Significance
HIV-1 requires integration for efficient gene expression, and the local chromatin environment significantly influences the level of HIV-1 transcription. Silent, integrated proviruses constitute the latent HIV reservoir. As HIV-1 commandeers cellular factors to dictate its preferred integration sites, these interactions consequentially influence latency. We examined the impact of polyadenylation specificity factor CPSF6, which binds HIV-1 capsid, and the integrase-binding chromatin reader LEDGF/p75 on viral infection and integration site distribution. Integration sites were determined in cells knocked down or knocked out for one or both host factors. Our data indicate that CPSF6 directs HIV-1 to transcriptionally active chromatin, where LEDGF/p75 predominantly directs the positions of integration within genes. These findings clarify the roles of cellular forces that dictate HIV-1 integration preferences and hence virus pathogenesis.
Keywords: HIV-1, integration, CPSF6, LEDGF, CFIm68
Abstract
Integration is vital to retroviral replication and influences the establishment of the latent HIV reservoir. HIV-1 integration favors active genes, which is in part determined by the interaction between integrase and lens epithelium-derived growth factor (LEDGF)/p75. Because gene targeting remains significantly enriched, relative to random in LEDGF/p75 deficient cells, other host factors likely contribute to gene-tropic integration. Nucleoporins 153 and 358, which bind HIV-1 capsid, play comparatively minor roles in integration targeting, but the influence of another capsid binding protein, cleavage and polyadenylation specificity factor 6 (CPSF6), has not been reported. In this study we knocked down or knocked out CPSF6 in parallel or in tandem with LEDGF/p75. CPSF6 knockout changed viral infectivity kinetics, decreased proviral formation, and preferentially decreased integration into transcriptionally active genes, spliced genes, and regions of chromatin enriched in genes and activating histone modifications. LEDGF/p75 depletion by contrast preferentially altered positional integration targeting within gene bodies. Dual factor knockout reduced integration into genes to below the levels observed with either single knockout and revealed that CPSF6 played a more dominant role than LEDGF/p75 in directing integration to euchromatin. CPSF6 complementation rescued HIV-1 integration site distribution in CPSF6 knockout cells, but complementation with a capsid binding mutant of CPSF6 did not. We conclude that integration targeting proceeds via two distinct mechanisms: capsid-CPSF6 binding directs HIV-1 to actively transcribed euchromatin, where the integrase-LEDGF/p75 interaction drives integration into gene bodies.
The integration of the DNA product of reverse transcription into the genome of an infected cell is an essential step in the retroviral replication cycle. Although integration strictly depends on the viral integrase (IN) enzyme, host factors play significant roles in determining where in the cellular genome the viruses integrate. As examples, the IN-binding proteins lens epithelium-derived growth factor (LEDGF)/p75 and bromo- and extraterminal (BET) domain proteins play important roles in determining sites of lentiviral and γ-retroviral integration, respectively (reviewed in ref. 1). The lentivirus HIV-1 preferentially integrates into gene-dense regions of chromosomes along the bodies of active genes (2). Depletion of LEDGF/p75 by RNA interference (RNAi) (3) or by knockout of the PSIP1 gene (4–6), which encodes for LEDGF/p75, significantly decreased HIV-1 integration into genes. However, because the frequency of gene-tropic integration remained significantly greater than random, other factors seem likely to contribute to HIV-1 integration site preferences. As subsets of integration sites become enriched in patients on suppressive antiretroviral therapy (7, 8), there is great interest to determine the mechanisms of HIV-1 integration targeting. However, the identity of host proteins other than LEDGF/p75 that direct the signature chromatin preferences of HIV-1 integration remains largely obscure (9, 10).
Active nuclear transport of the HIV-1 preintegration complex (PIC), which is required for virus replication, is principally mediated by the viral capsid (CA) protein (11). Several factors implicated in PIC nuclear import, including nucleoporins (NUPs) 153 and 358 and cleavage and polyadenylation specificity factor 6 (CPSF6), have been shown to bind HIV-1 CA (reviewed in ref. 12). Amino acid substitutions in CA that reduce binding to each of these factors can significantly alter the integration profile of HIV-1 (13, 14). In particular, the N74D change significantly reduced integration into genes and ablated the targeting of gene-dense regions of chromosomes (defined as gene number Mb−1 surrounding integration sites) (14). However, as this substitution reduced CA binding to both NUP358 and CPSF6 (13, 15, 16), the root cause of the altered N74D integration profile is unclear. Although knockdown of NUP153 (14, 17) or NUP358 (18) significantly reduced integration into genes and gene-dense regions, gene-tropic integration nevertheless remained significantly enriched over random under these conditions. The role of CPSF6 in HIV-1 integration targeting has not been reported.
CPSF6 is a component of the cleavage factor I (CFIm) complex (19) that regulates polyadenylation site selection and as a consequence the length of mRNA 3′ untranslated regions (UTRs) (20, 21). In addition to a potential role in PIC nuclear import (15), CPSF6 helps to suppress the activation of innate immunity in response to cytoplasmic HIV-1 nucleic acids in monocyte-derived macrophages (MDMs) (22). To address its potential role in integration site selection, a total of 698,561 unique HIV-1 sites in the human genome were mapped by Illumina sequencing following acute infection of control cells or cells depleted for CPSF6 and/or LEDGF/p75. Our results clarify a dominant role for CPSF6 in targeting HIV-1 to transcriptionally active chromatin.
Results
CPSF6 Knockdown Decreases Integration into Genes, Promoters, and Gene-Dense Regions.
To identify the relative contributions of CPSF6 and LEDGF/p75 to HIV-1 integration, each factor was initially knocked down in U2OS cells alone or in tandem. Whereas two different CPSF6-specific siRNAs depleted the protein to below the limit of detection of Western immunoblotting, LEDGF/p75 remained detectable at the time of virus infection in cells transfected with one of two siRNAs (siL21) (SI Appendix, Fig. S1A). Knockdown and control cells were infected with the single-round HIV-Luc reporter construct, which requires integration for efficient luciferase gene expression (4), and cells were lysed for luciferase assays at 48 h postinfection (hpi), the time of peak HIV-1 integration (23). As previously reported (24, 25), LEDGF/p75 depletion under these conditions did not discernibly affect luciferase expression (SI Appendix, Fig. S1B). Also consistent with prior reports (15, 26), CPSF6 knockdown increased viral gene expression by approximately twofold, compared with cells that received nontargeting control siRNA (siNON). Notably, infection of cells knocked down for both factors yielded approximately twofold increases in luminescence, a phenotype that was indistinguishable from those of sole CPSF6 knockdown (SI Appendix, Fig. S1B).
Sites of HIV-1 integration, which were amplified from cellular DNA by ligation-mediated PCR, were sequenced on the Illumina platform (27). The sequences were initially mapped with respect to RefSeq genes, surrounding gene density, and proximity to CpG islands and transcriptional start sites (TSSs). In siNON-treated cells, 67.3% of integrations were within genes; 4.9% and 4.1% were within 2.5 kb of a CpG island and TSS, respectively; and 16.3 genes Mb−1 on average surrounded the integration sites. Whereas these CpG and TSS values were largely similar to computationally processed matched random control (MRC) values (P = 4.8 × 10−5 and 0.85, respectively), genes and gene-dense regions were targeted to much greater extents than the MRC values of 44.7% and 8.7 Mb−1, respectively (SI Appendix, Tables S1 and S2; P < 2.2 × 10−302 for each metric). In line with the results of LEDGF/p75 immunoblotting, integration targeting was significantly altered only in cells transfected with siL3, though the corresponding values, for examples 65.4% and 15.6 genes Mb−1, were largely similar to those obtained using siNON. Consistent with prior findings (4), the reduction in gene-tropic integration was associated with significant upticks in promoter and CpG island-proximal integration (SI Appendix, Tables S1 and S2). Each CPSF6 siRNA unexpectedly decreased integration into genes to the level of the MRC; the ability to target CpG islands, TSSs, and gene-dense regions was moreover shifted to significantly below random values (SI Appendix, Fig. S1 C–E and Tables S1 and S2). Consistent with the results of virus infection, tandem factor knockdown in large part recapitulated the CPSF6 knockdown phenotype. Cotransfection with siL3 RNA, which on its own elicited noticeable integration retargeting, marginally shifted integration preferences back toward the values observed using siNON (SI Appendix, Fig. S1 C–E and Tables S1 and S2). We conclude from these data that CPSF6 plays a significant role in HIV-1 integration targeting.
CRISPR-Cas9 Knockout of CPSF6.
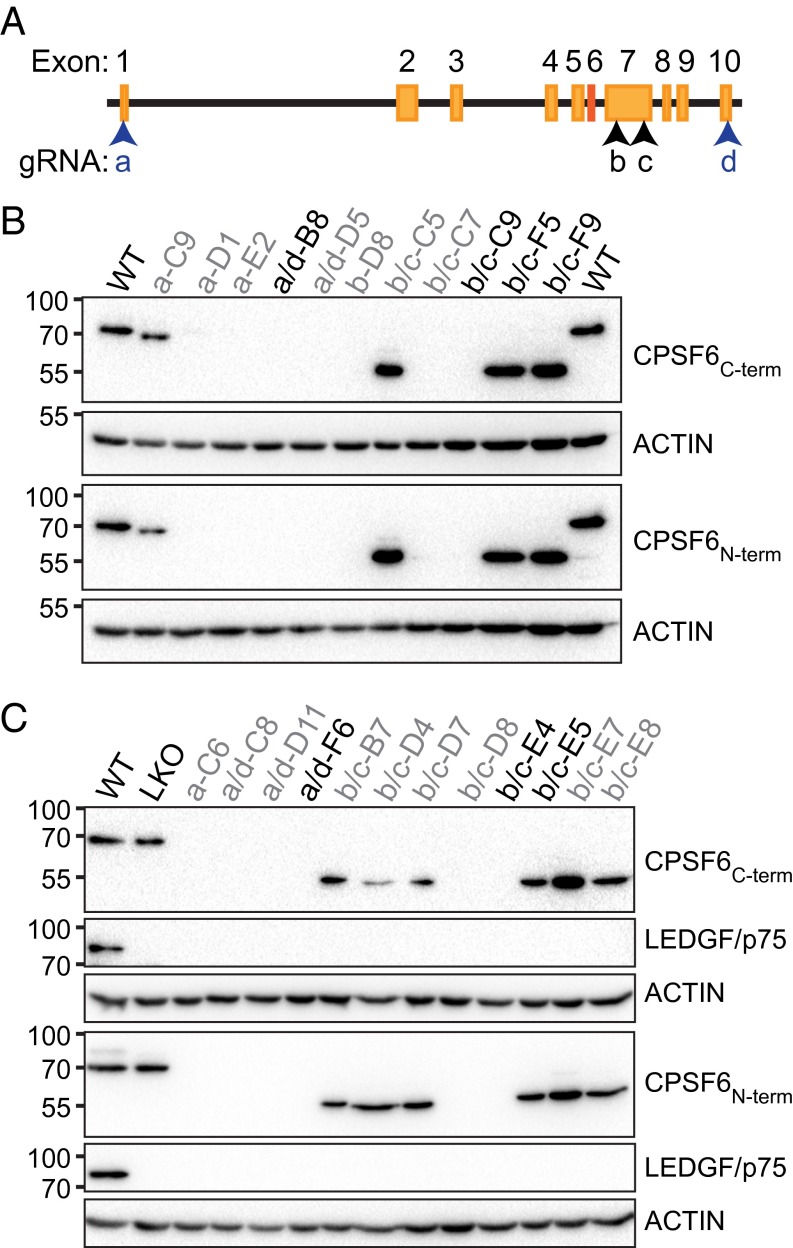
Because PSIP1 knockout (LKO) can yield greater integration and infectivity defects than those observed with LEDGF/p75 knockdown (4, 24, 25, 28), CPSF6 was knocked out in matched control and LKO HEK293T cells (28) using CRISPR-Cas9 to assess the contributions of CPSF6 versus LEDGF/p75 to integration targeting on an even playing field. Guide RNAs (gRNAs) complementary to proximal and distal CPSF6 exons 1 and 10, as well as internal exon 7, which encodes for the HIV-1 CA interacting portion of CPSF6 (29) (Fig. 1A), were introduced into cells with Cas9 (30) and an antibiotic selection marker, and cells were cloned by limiting dilution. Single cell clones were analyzed for evidence of CPSF6 knockout (CKO) by immunoblotting (Fig. 1 B and C) and by sequence analysis of PCR amplified genomic DNA fragments (SI Appendix, Fig. S2). The majority of gRNAs resulted in cells that lacked CPSF6 protein; the ∼55-kDa size of the truncated proteins observed in cells that expressed exon 7-targeting gRNAs is consistent with the internal deletion of the CA-binding region of CPSF6 (Fig. 1 B and C). Due to similar phenotypes in downstream assays (see below), cells that completely lack CPSF6 protein or express the internal truncation mutant are collectively referred to as CKO. Consistent with the known pseudotriploidy of HEK293T cells (31), upward of three unique knockout alleles were identified from sequencing cloned PCR fragments of CPSF6 exon 1, 7, and 10 regions (SI Appendix, Fig. S2). The DNA sequence analyses confirmed the CKO and double knockout (DKO) cell phenotypes observed by Western blotting.
Fig. 1.
CPSF6 knockout. (A) CPSF6 locus with gRNA targeted regions (arrowheads). Exon 6, which is excluded from the 551-residue isoform of CPSF6, is in red. (B) Immunoblots of clonal HEK293T cell lysates probed using the indicated antibodies. Numbers adjacent to the Left sides of the images indicate mass marker positions in kilodaltons. Cell line names and used gRNAs (a–d) are indicated atop the blots; names in black type were selected for downstream analyses. (C) Same as in B, except starting from LKO cells.
CKO Slows Cell Growth and Shortens 3′ UTRs.
Proliferative capacity was measured to assess potential effects of CKO on HEK293T cell physiology. Compared with WT cells, CKO cells displayed lower metabolic rates; LKO cells also displayed a slight, albeit significant decrease in metabolic rate compared with WT cells (SI Appendix, Fig. S3 A and B). The same pattern emerged from measuring rates of cell doubling (SI Appendix, Fig. S3C). Importantly, CKO did not effectively alter the fraction of HEK293T cells in interphase or mitosis (SI Appendix, Fig. S3D).
CPSF6 as part of CFIm influences polyadenylation site use and subsequent 3′ UTR length, and knockdown of either CPSF5, which is a constitutive member of CFIm, or CPSF6 results in preferential use of polyadenylation sites proximal to the translational stop codon (20, 32). To test the influence of CKO on CFIm function, RNA from three representative CKO cell lines (B8, C9, and F5) was subjected to RNA-seq analysis, and 3′ UTR length changes were assessed by percent distal polyadenylation usage index (PDUI) (33). PDUIs near zero indicate preferential use of proximal polyadenylation site(s), whereas PDUIs near 1.0 indicate preferential distal site use. Compared with WT cells, 10.0%, 14.3%, and 12.5% of transcripts from B8, C9, and F5 cells, respectively, had significant PDUI changes and of these, virtually all (97.5–99.5%) contained shortened 3′ UTRs (SI Appendix, Fig. S4 A–C, red dots in Lower Right quadrant). To correlate PDUI and transcriptional activity, ΔPDUI was calculated by subtracting WT PDUI from CKO PDUI and plotting ΔPDUI against fold change in expression for every gene with a detectable 3′ UTR. Negative ΔPDUIs indicate 3′ UTR shortening, whereas positive values indicate lengthening. Gene expression was not consistently affected by 3′ UTR shortening in CKO cells (SI Appendix, Fig. S4 D–F). Compared with WT cells, 0.82% and 14.9% of mRNAs in LKO and DKO cells had significantly altered PDUIs, respectively, with ∼46.2% and 97.4% of these exhibiting significant 3′ UTR shortening (SI Appendix, Fig. S4 G and H). Whereas 3′ UTR length did not significantly impact gene expression in LKO cells, the expression of messages with negative ΔPDUIs was down-regulated in DKO F6 cells (SI Appendix, Fig. S4 I and J). The observed changes in cellular proliferation and 3′ UTR lengths indicate that CKO profoundly affects cellular physiology.
CKO Increases HIV-1 Infectivity Independent of CA Residue Asn74.
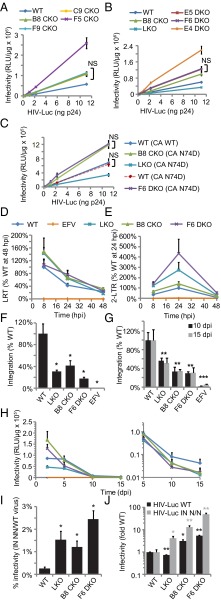
Similar to the results obtained using RNAi, CKO yielded increased levels of HIV-1 infection at 48 hpi. This increase hovered at ∼2-fold for the majority of CKO and DKO cell lines, whereas F5 CKO and E4 DKO cells supported somewhat more viral gene expression, ∼4.3- and 5.1-fold, respectively, compared with parental HEK293T cells (Fig. 2 A and B). By contrast, LKO cells supported about half as much HIV-Luc infection as control cells (Fig. 2B). As an additional control, cells were infected with the N74D CA mutant virus, which is deficient for CPSF6 binding (15, 16). The viral gene expression profiles of the N74D virus on WT, LKO, B8 CKO, and F6 DKO cells were similar to those of the WT virus (Fig. 2 A–C).
Fig. 2.
HIV-1 infectivity and DNA synthesis. (A) HIV-Luc infection of WT and indicated CKO cells. (B) Same as in A, including LKO and DKO cells. (C) Same as in A and B, except for the N74D mutant virus. (D–H) LRT products (D), 2-LTR circles (E), integration at 48 hpi by Alu-PCR (F), integration at 10 and 15 dpi by LRT PCR (G), and HIV-Luc infectivity (H). (I) NN infectivity relative to WT HIV-1. (J) Fold changes in WT (black bars) or NN (gray) infectivity, normalized to 1 (indicated virus in WT cells). Error bars indicate SE for two (H), three (D–G, I, and J), or six independent experiments (A–C). NS, not significant. Data from days 10 and 15 were averaged for the statistical analysis in G; *P < 0.05 **P < 0.01; ***P < 0.001. EFV, efavirenz.
Integration Is Defective in CKO and DKO Cells.
DNA species were quantified by real-time PCR (qPCR) to determine levels of DNA synthesis, PIC nuclear import, and integration during acute HIV-1 infection. Late reverse transcription (LRT) products represent all near-full-length HIV-1 DNA in the cell, whereas two long terminal repeat (2-LTR) containing circles and integrated proviruses are specific nuclear fractions (23, 34).
All cell types supported indistinguishable levels of reverse transcription throughout the 2-d infection time course (Fig. 2D). The cellular nonhomologous DNA end-joining (NHEJ) machinery converts a minor fraction of LRT products to 2-LTR circles (35), affording an indirect readout of viral nuclear import versus integration defects. Defective nuclear import reduces 2-LTR circles relative to total LRT (27, 36). By contrast, presumably due to the increased availability of substrate DNA for NHEJ, integration defects yield increased levels of 2-LTR circles (4, 9, 37). Expectedly (4, 9), LKO yielded an approximate 274% increase in 2-LTR circles at 24 hpi (Fig. 2E). B8 and F5 CKO cells supported 139% and 228% increases in 2-LTR circle formation, respectively (Fig. 2E and SI Appendix, Fig. S5B). The combined knockout of CPSF6 and LEDGF/p75 increased 2-LTR circles by about 435% and 443% in F6 and E4 cells, respectively (Fig. 2E and SI Appendix, Fig. S5B). HIV-1 integration was assessed by Alu-PCR at 48 hpi (23, 38). LKO expectedly decreased the level of HIV-1 integration (Fig. 2F). Integration in B8 CKO and F6 DKO cells was similarly decreased, by ∼2.5- and 4-fold, respectively, compared with WT cells (Fig. 2F). As decreased levels of integration seemed inconsistent with the increased levels of viral gene expression observed at 48 hpi in CKO and DKO cells, virus LRT product formation and infectivity were subsequently monitored over an extended time course.
The initial 48-hpi level of HIV-Luc expression in WT cells was maintained at 5 d postinfection (dpi); however, these values fell off dramatically by 10–15 dpi (Fig. 2H). LKO did not dramatically impact this kinetic profile. By contrast, the kinetic drop off in HIV-1 infection was significantly enhanced in the majority of cases by CKO, with the initial increases in virus expression observed in B8 CKO and E4 and F6 DKO cells at 48 hpi (Fig. 2H and SI Appendix, Fig. S5D) largely gone by 5 dpi. By 15 dpi, these cells supported reduced levels of HIV-1 infection, similar to the level observed in LKO cells (Fig. 2H, Right and SI Appendix, Fig. S5D). F5 CKO cells supported increased HIV-1 expression throughout the time course, with a similar decay rate as observed with WT cells (SI Appendix, Fig. S5D). After 2 dpi, relative levels of LRT products were reproducibly decreased in LKO, B8 CKO, and F6 DKO cells, whereas the level in F5 cells remained above those in WT cells throughout the time course (SI Appendix, Fig. S5A). LRT levels at 10–15 dpi, times at which unintegrated DNA has been diluted away and/or degraded (23), accordingly represent stable HIV-1 integration (5, 39). Consistent with the results of Alu-PCR, LRT levels in B8 CKO and F6 DKO cells were significantly reduced at 10 and 15 dpi (Fig. 2G). The outlier cell lines in initial infectivity measures, F5 CKO and E4 DKO (Fig. 2 A and B), supported an approximate 73% increase and an insignificant decrease in integration, respectively, relative to WT cells (SI Appendix, Fig. S5 A and C). Thus, integration is defective in the majority of CKO and DKO cells.
The IN D64N/D116N active site mutant virus (NN) was used to further probe the transient nature of HIV-1 gene expression in CKO and DKO cells. Although the IN NN virus is highly defective, it nevertheless supports sufficient levels of gene expression to be detected by the luciferase assay (38). IN active site mutant viruses were used previously to investigate the roles of host cell factors in integration and PIC nuclear import. Whereas WT and IN active site mutant viruses are similarly sensitive to nuclear import defects (36, 38), IN NN gene expression is relatively unperturbed by alterations that reduce WT virus integration (4, 27).
The IN NN virus was ∼0.3% as infectious as the WT virus in WT cells (Fig. 2I) and ∼0.9–2.5% as infectious as the WT in LKO, CKO, and DKO cells (Fig. 2I and SI Appendix, Fig. S5E). Expectedly, LKO decreased the level of WT virus infection, whereas CKO and DKO increased the extent of viral gene expression (Fig. 2J and SI Appendix, Fig. S5F, black bars). By contrast, in all knockout cell lines tested, the IN NN virus displayed increased levels of infectivity relative to the levels of infection it displayed with WT cells (Fig. 2J and SI Appendix, Fig. S5F, gray bars). Whereas LKO increased relative IN NN infectivity by fourfold, the relative levels of IN NN gene expression were increased by ∼13- and 15-fold in B8 and F5 CKO cells (Fig. 2J and SI Appendix, Fig. S5F), respectively. In F6 and E4 DKO cells, the relative levels of IN NN expression were increased to ∼50- and 13-fold of that observed in WT cells, respectively (Fig. 2J and SI Appendix, Fig. S5F).
Differential Effects of CPSF6 and LEDGF/p75 on HIV-1 Integration Targeting.
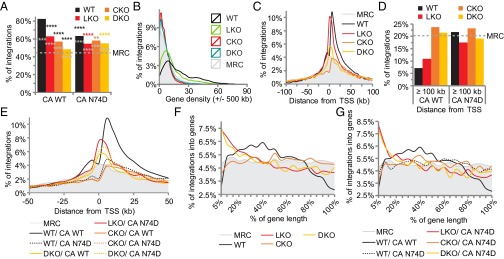
Preliminary experiments mapped WT HIV-1 and CA N74D viral integration sites in CKO lines B8, C9, and F5. As the results were largely similar across the different cells (SI Appendix, Tables S3 and S4), B8 CKO cells were compared with LKO and F6 DKO cells in subsequent infection experiments. Approximately 83% of integrations in WT cells fell within genes (Fig. 3A and SI Appendix, Tables S5 and S6). As expected (4–6), LKO significantly reduced the extent of gene targeting to 62.8% of all integrations (P < 2.2 × 10−305), with associated increases in CpG island and promoter proximal integration (P = 6.2 × 10−112 and 1.4 × 10−124, respectively). Consistent with the results of CPSF6 knockdown (SI Appendix, Fig. S1), CKO reduced integration into genes to a level (57%) that was significantly below the level observed by LKO (P = 4.1 × 10−26). CKO additionally reduced CpG island and promoter proximal integration to well below the MRC values. Thus, CPSF6 and LEDGF/p75 apparently promote gene-tropic integration via different mechanisms. Consistent with this interpretation, integration into genes was decreased to 48.3% by the DKO (Fig. 3A and SI Appendix, Table S5). The N74D mutation significantly reduced the extent of gene-tropic integration in WT and LKO cells. By contrast, gene targeting in CKO and DKO cells was significantly improved by the CA mutation (P = 0.01 and 10−18, respectively; Fig. 3A and SI Appendix, Tables S3–S6).
Fig. 3.
HIV-1 integration site distribution. Plots for integration into genes (A), as a function of gene density (B), and as a function of distance from TSS (C–E). (F and G) Integration graphed as a function of relative distance from TSSs (0%) and gene 3′ termini (100%) for the indicated cell lines. A statistical analysis shows black asterisks, versus WT virus in WT cells; gray asterisks, versus MRC; and colored asterisks, versus WT virus in indicated cell type. **P < 0.01; ****P < 0.0001. The MRC is shaded gray in B, C, and E–G to facilitate data comparison. See SI Appendix, Table S6 for full statistical analysis of A and B data and SI Appendix, Table S7 for F and G data.
The average gene density surrounding integration sites was 20.7 Mb−1 in WT cells (SI Appendix, Table S5; see Fig. 3B for distribution of all sites as function of gene density). Whereas LKO shifted the curve to the left and correspondingly decreased the average value to 13.7 genes Mb−1, CKO shifted the curve further to the left and reduced the average density to 5.8 genes Mb−1, which was well below the MRC value of 8.7. There was a slight reshift in the gene density curve with corresponding average value of 6.6 genes Mb−1 in DKO cells (Fig. 3B and SI Appendix, Table S5). Akin to the profiles of N74D mutant viral integration within genes, the CA change significantly reduced the targeting of gene-dense regions in WT and LKO cells, yet significantly increased the ability of the virus to target gene-dense regions in CKO and DKO cells (P = 0.04 and 8.7 × 10−97, respectively; SI Appendix, Tables S5 and S6).
HIV-1 displays local integration hotspots (2) and a hierarchy of genes that it prefers to target, which can reflect relative positioning with respect to the nuclear periphery (40). To assess if LEDGF/p75 or CPSF6 knockout affected the choice of which genes and chromosomal regions were targeted, integrations were plotted along chromosome lengths. Regions that were targeted in WT cells were also largely targeted in LKO cells (SI Appendix, Fig. S6 A and B, compare black and red lines). By contrast, fairly unique targeting preferences were evident in CKO and DKO cells. For example, the integration hotspot at ∼98 Mb from the origin of chromosome 7, which was maintained in LKO cells, was strongly disfavored in CKO and DKO cells. Moreover, the cold spot in WT and LKO cells at ∼53 Mb along chromosome 17 was strongly favored in CKO cells. The pattern of N74D integration in WT cells was similar to the WT virus pattern in CKO cells.
HIV-1 integration distribution positively correlates with histone modifications enriched in actively transcribed genes (e.g., H3K4me1) and negatively correlates with repressive epigenetic marks (e.g., H3K9me3) (41). Accordingly, integration in WT cells was enriched in regions containing acetylated histones and other marks associated with open chromatin, but was disfavored in areas linked with repressive histone marks (SI Appendix, Fig. S6C). Although LKO decreased integration near epigenetic marks associated with transcriptional activation, the corresponding values nevertheless remained significantly enriched compared with the MRC (SI Appendix, Fig. S6C). By contrast, integration near epigenetic marks associated with open chromatin were below the MRC in CKO and DKO cells. Integration in CKO and DKO cells was moreover enriched nearby repressive H3K9me3 marks. The correlation of integration sites and epigenetic marks was largely similar for the N74D virus in WT cells and the WT virus in CKO cells (SI Appendix, Fig. S6C).
Different Roles for LEDGF/p75 and CPSF6 in HIV-1 Integration Along Gene Bodies.
LKO significantly increases the percent of integrations near TSSs and CpG islands (4), whereas loss of CPSF6 has the opposite effect (SI Appendix, Table S5), indicating that LEDGF/p75 and CPSF6 might impact positional targeting along genes differently. The majority of integrations in WT cells occurred ∼2.5–50 kb downstream from TSSs, with a dip in activity that approached the MRC at the start sites (Fig. 3C). Consistent with gene-targeting preferences, integration at distances ≥100 kb from TSSs was highly disfavored in WT cells (Fig. 3D). Integration in LKO cells was highly enriched at TSSs, yet was disfavored at distances >15 kb upstream and ∼50 kb downstream from TSSs (Fig. 3C). CKO by contrast decreased integration at and adjacent to TSSs to below the MRC (Fig. 3C) and, consistent with the ablation of integration into gene-dense regions, increased integration at distances ≥100 kb from TSSs to levels that exceeded the MRC (Fig. 3D). The DKO yielded an intermediate level of TSS proximal integration that seemingly averaged the disparate LKO and CKO phenotypes; these frequencies approached or sank below the MRC at ∼5 kb upstream and 15 kb downstream from the start sites (Fig. 3 C and D). Infection of WT cells with N74D partially, but not fully, phenocopied the pattern of TSS proximal integration observed for the WT virus in CKO cells (Fig. 3 D and E). Integration near TSSs was marginally enhanced by the N74D mutation in CKO and DKO cells (Fig. 3E).
To ascertain the influence of integration within gene bodies, gene lengths were percentage normalized from 0% at TSSs to 100% at 3′ ends. HIV-1 preferentially targeted the midsections of genes that roughly encompassed 15–55% of their lengths. Relative to the MRC, integration was disfavored at approximately <10% and >75% of gene lengths (Fig. 3F). Consistent with a recent report (42), LKO changed the shape of the entire curve, with integration now highly favored within the initial 30% and disfavored to below the MRC for latter halves of genes (Fig. 3F). The shape of the curve in CKO cells was remarkably similar to that of the MRC (P = 0.03; SI Appendix, Table S7), with occasional blips of disfavored (e.g., <25%) and favored (∼50% and 80%) regions (Fig. 3F). The curve in DKO cells was strikingly similar to that observed in LKO cells (Fig. 3F, P = 0.81). Though N74D in large part reduced the significant preference to target the 15–55% midsection of genes and increased the targeting of gene 3′ regions in WT cells, the CA mutation did not seemingly impact the general shapes of the curves observed in the various knockout cells (compare Fig. 3F to 3G). Whereas CPSF6 is clearly important for gene-tropic integration, our data indicate that LEDGF/p75 plays a seemingly dominant role to determine where along gene lengths HIV-1 integrates. Whereas CPSF6 on average directs HIV-1 integration away from the 3′ regions of genes, LEDGF/p75 steers integration away from gene 5′ regions.
CPSF6 Targets Transcriptionally Active, Intron-Rich, and 3′ UTR-Containing Genes.
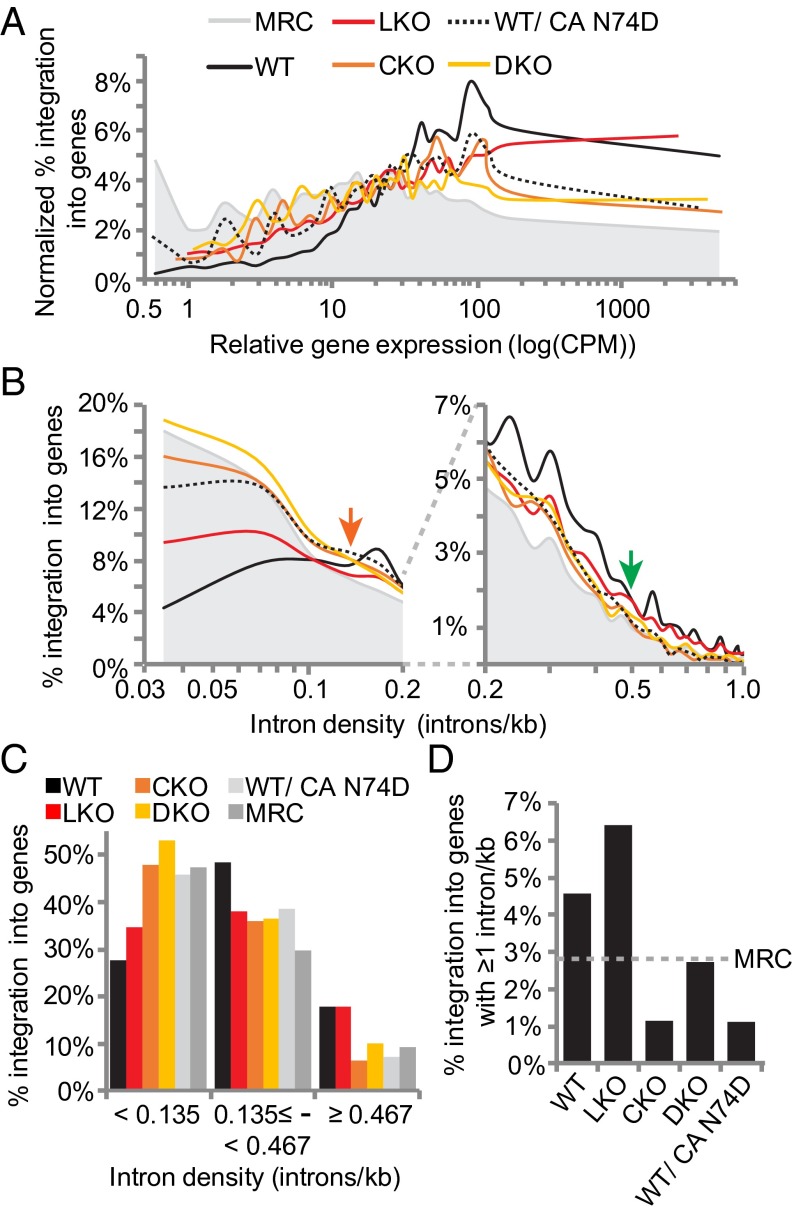
Genes were collated into 30 bins according to ascending expression level to investigate the influence of factor knockout on the targeting of active genes. In WT cells, HIV-1 avoided genes with expression levels of <10 log[counts per million (cpm)] and favored genes with >30 log(cpm). The sweet spot for HIV-1 integration hovered around genes with relative expression levels of ∼100 log(cpm) (Fig. 4A). As previously reported (4), LKO reduced but did not ablate the ability for HIV-1 to target active genes. Whereas the virus on average favored poorly expressed genes, genes expressed at very high levels, e.g., >400 log(cpm), were preferentially targeted. Although the LKO and CKO curves largely converged between 1 and 100 log(cpm), integration into genes expressed at >100 log(cpm) was preferentially crippled by CKO (Fig. 4A and SI Appendix, Table S8; P = 3.7 × 10−89). As integration into this relatively highly expressed gene subset remained suppressed in DKO cells, we infer that CPSF6 compared with LEDGF/p75 links integration to relatively highly expressed genes. The pattern of N74D integration in WT cells largely mimicked that of CKO (Fig. 4A).
Fig. 4.
CPSF6 is critical for integration into highly transcribed and spliced genes. (A) Integration as a function of gene expression. (B) Integration as a function of intron density; the graph was split to highlight on the Right differences between datasets at >0.2 introns kb−1. (C) Replot of B data to indicate results for relatively intron-sparse genes (left of the orange arrow in B), intermediary levels of intron density (between orange and green arrows in B), and relatively intron-dense genes (right of the green arrow). (D) Integration into genes with intron densities of ≥1 kb−1. See SI Appendix, Tables S8 and S9 for statistical analysis of A and C data, respectively.
The influence of CPSF6 and LEDGF/p75 on integration targeting into genes with 3′ UTRs was additionally investigated. Whereas the random calculated level of integration into such genes was 32.5%, 68.5% of targeted genes in WT cells harbored 3′ UTRs. LKO reduced this metric to 53.2%, whereas 46.6% and 46.8% of gene-tropic integrations were within 3′ UTR-containing genes in CKO and DKO cells, respectively (SI Appendix, Fig. S6D). The near-identical values in CKO and DKO cells (P = 0.81) suggest that CPSF6 plays a more dominant role than LEDGF/p75 in identifying 3′ UTR containing genes for integration. To address this link further, integration into genes with significant ΔPDUI values as defined in CKO cells was analyzed. Integration into genes with CPSF6-dependent 3′ UTRs did not significantly differ from random in WT cells, whereas LKO reduced this targeting to below the MRC (SI Appendix, Fig. S6E). By contrast, CKO and DKO yielded minor, albeit significant increases in the preference to integrate into genes that are regulated by CPSF6 (SI Appendix, Fig. S6E). Thus, although CPSF6 directs integration to genes that contain 3′ UTRs, the cellular CPSF6 function to regulate polyadenylation site use might not be necessary for HIV-1 to identify transcriptionally active genes.
LEDGF/p75 is implicated in mRNA splicing (42, 43), and CFIm components CPSF6 and CPSF7 interact with several spliceosome components (44–46). To investigate the relationship between splicing and integration targeting, frequencies of gene-tropic integration were correlated to intron density. Compared with the MRC, HIV-1 disfavored integration into genes with intron densities of <0.135 kb−1 (Fig. 4 B and C). LKO increased integration into genes with densities of <0.135 kb−1 (Fig. 4B, left of orange arrow) and decreased integration into genes with densities of ≥0.135 but <0.467 introns kb−1 (between the orange and green arrows in Fig. 4B; summarized in Fig. 4C). Although integration into genes with densities of ≥0.467 was similar in WT and LKO cells (P = 0.51; SI Appendix, Table S9), integration in genes with very high intron content, ≥1 kb−1, was actually favored in LKO cells (Fig. 4 C and D). CKO and DKO increased integration into genes with densities of <0.135 introns kb−1 to near the MRC (Fig. 4 B and C). HIV-1 targeted genes with intron densities between 0.135 and 0.467 kb−1 similarly in LKO, CKO, and DKO cells (Fig. 4 B and C and SI Appendix, Table S9). In contrast to LKO, integration into genes with densities of ≥0.467 was disfavored in CKO cells (Fig. 4 B–D). Integration into genes with relatively high intron content (≥0.467 kb−1) in DKO cells was similar to the MRC (Fig. 4 B–D). These data suggest that CPSF6 plays a greater role than LEDGF/p75 to direct integration to intron-dense genes.
CKO Does Not Affect Moloney Murine Leukemia Virus Integration Site Preferences.
Several controls were implemented in this study to address the specificity of the CKO phenotype. Moloney murine leukemia virus (MLV), which infects cells independent of LEDGF/p75 (4, 39) and CPSF6 (15), accordingly infected WT, LKO, CKO, and DKO cells similarly (SI Appendix, Fig. S7A). MLV integration sites were determined to assess potential affects from the various knockouts. Integration into genes was not significantly altered in LKO, CKO, or DKO cells (SI Appendix, Fig. S7B and Tables S10 and S11). MLV targets promoter regions and associated CpG islands to much greater extents than HIV-1 (47). Although LKO elicited relatively minor, albeit significant, effects on promoter-proximal integration, CKO did not affect the integration of MLV nearby CpG islands or TSSs (SI Appendix, Fig. S7 D and E). Moreover, neither LKO nor CKO significantly altered the targeting of gene dense regions by MLV (SI Appendix, Fig. S7C and Tables S10 and S11). Although the DKO did significantly reduce the targeting of gene-dense regions, CpG islands, and TSSs, we can infer that the dramatic alterations in HIV-1 integration targeting observed in CKO cells was not caused by global, nonspecific chromatin structural perturbations.
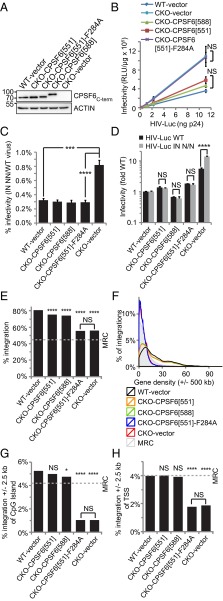
CA Binding Mutant CPSF6[551]-F284A Rescues Cellular but Not Viral CPSF6 Function.
CPSF6 expression was restored to CKO cells to further address the specificity of the HIV-1 integration defects. B8 CKO cells were transduced with MLV-derived vectors expressing the 588 amino acid isoform of CPSF6 (CPSF6[588]), CPSF6[551], which lacks exon 6 encoding sequences, or CPSF6[551] containing the F284A mutation that ablates binding to HIV-1 CA (16, 29). Backcomplemented CPSF6[551], CPSF6[551]-F284A, and CPSF6[588] were expressed at similar levels as endogenous CPSF6 in parental HEK293T cells (Fig. 5A). Notably, the larger CPSF6[588] isoform was not detected in WT HEK293T cells. Restoration of CPSF6 expression in large part restored proliferative capacity and 3′ UTR formation to CKO cells (SI Appendix, Fig. S8). Thus, the F284A change had no apparent effect on cellular CPSF6 function. CPSF6[551] and CPSF6[588] expression in large part negated the boost in HIV-1 infectivity that was observed at 48 hpi in CKO cells. By contrast, HIV-1 infected CPSF6[551]-F284A-expressing and control CKO cells indistinguishably (Fig. 5B). Thus, the increase in WT viral gene expression observed in CKO cells at 48 hpi results from the inability of CA to bind CPSF6. Interestingly, the additional boost in IN NN mutant gene expression relative to WT HIV-1 in CKO cells was similarly countermanded by CPSF6[551], CPSF6[588], or CPSF6[551]-F284A expression (Fig. 5 C and D).
Fig. 5.
The CA–CPSF6 interaction underlies gene-tropic HIV-1 integration. (A) Immunoblot of cell lines transduced with the indicated expression (or empty) vector. Mass standards in kilodaltons are to the Left. (B) HIV-1 infectivity as a function of input virus. (C) NN infectivity relative to WT HIV-1. (D) Fold changes in WT (black bars) or NN (gray) infectivity, normalized to 1 (WT/NN virus in WT cells). (E–H) HIV-1 integration into genes (E), as a function of gene density (F), and nearby CpG islands (G) and TSSs (H). Luciferase assays in B–D, which were performed at 48 hpi, show the average and SE from six independent experiments. *P < 0.05; ***P < 0.001, ****P < 0.0001; NS, not significant. See SI Appendix Tables S12 and S13 for E–H values and statistical analysis, respectively.
Proviral sites were determined in the backcomplemented cell lines to assess the role of CA-CPSF6 binding in integration targeting. WT and CKO cells that were transduced with the empty expression vector supported the expected frequencies of gene, gene-dense region, CpG island, TSS, intron density, and gene expression targeting preferences (Fig. 5 E–H and SI Appendix, Fig. S9 and Tables S12 and S13). Expression of CPSF6[551] or CPSF6[588] in large part restored integration targeting with respect to each of these genomic annotations. By contrast, integration preferences for genes, CpG islands, and TSSs were indistinguishable in control CKO cells and in cells that expressed CPSF6[551]-F284A (Fig. 5 E–H and SI Appendix, Tables S12 and S13; P value ranges from 0.08 to 0.86). Although a statistically significant difference in average density of genes Mb−1 occurred between control CKO and CPSF6[551]-F284A-expressing cells, we would note largely congruent integration as a function of gene density curves under these conditions (Fig. 5F, blue and red lines). We accordingly conclude that loss of CPSF6 binding to CA, and not loss of cellular CPSF6 function, underlies the failure of HIV-1 to identify euchromatin and active genes in CKO cells.
CPSF6 Knockdown Disrupts Integration Targeting in Primary Blood Cells.
As the work until now was performed in transformed cell lines, we next investigated the result of antagonizing CPSF6 and LEDGF/p75 expression levels in primary blood cells. Considering the relative difficulty to sufficiently suppress LEDGF/p75 expression levels by siRNA (SI Appendix, Fig. S1), and the fact that LEDGF/p75 expression is naturally lower in macrophages than in T cells (5), MDMs were transfected with control siNON or siRNA targeting CPSF6 or LEDGF/p75. Though Western blotting revealed moderately efficient knockdown at the time of HIV-1 infection, cellular CPSF6 levels partially recovered by 2 dpi (SI Appendix, Fig. S10A). Although disruption of integration targeting was expectedly partial under these conditions, CPSF6 and LEDGF/p75 knockdown significantly reduced integration into genes (P = 8 × 10−8 and 8.2 × 10−18, respectively; SI Appendix, Fig. S10B and Tables S14 and S15). Moreover, CPSF6 and LEDGF/p75 knockdown significantly decreased and increased promoter-proximal integration, respectively (SI Appendix, Fig. S10 C and D). Additionally, CPSF6 knockdown significantly altered the ability of HIV-1 to target gene-dense regions of chromosomes (SI Appendix, Fig. S10E; P < 1.9 × 10−320). We therefore conclude that LEDGF/p75 and CPSF6 play significant roles in HIV-1 integration targeting under physiologically relevant conditions.
Discussion
We show here that the interaction of CPSF6 with CA is critical to dictate HIV-1 integration site preferences. CPSF6 knockdown, knockout, or deletion of the CPSF6 CA binding domain disrupted HIV-1 integration targeting to gene-dense regions, transcriptionally active genes, intron-rich genes, and the positions of integration within genes (Figs. 3, 4, and 5; see Fig. 6 for summary model). LKO decreased integration into transcriptionally active genes, gene-dense regions, and intron-dense genes to lesser extents than did CKO (Figs. 3 A and B and 4). However, LEDGF/p75 was critical to target HIV-1 away from TSS proximal regions (Figs. 3 C–F and 6). In DKO cells, integration into genes was decreased below that observed in CKO cells (Fig. 3A), suggesting that CPSF6 and LEDGF/p75 orchestrate different aspects of HIV-1 integration targeting.
Fig. 6.
Model for the roles of CPSF6 and LEDGF/p75 in HIV-1 integration targeting. (Left column) Schematic of HIV-1 integration into genes, purported chromatin state (open or closed), and active genes in the various studied cell types. (Right column, boxes) Depiction of the effects of factor knockout on integration within genes. The Right sides of the boxes depict the generalized chromatin binding profiles of LEDGF/p75 (43, 52) and CPSF6 (51). LEDGF/p75 is additionally known to associate with the H3K36me3 histone modification (43, 61).
Does Integration into Gene-Dense Regions Promote HIV-1 Proviral Transcription?
Similar to results with CKO, the CA mutation N74D disrupted integration into transcriptionally active genes, gene-dense regions, and intron-rich genes (Figs. 3 and 4). However, we confirm that the effect of the CA N74D mutation on integration targeting is pleiotropic, as the patterns of the N74D and WT viruses largely differed from one another in CKO and DKO cells. Unexpectedly, the mutation partially restored defective targeting of genes and gene-dense regions in CKO and DKO cells (Fig. 3 A and B). The mutant virus additionally revealed the same increase in viral gene expression observed for the WT virus at 48 hpi in CKO cells (Fig. 2C). As the N74D change has been shown to reduce the affinity of the NUP358–CA interaction (13), it seems likely that disruption of multiple virus–host interactions could account for the nonidentical behavior of the mutant virus versus WT HIV-1 in CKO cells. We do note that TSS-proximal regions were for the most part similarly targeted by the WT and N74D viruses in CKO cells.
Our data imply that neither LEDGF/p75 nor CPSF6 is sufficient to identify the correct chromatin environment for integration (Fig. 6). Infectivity and overall provirus content are decreased by LKO as are, albeit to lesser extents than in CKO cells, integration into gene-dense regions and transcriptionally active genes (Figs. 2, 3, 4, and 6). Compared with WT, CKO and DKO cells supported increased HIV-1 gene expression that decreased over time (Fig. 2H), and integration into highly transcribed genes and gene-dense regions was greatly affected by CKO (Figs. 3, 4, and 6). The transient increases in HIV-1 infectivity observed in CKO and DKO cells were independent of integration, as the IN NN mutant virus that expresses its genes from unintegrated DNA also supported greater levels of HIV-1 expression in these knockout cells (Fig. 2 and SI Appendix, Fig. S5). As CPSF6[551]-F284A expression counteracted the fractional boost in IN NN mutant viral gene expression relative to the WT virus in CKO cells (Fig. 5 B and C), CKO apparently affords a transcriptional environment that specifically favors expression from unintegrated DNA templates. It is however noteworthy that normalized levels of WT and NN mutant viral gene expression remained similarly elevated in CPSF6[551]-F284A–expressing cells. One possibility that requires further investigation is that loss of CPSF6 localizes the PIC to a heterochromatin environment that on relatively short time frames supports increased levels of HIV-1 expression. Whatever this may be, the phenomenon is dependent on the CPSF6–CA interaction (Fig. 5).
CPSF6 Directs HIV-1 Integration to Active Genes and LEDGF/p75 Influences Positional Targeting Within Genes.
Various viral-binding cell factors, including cyclophilin A (13), NUP358 and transportin 3 (18), NUP153 (14, 17), LEDGF/p75 (3–6), and CPSF6 (results presented here) influence HIV-1 integration targeting. Binding of LEDGF/p75 or CPSF6 to IN and CA, respectively, targets integration not just to genes, but to a large extent to different regions within genes and the genome (Figs. 3 and 4 and SI Appendix, Fig. S6). We propose a model whereby nuclear CPSF6 binds CA before the engagement of LEDGF/p75 by IN. Consistent with a nuclear role for the CA–CPSF6 interaction, a small amount of CA is associated with the PIC inside the nucleus (48–50) and CFIm dictates the polyadenylation site on 3′ UTRs, a posttranscriptional, nuclear function (19). Furthermore, consistent with recent data that antagonized CPSF6 expression via RNAi (50), CKO did not obviously decrease HIV-1 nuclear import (Fig. 2E and SI Appendix, Fig. S5B), implying that CPSF6 does not directly function in PIC nuclear import.
We propose that CA binding to CPSF6 directs the PIC first to euchromatin, including regions of the genome enriched in genes, spliced mRNAs, and likely as a consequence, transcriptional activity (Fig. 6, WT cell). LKO was reported recently to decrease integration into genes enriched for intron content to near random (42). Interestingly, we found that CPSF6 more prominently affected integration into intron-dense genes than did LEDGF/p75 (Fig. 4 B–D). To control for the significant changes in gene targeting (Fig. 3A), we normalized our datasets to integrations within genes. Integration density kb−1 biases data with fewer gene-tropic integrations toward random because it fails to account for the decreased preference for genes upon LKO (42). Thus, although LEDGF/p75 influences integration targeting to genes, CPSF6 is the primary factor targeting HIV-1 to intron-rich genes.
Although both knockouts decreased integration into 3′ UTR containing genes, CPSF6 was more important than LEDGF/p75 to target this gene subset (SI Appendix, Fig. S6D). Somewhat unexpectedly, integration into genes with CPSF6-dependent changes in 3′ UTR length was not affected by CKO (SI Appendix, Fig. S6E), implying that CPSF6 function in CFIm might not underlie gene-tropic integration targeting. Importantly, integration correlated in a CPSF6-dependent manner with genomic sequences identified by CPSF6 chromatin-immunoprecipitation (ChIP)-seq. (51) (SI Appendix, Fig. S6F), suggesting that a CPSF6 binding partner(s) that links the factor to chromatin plays a role in HIV-1 integration site targeting. As LKO and DKO cells exhibited increased preferences for integration near promoters (Fig. 3 C–E), IN engagement of chromatin-bound LEDGF/p75 likely directs integration away from TSSs and into gene bodies (Fig. 3 C and F). The positional integration preferences within genes in LKO and CKO cells is similar to the chromatin binding profiles of CPSF6 (51) and LEDGF/p75 (43, 52), respectively (Fig. 6). Supporting the idea that LEDGF/p75 predominantly orchestrates integration targeting within genes, DKO and LKO cells demonstrated near identical preferences for this metric (Figs. 3 and 6).
DKO cells supported intermediate integration phenotypes, near MRC levels, for integration into gene dense regions and TSS-proximal regions (Fig. 3 B and C). HIV-1 integration hotspots have been identified near the nuclear envelope and adjacent to the nuclear pore (40, 53). Thus, the possibility exists that additional factors such as NUP153 and/or 358 could exert some influence on integration targeting at the nuclear pore. LEDGF/p75 knockdown cells supported increased integration near the center of the nucleus and decreased integration at regions adjacent to the nuclear pore (40). Conversely, CPSF6 knockdown or infection with mutant viruses that are deficient for CPSF6 binding increased integration in the peripheral region of the nucleus (50). Thus, similar to the effect of these factors on targeting the virus to gene-dense regions and TSSs, LEDGF/p75 and CPSF6 might exert opposing effects on the spatial distribution of integration within the nucleus. Our data moreover suggest a possible relationship between the sequestration of HIV-1 nucleic acids from innate immune signaling (22) and the downstream step of chromosomal DNA integration.
Materials and Methods
Plasmid Constructs.
Oligonucleotides for gRNAs were annealed and ligated with BbsI-digested pX330 (30) (Addgene) to generate pX330-CPSF6-a, pX330-CPSF6-b, pX330-CPSF6-c and pX330-CPSF6-d to express gRNA a, b, c, or d, respectively. Plasmids pRetroX-puro and pEGFP-C1 were from Clontech. The sequences of all oligonucleotides used in this work are listed in SI Appendix, Table S16.
Single-round HIV-1 expressing luciferase (HIV-Luc; WT or N74D) or green fluorescent protein (HIV-GFP) were expressed from pNLX.Luc.R-.ΔAvrII (14) and pNLENG1-ES-IRES (54), respectively. Single-round HIV-1 bearing IN mutations was expressed using pHP-dI-NA or pHP-dI-NN with pHI.Luc and pHCMV-VSV-G (38). Single-round MLV-Luc was expressed using pFb-Luc, pCG-MLV-gagpol, and pHCMV-VSV-G (4) whereas pSIV3+ and pCG-VPX were used to generate Vpx-containing virus like particles (55).
CPSF6[551] and CPSF6[588] cDNAs were amplified using AE6836 and AE6837 to add HindIII restriction sites. HindIII-digested DNAs were ligated with HindIII-digested pLB(N)CX (56). The F284A mutation was introduced into CPSF6[551] using PCR-directed mutagenesis. The coding regions of all plasmids synthesized by PCR were verified by sequencing.
Cells.
WT and LKO HEK293T cells were previously described (28). U2OS cells were purchased from America Type Culture Collection. HEK293T and U2OS cells were maintained in DMEM containing 10% (vol/vol) FBS, 100 IU penicillin, and 100 µg/mL streptomycin.
MDMs were isolated from human blood, which was purchased from Research Blood Components. The Dana-Farber Cancer Institute Office for Human Research Studies determined that the use of commercially obtained human blood, which lacks linking identifiers, was exempt from Institutional Review Board review. Blood was layered onto lymphocyte separation medium (GE Healthcare), and the interface containing peripheral blood mononuclear cells (PBMCs) was transferred to a new tube following centrifugation. PBMCs were washed three times with cold PBS–0.1% BSA. PBMCs were resuspended in RPMI 1640 media and cells were allowed to adhere to the plate. After washing four to five times with RPMI medium 1640, RPMI 1640 containing 10% (vol/vol) FBS, 100 IU penicillin, 100 µg/mL streptomycin, and 10 ng/mL macrophage colony stimulating factor (eBiosciences) (complete RPMI) was added. Media was replenished with complete RPMI every 2 d until day 7, when differentiated MDMs were transfected with siRNA.
Transfection, Virus Production, and Infection.
All siRNAs were generated by GE Dharmacon and contained ON-TARGET Plus modifications. U2OS cells were transfected with 20 nM total siRNA (10 nM targeting LEDGF/p75 and/or CPSF6, with 10 nM siNON for single factor knockdowns) using Lipofectamine RNAiMax (Life Technologies) 2 d before HIV-1 infection. MDMs were similarly transfected, except for using 50 nM siRNA and waiting 3 d for HIV-1 infection.
The following single-round viruses were pseudotyped with vesicular stomatitis virus G glycoprotein: HIV-GFP (SI Appendix, Fig. S10), MLV-Luc (SI Appendix, Fig. S7), and HIV-Luc (Figs. 2–5 and SI Appendix, Figs. S1, S5, S6, and S9). Plasmids were cotransfected using PolyJet (SignaGen Laboratories), and cell supernatants were concentrated as described (57, 58). HIV-Luc yield was quantified by p24 antigen capture assay (Advanced Bioscience Laboratories). To compensate for alterations resulting from CKO, HEK293T cells (WT and knockout) were infected within 2–3 h of plating to maintain similar multiplicities of infection across cell type. For infectivity assays, cells were infected using 0.0017, 0.0085, 0.017, and 0.085 pg p24 HIV-Luc per cell. Cells were infected with 1.67 and 0.125 pg p24 HIV-Luc per cell for Illumina sequencing and qPCR, respectively. See SI Appendix for luciferase and qPCR assay details.
Plasmid pLB(N)CX-based CPSF6 expression vectors (551 or 588 isoform) were cotransfected with pCG-MLVgagpol and pHCMV-VSV-G into WT HEK293T cells to make retroviruses expressing blasticidin resistance. WT or B8 CKO cells were infected for 24 h. At 48 hpi, cells were replated into media containing 7.5 µg/mL blasticidin (Invitrogen). Blasticidin concentration was lowered to 5 µg/mL after 14 d.
MDMs were pretreated with virus-like particles containing the simian immunodeficiency virus Vpx protein (55) for 6 h before infection with HIV-GFP at the approximate multiplicity of infection of 0.6 for 1 d. At 5 dpi, cells were processed for GFP expression by fluorescence-activated cell sorting (FACS) (FACS Canto, BD Biosciences) and for integration site sequencing.
Cas9/gRNA-Mediated CKO and Validation.
Plasmid pRetroX-puro was cotransfected with pX330-CPSF6-(a, b, c, a/d, or b/c) into WT or LKO HEK293T cells. After 2 d, 1.5 ng/µL puromycin (Life Technologies) was added for 3 d. Cells were then cloned by limiting dilution in 96-well plates.
Cells were lysed for immunoblotting as previously described (59) except that NaCl concentration was increased to 500 mM and 0.1% SDS was included in the lysis buffer. Lysate (15 µg protein) was fractionated through 8.5% (wt/vol) polyacrylamide gels under denaturing conditions. Gels were transferred at 17 V constant for 35 min using a Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (BioRad). The following antibodies were used for Western blotting: anti-CPSF6 (N-term) (ab175237, Abcam), anti-CPSF6 (C-term) (A301-358A, Bethyl Laboratories), anti-LEDGF/p75 (A300-848A, Bethyl Laboratories), and antiactin-HRP conjugated (A3854-200UL, Sigma).
Genomic DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen) and CPSF6 loci were amplified with EcoRI and BamHI tagged primers. EcoRI/BamHI-digested DNAs were ligated with BamHI/EcoRI-digested pEGFP-C1. DNA from eight bacterial colonies was sequenced from each ligation reaction.
Cell Growth and Metabolic Assays.
Cell metabolism was quantified by WST-1 assay (Roche) at 3 d postplating, using the manufacturer’s protocol. The growth rates of WT, LKO, CKO, and DKO cells were assessed by plating 250,000 cells into multiple wells of a six-well plate. At 2 and 3 d postplating, cells were counted using a Cellometer Auto T4 cell counter (Nexelcom Bioscience).
Cell Cycle Distribution.
Cells were detached, washed twice in PBS, and fixed at 4 °C for at least 24 h in ice-cold 70% (vol/vol) ethanol. Fixed cells were allowed to slowly equilibrate to room temperature and subsequently were washed twice with PBS. After staining at 37 °C for 30 min with propidium iodide staining solution [20 µg/mL propidium iodide (Sigma), 200 µg/mL RNase A (DNase-free, Sigma), 0.1% (vol/vol) Triton X-100 (Sigma) in PBS], cell cycle was quantified by FACS using the Watson (pragmatic) algorithm of FloJo version 10.
Integration Site Sequencing, RNA-Seq, and Bioinformatics.
Integration site sequencing, data analysis, and MRC generation were performed essentially as described (27) with some modifications in oligonucleotide design (SI Appendix, Table S16).
RNA was DNaseI treated and purified using the Quick-RNA Miniprep (Zymo Research). RNA prepped for 75-bp paired end Illumina HiSeq was sequenced on a NextSeq500 by the molecular biology core facilities at Dana-Farber Cancer Institute as described (60). See SI Appendix for bioinformatics and statistic protocols. Integration and RNA-seq sequences are accessible through the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) under accession no. SRP065607.
Supplementary Material
Acknowledgments
We thank David Levy for the gift of pNLENG1-ES-IRES and Jacek Skowronski for pSIN3+ and pCG-VPX plasmid DNAs. This work was supported by National Institutes of Health Grants T32 AI007245 (to G.A.S.), T32 AI007386 (to E.S.), R01 AI052014 (to A.N.E.), and P30 AI060354 (Harvard University Center for AIDS Research).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) (accession no. SRP065607).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1524213113/-/DCSupplemental.
References
- 1.Kvaratskhelia M, Sharma A, Larue RC, Serrao E, Engelman A. Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res. 2014;42(16):10209–10225. doi: 10.1093/nar/gku769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schröder AR, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110(4):521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 3.Ciuffi A, et al. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11(12):1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 4.Shun MC, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21(14):1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshall HM, et al. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One. 2007;2(12):e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schrijvers R, et al. LEDGF/p75-independent HIV-1 replication demonstrates a role for HRP-2 and remains sensitive to inhibition by LEDGINs. PLoS Pathog. 2012;8(3):e1002558. doi: 10.1371/journal.ppat.1002558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maldarelli F, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345(6193):179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner TA, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345(6196):570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, et al. HRP2 determines the efficiency and specificity of HIV-1 integration in LEDGF/p75 knockout cells but does not contribute to the antiviral activity of a potent LEDGF/p75-binding site integrase inhibitor. Nucleic Acids Res. 2012;40(22):11518–11530. doi: 10.1093/nar/gks913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schrijvers R, et al. HRP-2 determines HIV-1 integration site selection in LEDGF/p75 depleted cells. Retrovirology. 2012;9:84. doi: 10.1186/1742-4690-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamashita M, Emerman M. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol. 2004;78(11):5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matreyek KA, Engelman A. Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes. Viruses. 2013;5(10):2483–2511. doi: 10.3390/v5102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaller T, et al. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 2011;7(12):e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh Y, et al. Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. J Virol. 2013;87(1):648–658. doi: 10.1128/JVI.01148-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee K, et al. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe. 2010;7(3):221–233. doi: 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AJ, et al. CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS Pathog. 2012;8(8):e1002896. doi: 10.1371/journal.ppat.1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Nunzio F, et al. Nup153 and Nup98 bind the HIV-1 core and contribute to the early steps of HIV-1 replication. Virology. 2013;440(1):8–18. doi: 10.1016/j.virol.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ocwieja KE, et al. HIV integration targeting: a pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog. 2011;7(3):e1001313. doi: 10.1371/journal.ppat.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: Extent, regulation and function. Nat Rev Genet. 2013;14(7):496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 20.Martin G, Gruber AR, Keller W, Zavolan M. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Reports. 2012;1(6):753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Gruber AR, Martin G, Keller W, Zavolan M. Cleavage factor Im is a key regulator of 3′ UTR length. RNA Biol. 2012;9(12):1405–1412. doi: 10.4161/rna.22570. [DOI] [PubMed] [Google Scholar]
- 22.Rasaiyaah J, et al. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature. 2013;503(7476):402–405. doi: 10.1038/nature12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7(5):631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 24.Llano M, et al. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J Virol. 2004;78(17):9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandegraaff N, Devroe E, Turlure F, Silver PA, Engelman A. Biochemical and genetic analyses of integrase-interacting proteins lens epithelium-derived growth factor (LEDGF)/p75 and hepatoma-derived growth factor related protein 2 (HRP2) in preintegration complex function and HIV-1 replication. Virology. 2006;346(2):415–426. doi: 10.1016/j.virol.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 26.Henning MS, Dubose BN, Burse MJ, Aiken C, Yamashita M. In vivo functions of CPSF6 for HIV-1 as revealed by HIV-1 capsid evolution in HLA-B27-positive subjects. PLoS Pathog. 2014;10(1):e1003868. doi: 10.1371/journal.ppat.1003868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matreyek KA, et al. Host and viral determinants for MxB restriction of HIV-1 infection. Retrovirology. 2014;11:90. doi: 10.1186/s12977-014-0090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fadel HJ, et al. TALEN knockout of the PSIP1 gene in human cells: Analyses of HIV-1 replication and allosteric integrase inhibitor mechanism. J Virol. 2014;88(17):9704–9717. doi: 10.1128/JVI.01397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee K, et al. HIV-1 capsid-targeting domain of cleavage and polyadenylation specificity factor 6. J Virol. 2012;86(7):3851–3860. doi: 10.1128/JVI.06607-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin YC, et al. Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat Commun. 2014;5:4767. doi: 10.1038/ncomms5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masamha CP, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510(7505):412–416. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia Z, et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Commun. 2014;5:5274. doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munir S, Thierry S, Subra F, Deprez E, Delelis O. Quantitative analysis of the time-course of viral DNA forms during the HIV-1 life cycle. Retrovirology. 2013;10:87. doi: 10.1186/1742-4690-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, et al. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 2001;20(12):3272–3281. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Iaco A, et al. TNPO3 protects HIV-1 replication from CPSF6-mediated capsid stabilization in the host cell cytoplasm. Retrovirology. 2013;10:20. doi: 10.1186/1742-4690-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leavitt AD, Robles G, Alesandro N, Varmus HE. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J Virol. 1996;70(2):721–728. doi: 10.1128/jvi.70.2.721-728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matreyek KA, Engelman A. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J Virol. 2011;85(15):7818–7827. doi: 10.1128/JVI.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Llano M, et al. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314(5798):461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 40.Marini B, et al. Nuclear architecture dictates HIV-1 integration site selection. Nature. 2015;521(7551):227–231. doi: 10.1038/nature14226. [DOI] [PubMed] [Google Scholar]
- 41.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17(8):1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh PK, et al. LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1 integration to highly spliced genes. Genes Dev. 2015;29(21):2287–2297. doi: 10.1101/gad.267609.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012;8(5):e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Awasthi S, Alwine JC. Association of polyadenylation cleavage factor I with U1 snRNP. RNA. 2003;9(11):1400–1409. doi: 10.1261/rna.5104603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millevoi S, et al. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J. 2006;25(20):4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruepp MD, et al. The 68 kDa subunit of mammalian cleavage factor I interacts with the U7 small nuclear ribonucleoprotein and participates in 3′-end processing of animal histone mRNAs. Nucleic Acids Res. 2010;38(21):7637–7650. doi: 10.1093/nar/gkq613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300(5626):1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 48.Hulme AE, Kelley Z, Foley D, Hope TJ. Complementary assays reveal a low level of CA associated with viral complexes in the nuclei of HIV-1-infected cells. J Virol. 2015;89(10):5350–5361. doi: 10.1128/JVI.00476-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng K, et al. Quantitative microscopy of functional HIV post-entry complexes reveals association of replication with the viral capsid. eLife. 2014;3:e04114. doi: 10.7554/eLife.04114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chin CR, et al. Direct visualization of HIV-1 replication intermediates shows that capsid and CPSF6 modulate HIV-1 intra-nuclear invasion and integration. Cell Reports. 2015;13(8):1717–1731. doi: 10.1016/j.celrep.2015.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katahira J, et al. Human TREX component Thoc5 affects alternative polyadenylation site choice by recruiting mammalian cleavage factor I. Nucleic Acids Res. 2013;41(14):7060–7072. doi: 10.1093/nar/gkt414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Rijck J, Bartholomeeusen K, Ceulemans H, Debyser Z, Gijsbers R. High-resolution profiling of the LEDGF/p75 chromatin interaction in the ENCODE region. Nucleic Acids Res. 2010;38(18):6135–6147. doi: 10.1093/nar/gkq410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Primio C, et al. Single-cell imaging of HIV-1 provirus (SCIP) Proc Natl Acad Sci USA. 2013;110(14):5636–5641. doi: 10.1073/pnas.1216254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levy DN, Aldrovandi GM, Kutsch O, Shaw GM. Dynamics of HIV-1 recombination in its natural target cells. Proc Natl Acad Sci USA. 2004;101(12):4204–4209. doi: 10.1073/pnas.0306764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hrecka K, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kasper JS, Kuwabara H, Arai T, Ali SH, DeCaprio JA. Simian virus 40 large T antigen’s association with the CUL7 SCF complex contributes to cellular transformation. J Virol. 2005;79(18):11685–11692. doi: 10.1128/JVI.79.18.11685-11692.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jurado KA, et al. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc Natl Acad Sci USA. 2013;110(21):8690–8695. doi: 10.1073/pnas.1300703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matreyek KA, Yücel SS, Li X, Engelman A. Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog. 2013;9(10):e1003693. doi: 10.1371/journal.ppat.1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sowd GA, Li NY, Fanning E. ATM and ATR activities maintain replication fork integrity during SV40 chromatin replication. PLoS Pathog. 2013;9(4):e1003283. doi: 10.1371/journal.ppat.1003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H, Shun MC, Dickson AK, Engelman AN. Embryonic lethality due to arrested cardiac development in Psip1/Hdgfrp2 double-deficient mice. PLoS One. 2015;10(9):e0137797. doi: 10.1371/journal.pone.0137797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eidahl JO, et al. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 2013;41(6):3924–3936. doi: 10.1093/nar/gkt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.