Significance
C:G to T:A mutations constitute the largest class of spontaneous base substitutions in all organisms. These mutations are thought to be a result of cytosine deaminations, but what promotes these deaminations is unclear. We confirm here the hypothesis that they occur predominantly in single-stranded DNA (ssDNA) and identify the ssDNA in the lagging strand template as the preferred site of C:G to T:A mutations. As a consequence, replication creates a strand bias in these mutations, and this overwhelms any strand bias resulting from transcription. These results explain a long-recognized bias in base composition of microbial genomes called GC skew and predicts that C:G to T:A mutations created by the APOBEC3 family deaminases in cancer genomes should occur with the same strand bias.
Keywords: uracil-DNA glycosylase, APOBEC3A, APOBEC3B, kataegis, cancer genome mutations
Abstract
The rate of cytosine deamination is much higher in single-stranded DNA (ssDNA) than in double-stranded DNA, and copying the resulting uracils causes C to T mutations. To study this phenomenon, the catalytic domain of APOBEC3G (A3G-CTD), an ssDNA-specific cytosine deaminase, was expressed in an Escherichia coli strain defective in uracil repair (ung mutant), and the mutations that accumulated over thousands of generations were determined by whole-genome sequencing. C:G to T:A transitions dominated, with significantly more cytosines mutated to thymine in the lagging-strand template (LGST) than in the leading-strand template (LDST). This strand bias was present in both repair-defective and repair-proficient cells and was strongest and highly significant in cells expressing A3G-CTD. These results show that the LGST is accessible to cellular cytosine deaminating agents, explains the well-known GC skew in microbial genomes, and suggests the APOBEC3 family of mutators may target the LGST in the human genome.
Pairing of complementary DNA strands protects the DNA bases against modification by a number of hydrolytic, oxidizing, and alkylating chemicals (1–4). For example, water reacts with cytosine, creating uracil, and the rate of this reaction in single-stranded DNA (ssDNA) is more than 100-fold the rate in double-stranded DNA [dsDNA (5–7)]. Uracil-DNA glycosylase (Ung) excises uracils created by cytosine deamination in both ssDNA and dsDNA, resulting in abasic (AP) sites. In dsDNA, the AP sites are replaced with cytosines as a result of copying of the guanine in the complementary strand during repair by the base-excision repair (BER) pathway (8). In contrast, cytosine deaminations occurring in ssDNA are problematic because the complementary strand is not available to the BER pathway. Uracils that escape repair create C:G to T:A mutations, and incomplete repair of uracils can result in persistent AP sites and strand breaks that can destabilize the genome. Hence, identifying ssDNA regions that are susceptible to damage will increase our understanding of causes of mutations and genome instability.
The AID/APOBEC (activation-induced deaminase/apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) family of DNA–cytosine deaminases are specific for ssDNA (9, 10). They are found only in vertebrates and are good probes of ssDNA in cells because of their relatively small size (about 190-amino acid catalytic domain). They are active in heterologous hosts such as Escherichia coli (11, 12) and yeast (13–15) and cause mutations in the same sequence context as in their known targets, such as Ig genes and the DNA copy of HIV-1 genome (11, 16, 17). In particular, the catalytic domain of human APOBEC3G (A3G-CTD) was expressed in an engineered yeast strain lacking the UNG gene and was shown to target ssDNA generated through aberrant resection of telomeric ends (13).
To similarly probe ssDNA in E. coli, we expressed A3G-CTD on a plasmid (pA3G-CTD) in an ung mutant E. coli strain, and the resulting mutations were determined by whole-genome sequencing. These results were compared with results from similar experiments performed using ung mutant cells without a plasmid, ung mutant cells expressing catalytically inactive A3G-CTD (pA3G-CTDmut), and published results from the ung+ wild-type (WT) E. coli strain (18, 19).
Results and Discussion
C:G to T:A Mutations in the Absence of Ung.
About 200 independent lines of the four E. coli strains, PFM2, PFM16 (=PFM2 Δung), PFM275 (=PFM16 with plasmid pA3G-CTD), and PFM277 (=PFM16 with plasmid pA3G-CTDmut), were grown for more than 1,000 generations each, and the accumulated mutations in each line were determined by whole-genome sequencing (Table 1). More than 1,100 base-substitution mutations were identified in these lines and were classified according to the mutation type and whether they were in coding or noncoding DNA. These data are summarized in Table S1. A small number of the mutations (<10%) in each strain were insertion/deletions (Table S2), and these were not analyzed further.
Table 1.
Base-substitution mutations
| Strain | WT, no plasmid* | ung, no plasmid | ung, pA3G-CTD | ung, pA3G-CTDmut | ||||
| Independent lines | 61 | 50 | 48 | 46 | ||||
| Generations per line | 4,230 | 3,372 | 1,137 | 1,139 | ||||
| Mutations | Rate (×1010)† | Rate (×1010)† | Rate (×1010)† | Rate (×1010)† | ||||
| Total (n, %) | 246 (100) | 2.1 | 358 (100) | 4.6 | 411 (100) | 16.2 | 171 (100) | 7.0 |
| Transition | ||||||||
| Total (n, %) | 136 (55) | 1.1 | 280 (78) | 3.6 | 368 (90) | 14.5 | 127 (74) | 5.2 |
| C:G to T:A (n, %) | 86 (35) | 1.4 | 247 (69) | 6.2 | 349 (85) | 27.1 | 104 (61) | 8.4 |
| Transversion | 110 (45) | 0.9 | 78 (22) | 1.0 | 43 (10) | 1.7 | 44 (26) | 1.8 |
Table S1.
Details of base-substitution mutations
| Host | Wild type | ung | ung | ung | ||||
| Plasmid | None | None | pA3G-CTD | pA3G-CTDmut | ||||
| Independent lines | 61 | 50 | 48 | 46 | ||||
| Generation per line | 4,230 | 3,372 | 1,137 | 1,139 | ||||
| Mutations (n, %) | Mutations per nt per generation (×1010) | Mutations per nt per generation (×1010) | Mutations per nt per generation (×1010) | Mutations per nt per generation (×1010) | ||||
| Total | 246 (100) | 2.1 | 358 (100) | 4.6 | 411 (100) | 16.2 | 171 (100) | 7.0 |
| Transition | 136 (55) | 1.1 | 280 (78) | 3.6 | 368 (90) | 14.5 | 127 (74) | 5.2 |
| TA > CG | 50 (20) | 0.9 | 33 (9) | 0.9 | 19 (5) | 1.5 | 23 (13) | 1.9 |
| CG > TA | 86 (35) | 1.4 | 247 (69) | 6.2 | 349 (85) | 27.1 | 104 (61) | 8.4 |
| Transversion | 110 (45) | 0.9 | 78 (22) | 1.0 | 43 (10) | 1.7 | 44 (26) | 1.8 |
| AT > TA | 19 (8) | 0.3 | 19 (5) | 0.5 | 12 (3) | 1.0 | 9 (5) | 0.8 |
| GC > TA | 31 (13) | 0.5 | 26 (7) | 0.7 | 10 (2) | 0.8 | 9 (5) | 0.7 |
| AT > CG | 43 (17) | 0.7 | 27 (8) | 0.7 | 18 (4) | 1.4 | 16 (9) | 1.3 |
| GC > CG | 17 (7) | 0.3 | 6 (2) | 0.2 | 3 (1) | 0.2 | 10 (6) | 0.8 |
| Mutations in coding DNA | 189 (77) | 1.9 | 268 (75) | 4.0 | 315 (77) | 14.6 | 125 (73) | 6.0 |
| Mutations in noncoding DNA | 57 (23) | 3.2 | 90 (25) | 7.8 | 96 (23) | 25.6 | 46 (27) | 12.8 |
nt, nucleotides.
Table S2.
Insertion/deletion (Indel) mutations
| Strain | WT, no plasmid | ung, no plasmid | ung, pA3G-CTD | ung, pA3G-CTDmut |
| +1 A:T | 5 | 0 | 2 | 4 |
| +1 G:C | 1 | 2 | 1 | 3 |
| −1 A:T | 5 | 4 | 5 | 6 |
| −1 G:C | 11 | 5 | 1 | 2 |
| + >1 bp | 0 | 1 | 1 | 0 |
| − >1 bp | 2 | 0 | 4 | 1 |
| Runs of >1 bp | 22 | 9 | 13 | 15 |
| Total | 24 | 12 | 14 | 16 |
| Generations per line | 4,230 | 3,372 | 1,137 | 1,139 |
| Indel/generation (×103) | 0.09 | 0.07 | 0.26 | 0.31 |
| Indel/gen/nt (×1010) | 0.20 | 0.15 | 0.55 | 0.66 |
bp, base pair; nt, nucleotide.
In the absence of pA3G-CTD, the ung mutant strain had a twofold higher mutation rate compared with the WT strain (Table 1). This weak mutator phenotype contrasts with the 160-fold increase in the overall mutation rate that occurred when a pathway for the repair of 8-oxoguanines was blocked in the same genetic background (18) and shows that cytosine deamination is not the major form of endogenous DNA damage. The mutator phenotype of the ung mutant strain was almost entirely the result of a fourfold increase in the rate of C:G to T:A mutations, which increased from 35% of all of the mutations in the WT strain to 69% in the ung mutant strain, consistent with the lack of repair of U•G mispairs in the ung mutant strain. These mutation rates obtained using whole-genome sequencing are somewhat lower but are still consistent with previous studies using reporter genes that found loss of Ung in E. coli increased the frequencies of spontaneous mutations 1.5- to fivefold overall, and C:G to T:A mutations seven- to 50-fold (12, 20–24). When the plasmid pA3G-CTD was introduced in the ung mutant strain, the overall mutation rate increased a further 3.5-fold, principally as a result of a fourfold increase in C:G to T:A mutations, which constituted 85% of all of the mutations obtained (Table 1).
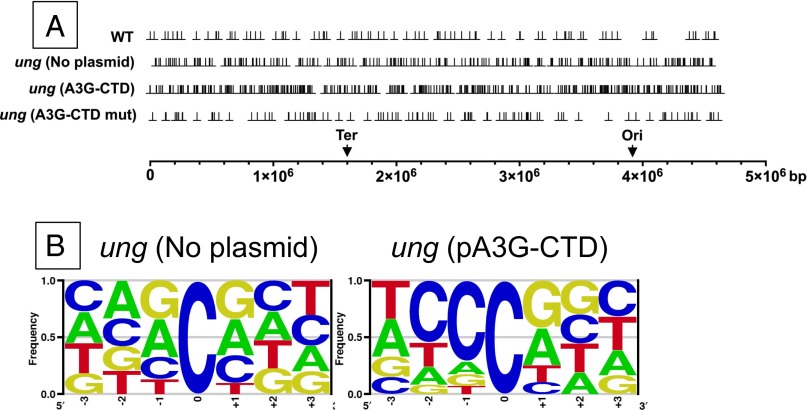
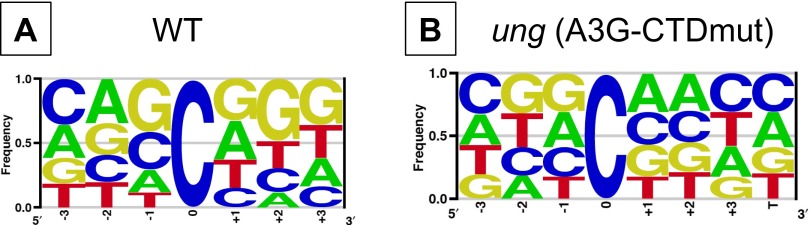
In all the strains tested, mutated cytosines were not localized to specific genomic regions, such as the origin or terminus of replication, but were distributed across the genome (Fig. 1A). This apparent lack of clustering may be because of the relatively small number of mutations obtained (several hundred), and a much larger number of mutations could reveal regional clustering (25). In the ung mutant strain both without a plasmid and carrying pA3G-CTDmut plasmid, there was no strong preference for flanking nucleotides (Fig. 1B and Fig. S1). However, when catalytically active A3G-CTD was expressed in the ung mutant strain, C:G to T:A mutations occurred preferentially in 5′CCCR3′/5′YGGG3′ sequences (target cytosine in bold and underlined; R is purine, Y is pyrimidine; Fig. 1B). Twenty-eight percent of the mutations occurred in this sequence, whereas only 2.5% were expected on the basis of the composition of E. coli genome (P ∼ 0 for the difference). This sequence preference of APOBEC3G in E. coli has been reported before (11) and is similar to that found in mammalian targets (12, 16, 26, 27).
Fig. 1.
Distribution and sequence context of C:G to T:A mutations. (A) Distribution of mutations. The positions of C:G to T:A mutations were plotted using the Graph Prism 6 for Mac software and are shown using the perpendicularity symbol (⊥); the strains in which the mutations occurred are indicated on the left. A straight line at the bottom is used to represent the E. coli genome, and downward arrows mark the positions of the replication origin (Ori) and terminus (Ter). (B) Sequence context of mutations. The three nucleotides on either side of the C:G pair that was mutated to T:A were used to create LOGOS plots using enoLOGOS software (63).
Fig. S1.
Sequence context of cytosines mutated to thymine in WT strain without a plasmid and in ung mutant strain with the plasmid pA3G-CTDmut. The three nucleotides on either side of the C:G pair that was mutated to T:A were used to create LOGOS plots using enoLOGOS software (63). Analysis of mutations in the WT strain (A) and the ung mutant strain containing plasmid pA3G-CTDmut (B).
Replication Strand Bias in Mutations Caused by APOBEC3G.
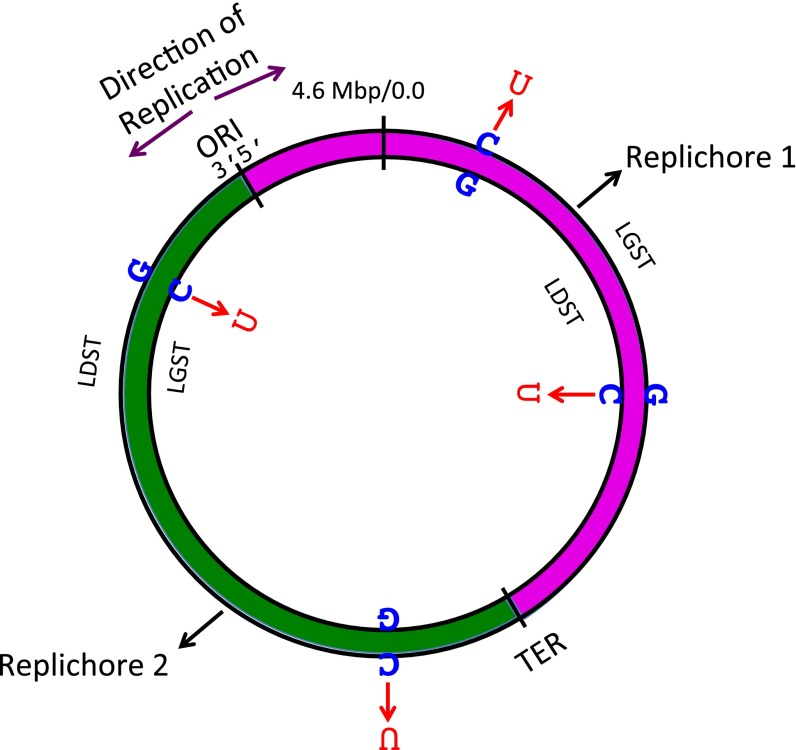
C:G to T:A mutations obtained in each E. coli strain were mapped to the two E. coli replichores and then separated into two groups on the basis of whether the mutated cytosine was in the lagging strand template (LGST) or in the leading strand template (LDST) for replication (Fig. S2). The numbers of C to T mutations in the two groups are presented in Table 2. Although in all four strains there were more mutations in the LGST than in the LDST, the LGST to LDST mutation ratio was highest, 3 to 4, when pA3G-CTD was present in the ung mutant strain (Table 2). This strong mutational strand bias was present in both E. coli replichores and was statistically highly significant (P << 0.0001). The presence of pA3G-CTD in the ung mutant strain increased the rate of C to T mutation in both LGST and LDST; the increase was sixfold when C was in the LGST and twofold when C was in the LDST (Tables 1 and 2). Thus, most of these strand-biased mutations must be a result of cytosine deaminations catalyzed by A3G-CTD.
Fig. S2.
Purple rightward arrows (→) show the overall direction of replication. Origin (ORI) and termination (TER) of replication are shown. Replication starts at ORI and proceeds in both clockwise (purple, Replichore 1) and anticlockwise (green, Replichore 2) fashion. The start of the E. coli sequence is shown at the top, and the strand directionality is shown at the ORI. The lagging and leading strand templates (LGST and LDST, respectively) are labeled for each replichore. Representative deaminations of cytosines in the LGST and LDST of each replichore are shown.
Table 2.
Replication strand bias in C:G to T:A mutations
| Replichore | C in the LGST scored mutation | C in the LDST scored mutation | Total | Ratio of C in LGST/LDST | P value* | Ratio normalized for C† | ||
| WT | ||||||||
| Right | C to T | 30 | G to A | 14 | 44 | 2.1 | 0.06 | 2.3 |
| Left | G to A | 31 | C to T | 11 | 42 | 2.8 | 0.02 | 3.0 |
| Total | C:G to T:A | 61 | C:G to T:A | 25 | 86 | 2.4 | 0.003 | 2.6 |
| ung | ||||||||
| Right | C to T | 64 | G to A | 55 | 119 | 1.2 | 0.41 | 1.2 |
| Left | G to A | 77 | C to T | 51 | 128 | 1.5 | 0.06 | 1.6 |
| Total | C:G to T:A | 141 | C:G to T:A | 106 | 247 | 1.3 | 0.05 | 1.4 |
| ung (pA3G-CTD) | ||||||||
| Right | C to T | 151 | G to A | 35 | 186 | 4.3 | 4x10−11 | 4.6 |
| Left | G to A | 126 | C to T | 37 | 163 | 3.4 | 6x10−08 | 3.6 |
| Total | C:G to T:A | 277 | C:G to T:A | 72 | 349 | 3.8 | 2x10−17 | 4.1 |
| ung (pA3G-CTDmut) | ||||||||
| Right | C to T | 37 | G to A | 17 | 54 | 2.2 | 0.03 | 2.3 |
| Left | G to A | 26 | C to T | 24 | 50 | 1.1 | 0.72 | 1.2 |
| Total | C:G to T:A | 63 | C:G to T:A | 41 | 104 | 1.5 | 0.08 | 1.6 |
P value is based on χ2 test of observed versus expected values. Expected values were calculated from the ratio of cytosines in LGST/LDST in each replichore.
Normalized for the number of cytosines in the LGST versus the LDST (ratio = 0.941 for the right replichore and 0.936 for the left replichore).
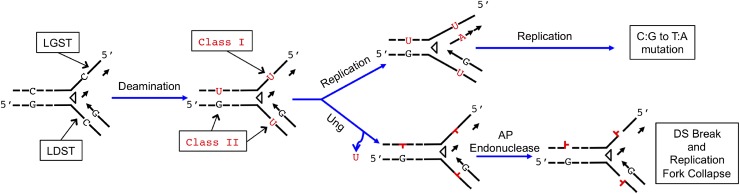
There are two reasons for the observed excess of C to T mutations in the LGST. First, A3G-CTD acts on ssDNA (28), and during replication, the LGST has longer stretches of this substrate than does the LDST (Fig. 2) (29). Second, once a cytosine in the LGST is deaminated to uracil (class I uracils in Fig. 2), it will be immediately copied by the lagging strand polymerase, creating a C:G to T:A mutation. This is likely to happen with or without A3G-CTD in cells in both ung mutant and ung+ genetic backgrounds (see following). Hence, the strand bias in mutations when A3G-CTD was expressed in the ung mutant strain strongly suggests that cytosines in LGST were much more accessible to the deaminase than cytosines in the LDST.
Fig. 2.
Consequences of cytosine deaminations at the replication fork. Deamination of three cytosines and the consequences of processing of the resulting uracils through replication or repair pathways are shown. Copying of the LGST is discontinuous, and two Okazaki fragments are shown. The open arrowhead represents the helicase DnaB and is pointed in the overall direction of replication. Class 1 and class 2 refer to two classes of uracils generated through cytosine deamination and have different biochemical consequences. The class 2 uracils are likely to be replaced with cytosines through BER, whereas the class 1 uracils lead to C:G to T:A mutations or double-strand (DS) breaks. The latter may be repaired through recombinational repair or result in the collapse of the replication fork.
Replication Strand Bias in Mutations in WT E. coli.
The WT strain lacking A3G-CTD also acquired twofold more C:G to T:A mutations when C was in the LGST than when it was in the LDST (P = 0.003; Table 2). This strand bias was reported previously, but it was not attributed to cytosine deaminations in that study (19). In light of the conclusion presented here that the A3G-CTD can access LGST, it is attractive to suggest that water, other small molecules, or proteins can access the LGST and deaminate cytosines in WT cells lacking A3G-CTD. Most uracils are likely to be copied immediately by DNA polymerase III, creating C:G to T:A mutations. However, occasionally, Ung may find the uracil before the polymerase and excise it, creating an AP site. This AP site cannot be replaced with cytosines by BER because of the lack of the complementary strand (Fig. 2). If the AP site is copied by one of the translesion synthesis DNA polymerases, it will generate a C:G to T:A mutation in about 50–70% of the cases, according to the “A” rule (30–32). Thus, both of these replicative pathways in ung+ cells will create more C:G to T:A mutations in the LGST than in the LDST.
Alternately, the AP site may be processed by an AP endonuclease, creating a double-strand break that will stop DNA replication (Fig. 2). If the break is not repaired through recombination, this will lead to replication fork collapse (29). This model further predicts that the strand breaks generated by AID/APOBEC enzymes should lead to cell death unless they are repaired through homology-directed repair or nonhomologous end-joining (in eukaryotes). These mutagenic and genome destabilizing effects of uracils created in the LGST contrast with the uracils created by deamination in dsDNA (class II uracils; Fig. 2). In WT cells, these uracils will be excised by Ung and repaired efficiently by BER, restoring the C:G pair.
Implications for Mutations in Cancer Genomes.
Many of the mutations in human tumors display “signatures” of APOBEC3 family deaminases (33, 34). In particular, a signature defined by C to T or C to G mutations in TCW context (W is A or T) is found in the genomes of a number of different cancers and is attributed to mutations caused by APOBEC3 enzymes (33, 35, 36). As in E. coli, C:G to T:A mutations in tumors would be caused when uracils created in LGST by one of the APOBEC3s are copied by the replicative DNA polymerases δ, or when AP sites created by the excision of uracils by UNG2 are copied by polymerase η, resulting in an insertion of adenines (37). Alternately, the AP sites may be copied by Rev1, creating C:G to G:C transversions (38, 39). One characteristic of cancer genome mutations is that they are found in clusters, suggesting that they occur in genomic regions that contain stretches of ssDNA (34). Although a number of cellular processes, including replication, transcription, and recombination, have been proposed as sources of ssDNA targets for APOBEC3s (13, 34, 40–42), experimental evidence for these ideas is limited. The data presented here suggest that the LGST at the replication forks of rapidly dividing cancer cells would be accessible to these enzymes, and hence the cytosines mutated in cancer genomes should be found preferentially in the LGST compared with the LDST. Recent results from whole-genome sequencing of mutations in yeast expressing APOBEC3A or APOBEC3B (43) and in 590 human tumors (44) found a strand bias in mutations consistent with this prediction.
Implications for the Microbial GC Skew.
Bacterial genomes contain a bias in base composition that is connected with replication. With few exceptions, these genomes have an excess of guanines over cytosines in the LGST [when normalized for the total G+C content (45)]. The hypotheses proposed to explain this “GC skew” include: the two DNA strands are replicated with different accuracy, the two strands are repaired with different frequency, and/or cytosines in the two strands deaminate at different rates (46–49). The data presented here strongly support the last of these hypotheses. As the LGST accumulates C to T mutations at least twice as frequently as the LDST, in the absence of any selection, there would be a progressive loss of cytosines from LGST creating the GC skew.
Mutations in ung Mutant Strain Lacking Active A3G-CTD.
The LGST/LDST ratio for C:G to T:A transitions in ung mutant cells without A3G-CTD or with inactive A3G-CTD was smaller than the ratio in the WT strain (Table 2). This was despite the fact that the overall C:G to T:A mutation rate was fourfold higher in the ung mutant strain without a plasmid and sixfold higher in the strain containing pA3G-CTDmut compared with the WT strain (Table 1). In the absence of Ung, class II U•G pairs will not be repaired, and both class I and class II U•G pairs will be replicated by DNA polymerases, increasing the overall mutation rate (Fig. 2). The class II uracils occur in dsDNA and have no strand bias. As a consequence, the addition of the resulting mutations to the overall C:G to T:A mutations reduces the LGST/LDST mutation ratio. Both the overall C:G to T:A mutation rate and LGST/LDST bias is slightly higher in the strain containing pA3G-CTDmut compared with the strain without a plasmid, suggesting the E259A substitution in A3G-CTD may have a residual ability to deaminate cytosines.
Lack of Strand Bias with Respect to Transcription.
As observed previously with the WT strain (18, 19), when normalized to the number of nucleotides in coding versus noncoding DNA, all three ung mutant strains showed a twofold bias against mutations occurring in coding sequences (Table S1). In ung+ strains, this bias was shown to be a result of better mismatch repair of the coding regions than of noncoding regions (19). Considering all the genes together, there was no significant bias in the frequency of C to T mutations in the transcribed versus the nontranscribed (coding) strand (Table S3). It is possible that a larger mutational harvest would reveal a significant transcription strand bias, especially in highly transcribed genes, but in our data, any such bias was overwhelmed by the observed replication strand bias. In contrast, Lada et al. found that most mutations caused by the lamprey cytosine deaminase, PmCDA1, in nondividing yeast were correlated with transcription (40). Together, these observations suggest that in dividing cells, the greatest source of C:G to T:A mutations is the deamination of cytosines in the template for the lagging strand synthesis, but transcription may play a larger role in nondividing cells.
Table S3.
Transcription-strand bias for C to T mutations in coding sequences
| Strain | Total | Number of C to T mutations | χ2 | P | |||||
| Observed | Expected | ||||||||
| T | NT | T/NT | T | NT | T/NT | ||||
| WT | 71 | 38 | 33 | 1.15 | 37 | 34 | 1.11 | 0.01 | 0.92 |
| ung | 186 | 90 | 96 | 0.94 | 98 | 88 | 1.11 | 0.68 | 0.41 |
| ung (pA3G-CTD) | 266 | 144 | 122 | 1.18 | 140 | 126 | 1.11 | 0.11 | 0.74 |
| ung (pA3G-CTDmut) | 75 | 36 | 39 | 0.92 | 40 | 35 | 1.11 | 0.33 | 0.57 |
NT, nontranscribed (coding) strand; T, transcribed strand.
Concluding Remarks
LGST is protected against digestion by nucleases by the ssDNA-binding protein (SSB) in bacteria and the replication protein A (RPA) in eukaryotes (50, 51). However, structural studies of both the proteins have found that both SSB and RPA wrap DNA around themselves in a conserved structure called an OB-fold, and the DNA bases largely lie on the outside of these complexes (52, 53). Thus, SSB or RPA are unlikely to prevent access to DNA bases by reactive small molecules or enzymes. This leaves the bases in LGST in both bacterial and eukaryotic replication forks highly susceptible to chemical and enzymatic damage. Therefore, the LGST is a chink in the armor with which the cell protects its DNA.
Experimental Procedures
Bacterial Media, Strains, and Plasmids.
Cultures were grown in liquid Miller Luria broth (LB), or on Miller LB agar plates (54). When appropriate, antibiotics were added to the growth media at the following concentrations: carbenicillin (Carb), 100 µg/mL; kanamycin (Kn), 50 µg/mL; and rifampicin (Rif), 100 µg/mL.
The ung mutant E. coli K12 strain used in this study, PFM16, was derived from WT strain PFM2 (18, 19). The Δung::Kn allele was moved into PFM2 from the Keio strain JW2564 (55) via P1 bacteriophage transduction, and the kanamycin-resistance gene was then removed using FLP recombination (56), leaving an in-frame scar sequence that encodes a 34-amino acid peptide.
The plasmid, pA3G-CTD, has been described previously (57), and the E259A mutant of A3G-CTD was constructed using the QuikChange site-directed mutagenesis strategy (Agilent Technologies). To remove restriction barriers, plasmid DNA was first transformed into E. coli strain DH5α, selecting for resistance to Carb (58). Plasmid DNA was then purified from DH5α, using Zyppy Plasmid Miniprep Kit (Zymo Research), and it was introduced into PFM16 via TSS transformation (59). PFM16/pA3G-CTD was designated PFM275, and PFM16/pA3G-CTDmut was designated PFM277.
Estimation of Mutation Rates by Fluctuation Tests.
Mutation rates to rifampicin-resistance (RifR) were estimated using fluctuation tests as described (60). Cultures of PFM275 and PFM277 were grown in LB broth containing Carb to maintain the presence of the plasmids. Because both A3G-CTD and A3G-CTDmut are under the control of lac promoter and inducible by IPTG (isopropyl β-d-1-thiogalactopyranoside), fluctuation assays with PFM275 and PFM277 were done both in the absence and presence of 1mM isopropyl β-d-1-thiogalactopyranoside. The addition of isopropyl β-d-1-thiogalactopyranoside made little difference to the mutation rates, so it was not added to the plates for the mutation accumulation (MA) procedure. Mutation rates from fluctuation tests were calculated using the Ma-Sandri-Sarkar maximum likelihood method (61) implemented using the FALCOR web tool found at www.mitochondria.org/protocols/FALCOR.html (62).
MA Protocol.
For PFM2 and PFM16, MA-line founders were generated by inoculating LB broth from a freezer stock, growing the culture at 37 °C overnight, and plating an appropriate dilution on LB agar plates (19). For PFM275 and PFM277, founders were generated by streaking from the freezer stock onto LB agar plates containing Carb and incubating the plates at 37 °C overnight. Two well-isolated colonies were then excised, soaked for 30 min in 0.01% gelatin solution in 0.85% NaCl, and vortexed for 60 s. Appropriate dilutions were then plated onto LB agar plates containing Carb to obtain at least 30 well-isolated colonies from each of the two parental colonies. These separate lines were then propagated for the duration of the MA procedure.
Each MA line was streaked for single colonies each day on an LB agar plate (PFM2 and PFM16) or LB agar plate with Carb (PFM275 and PFM277) and incubated overnight at 37 °C for 23–25 h. A well-isolated colony of each line was then picked and restreaked. After passage, plates were stored at 4 °C for a maximum of 2 d, to be used again if the original streaking did not yield well-isolated colonies. The number of required passages was determined from the mutation rates, as determined by fluctuation tests. The propagation of PFM2 was previously described (19). Both PFM275 and PFM277 were propagated for 40 passages. PFM16 was initially streaked for 55 passages, and then frozen stocks were made. Sequencing of the MA lines at that point revealed that the mutation rate had been overestimated. Lines were then reinoculated from the frozen stocks and streaked for another 65 passages, giving a total of 120 passages.
Estimation of Generations.
The number of generations between passages was estimated from the diameter of the colonies and the number of cells in colonies of a given diameter for each strain, as described (19). The average generation count was ∼27.5 per day. Generation counts for each line for the course of the MA experiment were totaled, and the average of this number of all of the lines for each strain multiplied by the number of lines was used to calculate the mutation rates.
Genomic DNA Preparation, Library Construction, and Whole-Genome Sequencing.
Genomic DNA was purified from 0.8 mL of overnight cultures in LB (PFM2 and PFM16) or LB broth with Carb (PFM275 and PFM277). DNA concentration and purity were assessed using an Epoch Microplate Spectrophotometer (BioTek Instruments, Inc.).
Before library construction, the ung deletion in each line was confirmed by visualizing the appropriate-sized PCR product of the genomic DNA, using primers ungFW (5′TGTCCAGCAGCCAGAAAGAG3′) and ungRV (5′ATAAATCAGCCGGGTGGCAA3′). To ensure presence of the appropriate plasmid, diagnostic restriction digestion were performed with each of the PFM275 and PFM277 MA lines. Plasmid DNA containing WT A3G-CTD has a unique Bgl1 restriction site, whereas plasmid containing A3G-CTDmut has a unique HaeII restriction site.
Libraries for PFM2 and PFM16 were made by the Beijing Genomics Institute and sequenced using the Illumina HiSeq2000 platform. Libraries for PFM275 and PFM277 were made by the Indiana University Center for Genomics and Bioinformatics and sequenced at the University of New Hampshire Hubbard Center for Genomic Studies, using the Illumina HiSeq2500 platform.
For quality control purposes, reads with any of the following characteristics were discarded, as described (19): ≥10% unreadable bases, ≥20% low-quality (≤Q20) bases, adapter contamination (≥15 bp overlap allowing up to 3 bp mismatch), and duplicate read-pairs. After this filtering, retained reads averaged 91.1%. Two MA lines from the PFM277 set had less than 30× sequence coverage and were eliminated.
Single-Nucleotide Polymorphism.
Procedures for single-nucleotide polymorphism calling were as described (18, 19). National Center for Biotechnology Information reference sequence NC_000913.2 was used as the reference genome sequence because it more accurately matches the sequence of PFM2 than the subsequent release. The sequences and mutations reported in this article have been deposited at the National Center for Biotechnology Information Sequence Read Archive (BioProject accession no. SPR013707) and in the IUScholarWorks Repository (URI TBA).
Some MA lines carried shared mutations arising either from mutations that occurred during the initial growth of founders or from cross-contamination during streaking. Such mutations were assigned to one MA line based on deduced lineage, if possible; otherwise, mutations were assigned randomly. Two MA lines of PFM275 that had many shared mutations but only one or zero unique mutations were eliminated.
Mutation Annotation.
Variants were annotated using custom scripts, as described (19).
Acknowledgments
A.S.B. thanks Gad Getz and Michael Lawrence (Broad Institute), and Steve Roberts (Washington State University) for sharing unpublished results and manuscripts. We thank the following current and past members of the P.L.F. laboratory for technical assistance: C. Coplen, J. Ferlmann, N. Ivers, D. Osiecki, S. Riffert, B. Souders, L. Whitson, N. Yahaya, A. Ying Yi Tan, and Ellen M. Popodi for useful discussions and B. Wanner and the National BioResource Project-E. coli at the National Institute of Genetics for providing bacterial strains and plasmids. This research was supported by US Army Research Office Multidisciplinary University Research Initiative (MURI) Award W911NF-09-1-0444 to P.L.F., H.T., M. Lynch, and S. E. Finkel; NIH Grant GM 57200 (to A.S.B.); and funds from Wayne State University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522325113/-/DCSupplemental.
References
- 1.Hayatsu H. Bisulfite modification of nucleic acids and their constituents. Prog Nucleic Acid Res Mol Biol. 1976;16:75–124. doi: 10.1016/s0079-6603(08)60756-4. [DOI] [PubMed] [Google Scholar]
- 2.Paleček E, Bartošík M. Electrochemistry of nucleic acids. Chem Rev. 2012;112(6):3427–3481. doi: 10.1021/cr200303p. [DOI] [PubMed] [Google Scholar]
- 3.Sinden RR. DNA Structure and Function. Academic Press; San Diego, CA: 1994. p. 398. [Google Scholar]
- 4.Singer B, Grunberger D. Molecular Biology of Mutagens and Carcinogens. Plenum Press; New York, NY: 1983. p. 347. [Google Scholar]
- 5.Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: Determination of rate constants and the activation energy. Biochemistry. 1990;29(10):2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 6.Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13(16):3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 7.Shen JC, Rideout WM, 3rd, Jones PA. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 1994;22(6):972–976. doi: 10.1093/nar/22.6.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- 9.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: An innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 10.Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- 11.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10(5):1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 12.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418(6893):99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 13.Chan K, et al. Base damage within single-strand DNA underlies in vivo hypermutability induced by a ubiquitous environmental agent. PLoS Genet. 2012;8(12):e1003149. doi: 10.1371/journal.pgen.1003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lada AG, et al. Genome-wide mutation avalanches induced in diploid yeast cells by a base analog or an APOBEC deaminase. PLoS Genet. 2013;9(9):e1003736. doi: 10.1371/journal.pgen.1003736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayorov VI, et al. Expression of human AID in yeast induces mutations in context similar to the context of somatic hypermutation at G-C pairs in immunoglobulin genes. BMC Immunol. 2005;6:10. doi: 10.1186/1471-2172-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beale RC, et al. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: Correlation with mutation spectra in vivo. J Mol Biol. 2004;337(3):585–596. doi: 10.1016/j.jmb.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 17.Rogozin IB, Pavlov YI. The cytidine deaminase AID exhibits similar functional properties in yeast and mammals. Mol Immunol. 2006;43(9):1481–1484. doi: 10.1016/j.molimm.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Foster PL, Lee H, Popodi E, Townes JP, Tang H. Determinants of spontaneous mutation in the bacterium Escherichia coli as revealed by whole-genome sequencing. Proc Natl Acad Sci USA. 2015;112(44):E5990–E5999. doi: 10.1073/pnas.1512136112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci USA. 2012;109(41):E2774–E2783. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncan BK, Rockstroh PA, Warner HR. Escherichia coli K-12 mutants deficient in uracil-DNA glycosylase. J Bacteriol. 1978;134(3):1039–1045. doi: 10.1128/jb.134.3.1039-1045.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duncan BK, Weiss B. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J Bacteriol. 1982;151(2):750–755. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster PL. Escherichia coli strains with multiple DNA repair defects are hyperinduced for the SOS response. J Bacteriol. 1990;172(8):4719–4720. doi: 10.1128/jb.172.8.4719-4720.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutsenko E, Bhagwat AS. Principal causes of hot spots for cytosine to thymine mutations at sites of cytosine methylation in growing cells. A model, its experimental support and implications. Mutat Res. 1999;437(1):11–20. doi: 10.1016/s1383-5742(99)00065-4. [DOI] [PubMed] [Google Scholar]
- 24.Nordman J, Wright A. The relationship between dNTP pool levels and mutagenesis in an Escherichia coli NDP kinase mutant. Proc Natl Acad Sci USA. 2008;105(29):10197–10202. doi: 10.1073/pnas.0802816105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foster PL, Hanson AJ, Lee H, Popodi EM, Tang H. On the mutational topology of the bacterial genome. G3 (Bethesda) 2013;3(3):399–407. doi: 10.1534/g3.112.005355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300(5622):1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424(6944):94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris RS, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113(6):803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 29.Langston LD, O’Donnell M. DNA replication: Keep moving and don’t mind the gap. Mol Cell. 2006;23(2):155–160. doi: 10.1016/j.molcel.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence CW, Borden A, Banerjee SK, LeClerc JE. Mutation frequency and spectrum resulting from a single abasic site in a single-stranded vector. Nucleic Acids Res. 1990;18(8):2153–2157. doi: 10.1093/nar/18.8.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274(45):31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 32.Tang M, et al. UmuD′(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci USA. 1999;96(16):8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexandrov LB, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nik-Zainal S, et al. Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149(5):979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: Footprints and mechanisms. Nat Rev Cancer. 2014;14(12):786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC Enzymes: Mutagenic Fuel for Cancer Evolution and Heterogeneity. Cancer Discov. 2015;5(7):704–712. doi: 10.1158/2159-8290.CD-15-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J. 2000;19(12):3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrence CW, Hinkle DC. DNA polymerase zeta and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surv. 1996;28:21–31. [PubMed] [Google Scholar]
- 39.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 40.Lada AG, et al. Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes. PLoS Genet. 2015;11(5):e1005217. doi: 10.1371/journal.pgen.1005217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts SA, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46(4):424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor BJ, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoopes J, et al. APOBEC3A and APOBEC3B deaminate the lagging strand template during DNA replication. Cell Reports. 2016 doi: 10.1016/j.celrep.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haradhvala NJ, et al. Mutational strand asymmetries across cancer reveal mechanisms of DNA damage and repair. Cell. 2016 doi: 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13(5):660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 46.Francino MP, Ochman H. Strand asymmetries in DNA evolution. Trends Genet. 1997;13(6):240–245. doi: 10.1016/S0168-9525(97)01118-9. [DOI] [PubMed] [Google Scholar]
- 47.Frank AC, Lobry JR. Asymmetric substitution patterns: A review of possible underlying mutational or selective mechanisms. Gene. 1999;238(1):65–77. doi: 10.1016/s0378-1119(99)00297-8. [DOI] [PubMed] [Google Scholar]
- 48.Rocha EP. The replication-related organization of bacterial genomes. Microbiology. 2004;150(Pt 6):1609–1627. doi: 10.1099/mic.0.26974-0. [DOI] [PubMed] [Google Scholar]
- 49.Karlin S. Bacterial DNA strand compositional asymmetry. Trends Microbiol. 1999;7(8):305–308. doi: 10.1016/s0966-842x(99)01541-3. [DOI] [PubMed] [Google Scholar]
- 50.Alani E, Thresher R, Griffith JD, Kolodner RD. Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J Mol Biol. 1992;227(1):54–71. doi: 10.1016/0022-2836(92)90681-9. [DOI] [PubMed] [Google Scholar]
- 51.Molineux IJ, Gefter ML. Properties of the Escherichia coli DNA-binding (unwinding) protein interaction with nucleolytic enzymes and DNA. J Mol Biol. 1975;98(4):811–825. doi: 10.1016/s0022-2836(75)80012-x. [DOI] [PubMed] [Google Scholar]
- 52.Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21(7):1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol. 2000;7(8):648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 54.Miller JH. A short course in bacterial genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1992. [Google Scholar]
- 55.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carpenter MA, Rajagurubandara E, Wijesinghe P, Bhagwat AS. Determinants of sequence-specificity within human AID and APOBEC3G. DNA Repair (Amst) 2010;9(5):579–587. doi: 10.1016/j.dnarep.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grant SG, Jessee J, Bloom FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87(12):4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chung CT, Niemela SL, Miller RH. One-step preparation of competent Escherichia coli: Transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86(7):2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foster PL. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006;409:195–213. doi: 10.1016/S0076-6879(05)09012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarkar S, Ma WT, Sandri GH. On fluctuation analysis: A new, simple and efficient method for computing the expected number of mutants. Genetica. 1992;85(2):173–179. doi: 10.1007/BF00120324. [DOI] [PubMed] [Google Scholar]
- 62.Hall BM, Ma CX, Liang P, Singh KK. Fluctuation analysis CalculatOR: A web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009;25(12):1564–1565. doi: 10.1093/bioinformatics/btp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Workman CT, et al. enoLOGOS: A versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005;33(Web Server issue):W389–W392. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]