Significance
Extensive studies of the structure–function relationship of antibodies have established that conventional immunoglobulins contain two copies of the antigen-binding fragment (Fab), each of which serves as an autonomous and complete unit for recognizing an antigen. In this paper, we report a previously unidentified mode of antibody–antigen recognition, dubbed “antigen clasping,” where two antigen-binding sites cooperatively clasp one antigen, and the design of a long-neck antibody format that facilitates antigen clasping. Antigen clasping led to recombinant antibodies for histone posttranslational modifications with extraordinarily high specificity, valuable tools for epigenetic research. This study substantially broadens the long-standing paradigm for antibody–antigen recognition.
Keywords: antibody engineering, epigenetics, antibody validation, protein–protein interaction, data reproducibility
Abstract
Antibodies have a well-established modular architecture wherein the antigen-binding site residing in the antigen-binding fragment (Fab or Fv) is an autonomous and complete unit for antigen recognition. Here, we describe antibodies departing from this paradigm. We developed recombinant antibodies to trimethylated lysine residues on histone H3, important epigenetic marks and challenging targets for molecular recognition. Quantitative characterization demonstrated their exquisite specificity and high affinity, and they performed well in common epigenetics applications. Surprisingly, crystal structures and biophysical analyses revealed that two antigen-binding sites of these antibodies form a head-to-head dimer and cooperatively recognize the antigen in the dimer interface. This “antigen clasping” produced an expansive interface where trimethylated Lys bound to an unusually extensive aromatic cage in one Fab and the histone N terminus to a pocket in the other, thereby rationalizing the high specificity. A long-neck antibody format with a long linker between the antigen-binding module and the Fc region facilitated antigen clasping and achieved both high specificity and high potency. Antigen clasping substantially expands the paradigm of antibody–antigen recognition and suggests a strategy for developing extremely specific antibodies.
The antigen-binding site of conventional immunoglobulins (Igs) is primarily composed of six complementarity-determining regions (CDRs) located in the VH and VL domains (Fig. 1A). Antibody fragments such as Fab and Fv are viewed as an autonomous unit containing a single, complete site for antigen recognition (1). The 1:1 stoichiometry of the antigen and Fab (or Fv) is conserved among known antibody structures and isotypes, including the “two-in-one” antibodies whose Fab specifically binds to two distinct antigens, but one at a time (2). This paradigm has been a guiding principle in the engineering of diverse antibody formats such as bispecific antibodies (3).
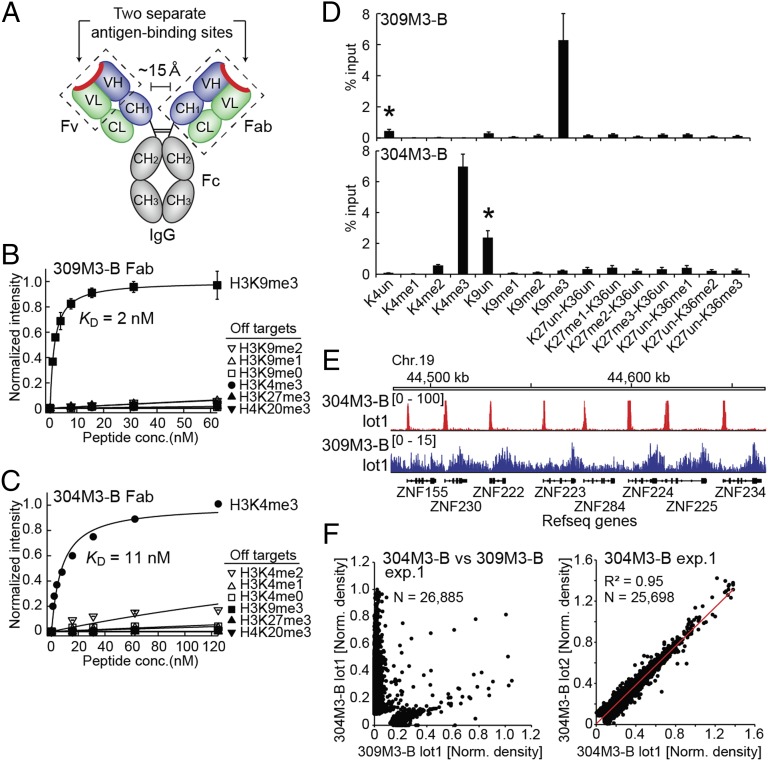
Fig. 1.
Exquisite specificity and high affinity of recombinant antibodies to H3K4me3 and H3K9me3. (A) Schematic structure of the IgG. (B and C) Binding titration curves of the 309M3-B (B) and 304M3-B (C) antibodies to their cognate peptide and off-target peptides measured with the peptide IP assay. The calculated KD value to the cognate peptide is shown. (D) Equal molar amounts of synthetic peptides harboring different PTMs were mixed, and then peptides were captured with 309M3-B (Top) or 304M3-B (Bottom) were quantified with MS. The ratio to the input for each peptide is shown. *, The apparent enrichment of unmodified peptides is derived from enrichment of the input peptides (residues 1–21) harboring H3K9me3 or H3K4me3 that also produce the peptides for H3K4un (residues 3–8) or H3K9un (residues 9–17), respectively, after trypsin digestion. (E) Comparison of ChIP-seq data at the same loci obtained with 304M3-B lot1 and 309M3-B lot1. (F) Scatter plots comparing the normalized read densities (reads per base pair per million mapped reads) for called peaks of the dataset with 304M3-B and 309M3-B (Left) and with different lots of 304M3-B (Right). The square of the Pearson product-moment correlation coefficient (R2) and the total number of peaks compared (N) are indicated.
The terminal regions of histone proteins (“histone tails”) are unstructured and contain many posttranslational modification (PTM) sites that are recognized by epigenetic regulatory machineries involved in transcriptional regulation (4, 5). Antibodies to histone PTMs are essential tools for epigenetics research, but limited validation and large lot-to-lot variation of currently available anti-histone PTM antibodies are major sources of low reproducibility (6–9). The challenge in achieving high specificity and affinity can be reasoned by minute differences among chemical moieties of PTMs, as small as a single methyl group, and sequence similarity surrounding modification sites (e.g., those encompassing H3K9 and H3K27) (SI Appendix, Fig. S1) and by the fundamental challenge in recognizing flexible polypeptides due to unfavorable entropic changes associated with binding. Highly specific recombinant antibodies to histone PTMs, with their essentially infinite renewability, could fundamentally eliminate this major limitation (9).
The limited understanding of the molecular mechanisms underlying the recognition of histone PTMs has severely limited our ability to apply mechanism-based designs to the generation of recombinant antibodies to a wider range of histone PTMs. For example, it is unknown whether existing anti-histone PTM antibodies and natural “reader” proteins use similar mechanisms. Crystal structures of antibody–antigen complexes are critical information for structure-guided design and engineering of antibody affinity and specificity (2, 10). In this study, we isolated highly specific and potent antibodies to trimethylated Lys4 and Lys9 on histone H3 (abbreviated H3K4me3 and H3K9me3, respectively) that are exquisitely specific, potent, and fully validated in standard epigenetics applications. Further, we describe the crystal structures of both antibodies in complex with their respective targets. Their structural and functional analyses revealed an unprecedented mechanism of antigen recognition where two antigen-binding sites cooperatively recognize one antigen.
Results
Generation of Recombinant Antibodies to Trimethylated Histone H3.
We generated recombinant antibodies to H3K4me3 and H3K9me3 by using directed evolution methods that mimic the processes underlying the natural immune system: i.e., generation of low affinity and specificity clones followed by maturation. We have previously identified an antibody that weakly but specifically recognizes trimethyl Lys, termed “clone 4-5”, from a naive human antibody library (9). Then, we generated a single chain Fv (scFv) phage-display library where we diversified a subset of CDR residues (SI Appendix, Fig. S1) (9). After procedures that led to our initial success in targeting H3K9me3 (9) that involved rounds of stringent selection for specific binding to histone peptides, we isolated new antibodies to H3K9me3 and H3K4me3, termed 309M3-B and 304M3-B, respectively.
We then produced these antibodies in the form of Fab and subjected them to extensive validation. They showed apparent dissociation constants (KDs) of 2 nM and 11 nM to their cognate peptide, respectively, as measured with the peptide immunoprecipitation (IP) assay (8). The reason for using the term “apparent” is described in the next section. They had almost no detectable binding to most other peptides tested, indicating high affinity and exquisite specificity (Fig. 1 B and C). 309M3-B was more sensitive to modifications of adjacent residues than our first recombinant antibody to H3K9me3, termed 309M3-A, which we reported previously (9), indicating that 309M3-B recognizes many features of the H3K9me3 peptides outside the K9me3 site itself (SI Appendix, Fig. S1). Similarly, 304M3-B preferentially recognized unmodified forms of R2, T3, and T6 residues (SI Appendix, Fig. S1). Characterization by IP-mass spectrometry (IP-MS) demonstrated that both antibodies specifically enriched their cognate targets from a mixture of synthetic peptides (Fig. 1D) (note that the apparent enrichment of unmodified H3K4 and H3K9 was an artifact due to experimental limitations) and from natural histone extracts (SI Appendix, Fig. S2 and Table S1). The exquisite specificity of both antibodies was further validated by using internal standard calibrated chromatin immunoprecipitation followed by sequencing (ICeChIP) that allows quantitative assessment of specificity in the ChIP format using semisynthetic nucleosomes (11) (SI Appendix, Fig. S2).
Both antibodies performed well in common epigenetics applications. They detected histone H3 in Western blotting of the whole cell extracts, and, in the case of 304M3-B, the intense histone H3 band was absent in the blot for the extracts from a Set1-deleted yeast strain (SI Appendix, Fig. S3). In ChIP-seq of HEK293 cells, 304M3-B produced sharp peaks localized in the promotor regions and 309M3-B produced diffused peaks, and their locations were mutually exclusive (Fig. 1 E and F, Left), patterns consistent with the known distribution of these histone PTM marks (9, 12, 13). As expected for recombinant proteins, results using different batches of these antibodies were highly reproducible (Fig. 1F, Right and SI Appendix, Fig. S3). Therefore, these recombinant antibodies represent extensively validated, high-performance tools for achieving accurate and reproducible results.
Formation of Head-to-Head Dimers That Sandwich the Antigen.
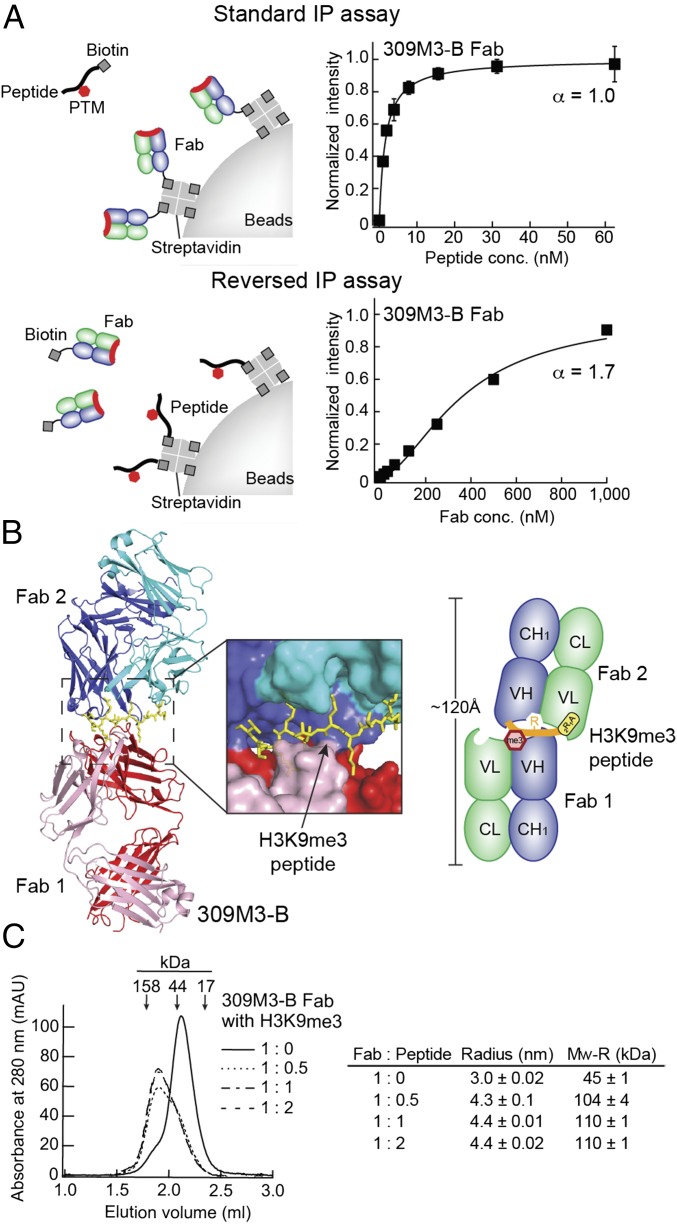
Unexpectedly, the binding properties of these antibodies differed substantially, depending on the orientation of the IP assay (Fig. 2A and SI Appendix, Fig. S4). In our standard IP assay, which mimics the format of the standard immunoprecipitation method (8), a biotinylated Fab is immobilized on streptavidin-coated beads first, and a peptide captured by the antibody is quantified using flow cytometry. In this format, we observed hyperbolic curves consistent with a simple 1:1 binding model. Titrations in the reversed orientation (i.e., soluble antibodies to a peptide immobilized on beads) showed sigmoidal curves with a Hill coefficient substantially greater than unity, indicating cooperating binding, and with much higher half-saturation concentrations than those from the standard format (Fig. 2A and SI Appendix, Fig. S4).
Fig. 2.
Cooperative dimerization of two antigen-binding sites. (A) Effects of the assay orientation on the binding reactions of 309M3-B. (Top) Scheme and titration of a soluble peptide to immobilized antibodies: i.e., the standard IP assay. (Bottom) Scheme and titration in the reversed orientation where a soluble Fab protein is added to immobilized peptide. The titration data in the Top panel is the same as that shown in Fig. 1B. The curves show the best fit of the Hill equation, with the values of the Hill coefficient, α, indicated. Note the different concentration ranges on the horizontal axis. Data shown are from triplicate measurements. (B) The overall structures of 309M3-B in complex with the H3K9me3 peptide. The scheme depicts the binding mechanism of the 309M3-B antibody that forms asymmetric homodimerization with the H3K9me3 peptide. The heavy and light chains in Fab 1 are shown in red and pink, respectively, and those in Fab 2 (the dimerization partner of Fab1) are shown in blue and cyan, respectively. (C) Dimerization of 309M3-B in solution analyzed using gel filtration chromatography (graph) and dynamic light scattering (table). The Fab sample was mixed with the peptides with the indicated ratios and analyzed. Arrows indicate the peak positions of calibration proteins (γ-globulin, ovalbumin, and myoglobin, from left to right). The complete dataset for dynamic light scattering is in SI Appendix, Fig. S5.
Strikingly, the crystal structures of 309M3-B and 304M3-B in complex with peptides corresponding to their respective antigens revealed that both antibodies formed head-to-head Fab dimers (Fig. 2B and SI Appendix, Fig. S4). Two 309M3-B Fab molecules bound one H3K9me3 peptide, with no internal symmetry (Fig. 2B). Similarly, the 304M3-B Fab dimer bound two H3K4me3 peptides, but with the two molecules related by an approximately twofold rotational symmetry operation (SI Appendix, Fig. S4). In contrast, the crystal structure of 309M3-B in complex with trimethylated Lys (i.e., a single amino acid without a polypeptide chain) did not show dimerization, suggesting that its dimerization requires peptide residues adjacent to trimethyl Lys (SI Appendix, Fig. S4).
We then confirmed the occurrence of this unusual binding mode in solution by using size-exclusion chromatography and dynamic light scattering. The sizes of the antibody–peptide complexes were about twice as large as the antibodies alone (Fig. 2C and SI Appendix, Fig. S5). A 2:1 molar ratio of antibody to peptide was sufficient to fully induce the size shift of 309M3-B, whereas a 1:1 ratio was required for 304M3-B. The binding stoichiometries in solution are consistent with those observed in crystals. Also consistent with the crystal data, trimethyl Lys did not induce the size shift of 309M3-B (SI Appendix, Fig. S5). These results conclusively demonstrated antigen-induced dimerization of 309M3-B and 304M3-B. We term this mode of antigen recognition “antigen clasping” hereafter.
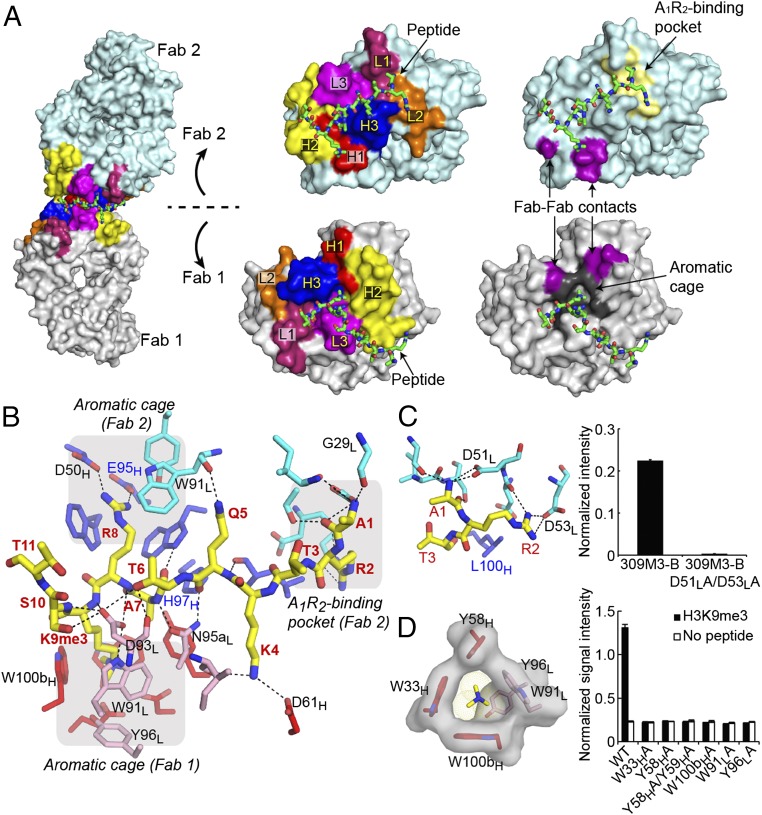
Anti-peptide or anti-small compound antibodies typically have a deep cleft created by long CDRs (14, 15) (SI Appendix, Fig. S6). In contrast, the antigen-binding sites of our antigen-clasping antibodies are flat, consistent with their short CDRs (Fig. 3A and SI Appendix, Fig. S6), a topography similar to that found for antibodies to large spherical antigens such as structured proteins. Antigen clasping by two copies of a flat antigen-binding site created large interaction surfaces. The interfaces (1,177 Å2 for the H3K9me3 peptide and 987 Å2 for the H3K4me3 peptide) (SI Appendix, Table S2) were nearly twice as large as typical peptide–protein binding interfaces (16). 309M3-B used a total of 8 CDRs to recognize the H3K9me3 peptide, and, remarkably, 304M3-B used 12 CDRs (i.e., all CDRs available in the Fab dimer) in binding to two H3K4me3 peptide molecules (Fig. 3A and SI Appendix, Fig. S6). Therefore, antigen clasping enables antibodies to use most CDRs and create large interacting surfaces, leading to high affinity and specificity toward peptide antigens.
Fig. 3.
Recognition of a single H3K9me3 peptide at the interface of two antigen-binding sites. (A) Interactions of the H3K9me3 peptide with the Fab dimer. (Center and Right) The peptide-recognition interface in an open book manner. CDRs that interact with the peptide are labeled in yellow. (Right) Key elements in Fab1 and Fab2. (B) Details of the antibody–peptide interaction interface. The amino acid residues are colored in the same manner as in Fig. 2B. Peptide residue numbers are in red. The A1R2-binding pocket and the aromatic cage are enclosed in a gray shade. Polar interactions are marked as dashed lines. Residues mutated with respect to the lead antibody, 4-5, are labeled in blue. (C) A close-up view of the recognition of the A1R2 motif with its binding pocket in 309M3-B (Left). The D51LA/D53LA double mutant abolishes binding as tested with the peptide IP assay (Right). (D) The aromatic cage in 309M3-B. (Left) The side chains that create the aromatic cage and trimethylated Nε of Lys9. (Right) The effects of alanine mutations of these residues tested with phage ELISA. Binding data shown here are from triplicate measurements. See SI Appendix, Fig. S8 for additional data.
Recognition of Trimethylated Lys and Histone N Terminus with Two Distinct Pockets.
Close inspection of the antibody–peptide interfaces revealed two common features between the two structures. First, there is an “aromatic cage” located at the interface between the heavy and light chains that perfectly fit the trimethylated quaternary ammonium cation of Lys Nε (Fig. 3 B and D and SI Appendix, Fig. S6). Aromatic cages are known binding pockets for methylated Lys moieties in natural histone reader proteins, in which methylated ammonium groups interact with the aromatic side chains mediated by the cation–π interaction (17, 18). The aromatic cage in our antibodies is more extensive and optimal for trimethylated Lys than natural counterparts, rationalizing the much higher ability of our antibodies to distinguish trimethylated Lys from dimethylated Lys (SI Appendix, Fig. S7). Interestingly, Arg8 of the H3K9me3 peptide fit in the aromatic cage in the second Fab molecule and made electrostatic interactions with acidic residues located at the base of the cavity (Fig. 3B). Occupation of an aromatic cage by the Arg side chain has been observed for the SET domain of Ezh2 (19), suggesting that aromatic cages have inherent affinity to Arg.
Second, a binding pocket formed by residues in CDRL1 and L2 recognized the N-terminal two residues (Ala1 and Arg2) of the histone (Fig. 3 B and C and SI Appendix, Fig. S6). Mutation studies confirmed the importance of both the aromatic cage and A1R2-binding pocket for antigen recognition (Fig. 3 C and D and SI Appendix, Figs. S6 and S8).
Antigen Clasping Enables Fine Control of the Distance Between the Two Pockets.
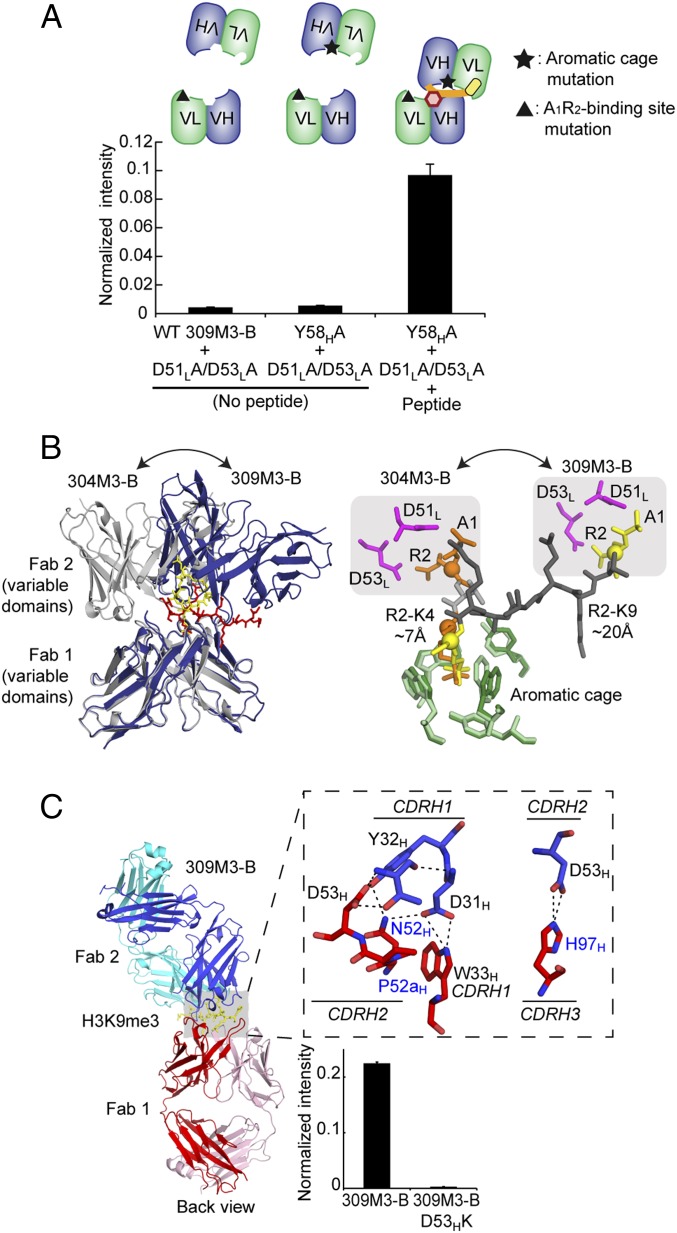
Remarkably, the peptides bound to the A1R2-binding pocket in one Fab and to the aromatic cage in the other in both structures, thereby bridging the two Fab molecules across the dimer interface (Fig. 3 A and B and SI Appendix, Fig. S6). We confirmed this binding mode by constructing an active heterodimer with two complementary “half-site” mutants, one containing a mutation in the aromatic cage and the other containing a mutation in the A1R2-binding pocket (Fig. 4A). Although the homodimers containing mutation in the aromatic cage (Y58HA) or in the A1R2-binding site (D51LA/D53LA) were inactive, the heterodimer of the two mutants regained binding. Together, these features rationalize the unusual cooperative binding and high functionality of these antibodies.
Fig. 4.
Structural basis for the exquisite sequence specificity of the antibodies. (A) Two complementary half-site mutants retained antigen-binding function by forming a heterodimer. The addition of the 309M3-B A1R2-binding site mutant to yeast cells expressing the 309M3-B aromatic cage mutant resulted in “heterodimer” formation only in the presence of H3K9me3 peptide. The binding of the peptide was detected in the same manner as the peptide IP assay. (B) Superposition of 309M3-B (dark purple) and 304M3-B (gray) using the variable domains of Fab 1 as the common element (Left). Only the variable domains are shown for clarity. The H3K9me3 and H3K4me3 peptides are shown as stick models in red and yellow, respectively. Only one molecule for H3K4me3 peptide is shown for clarity. (Right) A comparison of the bound peptides using superposition of the aromatic cages. The aromatic cages of 304M3-B and 309M3-B are shown in light and dark green, respectively, and residues in the A1R2-binding pockets are shown in magenta. A1, R2, and trimethylated lysine residues in the H3K4me3 peptide are shown in orange, and their counterparts in the H3K9me3 peptide are shown in yellow. The Cα atoms of the R2 and Kme3 residues are shown as spheres, and the distances between these atoms in the two peptides are indicated. (C) Antibody–antibody contacts between two 309M3-B molecules. Polar interactions are marked as dashed lines. The color scheme is the same as in Fig. 2B. The residues that are different from clone 4-5 are labeled in blue. See SI Appendix, Fig. S9 for more detail. Mutation of D53H abolishes antigen biding as tested with the peptide IP assay (Right).
304M3-B and 309M3-B achieved their high sequence specificity to different Kme3 marks using a set of common recognition motifs: i.e., the aromatic cage and the A1R2-binding pocket. Curiously, although these motifs are conserved in these antibodies (and the starting clone) because we did not mutate residues forming these pockets (SI Appendix, Fig. S1), their sequence specificity profiles are essentially mutually exclusive (Fig. 1 B and C). Superposition of the two structures revealed a large difference in the relative position of the two Fab molecules (Fig. 4B), with a concomitant alteration in the spacing between the aromatic cages and the A1R2-binding pockets. The span between the two pockets seems optimal for the H3K9me3 peptide in the 309M3-B complex (∼20 Å) and for the H3K4me3 peptide in the 304M3-B complex (∼7 Å), but much too short for other major Lys modification sites (e.g., H3K27, H3K36, and H4K20) (Fig. 4B), explaining their specificity profiles. Likewise, the need for optimizing the distance between the two pockets rationalizes why the peptide is bound across two Fab molecules because this distance within a single Fab molecule (∼13 Å) is invariant and too large for H3K4me3 and too small for H3K9me3. Therefore, we propose that the dimerization and the strategically positioned two recognition motifs in these antibodies play important roles in highly specific recognition of the histone marks.
The structures suggest that antigen clasping is obligatory for these antibodies to achieve high affinity and high specificity. Mutation of a single residue (two for the Fab dimer) in the Fab–Fab interface abolished antigen binding, indicating the importance of this contact in achieving the observed mode of peptide recognition (Fig. 4C and SI Appendix, Fig. S9). Interestingly, the ability to form a peptide-induced dimer emerged in the directed evolution process. The Fab–Fab contacts in the two antibodies are different and involve residues in CDRH2 that were altered with respect to the starting clone (Fig. 4C and SI Appendix, Fig. S9). In contrast to these two antibodies, the previously developed antibody for H3K9me3, 309M3-A, did not exhibit antigen clasping although it was isolated from the same phage-display library and it differs in only 10 CDR positions from 309M3-B (SI Appendix, Figs. S1 and S5). It is notable that mutations required for antigen clasping are within CDRs, regions that are highly varied in natural antibodies, suggesting that these antibodies and other yet-to-be-identified dimerizing Fab molecules could exist in the natural immune system.
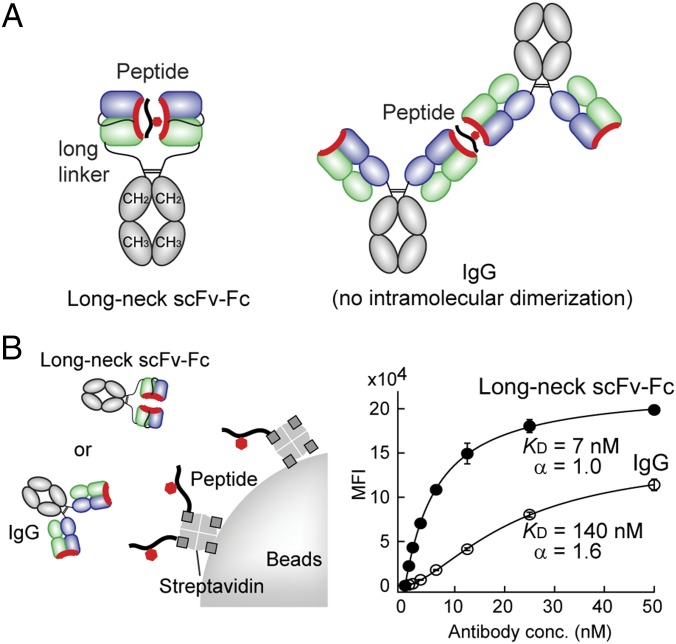
A “Long-Neck” Format Promotes Antigen Clasping.
We then designed an antibody format, termed “long-neck antibody,” to facilitate antigen clasping. We reasoned that the format of our phage display system, where scFv molecules are tethered to a phage coat protein via a long linker, enabled us to identify these unusual antibodies. Thus, we connected an antigen-binding module (scFv in this case) to Fc with a 17-residue linker (Fig. 5A). We chose scFv over Fab because the use of Fab would require a much longer linker. As expected, the long-neck scFv-Fc antibodies did not show cooperativity in the reversed IP assay, demonstrating that this format promotes antigen clasping within a single antibody molecule (Fig. 5B and SI Appendix, Fig. S10). In contrast, the same antigen-binding sites in the standard IgG format showed weaker binding and strong cooperativity. The long-neck scFv-Fc antibodies worked well in Western blotting and immunostaining analyses at high dilutions (SI Appendix, Fig. S10), further supporting the potency of the long-neck scFv-Fc format.
Fig. 5.
An antibody format that promotes head-to-head dimerization. (A) Schematic structure of the long-neck scFv-Fc format and the IgG format of an antibody exhibiting antigen clasping. (B) The reversed IP assay of the long-neck scFv-Fc and the IgG formats of 309M3-B. The long-neck scFv-Fc did not show strong cooperativity whereas the IgG did. Compare these results with Fig. 2A. The curves show the best fit of the Hill equation with the values of the Hill coefficient, α, indicated. All binding data shown here are from triplicate measurements.
Discussion
In this study, we generated recombinant antibodies to H3K4me3 and H3K9me3 that performed well in epigenetics applications, successfully determined the crystal structures of antibody–peptide complexes, and elucidated how these antibodies achieved high specificity and high affinity to challenging targets, histone tails containing Kme3 marks. These highly validated recombinant antibodies, as well as future recombinant antibodies engineered based on the mechanistic insights gained in this work, will provide the epigenetic research community with a set of standard reagents that will improve data accuracy and reproducibility.
Antigen clasping is highly unusual among known antibody structures. Dimerization of Fab molecules in crystals is observed in cases where the antigen itself has internal twofold symmetry, as expected (20, 21), but histone peptides described here do not form dimers or have internal symmetry. Antigen clasping is also distinct from the binding mode of the bispecific, chelating recombinant antibody (CRAb) because a CRAb is constructed by linking two different scFv molecules (22) and the two scFv units bind to nonoverlapping epitopes of a protein. Among ∼1,200 structures of antibody–antigen complexes in the Protein Data Bank, we found a single example, anti–C-myc antibody 9E10, that seems to exhibit antigen clasping. However, the binding mode of 9E10 conforms to the convention of anti-peptide antibodies because one Fab dominates peptide recognition by capturing the peptide into a deep cleft created with long CDRs, and this antibody showed no cooperative binding (23). By contrast, in our antibodies, two Fab molecules synergistically contribute to peptide recognition. Nevertheless, the 9E10 structure supports the view that natural antibodies have an inherent capacity to clasp an antigen.
What factor restricts the immune systems from producing antibodies exhibiting antigen clasping? Or have such antibodies been simply overlooked because the existing paradigm of antibody–antigen interaction is so well established? In conventional antibodies, two Fab segments are connected with disulfide bonds in the hinge region located between the Fab and Fc regions (Fig. 1A). This architecture makes the antigen-binding sites in the two Fab regions face opposite directions to each other. The distance between the C termini of two Fab molecules in IgG (∼15 Å between the C termini of the CH1 domains) (Fig. 1A) is much too short for forming a head-to-head dimer (∼120Å) (Fig. 2B) (24, 25). Thus, it is sterically difficult for the two Fab arms of a single IgG molecule to form a head-to-head dimer. Indeed, 309M3-B in the IgG format still shows cooperative binding with low affinity in the reverse IP assay, indicating dimer formation between two IgG molecules (Fig. 5 A and B). Such intermolecular dimerization leads to low efficacy, diminishing the likelihood of isolating this type of antibodies. Uncontrolled interaction between antigen-binding sites may promote aggregation, perhaps leading to low probability of generating functional antibodies. In addition to the geometrical restrictions described above, a clasping antibody requires a single antigen-binding unit with binding interfaces that recognize different parts of the peptide, as well as the antibody/antibody contacts that are not energetically unfavorable. These considerations rationalize why antibodies exhibiting antigen clasping are rare.
Whereas generation of homodimeric clasping antibodies like those reported here may be challenging, one can envision constructing clasping antibodies with two different antigen-binding units. One could iteratively engineer a first unit binding to the antigen and then a second unit binding to the complex of the first unit and the antigen to achieve clasping. Indeed, our group has generated a class of synthetic binding proteins termed “affinity clamps” by using a natural peptide-binding domain as the first unit and a synthetic binding protein (“monobody”) as the second unit (26). The successes of affinity clamp engineering (26, 27) support the feasibility of generating heterodimeric clasping antibodies.
This study has expanded the paradigm for antibody–antigen recognition and identified an evolutionary restriction contributing to the rarity of antibodies that form Fab dimers. Antigen clasping doubles the size of the antigen recognition interface and allows for the formation of extensive interactions that completely surround a small antigen. We anticipate that antibody formats enabling antigen clasping (e.g., the long-neck format) and iterative selection strategies will have a strong impact on unleashing molecular recognition potentials of antibodies toward currently challenging targets, including histone PTMs and small compounds.
Materials and Methods
Selection, purification, and characterization of recombinant antibodies to histone PTMs were performed essentially as described previously (9). IP-MS, ICeChIP, and ChIP-seq were performed following published methods (11, 28, 29). Further details on the materials and methods used in this study are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank J. Osipiuk for assistance with data collection at the Advanced Photon Source, Drs. A. Gupta and S. Tanaka for assistance with X-ray structure determination, Dr. D. Kovar for access to a cell homogenizer, and Drs. A. Kossiakoff and M. Lugowski for access to cell culture equipment. This work was supported by National Institutes of Health (NIH) Grants R21 DA025725 and RC1 DA028779 (to S.K.) and GM067193 (to N.L.K.). B.D.S. acknowledges funding from the W. M. Keck Foundation. S.K., A.J.R., and N.L.K. acknowledge funding from the Chicago Biomedical Consortium, with support from the Searle Funds at the Chicago Community Trust. We acknowledge the use of the University of Chicago Genomics, Flow Cytometry, and Biophysics core facilities that are supported by the University of Chicago Comprehensive Cancer Center under NIH Grant P30 CA014599. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
Footnotes
Conflict of interest statement: T.H., A.K., and S.K. are named as inventors in a patent application filed by the University of Chicago on the described materials.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4YHP (309M3-B with the H3K9me3 peptide), 4YHY (309M3-B with Kme3), and 4YHZ (304M3-B with the H3K4me3 peptide). ChIP-seq data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE66530).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522691113/-/DCSupplemental.
References
- 1.Porter RR. Separation and isolation of fractions of rabbit gamma-globulin containing the antibody and antigenic combining sites. Nature. 1958;182(4636):670–671. doi: 10.1038/182670a0. [DOI] [PubMed] [Google Scholar]
- 2.Bostrom J, et al. Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site. Science. 2009;323(5921):1610–1614. doi: 10.1126/science.1165480. [DOI] [PubMed] [Google Scholar]
- 3.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23(9):1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 4.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 5.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Egelhofer TA, et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18(1):91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuchs SM, Strahl BD. Antibody recognition of histone post-translational modifications: Emerging issues and future prospects. Epigenomics. 2011;3(3):247–249. doi: 10.2217/epi.11.23. [DOI] [PubMed] [Google Scholar]
- 8.Nishikori S, et al. Broad ranges of affinity and specificity of anti-histone antibodies revealed by a quantitative peptide immunoprecipitation assay. J Mol Biol. 2012;424(5):391–399. doi: 10.1016/j.jmb.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hattori T, et al. Recombinant antibodies to histone post-translational modifications. Nat Methods. 2013;10(10):992–995. doi: 10.1038/nmeth.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diskin R, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334(6060):1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grzybowski AT, Chen Z, Ruthenburg AJ. Calibrating ChIP-seq with nucleosomal internal standards to measure histone modification density genome wide. Mol Cell. 2015;58(5):886–899. doi: 10.1016/j.molcel.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121(6):859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 13.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442(7098):86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 14.Vargas-Madrazo E, Lara-Ochoa F, Almagro JC. Canonical structure repertoire of the antigen-binding site of immunoglobulins suggests strong geometrical restrictions associated to the mechanism of immune recognition. J Mol Biol. 1995;254(3):497–504. doi: 10.1006/jmbi.1995.0633. [DOI] [PubMed] [Google Scholar]
- 15.Collis AV, Brouwer AP, Martin AC. Analysis of the antigen combining site: Correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J Mol Biol. 2003;325(2):337–354. doi: 10.1016/s0022-2836(02)01222-6. [DOI] [PubMed] [Google Scholar]
- 16.London N, Movshovitz-Attias D, Schueler-Furman O. The structural basis of peptide-protein binding strategies. Structure. 2010;18(2):188–199. doi: 10.1016/j.str.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14(11):1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes RM, Wiggins KR, Khorasanizadeh S, Waters ML. Recognition of trimethyllysine by a chromodomain is not driven by the hydrophobic effect. Proc Natl Acad Sci USA. 2007;104(27):11184–11188. doi: 10.1073/pnas.0610850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015;350(6258):aac4383. doi: 10.1126/science.aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuh G, et al. Structure-function studies of two synthetic anti-vascular endothelial growth factor Fabs and comparison with the Avastin Fab. J Biol Chem. 2006;281(10):6625–6631. doi: 10.1074/jbc.M507783200. [DOI] [PubMed] [Google Scholar]
- 21.Hillig RC, et al. Fab MOR03268 triggers absorption shift of a diagnostic dye via packaging in a solvent-shielded Fab dimer interface. J Mol Biol. 2008;377(1):206–219. doi: 10.1016/j.jmb.2007.12.071. [DOI] [PubMed] [Google Scholar]
- 22.Neri D, Momo M, Prospero T, Winter G. High-affinity antigen binding by chelating recombinant antibodies (CRAbs) J Mol Biol. 1995;246(3):367–373. doi: 10.1006/jmbi.1994.0091. [DOI] [PubMed] [Google Scholar]
- 23.Krauss N, et al. The structure of the anti-c-myc antibody 9E10 Fab fragment/epitope peptide complex reveals a novel binding mode dominated by the heavy chain hypervariable loops. Proteins. 2008;73(3):552–565. doi: 10.1002/prot.22080. [DOI] [PubMed] [Google Scholar]
- 24.Saphire EO, et al. Crystal structure of a neutralizing human IGG against HIV-1: A template for vaccine design. Science. 2001;293(5532):1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 25.Harris LJ, Skaletsky E, McPherson A. Crystallographic structure of an intact IgG1 monoclonal antibody. J Mol Biol. 1998;275(5):861–872. doi: 10.1006/jmbi.1997.1508. [DOI] [PubMed] [Google Scholar]
- 26.Huang J, Koide A, Makabe K, Koide S. Design of protein function leaps by directed domain interface evolution. Proc Natl Acad Sci USA. 2008;105(18):6578–6583. doi: 10.1073/pnas.0801097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasui N, et al. Directed network wiring identifies a key protein interaction in embryonic stem cell differentiation. Mol Cell. 2014;54(6):1034–1041. doi: 10.1016/j.molcel.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng Y, Thomas PM, Kelleher NL. Measurement of acetylation turnover at distinct lysines in human histones identifies long-lived acetylation sites. Nat Commun. 2013;4:2203. doi: 10.1038/ncomms3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brand M, Rampalli S, Chaturvedi CP, Dilworth FJ. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation of nucleosomes isolated using hydroxyapatite chromatography. Nat Protoc. 2008;3(3):398–409. doi: 10.1038/nprot.2008.8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.