) for 18 h. The cells then were transfected with biotinylated pIC (200 ng/m total). After 4 h the cells were harvested in lysis buffer [50 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 2.5 mM/mg Cl2, 0.5% Nonidet P-40, protease inhibitor mixture], and 1.5 mg/mL of total cell lysates were incubated with streptavidin beads (100 µL) for 18 h at 4 °C. The beads then were washed five times with wash buffer [50 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 2.5 mM/mg Cl2, 0.5 mM EDTA, 0.5%Nonidet P-40, and protease inhibitor mixture), centrifuged at 12,000 × g for 5 min at 4 °C, and dissolved in 100 µL of sample dye. Thirty percent (30 µL) of each sample was separated in 10% SDS PAGE. One percent of the input (15 µg of total protein) of each sample also was resolved in the same gel. Three membranes were probed separately with OAS1, OAS2, and OAS3 antibodies at same time under identical conditions.
mRNA Quantification by Quantitative RT-PCR.
A549 cells were infected with SINV (MOI = 5) in triplicate in six-well dishes. Cells were lysed 2, 6, 12, and 24 hpi in RLT Plus RNA lysis buffer (Qiagen), and RNA was isolated using the RNeasy Plus Mini Kit (Qiagen). Quantitative RT-PCR (qRT-PCR) was performed using an iQ5 iCycler (Bio-Rad), and cycle threshold (CT) values were recorded, all as described previously (49). CT values were normalized to β-actin. OAS mRNA levels were calculated relative to β-actin mRNA and expressed as fold over levels at 2 hpi with the equation 2-Δ(ΔCT) (ΔCT = CTgene of interest – CTactin). Primer sequences are listed in Table S4.
Acknowledgments
We thank Dr. Paul Bates and Stephen Bart for plasmids psPAX2 and pCMV-VSV-G and for advice on CRISPR-Cas9 technology; Dr. Babal Kant Jha for the 2-5A and RNase L used in the FRET assays; and Danika Baskar for help with the Western blots. This work was supported by NIH Grants R01-AI104887 (to S.R.W. and R.H.S.), R01-NS081008 (to S.R.W.), and R01-CA044059 (to R.H.S.) and by the Mal and Lea Bank Chair Fund (R.H.S.). Y.W. was supported in part by the China Scholarship Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519657113/-/DCSupplemental.
References
- 1.Silverman RH. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol. 2007;81(23):12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kristiansen H, Gad HH, Eskildsen-Larsen S, Despres P, Hartmann R. The oligoadenylate synthetase family: An ancient protein family with multiple antiviral activities. J Interferon Cytokine Res. 2011;31(1):41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- 3.Ibsen MS, et al. Structural and functional analysis reveals that human OASL binds dsRNA to enhance RIG-I signaling. Nucleic Acids Res. 2015;43(10):5236–5248. doi: 10.1093/nar/gkv389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hornung V, Hartmann R, Ablasser A, Hopfner KP. OAS proteins and cGAS: Unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol. 2014;14(8):521–528. doi: 10.1038/nri3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silverman RH, Weiss SR. Viral phosphodiesterases that antagonize double-stranded RNA signaling to RNase L by degrading 2-5A. J Interferon Cytokine Res. 2014;34(6):455–463. doi: 10.1089/jir.2014.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drappier M, Michiels T. Inhibition of the OAS/RNase L pathway by viruses. Curr Opin Virol. 2015;15:19–26. doi: 10.1016/j.coviro.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee MS, Kim B, Oh GT, Kim YJ. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat Immunol. 2013;14(4):346–355. doi: 10.1038/ni.2535. [DOI] [PubMed] [Google Scholar]
- 8.Zhu J, et al. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity. 2014;40(6):936–948. doi: 10.1016/j.immuni.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim JK, et al. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog. 2009;5(2):e1000321. doi: 10.1371/journal.ppat.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandal S, Abebe F, Chaudhary J. 2′-5′ oligoadenylate synthetase 1 polymorphism is associated with prostate cancer. Cancer. 2011;117(24):5509–5518. doi: 10.1002/cncr.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Field LL, et al. OAS1 splice site polymorphism controlling antiviral enzyme activity influences susceptibility to type 1 diabetes. Diabetes. 2005;54(5):1588–1591. doi: 10.2337/diabetes.54.5.1588. [DOI] [PubMed] [Google Scholar]
- 12.Fedetz M, et al. OAS1 gene haplotype confers susceptibility to multiple sclerosis. Tissue Antigens. 2006;68(5):446–449. doi: 10.1111/j.1399-0039.2006.00694.x. [DOI] [PubMed] [Google Scholar]
- 13.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silverman RH, Skehel JJ, James TC, Wreschner DH, Kerr IM. rRNA cleavage as an index of ppp(A2'p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J Virol. 1983;46(3):1051–1055. doi: 10.1128/jvi.46.3.1051-1055.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schroeder A, et al. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thakur CS, Xu Z, Wang Z, Novince Z, Silverman RH. A convenient and sensitive fluorescence resonance energy transfer assay for RNase L and 2′,5′ oligoadenylates. Methods Mol Med. 2005;116:103–113. doi: 10.1385/1-59259-939-7:103. [DOI] [PubMed] [Google Scholar]
- 18.Min JY, Krug RM. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc Natl Acad Sci USA. 2006;103(18):7100–7105. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper DA, et al. RNase L targets distinct sites in influenza A virus RNAs. J Virol. 2015;89(5):2764–2776. doi: 10.1128/JVI.02953-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-Sastre A, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252(2):324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 21.Beattie E, et al. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J Virol. 1995;69(1):499–505. doi: 10.1128/jvi.69.1.499-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiang Y, et al. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J Virol. 2002;76(10):5251–5259. doi: 10.1128/JVI.76.10.5251-5259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kjær KH, et al. Mitochondrial localization of the OAS1 p46 isoform associated with a common single nucleotide polymorphism. BMC Cell Biol. 2014;15:33. doi: 10.1186/1471-2121-15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasheed S, Nelson-Rees WA, Toth EM, Arnstein P, Gardner MB. Characterization of a newly derived human sarcoma cell line (HT-1080) Cancer. 1974;33(4):1027–1033. doi: 10.1002/1097-0142(197404)33:4<1027::aid-cncr2820330419>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 25.Donovan J, Whitney G, Rath S, Korennykh A. Structural mechanism of sensing long dsRNA via a noncatalytic domain in human oligoadenylate synthetase 3. Proc Natl Acad Sci USA. 2015;112(13):3949–3954. doi: 10.1073/pnas.1419409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin RJ, et al. Distinct antiviral roles for human 2′,5′-oligoadenylate synthetase family members against dengue virus infection. J Immunol. 2009;183(12):8035–8043. doi: 10.4049/jimmunol.0902728. [DOI] [PubMed] [Google Scholar]
- 27.Zheng S, et al. 2015. Porcine 2', 5′-oligoadenylate synthetases inhibit Japanese encephalitis virus replication in vitro. J Med Virol, 10.1002/jmv.24397.
- 28.Bréhin AC, et al. The large form of human 2′,5′-Oligoadenylate Synthetase (OAS3) exerts antiviral effect against Chikungunya virus. Virology. 2009;384(1):216–222. doi: 10.1016/j.virol.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 29.Wells JA, Swyryd EA, Stark GR. An improved method for purifying 2′,5′-oligoadenylate synthetases. J Biol Chem. 1984;259(2):1363–1370. [PubMed] [Google Scholar]
- 30.Zhao L, et al. Cell-type-specific activation of the oligoadenylate synthetase-RNase L pathway by a murine coronavirus. J Virol. 2013;87(15):8408–8418. doi: 10.1128/JVI.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ibsen MS, et al. The 2′-5′-oligoadenylate synthetase 3 enzyme potently synthesizes the 2′-5′-oligoadenylates required for RNase L activation. J Virol. 2014;88(24):14222–14231. doi: 10.1128/JVI.01763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donovan J, Dufner M, Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc Natl Acad Sci USA. 2013;110(5):1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ilson DH, Torrence PF, Vilcek J. Two molecular weight forms of human 2′,5′-oligoadenylate synthetase have different activation requirements. J Interferon Res. 1986;6(1):5–12. doi: 10.1089/jir.1986.6.5. [DOI] [PubMed] [Google Scholar]
- 34.Zhou A, et al. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 1997;16(21):6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castelli JC, et al. The role of 2′-5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ. 1998;5(4):313–320. doi: 10.1038/sj.cdd.4400352. [DOI] [PubMed] [Google Scholar]
- 36.Huang H, et al. Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity. Mol Cell. 2014;53(2):221–234. doi: 10.1016/j.molcel.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gack MU, et al. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5(5):439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall RA, Scherret JH, Mackenzie JS. Kunjin virus: An Australian variant of West Nile? Ann N Y Acad Sci. 2001;951:153–160. [PubMed] [Google Scholar]
- 39.Frolova EI, et al. Roles of nonstructural protein nsP2 and Alpha/Beta interferons in determining the outcome of Sindbis virus infection. J Virol. 2002;76(22):11254–11264. doi: 10.1128/JVI.76.22.11254-11264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oleszak EL, Leibowitz JL, Rodriguez M. Isolation and characterization of two plaque size variants of Theiler’s murine encephalomyelitis virus (DA strain) J Gen Virol. 1988;69(Pt 9):2413–2418. doi: 10.1099/0022-1317-69-9-2413. [DOI] [PubMed] [Google Scholar]
- 41.Hoskins JM, Sanders FK. Propagation of mouse encephalomyocarditis virus in ascites tumour cells maintained in vitro. Br J Exp Pathol. 1957;38(3):268–272. [PMC free article] [PubMed] [Google Scholar]
- 42.Basler CF, et al. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol. 2003;77(14):7945–7956. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson WH, Kalfayan B, Anslow RO. Isolation of California Encephalitis Group Virus from a Fatal Human Illness. Am J Epidemiol. 1965;81:245–253. doi: 10.1093/oxfordjournals.aje.a120512. [DOI] [PubMed] [Google Scholar]
- 44.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003;77(8):4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong B, Silverman RH. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J Biol Chem. 1995;270(8):4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 46.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elbahesh H, Jha BK, Silverman RH, Scherbik SV, Brinton MA. The Flvr-encoded murine oligoadenylate synthetase 1b (Oas1b) suppresses 2-5A synthesis in intact cells. Virology. 2011;409(2):262–270. doi: 10.1016/j.virol.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao L, et al. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe. 2012;11(6):607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]