Significance
The evolutionary diversification of brain morphology is one of the most prominent features of the primate adaptive radiation and a likely determinant of primate evolutionary success. However, the ecological factors responsible for the diversification of the primate brain are largely unknown. In this work, we use a comparative approach to study brain diversification during the adaptive radiation of a major primate clade—the New World monkeys. We show that brain morphology evolved in association with the occupation of several ecological niches, and that convergence in brain morphology among clades may be associated with an evolutionary increase in the complexity of social behaviors.
Keywords: comparative method, primates, adaptive evolution, geometric morphometrics, Platyrrhini
Abstract
Primates constitute one of the most diverse mammalian clades, and a notable feature of their diversification is the evolution of brain morphology. However, the evolutionary processes and ecological factors behind these changes are largely unknown. In this work, we investigate brain shape diversification of New World monkeys during their adaptive radiation in relation to different ecological dimensions. Our results reveal that brain diversification in this clade can be explained by invoking a model of adaptive peak shifts to unique and shared optima, defined by a multidimensional ecological niche hypothesis. Particularly, we show that the evolution of convergent brain phenotypes may be related to ecological factors associated with group size (e.g., social complexity). Together, our results highlight the complexity of brain evolution and the ecological significance of brain shape changes during the evolutionary diversification of a primate clade.
Adaptive radiation, defined as the rapid and exceptional adaptive diversification of a single phylogenetic lineage into a variety of different ecological niches (1, 2), is thought to be one of the main evolutionary processes generating biodiversity on Earth (3). Although several adaptive radiations have now been thoroughly studied (e.g., African cichlids, Caribbean anoles, Galapagos finches, etc.), we still need more studies from which generalizations on this process can be drawn (2). Particularly, under what conditions a lineage will undergo adaptive radiation has long been debated, although both empirical and theoretical models point to the existence of ecological opportunity as a major factor (2, 3). This opportunity may appear, among others, through the colonization of a new area, the extinction of a strong ecological competitor, or the evolution of a new trait—a “key innovation”—that allows the utilization of resources in ways that were not previously possible (3, 4). Therefore, a significant dimension of adaptive radiations is the diversification of ecologically relevant phenotypic traits (2, 5). Among the studied cases of adaptive radiations, several ecologically relevant traits have been identified. For example, relative limb size in anoles lizards (6), beak shape in Darwin’s finches (7), or overall body shape in cichlid fishes (8).
Remarkably, a trait that has received less attention in the study of adaptive radiations among vertebrate clades is brain morphology. Brains have substantial ecological and adaptive importance because they underlie the behavior that allows an animal to successfully interact with its environment. In this sense, primates constitute a notable example, as the evolution of brain morphology is one of the most prominent features of their diversification (9). Primates generally engage in complex foraging and social behaviors (10), and therefore, the evolution of enhanced cognitive capacities associated with enlarged and/or more complex brains may constitute a major axis of their adaptive ecological diversification. For example, possessing a large brain is perhaps the single most relevant phenotypic trait of our own species, Homo sapiens. Specifically, previous works have pointed out that the evolution of enlarged brains could be important ecologically because these changes are related to the acquisition of the cognitive abilities required to sustain complex social interactions—a behavioral trait probably involved in the origin and maintenance of the evolutionary success of primates (the social brain hypothesis) (11).
However, although brain size has been traditionally the preferred measured trait in evolutionary studies, brains are not uniform structures but are constituted by several anatomically distinct functional systems or modules (e.g., neocortex, cerebellum, etc.). Previous work has suggested that these modules can vary in their relative size among species of some mammalian clades (including primates) (12–14) and, moreover, that these mosaic changes can better explain neural diversity than total—absolute or relative—brain size changes alone (e.g., among primates) (14). Additionally, in several vertebrate clades, part of this mosaic variation has been related to particular behavioral capacities (e.g., refs. 15 and 16). Thus, brain diversification is probably a complex process involving several dimensions of change occurring during the divergence of the species and along multiple ecological axes. Moreover, as in previous adaptive radiation studies where shape is the most ecologically relevant trait (6, 7), brain shape (i.e., the relative position and size of individual brain components) is probably a more important aspect of brain evolution than its total relative size. However, despite the potential ecological relevance of brain shape variation for primates, its evolutionary dynamics have been less studied in macroevolution (14).
New World monkeys or platyrrhines, one of the three major primate clades, constitute an example of a major mammalian adaptive radiation that unfolded in isolation in Central and South America during the last 25–35 Ma, resulting in broad ecological and morphological diversity (17, 18). Noticeably, previous works have pointed to the putative occurrence of several evolutionary independent processes of increase and decrease in relative brain size (19–21) in this clade, indicating an extensive diversification of brain morphology. However, it is unknown how brain shape has evolved in the context of this primate adaptive radiation.
In this study, we investigate the process of brain shape diversification of New World monkeys during their adaptive radiation in relation to different ecological dimensions. We first quantify brain shape variation using virtual reconstructions of cranial endocasts and geometric morphometrics methods, and then explore the evolutionary processes underlying brain diversification using phylogenetic comparative methods. We hypothesized that, if brain shape is an ecologically relevant trait for platyrrhines, a pattern of variation that departs from neutral models of evolution would be expected. To address this hypothesis, we analyzed the relationship between phenotypic variation and the branching process of the species based on a Brownian (random) model of evolution. Next, we investigated whether a model involving changes in adaptive peaks in a macroevolutionary landscape for brain shape can better account for the observed diversity. Moreover, considering that a likely outcome of an adaptive radiation, given the importance of ecological factors in shaping diversity, is the repeated evolution of similar phenotypes (22), we explicitly test for the existence of brain morphological convergence in the platyrrhine radiation. Finally, we explore previously hypothesized ecological factors (e.g., group size) (11) driving brain shape diversification and convergence in this primate clade.
Results
Brain Shape Variation.
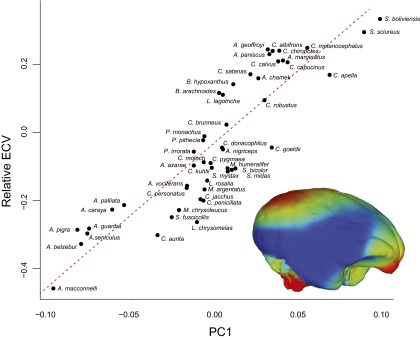
To study the pattern of variation in brain shape in the platyrrhine clade we performed a principal-component analysis (PCA) on the Procrustes shape coordinates of 399 landmarks and semilandmarks measured on 49 species (Table S1) that describe each species external brain morphology (Fig. 1A and Table S2). Fig. 1A shows the main trends in shape variation, as represented by the first two principal components (PCs), which together account for ∼61% of the total variation, along a projection of the phylogeny on the morphospace. A general separation among families, and a largely phylogenetically structured occupation of the morphospace can be observed, although remarkable shape proximity occurs among members from the three families (the atelids Ateles, Brachyteles, and Lagothrix; the pithecids Chiropotes and Cacajao; and the cebid Cebus) showing high and similar encephalization (i.e., total relative brain size) values [relative endocranial volume (rECV)] (Figs. S1 and S2), suggesting the existence of brain shape convergence among these clades. Furthermore, Alouatta and Saimiri species, extremes on the encephalization scale, also appear as extremes in shape space. Although these results may suggest that shape and total relative size describe the same morphological properties of the brain, only a fraction of the total shape variation can be accounted by encephalization [phylogenetic generalized least-squares regression (PGLS): λ = 0.74; R2 = 0.11; P = 0.010]. Brain shape differences along PC1 (Fig. 1B and Movie S1), which explains ∼43% of variation, are mainly related to general enlargement of the neocortex area with respect to the stem and cerebellum areas; and the relative position of the stem, which contrasts the posterior location in Alouatta with the more ventral position in Saimiri. Specifically, shape changes in the neocortex are concentrated mainly in the frontal, parietal, and occipital lobes, with the largest variation observed in the occipital lobe, which expands posteriorly and in a dorsoventral direction. These changes contribute to an overall more globular brain shape for positive scores. Additionally, the brain base exhibits a more ventrally flexed morphology among species with more positive scores. Furthermore, although PC1 is highly correlated with rECV (PGLS: λ = 0.87; R2 = 0.77, P = 0.001; Fig. S3), our results clearly show that PC1 describes complex morphological changes that cannot be characterized merely by relative total brain volume measurements.
Table S1.
Species included in this study
| Species | n | ECV,* cm3 | CS* | Base CS* | BM, g† | GS‡ |
| Alouatta belzebul | 3 | 58.54 | 506.43 | 168.00 | 6,395 | 7 |
| Alouatta caraya | 3 | 49.27 | 475.41 | 152.78 | 5,383 | 8.29 |
| Alouatta guariba | 3 | 56.46 | 501.10 | 163.33 | 5,175 | 7 |
| Alouatta macconnelli | 3 | 51.78 | 489.24 | 168.48 | 5,350 | 9.01 |
| Alouatta palliata | 5 | 47.37 | 468.82 | 150.01 | 6,250 | 14.4 |
| Alouatta pigra | 2 | 56.73 | 498.22 | 163.78 | 6,717 | 6.13 |
| Alouatta seniculus | 3 | 55.09 | 493.09 | 162.75 | 5,950 | 8.99 |
| Aotus azarae | 3 | 16.32 | 327.75 | 99.50 | 1,205 | 3.27 |
| Aotus nigriceps | 5 | 16.67 | 329.20 | 98.60 | 957 | 4.1 |
| Aotus vociferans | 1 | 15.32 | 323.39 | 99.37 | 703 | 3.3 |
| Ateles chamek | 3 | 111.18 | 611.00 | 177.26 | 9,370 | 38.5 |
| Ateles geoffroyi | 4 | 102.79 | 588.74 | 167.48 | 7,980 | 42.9 |
| Ateles marginatus | 4 | 122.25 | 625.09 | 179.91 | 8,608 | 33.13§ |
| Ateles paniscus | 4 | 115.63 | 614.71 | 175.32 | 8,280 | 18 |
| Brachyteles arachnoides | 6 | 108.03 | 603.88 | 178.15 | 8,840 | 36.5 |
| Brachyteles hypoxanthus | 2 | 115.00 | 619.66 | 180.45 | 8,865 | 36.5§ |
| Cacajao calvus | 4 | 69.61 | 518.04 | 148.07 | 3,165 | 21.7 |
| Cacajao melanocephalus | 3 | 68.20 | 513.89 | 145.01 | 2,935 | 30 |
| Callicebus brunneus | 3 | 15.46 | 317.78 | 93.67 | 829 | 2.5 |
| Callicebus donacophilus | 4 | 13.80 | 309.40 | 92.18 | 950 | 4§ |
| Callicebus moloch | 5 | 16.97 | 329.73 | 100.53 | 988 | 3.5 |
| Callicebus personatus | 5 | 20.54 | 353.82 | 110.27 | 1,325 | 6 |
| Callimico goeldii | 1 | 10.89 | 289.90 | 84.87 | 483 | 6.31 |
| Callithrix aurita | 1 | 7.50 | 254.50 | 81.54 | 429 | 3.1 |
| Callithrix jacchus | 5 | 8.11 | 261.27 | 80.78 | 320 | 8.5 |
| Callithrix kuhlii | 4 | 8.21 | 258.80 | 78.56 | 375 | 5 |
| Callithrix penicillata | 5 | 7.19 | 250.93 | 77.59 | 328 | 13 |
| Cebuella pygmaea | 2 | 4.20 | 205.99 | 61.92 | 116 | 5.36 |
| Cebus apella | 3 | 68.84 | 520.91 | 150.59 | 3,085 | 16 |
| Cebus albifrons | 4 | 70.24 | 517.81 | 145.94 | 2,735 | 19.8 |
| Cebus capucinus | 4 | 72.42 | 524.56 | 150.27 | 3,110 | 17.2 |
| Cebus robustus | 4 | 59.76 | 496.46 | 146.10 | 2,731 | 3.3 |
| Chiropotes | 3 | 56.06 | 477.56 | 135.86 | 2,740 | 22.5¶ |
| Chiropotes satanas | 2 | 51.51 | 466.78 | 135.12 | 3,030 | 22.5¶ |
| Lagothrix lagotricha | 4 | 93.28 | 576.65 | 169.62 | 7,150 | 34.18 |
| Leontopithecus chrysomelas | 4 | 12.55 | 304.69 | 96.23 | 577 | 5 |
| Leontopithecus rosalia | 5 | 12.31 | 297.39 | 91.61 | 609 | 5.43 |
| Mico argentatus | 4 | 8.07 | 260.21 | 79.83 | 345 | 7.8 |
| Mico chrysoleucus | 3 | 8.04 | 260.93 | 81.45 | 409 | 7.15§ |
| Mico humeralifer | 1 | 8.42 | 264.04 | 79.30 | 473.5 | 6.5 |
| Pithecia irrorata | 5 | 34.33 | 417.47 | 127.02 | 2,160 | 2.58 |
| Pithecia monachus | 4 | 33.02 | 410.20 | 123.36 | 2,360 | 4 |
| Pithecia | 4 | 31.14 | 400.70 | 121.36 | 1,760 | 4.02 |
| Saguinus bicolor | 3 | 10.18 | 281.35 | 84.73 | 429 | 5 |
| Saguinus fuscicollis | 5 | 7.82 | 258.65 | 81.25 | 401 | 6.07 |
| Saguinus midas | 5 | 10.95 | 287.49 | 86.99 | 563 | 5.8 |
| Saguinus mystax | 4 | 11.07 | 289.36 | 87.49 | 584 | 5.06 |
| Saimiri boliviensis | 4 | 24.12 | 362.83 | 98.13 | 811 | 37.5 |
| Saimiri sciureus | 8 | 21.55 | 350.33 | 95.64 | 799 | 33.5 |
| Total | 179 |
Base CS, brain base centroid size; BM, body mass; CS, brain centroid size; ECV, measured mean endocranial volume; GS, mean group size; n, number of specimens analyzed. Species were represented by one to eight individuals each (mode = 4), with smaller sample sizes belonging to species that are scarce in nature and/or museum collections. Taxonomy was reassessed using geographic location and distribution maps following International Union for Conservation of Nature red lists (34).
This work.
Data from ref. 18 and references therein.
Data from ref. 58 and references therein.
Genus mean.
Data from C. albinasus.
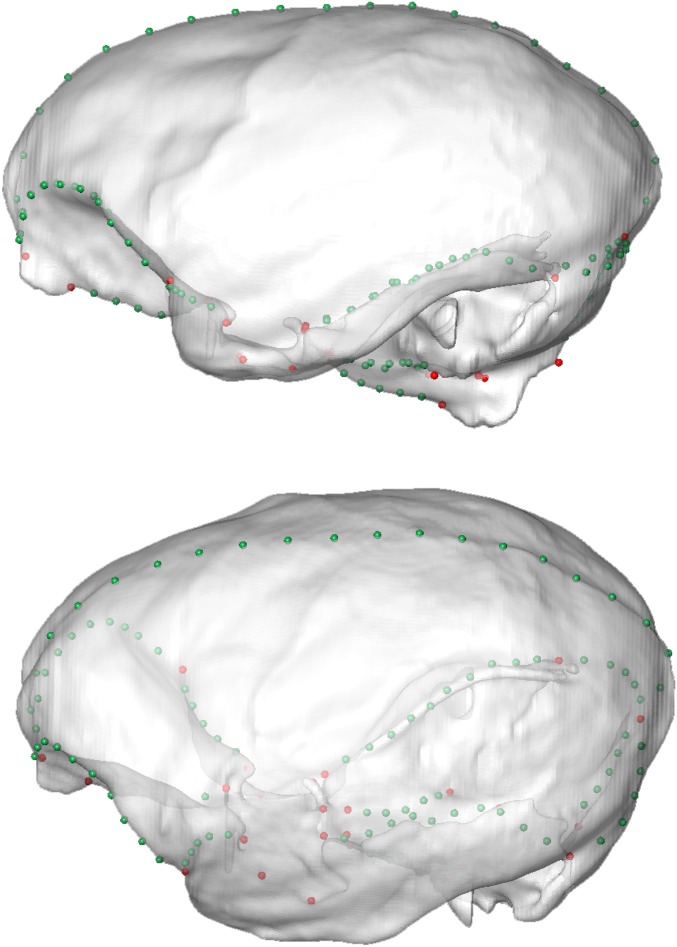
Fig. 1.
Morphometric analysis of New World monkey’s brain shape. (A) Ordination of 49 platyrrhine species in the morphospace defined by the first two principal components (PCs) of brain shape variation, which together account for ∼61% of total variance. Endocast at Left shows the measured landmarks (red), and curves (green) and surfaces (blue) semilandmarks on each individual (see also Fig. S6 and Table S2). Encephalization values (relative ECV) for each species are depicted by the color of each data point. Additionally, the three platyrrhine families are indicated, along with the genus Aotus, whose position in the platyrrhine tree is contentious. (B) Brain shape changes associated with the main axes of variation. Models were obtained by warping a surface model of the mean platyrrhine shape along PC1 and PC2 scores. See also Movies S1 and S2.
Table S2.
Landmarks and curves used in this study
| Landmarks/curves | Definition |
| Landmarks | |
| Basion | Midpoint of the anterior margin of the foramen magnum |
| Opisthion | Midpoint of the posterior margin of the foramen magnum |
| Middle spheno-occipital | Spheno-occipital suture at the midline |
| Dorsum sellae | Anterior edge at the midline |
| Sphenoidale | Most posterior, superior midline point of planum sphenoideum |
| Posterior “olfactory pit” | Posterior edge of the olfactory pit at the midline |
| Anterior olfactory pit | Anterior edge of the olfactory pit at the midline |
| Internal occipital protuberance | Superior edge of transverse sinus at the midline |
| Left/right lateral foramen magnum | Widest point, immediately above left/right hypoglossal canal |
| Left/right superior orbital fissure | Lateral vertex |
| Left/right foramen ovale | Most posterior and lateral point |
| Left/right foramen rotundum | Lateral vertex |
| Left/right jugular foramen | Most anterior and inner point |
| Left/right spheno-occipital | Lateral spheno-occipital suture |
| Left/right spheno-parietal | At the junction |
| Left/right petrous-occipital | Maximum curvature point at the junction |
| Left/right petrous | Petrous apex |
| Curves | |
| Midline occipital | Along the midline from foramen magnum to spheno-occipital suture |
| Middle sphenoid | Along the midline in anterior cranial fossa |
| Left/right lateral occipital | From jugular foramen to spheno-occipital suture. Delimiting brainstem |
| Left/right sphenoid | From superior orbital fissure to spheno-parietal junction. Delimiting anterior and middle cranial fossa |
| Left/right petrous | Delimiting posterior and middle cranial fossa |
| Transverse sinus | Delimiting vault from posterior cranial fossa |
| Left/right anterior cranial fossa | From olfactory pit to spheno-parietal junction. Delimiting frontal lobe/anterior cranial fossa ventral and dorsal regions |
| Midsagittal | From anterior olfactory pit to internal occipital protuberance |
| Basioccipital | From internal occipital protuberance to opisthion, at the midline |
Fig. S1.
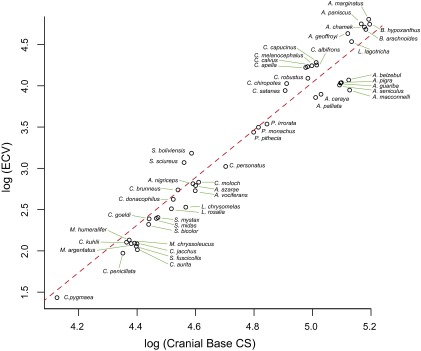
PGLS regression of log endocranial volume (ECV) on the log of the cranial base centroid size (CS) (λ = 0.96; R2 = 0.88; P = 0.0001). Residuals of this regression were used as a measure of relative endocranial—or brain—volume or encephalization (rECV).
Fig. S2.
Measured rECV (encephalization) values for each species mapped on the phylogeny. Values at nodes and branches were reconstructed using a maximum-likelihood ancestral character estimation method based on a Brownian motion model of evolution (57). Although ancestral reconstructions can have a large uncertainty, this tool is useful to visualize general trends in the evolution of a character.
Fig. S3.
PGLS regression of encephalization (rECV) on the main axis of platyrrhine brain shape variation (PC1). Colors in the brain model shows the areas in which shape change is concentrated along PC1. Red colors indicate the most shape change, whereas blue colors indicate less change. Colors were assigned based on a Procrustean superimposition of the extremes of variation along PC1.
Shape variation along PC2 (18.5% of total variation) is mainly related to changes in the prefrontal area, which becomes less expanded, particularly along the brain base midline, in the positive scores direction; and with a general increase in globularity for negative scores (Fig. 1B and Movie S2). Importantly, although New World monkeys exhibit a remarkable body size diversity (∼0.01–10 kg.), brain shape variation cannot be merely attributed to evolutionary allometric effects (PGLS: λ = 0.69; R2 = 0.15; P = 0.003).
Brain Shape Convergence.
Next, we explicitly test whether the pattern of shape proximity exhibited by members of the three families departs from what can be expected by chance. We calculated two pattern-based measures of morphological convergence (C1 and C5) using the first two PCs and compared them against a null distribution obtained using simulations under a random model of evolution (Brownian motion). Although only the first measure showed a strong convergence signal (C1 = 0.63; P = 0.01; C5 = 5; P = 0.22), these analyses point to the existence of nonrandom evolutionary processes generating brain shape similarity among these clades.
The Evolution of Brain Shape.
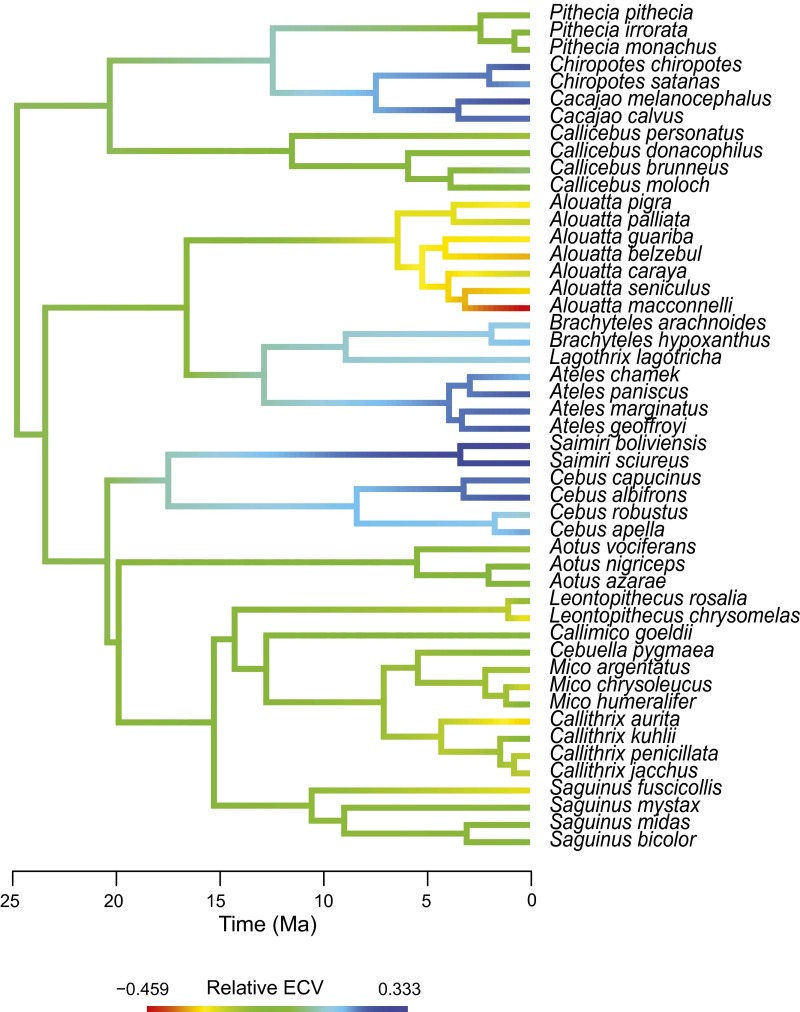
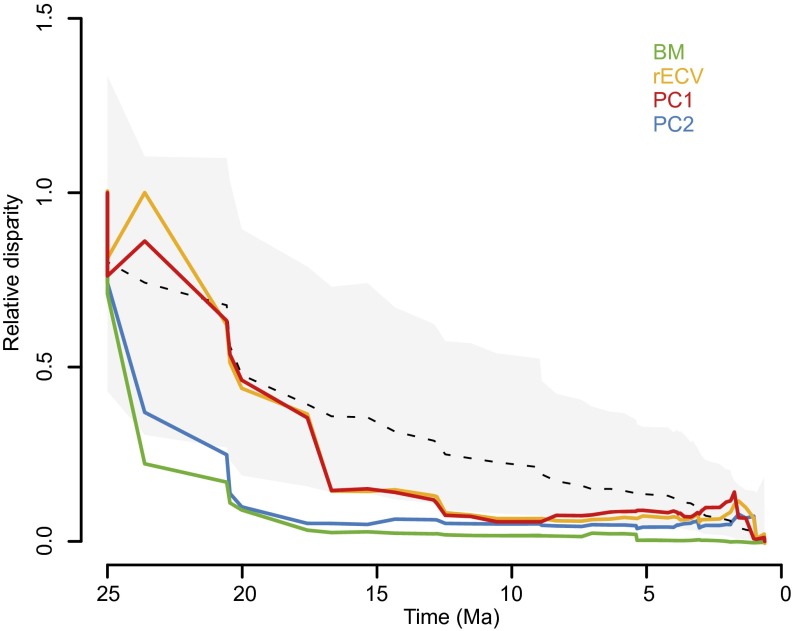
Phylogenetic signal analyses allowed to further unravel the evolutionary dynamics of brain shape diversification. Complementing the observed cladewise convergence, K statistic values both for PC1 (K = 1.30, P = 0.001) and PC2 (K = 2.32, P = 0.001) showed that variance was concentrated among clades rather than within clades, indicating, besides a considerable phylogenetic signal, a pattern of early brain morphological diversification that departs from neutral expectations. Disparity-through-time (DTT) plots further support this view: an early fast diversification of the morphological aspects represented by PC2 is observed (Fig. 2); whereas, for PC1, a drop in disparity beyond the neutral expectation can be seen between ∼17 and 12 Ma, pointing to a burst of brain shape evolution approximately associated with the origin of the extant subfamilies (Fig. 2 and Fig. S2), followed by a slowdown in disparity changes until present times. Moreover, these results also point to the existence of separate bursts of brain evolution during the platyrrhine radiation, each affecting different aspects of brain morphology.
Fig. 2.
Disparity-through-time (DTT) plots for brain shape, encephalization (rECV), and log body mass (BM). Relative disparity at each point indicates the average extant disparity of the subclades that had an ancestor at that time with respect to the whole clade disparity. Dashed line and gray shadow represent the expectation under a BM model of evolution, estimated via simulations, and its 95% confidence interval, respectively.
Finally, we explicitly asked whether a model of adaptive evolution could be invoked to explain brain shape diversification in platyrrhine monkeys, and particularly, to explain the observed pattern of shape convergence. To do this, we implement a model selection approach, in which the likelihoods of several alternative evolutionary scenarios are compared using an information content criterion [corrected Akaike information criterion (AICc)] to find the best-fitting model. The explored models were a random walk [modeled as Brownian motion (BM)]; early burst (where the rate of Brownian evolution decays exponentially through time); and several adaptive evolution scenarios modeled as Ornstein–Uhlenbeck (OU) processes (Fig. S4). In OU models, nodes and branches of the phylogeny are assigned to different selective regimes representing phenotypic adaptive peaks on a Simpsonian macroevolutionary landscape. This way, directional and stabilizing selection can be modeled. To construct these scenarios, we combined a priori biological hypotheses with the use of the SURFACE method (Supporting Information and Figs. S4 and S5), which, using a data-driven algorithm, finds the model that has the best statistical fit. By combining these approaches, we generated a likely biological hypothesis with a high absolute statistical support, excluding the possibility of choosing only the best among several bad biological hypotheses (see Supporting Information for details). Our results support as the best fit an OU model based on a multidimensional ecological niche hypothesis (AICc weight = 0.94; Fig. 3 and Table S3) in which diet composition, locomotion strategy, and group size are the main adaptive landscape-defining variables. Moreover, this suggests that ecological factors represented by group size could be responsible for the observed cladewise convergence in platyrrhines, as further indicated by significant shape differences between species with large (i.e., more than 15 individuals) and small groups (phylogenetic multivariate ANOVA for PC1 and PC2; P = 0.001, R2 = 0.74). Overall, our results indicate that adaptive brain shape diversification in platyrrhines can be invoked as a plausible explanation for the observed patterns, and particularly, to account for brain shape convergence in the clade.
Fig. S4.
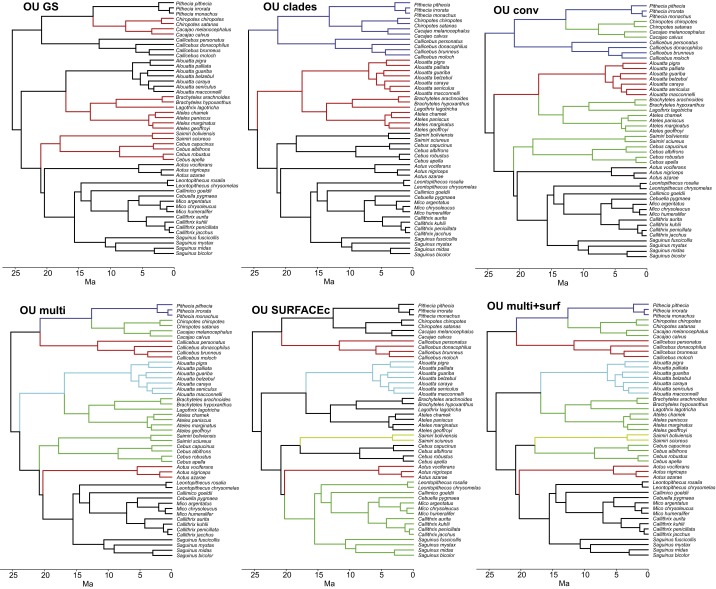
Alternative multivariate OU hypotheses for the evolution of platyrrhine brain shape. The trees depict the different multiregime OU hypotheses included in the model selection analyses performed in mvMORPH. The first model (OU GS) hypothesizes group size (GS) as the only defining dimension of the adaptive landscape for brain shape [large GS species (i.e., 15 or more individuals) vs. small GS species]. The second model (OU clades) considers a fully phylogenetic hypothesis where each platyrrhine family occupies a separate adaptive peak. The third model (OU conv) is a more complex model (five regimes) where each family occupies an adaptive peak but GS drives morphological convergence. “OU multi” model is equally complex as “OU conv” model but hypothesizes an adaptive landscape defined by three ecological dimensions (diet composition, locomotion strategy, and GS; Supporting Information). In this model, GS is also the ecological dimension driving brain shape convergence. The other two ecological dimensions were previously used to explain phenotypic diversification (e.g., in body size) in the platyrrhine adaptive radiation (17, 18). The “OU SURFACEc” model is a slightly modified version of the best-fitting model returned by the SURFACE analysis. The last model (“OU multi + surf”) is a modified version of the OU multi model after the SURFACE results, in which Saimiri is assigned a different adaptive regime. Additionally to these multiregime OU models, we also fitted a single-peak OU, Brownian motion, and early-burst models.
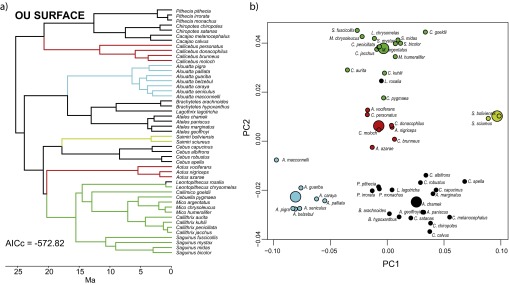
Fig. S5.
Results of the SURFACE analysis. Adaptive regimes for the best-fitting model found by SURFACE are mapped on the platyrrhine tree (A) and in the morphospace of PC1 and PC2 (B). Large circles in B represent the position of the estimated optima for each regime.
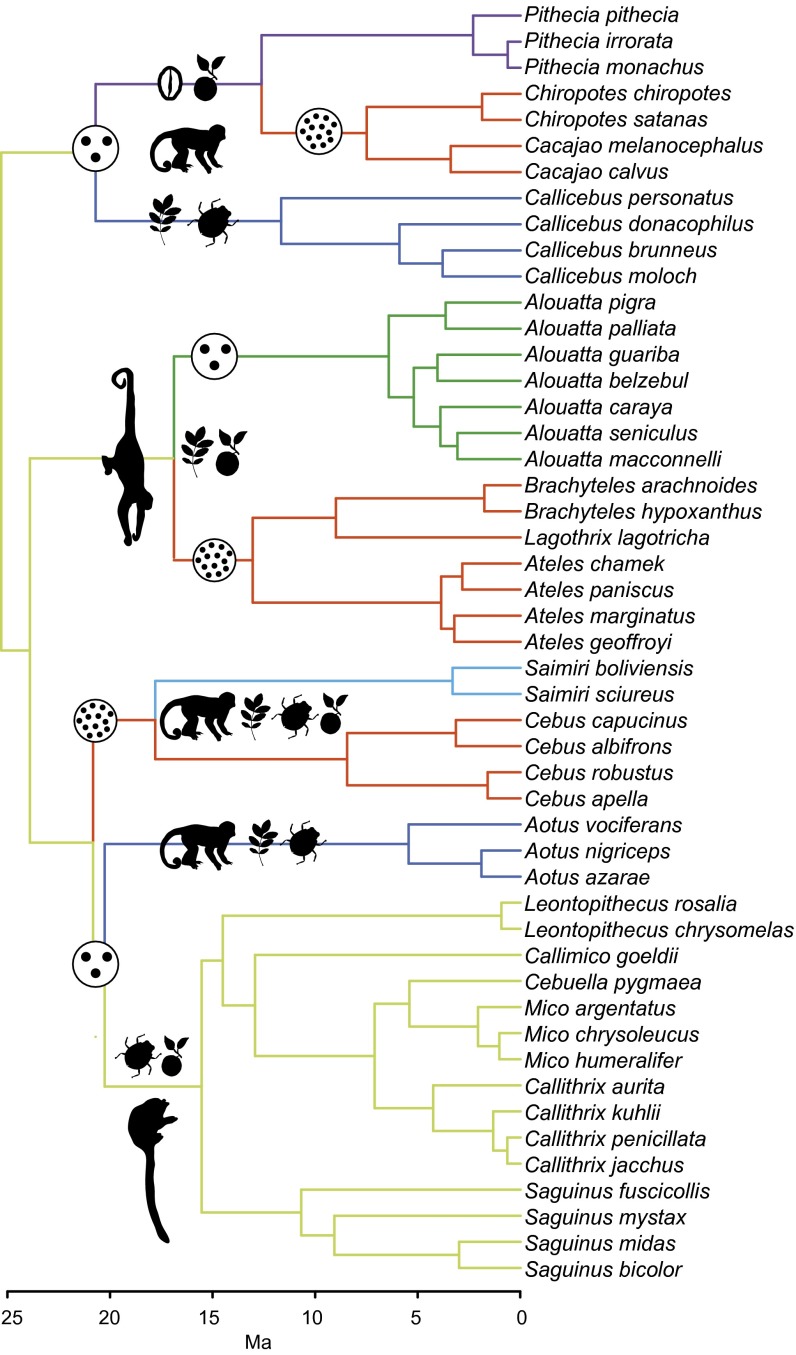
Fig. 3.
Time-calibrated phylogenetic tree for the studied New World monkey species showing adaptive regimes for the best-fitting model of brain shape evolution. Regimes were assigned based on diet composition, locomotion strategy, and group size with exception of Saimiri, which was assigned based on the SURFACE results (Supporting Information). Drawings broadly depict the ecological categories that define these regimes and are not intended to represent ancestral states. Diets are represented by niche-defining food items (17). Locomotion is represented by typical behaviors. Group size (small and large) is represented by encircled dots. Convergent regimes are defined by having large group size. Aotus and Callicebus are considered by some workers to be sister clades.
Table S3.
Results of the multivariate model-fitting analyses for brain shape
| Model | P | AIC | AICc | ΔAICc | AICc weight |
| OU multi + surf | 18 | −594.44 | −571.64 | 0 | 0.94 |
| SURFACEc | 16 | −583.29 | −566.29 | 5.35 | 0.06 |
| GS | 10 | −548.12 | −542.33 | 29.31 | 0.00 |
| OU multi | 16 | −549.41 | −532.41 | 39.23 | 0.00 |
| BM | 5 | −530.20 | −528.81 | 42.83 | 0.00 |
| EB | 6 | −530.17 | −528.17 | 43.47 | 0.00 |
| OU1 | 8 | −525.44 | −521.84 | 49.80 | 0.00 |
| Conv | 14 | −523.13 | −510.77 | 60.87 | 0.00 |
| Clades | 12 | −467.58 | −458.92 | 112.72 | 0.00 |
For each model, the number of parameters (P), the Akaike information criterion (AIC), the small sample corrected AIC (AICc), and the relative fit (ΔAICc) and support (AICc weight) are shown. The best model has the lowest ΔAICc. See Fig. S4 and Supporting Information for detailed descriptions of OU models.
Discussion
The evolution of relatively large and complex brains is perhaps one of the key determinants of primate evolutionary success. Additionally, several extant and extinct primate clades constitute putative examples of adaptive radiations (10), where one of the expected outcomes is the marked adaptive diversification of ecologically relevant phenotypic traits. However, brain morphology has received little attention in the study of primate adaptive radiations despite its manifest ecological importance. Moreover, most previous comparative studies of primate brain evolution have mainly explored the diversification of relative brain size, having generally overlooked the evolutionary and ecological importance of variation in other phenotypic dimensions of the process of brain evolution, e.g., variation in the relative size and position—shape—of the different brain modules.
Our results indicate that New World monkeys present an extensive variation in brain shape and, importantly, that only a fraction of this variation is related to total relative brain size, indicating that a significant amount of information is not captured by this variable, as other workers have pointed out (23). Relative brain size changes are mostly associated with a relative enlargement of the neocortex, with most localized shape changes concentrated on the frontal and occipital areas, and correlated changes in other brain modules (Fig. S3). Additionally, in species exhibiting this neocortex enlargement, the brainstem shifts its relative position to a more downwardly oriented location (Fig. 1B and Movie S1). Therefore, although our approach (i.e., virtual endocasts and geometric morphometrics methods) only allowed quantification of external brain morphology, it can contribute valuable information that may be otherwise overlooked, as the conjoint changes in relative size and position of brain components or structures—as described above—cannot be measured only with volumetric assessments of the whole brain (i.e., its absolute or relative volume) or brain parts.
Our results also show that platyrrhines display a significantly high phylogenetic structure in brain shape variation, as in other previously studied phenotypic traits (e.g., refs. 21 and 24). Although this is not unexpected at macroevolutionary scales (25), our results additionally show that several clades exhibit significantly convergent brain morphologies, suggesting the existence of ecological or nonneutral evolutionary factors structuring brain shape variation. Moreover, our model selection results indicate that, beyond this clear phylogenetic structure, brain diversification among platyrrhines can be explained invoking a Simpsonian model of adaptive peak shifts to unique and shared optima, mainly defined by a multidimensional ecological hypothesis. Particularly, our results suggest that brain shape diversification in the platyrrhine radiation may have unfolded in at least two stages (Fig. 3): an early differentiation (i.e., among families) within an ecological adaptive landscape mainly defined by diet and locomotion strategy axes, followed by several shifts at the subfamily level to a shared adaptive peak defined by social group size, generating shape convergence. The first stage may be related—although not exclusively—to the changes represented by PC2, which shows a strong early diversification pattern, strikingly similar to that of body mass (Fig. 2), which previous works proposed to have diversified within an adaptive landscape associated with diet and locomotion strategies (17, 18). This similarity in the patterns of diversification of body mass and a component of brain shape may point to the action of common selective factors on both traits, or alternatively, to the existence of developmental constraints. Interestingly, we only found evidence for a moderate evolutionary allometry linking variation in both traits (PGLS: λ = 0.92; R2 = 0.25; P = 0.001); thus, modular over integrated explanations may be favored. However, we cannot rule out completely the existence of integrated changes affecting brain morphology. For example, correlated structural changes with the face—which may reflect dietary adaptations—could be a major source of the ecological signal in brain shape.
Following this initial divergence process, our results suggest that ensuing shifts to a shared adaptive peak (Fig. 3), generating cladewise convergence at the subfamily level in particular aspects of brain morphology (e.g., expansion of the neocortex and its correlated increase in relative brain size; Fig. S3), could be a significant mode of brain shape change in platyrrhine evolution. Particularly, a model explicitly incorporating brain shape convergence (Fig. 3) in relation to the several ecological factors that are likely represented by group size (e.g., social complexity; see below) (16) was strongly favored over several nonconvergent models (Fig. S4). Recent studies have suggested that phenotypic convergence could be a frequent phenomenon in macroevolutionary radiations (26–28), although this is typically observed in radiations where the convergent species evolved in geographic isolation from each other. Although some theoretical models (29) and empirical examples (8) describe the emergence of morphological convergence in the same geographic area, these cases refer to communities where the number of species largely exceeds the number of available ecological niches. Alternatively, our results suggest that brain shape convergence was likely attained at a late stage of the radiation, as described above, only after divergence in several other dimensions of the niche space was achieved.
This scenario may be in agreement with proposed general models and examples of vertebrate adaptive radiations (4, 30), in which clades diversify in stages, for example, first diverging in traits allowing differential exploitation of the macrohabitat [e.g., body size for platyrrhines (17, 18)], followed by more narrow sequential partitioning of niche dimensions (e.g., microhabitat, diet specializations, behavior, communication, etc.). In the case of platyrrhines, one of this late niche dimensions could be represented by the complexity of social interactions (i.e., a cognitive dimension of the niche). Convergent phenotypes exhibit, although not exclusively, encephalization-related shape changes. Particularly, the neocortex is the main structure exhibiting a relative enlargement. Because brain size in primates scales almost isometrically with cell number (31), relatively larger brain structures would have relatively higher computational power (14), indicating an increase in the cognitive abilities related with the enlarging neocortex. In this sense, our results suggests that the social brain hypothesis, which posits that the large neocortex of primates evolved in response to the cognitive demands arising from living in complex social groups (16, 32), can be invoked to better understand the evolution at macroevolutionary scales of brain shape and encephalized phenotypes in an ecological context, and particularly, the factors generating brain morphological convergence among primate species. In the case of Saimiri, which is the most encephalized genus and which our results showed to occupy its own adaptive peak, additional factors beyond those considered here may be driving its evolution.
Summarizing, we show that brain shape constitutes an ecologically relevant phenotypic trait in the platyrrhine adaptive radiation, and particularly, that the evolution of specific aspects of brain shape probably allowed the exploitation of additional dimensions of the platyrrhine ecological niche space. Overall, this further indicates that brain shape evolution could be of relevance for understanding other cases of primate and, more generally, vertebrate adaptive radiations.
Materials and Methods
Sample.
Our sample consisted of 179 adult skulls of both sexes from 49 platyrrhine species belonging to all 17 broadly recognized genera (Table S1). This represents a large sample of platyrrhine phylogenetic diversity (∼40–60% of extant species) (33, 34).
Morphological Analyses.
For each specimen X-ray computed tomography or micro-computed tomography scans were acquired or downloaded from public repositories (21). Three-dimensional virtual endocasts were generated from these data following a threshold-based 2D segmentation procedure (21, 35). From the segmented images, a surface 3D model—the virtual endocast—was generated, and the endocranial volume (ECV) was measured as the volume enclosed by this surface.
A total of 26 anatomical landmarks (Table S2) and 373 semilandmarks along curves and surfaces were digitized on each endocast (Fig. 1A and Fig. S6). To digitize the surface semilandmarks, a mesh of 268 roughly equidistant points was generated automatically on one specimen and then projected onto every other endocast, using the landmarks and curve semilandmarks as reference points for alignment and a thin-plate spline interpolation (36), as implemented in the geomorph package for R (37). To remove nonshape variation, a generalized Procrustes analysis (GPA) was performed. GPA also returns the centroid size (CS) of each configuration, a measure of the size of the structure. Additionally, semilandmarks were allowed to slide along tangents and tangent planes to the curves and surfaces, respectively, minimizing the bending energy distance between semilandmarks in each specimen and the consensus configuration (38). The resulting Procrustes shape coordinates were extracted.
Fig. S6.
Landmarks and curves semilandmarks used in this study. Curves were placed to delimitate different anatomical regions of the brain, such as the frontal from the temporal lobe or the cerebellum and stem areas from the rest of the brain. Also, the left and right hemispheres are separated by a curve (Table S2).
Variability in brain shape was characterized by means of a PCA of the species-averaged Procrustes coordinates (Dataset S1). This reduces the dataset dimensionality and generates uncorrelated axes (PCs) describing the main trends in shape variation among species. Subsequently, PC scores were used in the following analyses as brain shape variables.
We measured encephalization (or relative brain size) as the residual values (rECV) from a phylogenetic regression of ECV on basicranial CS (21). Encephalization has been measured in many different ways in primate studies, and the choice mainly depends on the question being asked as all present strengths and weaknesses (39). Our measure is conceptually different from that of other platyrrhine studies, which mainly corrected for body mass (e.g., refs. 19 and 20) but is similar to that used by other workers (e.g., refs. 40 and 41). Using this measurement, we have a more structural encephalization definition, avoiding the use of body mass data, which can be subjected to different selective pressures to that experienced by brain size, affecting evolutionary interpretations (42).
Comparative Analyses.
The phylogeny for the platyrrhine species was obtained from the fossil-calibrated Bayesian molecular tree in Aristide et al. (18) and was pruned to match the species in our morphometric dataset (Dataset S2).
PGLSs were used to assess the association between the various analyzed variables. PGLS accounts for the expected lack of independence among samples arising from phylogenetic structure (43) by modeling residual variation assuming Brownian evolution. This assumption can be ameliorated by incorporating an additional parameter to the regression, λ [ranging from 0 to 1 and estimated by maximum likelihood (44)], which measures the phylogenetic signal in the regression residuals.
Convergence was assessed using two pattern-based test statistics (C1 and C5), which quantify independently evolved similarity without making assumptions about the processes generating this similarity (45). This way, investigation of the processes driving convergent evolution can be separated from its identification. C1 quantifies the degree to which the putatively convergent taxa have evolved to be more similar than their reconstructed ancestors were. C5 simply counts the number of lineages entering the region of the morphospace occupied by the hypothesized convergent taxa. Both measures were compared against a null expectation generated using 999 BM trait simulations along the phylogeny (45).
Phylogenetic signal in trait data were estimated by Blomberg’s K statistic (25). K measures the association between the pattern of variation in the data and the structure of the phylogenetic tree, considering a BM model of evolution as a null expectation. Values of K near 1 indicate a strong phylogenetic signal, whereas values near 0 indicate a decoupling of phylogenetic and phenotypic divergence. K values above 1 indicate that species are more similar than expected under BM, a pattern that may be associated with early niche-filling scenarios (46).
DTT plots (47) were generated in the GEIGER package for R (48) to examine the temporal pattern of change in relative phenotypic disparity along the platyrrhine phylogeny. DTT analyses allow comparing the observed pattern of intraclade versus among-clades disparity through time with an expectation under BM. High relative disparity values are indicative of extensive within-clade diversification and among-clades overlap, whereas values near 0 suggest that variation is mainly partitioned among clades (47).
Model selection analyses were performed with the mvMORPH package for R (49), which allows fitting several evolutionary models to trait data and a phylogeny in a multivariate framework. For each model, the relative fit was assessed using the sample size AICc (50). Several models were evaluated, with BM evolution as the simplest. More complex models included early burst (51), were the rates of Brownian evolution decays exponentially with time, mimicking niche-filling scenarios; and several adaptive OU models (Fig. S4). The SURFACE method (52) was used to explore the OU model space to corroborate that our best-fitting biological hypothesis is not markedly different from the best possible statistical hypothesis (Supporting Information and Fig. S5). OU models describe processes where traits values are constrained around one or several optima that can be considered adaptive peaks in a macroevolutionary landscape. These models incorporate to the stochastic BM model a deterministic term representing the action of selection toward an optimum value, and constitute the simplest mathematical expression of an evolutionary model with selection (53). Each OU model is constructed by assigning hypothesized adaptive regimes to each branch and node of the phylogeny following alternative evolutionary scenarios for the traits under study.
Testing for Adaptive Brain Shape Evolution in New World Monkeys
We constructed several OU models to test whether adaptive evolution could be invoked to explain brain diversification. Each model represents alternative biological hypotheses about the factors that may structure the adaptive landscape for platyrrhines. For each model, we calculated a measure of explanatory power or statistical fit. Importantly, the purpose of such evolutionary models is to aid in the reasoning about the factors and mechanisms underlying the evolution of the studied traits (54) and are not, therefore, definite or complete explanations. We initially constructed four adaptive hypotheses based on different biological criteria (Fig. S4) and tested their relative fit using AICc scores. This way we obtained a measure of the relative explanatory power of each hypothesis (ΔAICc). We followed Rosenberger (17) criteria to assign each primate species to dietary and locomotion categories in the case of our “multidimensional” models (Fig. S4; “OU multi” and “OU multi + surf”). Rosenberger proposed that the platyrrhine radiation unfolded as the lineages diversified into several dietary adaptive zones, which are associated with the evolution of locomotor and body size adaptations. Starting with this model, we hypothesize group size as an additional dimension structuring the adaptive landscape for brain shape (Fig. 3). We created two categories [small and large (more than 15 individuals)] and treated group size as a discrete variable. Ancestral states were reconstructed following a maximum-likelihood criterion, with the exception of the states at the root and basal branches. For these, the states were assigned based on the fossil record that indicates a generalized locomotion mode, a more insectivorous diet, and a small size for the oldest platyrrhines (55), most similar to the smaller members of the Cebidae family (Callitrichinae). Ancestral states in other models were assigned manually, considering shared states of descending branches for each node.
As these tested hypotheses constitute only a limited set among all possible alternatives, it may well be that none of them represent an accurate description of the actual macroevolutionary landscape. In other words, we may be only choosing the best among very bad hypotheses. To overcome this potential problem, we additionally used the SURFACE (52) method. This method uses a stepwise algorithm to locate selective regime shifts along the phylogeny without resorting to any a priori assignation of such regimes. Briefly, the SURFACE algorithm sequentially adds regime changes to the tree wherever they produce an improvement in the AICc score of the model (“forward” phase) until no further improvement is achieved (i.e., ΔAICc < 2). Next, the method assesses whether collapsing regimes in different branches to the same adaptive peak (implicating convergence) can further improve the model obtained in the forward phase. This “backward” phase proceeds step by step until, again, no further improvement is achieved. The SURFACE method thereby can explore several hundred OU models, obtaining as a result the best possible model in statistical terms; or, at least, a model with a high absolute statistical support because not all of the possible models are explored by the algorithm. Last, if the best model found by SURFACE is markedly different from our biological model, this may indicate that our model is probably not any good at describing the macroevolutionary landscape for brain shape in the platyrrhines. Because SURFACE cannot fit data in a multivariate fashion (i.e., considering evolutionary correlations among variables), the model found by SURFACE was translated into the mvMORPH package and tested along the alternative hypotheses. The best model found by SURFACE (Fig. S5) is broadly similar to our OU multi model (Fig. S4), with a few differences. Noticeably, the SURFACE model reconstructs as the ancestral regime for platyrrhines the regime that we hypothesize as convergent (Fig. S4). This would imply that callitrichines (Callitrichinae subfamily; black regime in the OU multi model; Fig. S4) are derived with respect to the ancestral platyrrhines. However, this is in disagreement with the fossil record, which shows that the oldest platyrrhine representatives were probably similar (e.g., in body size and diet) to members of this subfamily (55) and, most importantly, that oldest platyrrhines had small brain size and a reduced frontal lobe (56), contrary to the species assigned to the ancestral regime by SURFACE. Still, SURFACE coincides with our biological model in the assignment of a common regime to these large-brained species, whether it is ancestral or not. On the other hand, in the SURFACE model, Leontopithecus rosalia is assigned to this same regime, which makes no biological sense because L. rosalia is very similar to its sister species both from morphological and ecological viewpoints. Additionally, Saimiri species occupy their own adaptive peak, which is not surprising given our morphometric results (Fig. 1 and Fig. S5). Given this last difference, we decided to incorporate a new regime for Saimiri in our OU multi model (OU multi + surf; Fig. S4). Noticeably, this lead to a high increase in the fit of our model, making it statistically indistinguishable from the SURFACE result (ΔAICc < 2). This indicates not only that our corrected a priori hypothesis is the best among several others (Table S3) but also that it has a high absolute statistical support. Moreover, taking into account the SURFACE results for L. rosalia, we constructed a modified version of this model (“SURFACEc”; Fig. S4) in which this species is assigned to the same regime as its sister species, thus being more biologically realistic. This minor change led to a marked drop in the statistical support of this model (Table S3), making our a priori biological hypothesis, with the addition of the Saimiri regime, the preferred one (AICc weight = 0.94). Thus, by combining our a priori expectations with the SURFACE results, we constructed a likely biological model with a high absolute statistical support for the evolution of brain shape in the New World monkeys.
Supplementary Material
Acknowledgments
We thank J. de Oliveira (Museu Nacional do Rio de Janeiro), D. Flores (Museo Argentino de Ciencias Naturales), M. de Vivo (Museu de Zoologia da Universidade de São Paulo), and the Digital Morphology Museum (Kyoto University) for granting us access to the collections under their care. We are grateful to A. Rosenberger and two anonymous reviewers for useful comments. This research was supported by grants from the Fondo para la Investigación Científica y Tecnológica, Conselho Nacional de Desenvolvimento Científico e Tecnológico, and Fundação de Amparo à Pesquisa do Estado de São Paulo.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.C.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514473113/-/DCSupplemental.
References
- 1.Futuyma DJ. 1998. Evolutionary Biology (Sinauer Associates, Sunderland, MA)
- 2.Gavrilets S, Losos JB. Adaptive radiation: Contrasting theory with data. Science. 2009;323(5915):732–737. doi: 10.1126/science.1157966. [DOI] [PubMed] [Google Scholar]
- 3.Simpson GG. The Major Features of Evolution. Columbia Univ Press; New York: 1953. [Google Scholar]
- 4.Losos JB. Adaptive radiation, ecological opportunity, and evolutionary determinism. American Society of Naturalists E. O. Wilson Award Address. Am Nat. 2010;175(6):623–639. doi: 10.1086/652433. [DOI] [PubMed] [Google Scholar]
- 5.Jønsson KA, et al. Ecological and evolutionary determinants for the adaptive radiation of the Madagascan vangas. Proc Natl Acad Sci USA. 2012;109(17):6620–6625. doi: 10.1073/pnas.1115835109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Losos JB. Lizards in an Evolutionary Tree: Ecology and Adaptive Radiation of Anoles. Univ of California Press; Berkeley, CA: 2009. [Google Scholar]
- 7.Soons J, et al. Is beak morphology in Darwin’s finches tuned to loading demands? PLoS One. 2015;10(6):e0129479. doi: 10.1371/journal.pone.0129479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muschick M, Indermaur A, Salzburger W. Convergent evolution within an adaptive radiation of cichlid fishes. Curr Biol. 2012;22(24):2362–2368. doi: 10.1016/j.cub.2012.10.048. [DOI] [PubMed] [Google Scholar]
- 9.Barton RA. Primate brain evolution: Integrating comparative, neurophysiological and ethological data. Evol Anthropol. 2006;15(6):224–236. [Google Scholar]
- 10.Fleagle JG. Primate Adaptation and Evolution. Academic; San Diego: 2013. [Google Scholar]
- 11.Dunbar RIM. The social brain hypothesis. Evol Anthropol. 1998;6(5):178–190. [Google Scholar]
- 12.Barton RA, Harvey PH. Mosaic evolution of brain structure in mammals. Nature. 2000;405(6790):1055–1058. doi: 10.1038/35016580. [DOI] [PubMed] [Google Scholar]
- 13.de Winter W, Oxnard CE. Evolutionary radiations and convergences in the structural organization of mammalian brains. Nature. 2001;409(6821):710–714. doi: 10.1038/35055547. [DOI] [PubMed] [Google Scholar]
- 14.Smaers JB, Soligo C. Brain reorganization, not relative brain size, primarily characterizes anthropoid brain evolution. Proc Biol Sci. 2013;280(1759):20130269. doi: 10.1098/rspb.2013.0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huber R, van Staaden MJ, Kaufman LS, Liem KF. Microhabitat use, trophic patterns, and the evolution of brain structure in African cichlids. Brain Behav Evol. 1997;50(3):167–182. doi: 10.1159/000113330. [DOI] [PubMed] [Google Scholar]
- 16.Dunbar RIM, Shultz S. Evolution in the social brain. Science. 2007;317(5843):1344–1347. doi: 10.1126/science.1145463. [DOI] [PubMed] [Google Scholar]
- 17.Rosenberger AL. Evolution of feeding niches in New World monkeys. Am J Phys Anthropol. 1992;88(4):525–562. doi: 10.1002/ajpa.1330880408. [DOI] [PubMed] [Google Scholar]
- 18.Aristide L, Rosenberger AL, Tejedor MF, Perez SI. Modeling lineage and phenotypic diversification in the New World monkey (Platyrrhini, Primates) radiation. Mol Phylogenet Evol. 2015;82(Pt B):375–385. doi: 10.1016/j.ympev.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Hartwig W, Rosenberger AL, Norconk MA, Owl MY. Relative brain size, gut size, and evolution in New World monkeys. Anat Rec (Hoboken) 2011;294(12):2207–2221. doi: 10.1002/ar.21515. [DOI] [PubMed] [Google Scholar]
- 20.Allen KL, Kay RF. Dietary quality and encephalization in platyrrhine primates. Proc Biol Sci. 2012;279(1729):715–721. doi: 10.1098/rspb.2011.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aristide L, et al. Encephalization and diversification of the cranial base in platyrrhine primates. J Hum Evol. 2015;81:29–40. doi: 10.1016/j.jhevol.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Schluter D. The Ecology of Adaptive Radiation. Oxford Univ Press; Oxford: 2000. [Google Scholar]
- 23.Healy SD, Rowe C. A critique of comparative studies of brain size. Proc Biol Sci. 2007;274(1609):453–464. doi: 10.1098/rspb.2006.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez SI, Klaczko J, Rocatti G, Dos Reis SF. Patterns of cranial shape diversification during the phylogenetic branching process of New World monkeys (Primates: Platyrrhini) J Evol Biol. 2011;24(8):1826–1835. doi: 10.1111/j.1420-9101.2011.02309.x. [DOI] [PubMed] [Google Scholar]
- 25.Blomberg SP, Garland T, Jr, Ives AR. Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution. 2003;57(4):717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 26.Mahler DL, Ingram T, Revell LJ, Losos JB. Exceptional convergence on the macroevolutionary landscape in island lizard radiations. Science. 2013;341(6143):292–295. doi: 10.1126/science.1232392. [DOI] [PubMed] [Google Scholar]
- 27.Moen DS, Irschick DJ, Wiens JJ. Evolutionary conservatism and convergence both lead to striking similarity in ecology, morphology and performance across continents in frogs. Proc Biol Sci. 2013;280(1773):20132156. doi: 10.1098/rspb.2013.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grundler MC, Rabosky DL. Trophic divergence despite morphological convergence in a continental radiation of snakes. Proc Biol Sci. 2014;281(1787):20140413. doi: 10.1098/rspb.2014.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheffer M, van Nes EH. Self-organized similarity, the evolutionary emergence of groups of similar species. Proc Natl Acad Sci USA. 2006;103(16):6230–6235. doi: 10.1073/pnas.0508024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Streelman JT, Danley PD. The stages of vertebrate evolutionary radiation. Trends Ecol Evol. 2003;18:126–131. [Google Scholar]
- 31.Herculano-Houzel S, Collins CE, Wong P, Kaas JH. Cellular scaling rules for primate brains. Proc Natl Acad Sci USA. 2007;104(9):3562–3567. doi: 10.1073/pnas.0611396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunbar RIM. Neocortex size as a constraint on group size in primates. J Hum Evol. 1992;22(6):469–493. [Google Scholar]
- 33.Groves CP. Primate Taxonomy. Smithsonian Institution Press; Washington, DC: 2001. [Google Scholar]
- 34. International Union for Conservation of Nature (2014) IUCN Red List of Threatened Species, Version 2015.2. Available at www.iucnredlist.org. Accessed March 17, 2015.
- 35.Neubauer S, Gunz P, Hublin J-J. The pattern of endocranial ontogenetic shape changes in humans. J Anat. 2009;215(3):240–255. doi: 10.1111/j.1469-7580.2009.01106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitteroecker P, Gunz P. Advances in geometric morphometrics. Evol Biol. 2009;36:235–247. [Google Scholar]
- 37.Adams DC, Otarola-Castillo E. geomorph: An R package for the collection and analysis of geometric morphometric shape data. Methods Ecol Evol. 2013;4(4):393–399. [Google Scholar]
- 38.Gunz P, Mitteroecker P, Bookstein FL. Semilandmarks in three dimensions. In: Slice DE, editor. Modern Morphometrics in Physical Anthropology. Kluwer Academic/Plenum Publishers; New York: 2005. pp. 73–98. [Google Scholar]
- 39.Lefebvre L. Primate encephalization. In: Hofman MA, Falk D, editors. Progress in Brain Research. Vol 195. Elsevier; Amsterdam: 2012. pp. 393–412. [DOI] [PubMed] [Google Scholar]
- 40.Gould SJ. Ontogeny and Phylogeny. Harvard Univ Press; Cambridge, MA: 1977. [Google Scholar]
- 41.Ross CF, Ravosa MJ. Basicranial flexion, relative brain size, and facial kyphosis in nonhuman primates. Am J Phys Anthropol. 1993;91(3):305–324. doi: 10.1002/ajpa.1330910306. [DOI] [PubMed] [Google Scholar]
- 42.Smaers JB, Dechmann DK, Goswami A, Soligo C, Safi K. Comparative analyses of evolutionary rates reveal different pathways to encephalization in bats, carnivorans, and primates. Proc Natl Acad Sci USA. 2012;109(44):18006–18011. doi: 10.1073/pnas.1212181109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martins EP, Hansen TF. Phylogenies and the comparative method: A general approach to incorporating phylogenetic information into the analysis of interspecific data. Am Nat. 1997;149(4):646–667. [Google Scholar]
- 44.Freckleton RP, Harvey PH, Pagel M. Phylogenetic analysis and comparative data: A test and review of evidence. Am Nat. 2002;160(6):712–726. doi: 10.1086/343873. [DOI] [PubMed] [Google Scholar]
- 45.Stayton CT. The definition, recognition, and interpretation of convergent evolution, and two new measures for quantifying and assessing the significance of convergence. Evolution. 2015;69(8):2140–2153. doi: 10.1111/evo.12729. [DOI] [PubMed] [Google Scholar]
- 46.Revell LJ, Harmon LJ, Collar DC. Phylogenetic signal, evolutionary process, and rate. Syst Biol. 2008;57(4):591–601. doi: 10.1080/10635150802302427. [DOI] [PubMed] [Google Scholar]
- 47.Harmon LJ, Schulte JA, 2nd, Larson A, Losos JB. Tempo and mode of evolutionary radiation in iguanian lizards. Science. 2003;301(5635):961–964. doi: 10.1126/science.1084786. [DOI] [PubMed] [Google Scholar]
- 48.Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. GEIGER: Investigating evolutionary radiations. Bioinformatics. 2008;24(1):129–131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
- 49.Clavel J, Escarguel G, Merceron G. mvMORPH: An R package for fitting multivariate evolutionary models to morphometric data. Methods Ecol Evol. 2015;6(11):1311–1319. [Google Scholar]
- 50.Burnham KP, Anderson DR. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. Springer; New York: 2002. [Google Scholar]
- 51.Harmon LJ, et al. Early bursts of body size and shape evolution are rare in comparative data. Evolution. 2010;64(8):2385–2396. doi: 10.1111/j.1558-5646.2010.01025.x. [DOI] [PubMed] [Google Scholar]
- 52.Ingram T, Mahler DL. SURFACE: Detecting convergent evolution from comparative data by fitting Ornstein–Uhlenbeck models with stepwise Akaike information criterion. Methods Ecol Evol. 2013;4(5):416–425. [Google Scholar]
- 53.Butler MA, King AA. Phylogenetic comparative analysis: A modeling approach for adaptive evolution. Am Nat. 2004;164:683–695. doi: 10.1086/426002. [DOI] [PubMed] [Google Scholar]
- 54.Cressler CE, Butler MA, King AA. Detecting adaptive evolution in phylogenetic comparative analysis using the Ornstein–Uhlenbeck model. Syst Biol. 2015;64(6):953–968. doi: 10.1093/sysbio/syv043. [DOI] [PubMed] [Google Scholar]
- 55.Bond M, et al. Eocene primates of South America and the African origins of New World monkeys. Nature. 2015;520(7548):538–541. doi: 10.1038/nature14120. [DOI] [PubMed] [Google Scholar]
- 56.Kay RF, et al. Paleobiology of Santacrucian primates. In: Vizcaíno S, Kay RF, Bargo M, editors. Early Miocene Paleobiology in Patagonia: High-Latitude Paleocommunities of the Santa Cruz Formation. Cambridge Univ Press; Cambridge, UK: 2012. pp. 306–330. [Google Scholar]
- 57.Revell LJ. Two new graphical methods for mapping trait evolution on phylogenies. Methods Ecol Evol. 2013;4:754–759. [Google Scholar]
- 58.Santana SE, Alfaro JL, Alfaro ME. Adaptive evolution of facial colour patterns in Neotropical primates. Proc Biol Sci. 2012;279(1736):2204–2211. doi: 10.1098/rspb.2011.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.