Significance
X-ray crystallography provides a structural basis for enzyme mechanisms by elucidating information about the chemical reaction occurring within the active site. Crystallographic structures can also aid in rational drug design. A highly conserved family of methionine adenosyltranferases (MATs) produces S-adenosylmethionine (SAMe) via an ATP-driven process. Dysregulation of MAT enzymes has been tightly linked to liver and colon cancer. Here we present crystal structures of human MATα2 proteins containing different ligands within the active site, allowing for a step change in our understanding of how this enzyme uses its substrates, methionine and adenosine, to produce the product SAMe.
Keywords: methionine adenosyltransferase, cell growth, liver cancer, X-ray crystallography, methylation
Abstract
The principal methyl donor of the cell, S-adenosylmethionine (SAMe), is produced by the highly conserved family of methionine adenosyltranferases (MATs) via an ATP-driven process. These enzymes play an important role in the preservation of life, and their dysregulation has been tightly linked to liver and colon cancers. We present crystal structures of human MATα2 containing various bound ligands, providing a “structural movie” of the catalytic steps. High- to atomic-resolution structures reveal the structural elements of the enzyme involved in utilization of the substrates methionine and adenosine and in formation of the product SAMe. MAT enzymes are also able to produce S-adenosylethionine (SAE) from substrate ethionine. Ethionine, an S-ethyl analog of the amino acid methionine, is known to induce steatosis and pancreatitis. We show that SAE occupies the active site in a manner similar to SAMe, confirming that ethionine also uses the same catalytic site to form the product SAE.
Transmethylation, the transfer of a methyl group from one molecule to another, is a fundamental chemical reaction that plays a central role in important biological processes such as gene expression, cell growth, and apoptosis (1). The highly conserved methionine adenosyltransferase (MAT) enzymes synthesize the main source of methyl groups in all living organisms in the form of S-adenosylmethionine (SAMe) (2). The MAT family of enzymes is conserved throughout the kingdoms, emphasizing both their importance and essential role in maintaining appropriate levels of SAMe.
Mammals express three MAT genes: MAT I alpha (MAT1A), MAT II alpha (MAT2A), and MAT II beta (MAT2B). MAT1A and MAT2A encode for the catalytic subunits, MATα1 and MATα2, which share an 84% sequence similarity. MAT2B encodes for MATβ, the regulatory subunit that has low sequence similarity (7%) to the catalytic subunits. MATα and MATβ subunits can form several MATαβ complexes (3, 4). There are two major splicing forms of the MAT2B gene that encode for MATβV1 and MATβV2, which share very high sequence similarity and differ by only 20 residues in the N terminus (5). We recently reported that MATβ isoforms interact through their C terminus with MATα2 to form the MATαβ complexes MATαβV1 and MATαβV2 (4). MATα1 can exist in two oligomeric states, dimer and tetramer; however, there was little information on the oligomeric state of MATα2 until recently, when the structure of MATα2βV2 showed that MATα2 can exist and function as a tetramer in the presence of MATβ (4).
Several diseases are known to arise from dysregulation of MAT1A, MAT2A, and MAT2B. For example, mutations involving the MAT1A gene have links with hepatic MAT deficiency which leads to hypermethioninemia (6, 7). Expression of MATα2 confers a growth advantage in cells and is important for differentiation and apoptosis (1) in diseases such as human hepatocellular carcinoma (5), colon cancer (8), and leukemia (9).
A diverse series of structures are available for MAT enzymes from bacteria, rat, and human (4, 10–12), which, supported by a range of biochemical evidence, have provided significant insight into the enzymatic mechanism. SAMe synthesis follows an SN2 catalytic mechanism (13, 14) in which the reaction is initiated through a nucleophilic attack by the sulfur atom of methionine on the C5′ atom of ATP, which produces the intermediate tripolyphosphate (PPPi). Hydrolysis of the PPPi into pyrophosphate (PPi) and orthophosphate (Pi) then occurs (Fig. S1).
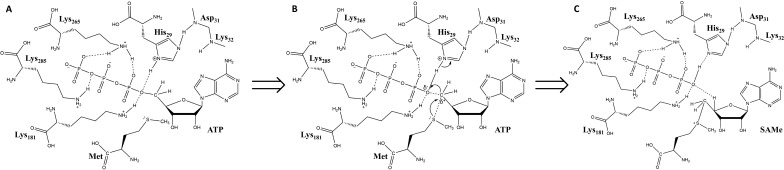
Fig. S1.
Schematic of the enzymatic reaction of SAMe synthesis. Residues of MATα2 that interact with ATP within the active site are shown; dashed lines indicate hydrogen bonds. (A) The interaction of the amide groups of Asp31 and Lys32 with His29 polarizing its imidazol ring and favouring the N-proton release. (B) The sulphur atom of methionine performs a nucleophilic attack on the C5′ atom of ATP. Arrows indicate the movement of electron pairs. (C) Through a nucleophilic substitution reaction the sulphur atom and the C5′ atom form a new bond creating SAMe as the C5′–O5′ of ATP is cleaved.
A common feature of MAT enzymes is a gating loop that flanks the active site (in human MATα2 residues 113–131), which has been postulated to act in a dynamic way to allow access to the active site. It has been suggested that when the active site is occupied, the loop is closed like a gate, but when the active site is empty, the loop becomes invisible in the structure, presumably resulting from an open dynamic gate (11). Whether the opening of the loop/gate is required for the entry of substrate and release of products remains unclear. It has been shown that MAT enzymes not only metabolize methionine, but also, depending on species, can process many methionine analogs (15). Ethionine, an S-ethyl analog of the amino acid methionine, is known to induce steatosis and pancreatitis in rats (16). MAT enzymes are able to produce S-adenosylethionine (SAE) from ethionine (17), which in turn acts as a competitor of methionine. As a result, insufficient levels of SAMe are produced within the cell, leading to disruption of SAMe-dependent processes, such as nicotinamide catabolism (18). SAE can be used to ethylate targets by donating its ethyl group in direct competition with methylation, which results in abnormal alkylation of nucleic acids (19, 20).
Here, for the first time to our knowledge, we report several ligand-bound structures of human MATα2 at 1.1 Å (SAMe+ADO+MET+PPNP), 1.85 Å (SAE), and 2.3 Å [p-nitrophenyl phosphate (PPNP)], providing information on the formation of SAMe and SAE in human MATα2 and supporting the common mechanism that exists among MAT enzymes. A detailed view of the position of the triple phosphate group within the active site clearly reveals the orientation of the three phosphoryl moieties, as well as the placement, coordination, and identity of ions within the active site. The resulting insights into the movement of methionine during catalysis elucidate some of the key catalytic events that lead to product formation. We also report the first, to our knowledge, nonarchaeal MAT enzyme structure containing a nonnative product, SAE, that occupies the active site in a similar manner as SAMe, suggesting a similar mechanism for the utilization of ethionine to form the product SAE.
Results
From Substrate Binding to SAMe Formation.
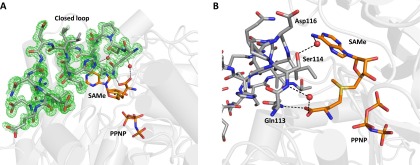
We obtained the atomic resolution structure (SAMe+ADO+MET+PPNP) at 1.1 Å (Table 1) by incubation with AMP-PNP and methionine (Table S1). The atomic resolution allows us to resolve multiple ligands with partial occupancies, including substrates and products within the active site; methionine (occupancy, 0.3), adenosine (occupancy, 0.5), SAMe (occupancy, 0.5), and PPNP (occupancy, 0.7) can be seen. An omit map clearly shows the substrate methionine in a position not previously observed. It sits within the active site, so that its functional group is directed away from its final location as part of the product SAMe (Fig. 1A).
Table 1.
Data collection and refinement statistics
| Variable | SAME+ADO+MET+PPNP | SAE bound | PPNP bound |
| Wavelength, Å | 0.92 | 0.92 | 0.92 |
| Detector | Pilatus | Pilatus | Pilatus |
| Space group | I222 | I222 | I222 |
| Unit-cell dimensions (a,b,c), Å | 67.92, 94.07, 117.22 | 68.39, 94.39, 117.39 | 66.11, 95.33, 117.51 |
| Resolution, Å | 73.37–1.10 (1.14–1.10) | 58.7–1.85 (1.90–1.85) | 74.04–2.34 (2.40–2.34) |
| Rmerge, % (last shell) | 8.9 (91.0) | 10.3 (81.4) | 11.6 (59.7) |
| I/σ (last shell) | 24.5 (3.4) | 21.3 (3.4) | 20.7 (3.9) |
| Completeness, % | 99.1 (99.0) | 99.5 (97.5) | 99.9 (99.9) |
| Redundancy | 12.3 (9.7) | 13.3 (12.5) | 13.0 (13.7) |
| No. of reflections | 176,512 | 33,901 | 16,107 |
| Rwork/Rfree | 10.5/12.6 | 13.4/17.7 | 14.8/20.1 |
| No. of atoms | |||
| Protein | 3,290 | 2,972 | 2,876 |
| Ligand/ion | 68/4 | 41/3 | 13/3 |
| Water | 447 | 265 | 161 |
| B factors, Å2 | |||
| Protein | 11.2 | 20.0 | 29.1 |
| Ligands/ions | SAMe/ADO/MET/PPNP/Mg/K | SAE/PPNP/Mg/K | PPNP/Mg/K |
| 7.5/13.3/14.5/10.3/8.5/10 | 22.5/22/21/27 | 32.8/28.5/62 | |
| Water | 24.0 | 29.1 | 32.4 |
| Ramachandran statistics | |||
| Residues in preferred regions | 422 (98%) | 369 (97%) | 354(97%) |
| Residues in allowed regions | 11 (2%) | 12 (3%) | 12 (3%) |
| Outliers | 0 (0%) | 0 (0%) | 0 (0%) |
| rmsd | |||
| Bond length, Å | 0.014 | 0.020 | 0.015 |
| Bond angles, ° | 1.79 | 1.83 | 1.66 |
| PDB ID code | 5A1I | 5A1G | 5A19 |
Values in parentheses are for the highest resolution shell. Each structure was solved using molecular replacement using of human MATα2 as model (PDB ID code 2P02).
Table S1.
Summary of incubation conditions used for all structures referred to in this study
| Crystal name | PBD ID code | Pregel filtration incubation* | Precrystallization incubation | Cryoprotectant |
| SAMe+ADO+MET+PPNP bound | 5A1I | 100 μM AMP-PNP, 10 mM methionine | 250 μM AMP-PNP | Reservoir solution |
| PPNP bound | 5A19 | 100 μM AMP-PNP | 250 μM SeMet | Reservoir solution |
| 10 mM methionine | ||||
| SAE bound | 5A1G | 100 μM AMP-PNP | 250 μM ethionine | Reservoir solution + 20% ethylene glycol |
| 10 mM ethionine | ||||
| MAT(α2)4(βV2)2 | 4NDN | 300 mM NADP | 250 μM AMP-PNP + 1 mM methionine | Reservoir solution + 20% ethylene glycol |
The proteins were incubated in 50 mM Hepes buffer pH 7.5, 10 mM MgCl2, 50 mM KCl, and 10 mM DTT with the ligands shown in the table, after which gel filtration was performed using 25 mM Hepes pH 7.5, 200 mM NaCl, 1 mM MgCl2, 5 mM KCl, and 1 mM TCEP as a buffer.
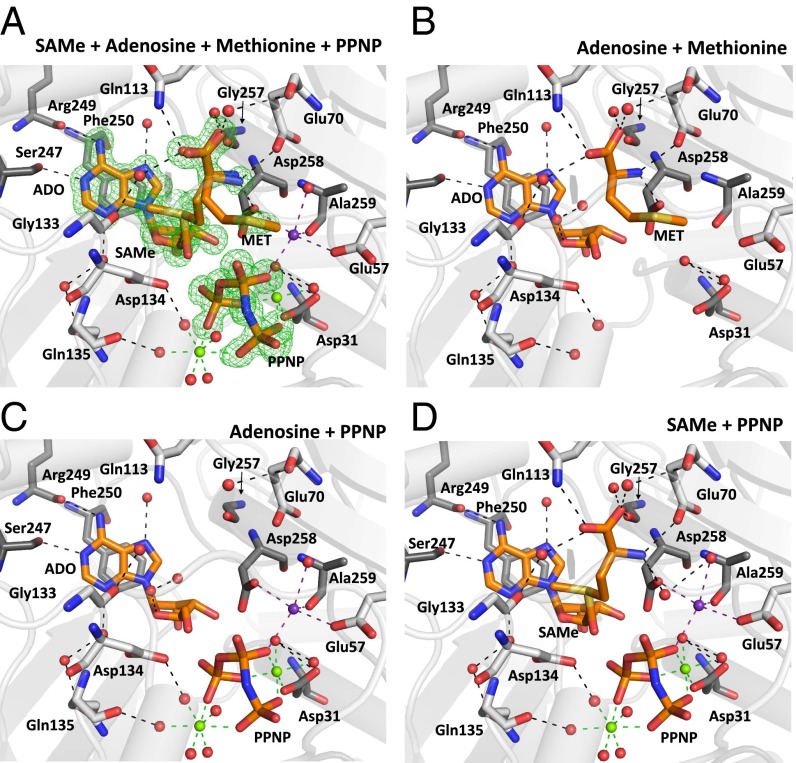
Fig. 1.
SAMe synthesis in human MATs. (A) Active site of SAMe+ADO+MET+PPNP MATα2 showing low-occupancy substrate methionine (0.3), adenosine (0.5), SAMe (0.5), and PPNP (0.7). The omit map (Fo-Fc) is colored green and contoured at the three sigma level. (B) Novel positions of substrate methionine and adenosine, both hydrogen-bonded to MATα2 residues and water molecules. (C) Adenosine and PPNP occupying the active site, hydrogen-bonded to MATα2 residues, ions, and water molecules. (D) Product SAMe and PPNP shown hydrogen-bonded by the same residues that secure both substrates. Two MATα2 monomers form a dimer, residues of which are shown in gray and dark gray. Substrates and residues are shown as sticks, color-coded according to the nature of the atom (blue, N; red, O). Hydrogen bonds are shown as black dotted lines. Green and purple dotted lines show magnesium and potassium ion coordination, respectively. Water molecules, magnesium ions, and potassium ions are represented as red, green, and purple spheres, respectively.
To clearly illustrate the components of the mixed active site, we present three snapshots representing different states in Fig. 1 B–D. In the first step, the orientation of methionine before SAMe formation (Fig. 1B) and the hydrolysis of the AMP-PNP into adenosine (ADO) and PPNP (Fig. 1C) are observed, whereas in the second step, the formation of SAMe and the placement of PPNP before its hydrolysis into pyrophosphate (PPi) and orthophosphate (Pi) (Fig. 1D) is seen. The substrate methionine is hydrogen-bonded only via its nitrogen with OE1 atoms of Glu70 (2.8 Å) and OD1 atoms of Asp258 (2.7 Å) and via its two terminal oxygens with NE2 atoms of Gln113 (3.0 Å) and water ligands (Fig. 1B), which allows its side chain extra flexibility. The position of ADO is maintained through π-π stacking between the purine base adenine and Phe250, as well as several hydrogen bonds with the side chain O of Ser247 (2.8 Å), main chain oxygen of Arg249 (2.9 Å), and water (Fig. 1 A–D). One water molecule forms a hydrogen bond with another water molecule and the O4 of adenosine, and occupies the position in which eventually the sulfur of SAMe lies (Fig. 1D).
The triple phosphate can be modeled precisely owing to the high quality of the omit map at atomic resolution (Figs. 1 A, C, and D and 2A). The PPNP is secured in place through multiple interactions with residues His29, Lys181, Lys265, Ap291, and Ala281; water; two hexacoordinated magnesium ions; and a pentacoordinated potassium ion (Fig. S2). The product SAMe lies within the active site, interacting with the same residues that secure the methionine main chain and the ADO molecule, identical to those in the first step, whereas the S atom of SAMe occupies the position previously occupied by a water molecule.
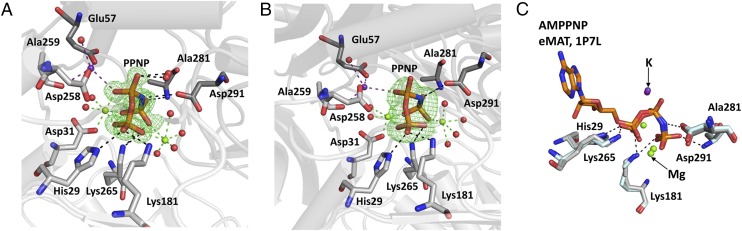
Fig. 2.
Comparison of phosphate moiety conformations in human MAT enzymes. (A) Omit (Fo-Fc) electron density map, contoured at the three sigma level around the PPNP from the SAMe+ADO+MET+PPNP MATα2 structure. (B) Coordination of PPNP from rerefined MAT(α2)4(βV2)2 (PDB ID code 4NDN), showing that the position of PPNP is the same as in SAMe+ADO+MET+PPNP. Omit map and color cording are the same as in A. (C) Superposition of AMP-PNP from eMAT (PDB ID code 1P7L) and PPNP from SAMe+ADO+MET+PPNP MATα2 shown in stick representation. Residues from MATα2 monomers are shown as sticks in gray or dark gray, with hydrogen bonds as black dotted lines. Magnesium ions are shown as green spheres; potassium ions, as purple spheres. Coordination of magnesium and potassium ions is indicated by green and purple dotted lines, respectively. All phosphate moieties are shown as sticks, color-coded by atom (orange, P; red, O; blue, N).
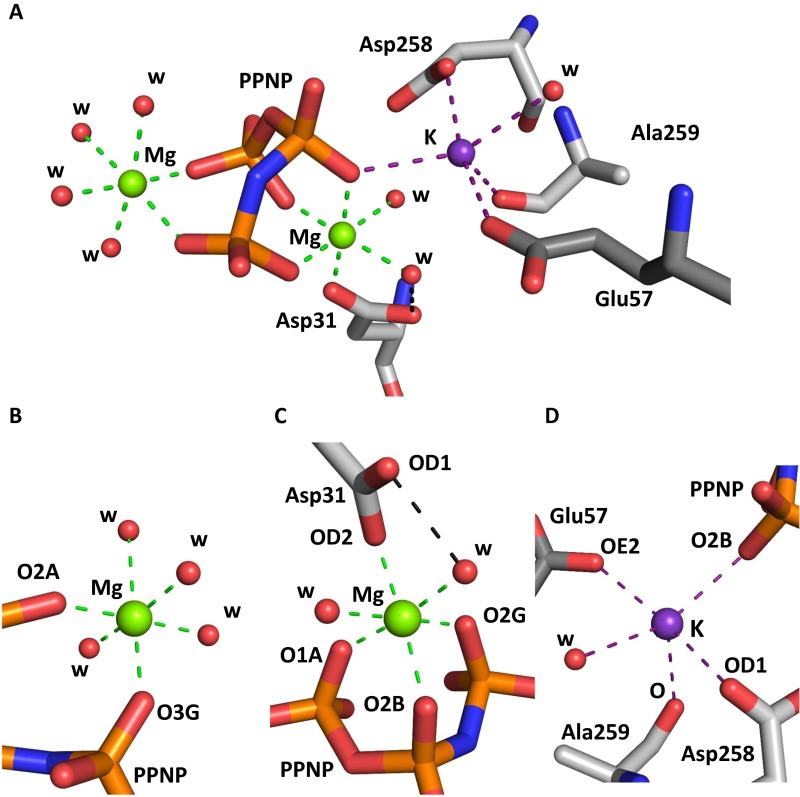
Fig. S2.
Magnesium and potassium ion binding in ligand-bound MATα2. (A) Active site of MATα2 (SAMe+ADO+MET+PPNP) showing two magnesium ions (green) and one potassium ion (purple). (B) Closer view of the first magnesium ion, hexacoordinated by four water molecules and two oxygen atoms, O3G and O2A, from PPNP. (C) Closer view of the second magnesium ion, hexacoordinated by two water molecules and four oxygen atoms, OD2 Asp31, O1A, O2B, and O2G, from PPNP. (D) Potassium ion pentacoordinated by water and four oxygen atoms originating from PPNP (O2B) and residues Asp258 (OD1), Ala259 (O), and Glu57 (OE2). PPNP and MATα2 residues are shown as sticks color-coded according to atom type (gray, C; red, O); water molecules are shown as red spheres. Coordination bonds for magnesium ions are indicated by green dotted lines; those for potassium, by purple dotted lines. Black dotted lines represent hydrogen bonds.
Mode of PPNP Binding Is Persevered in the Absence of the Regulatory Subunit.
Reexamination of the PPNP conformation in the MAT(α2)4(βV2)2 complex structure [Protein Data Bank (PDB) ID code 4NDN] using this group from the 1.1-Å SAMe+ADO+MET+PPNP structural model suggests that the PPNP was not correctly positioned, given that lower-resolution MATα2β structures did not allow identification of Mg ions. Positioning of these phosphates and three metal ions from the current atomic resolution structure of MATα2 into the lower-resolution electron density map of the MATα2β complexes gives a much improved fit to the electron density (Fig. 2B). The conformation of the triple phosphate is preserved in both the MATα2βV2 and SAMe+ADO+MET+PPNP structures. The position is also the same in AMP-PNP from Escherichia coli MAT (PDB ID code 1P7L), despite cleavage from the adenosine moiety (Fig. 2C).
PPNP Does Not Provide the Energy to Open the Gating Loop.
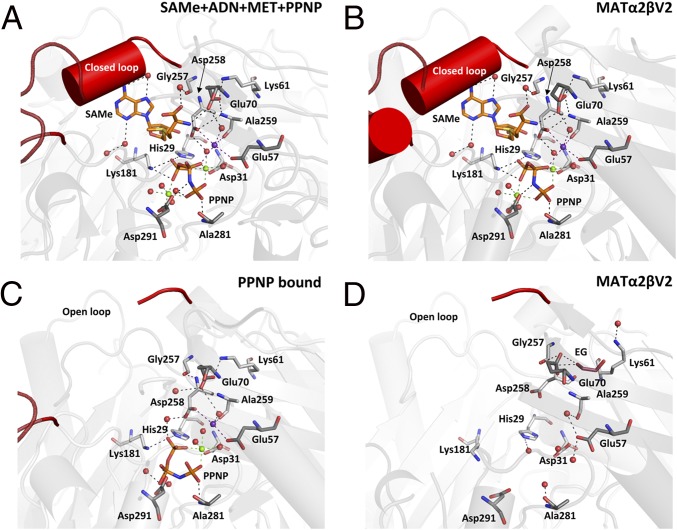
The structure of (PPNP-bound) MATα2 has a disordered gating loop, providing direct evidence against the idea that it provides the energy for the opening of the gate. The complex MAT(α2)4(βV2)2 (PDB ID code 4NDN) contains four active sites, two that reveal ordered gating loops and two that do not show a gating loop, presumably owing to a highly flexible, disordered nature (4). The two ordered sites contain SAMe, PPNP, Mg2+, and K+, whereas the disordered sites contain only an ethylene glycol molecule as a result of the cryoprotection. The structure of (PPNP-bound) MATα2 has a disordered gating loop, and a comparison of the PPNP-bound, MAT(α2)4(βV2)2, and SAMe+ADO+MET+PPNP structures shows that the gate is open when active site is occupied by PPNP. In contrast, the gate is shut like a lid when the active site is occupied by SAMe or adenosine (Fig. S3A).
Fig. S3.
Ordered gating loop of holo MATα2. (A) The omit map (Fo-Fc) is colored green around the gating loop from the SAMe+ADO+MET+PPNP-bound MATα2 structure and contoured at the three sigma level. (B) Interactions between the gating loop and the product SAMe. Gating loop residues are shown in stick representation. Two MATα2 monomers form a dimer colored in cartoon representation. Substrates and residues are shown as sticks, color-coded according to the nature of the atom (blue, N; red, O). Hydrogen bonds are shown as black dotted lines; water molecules, as red spheres.
A comparison of SAMe+ADO+MET+PPNP structure with holo rerefined MATα2 from the MAT(α2)4(βV2)2 complex (Fig. 3 A and B) shows that the residues in the active site do not differ in position, and that the nature of the active site in the catalytic subunit is preserved in the absence of the regulatory subunit MATβ. Gln113 is the sole residue of the gating loop that interacts with SAMe directly through its NE2 atom, forming a hydrogen bond with the O of SAMe. Ser114 interacts with a water molecule that also interacts with SAMe (Fig. S3B). The gating loop in the PPNP-bound (Fig. 3C) and apo subunit of the MAT(α2)4(βV2)2 complex (Fig. 3D) are invisible, demonstrating that the opening of the gate to release product is not driven by hydrolysis of the triple phosphate. What drives the opening of the gate and release of product remains an open question.
Fig. 3.
Comparison of the holo and apo active sites in MATα2 and MATαβ. (A) Holo SAMe+ADO+MET+PPNP MATα2 active site. (B) Holo MATαβ (PDB ID code 4NDN) active site. (C) Holo PPNP-bound MATα2 active site with disordered gating loop. (D) Apo MATαβ (PDB ID code 4NDN). Gating loop is highlighted in red. Two MATα2 monomers form a dimer colored gray and dark gray with residues of interest shown as sticks. SAMe is shown in orange; PPNP, as orange sticks. Magnesium ions are shown as green spheres; potassium ions, as purple spheres; and water molecules, as red spheres. Hydrogen bonds are indicated by black dotted lines. Green dotted lines indicate interaction with magnesium ions; purple dotted lines, interaction with potassium ions.
In the SAMe+ADO+MET+PPNP structure, OE1 atoms of Glu70 interact with the nitrogen atoms of Lys61 (NZ) and Ala259 (N), whereas its OE2 oxygen forms a hydrogen bond to the substrate methionine or SAMe (2.8 Å). Glu70 maintains the same position in all holo active sites (Fig. 3 A–C). In the apo MATα2 subunit of the MAT(α2)4(βV2)2 complex (Fig. 3D) significant movement of the residues occurs; for example, Lys61 adopts a different conformation than the residues in all other MATs, allowing Glu70 to hydrogen-bond to an ethylene glycol molecule.
The change in lysine conformation also enables movement of other residues in the active site; for example, Gly257 and Asp258 both move away from the active site. The oxygen atom, OD1, of Asp258 can no longer participate in the interaction with the potassium ion, but the movement of Asp258 enables it to interact with N of Ala259. The oxygen atom, OE1, of Glu70 is unable to interact with Lys61 or Ala259, but can interact with the N of Asp258, which alters its conformation as a result of the movement of Lys61.
In the MATα2 structure SAMe+ADO+MET+PPNP (Fig. 3A), the potassium ion is present and interacts with Asp258 and PPNP. In the PPNP-bound MATα2 (Fig. 3C), the PPNP adopts a different position in which the central phosphate PB rotates onto the side. The orientation of the residues around the PPNP remains the same as in the SAMe+ADO+MET+PPNP structure, and interaction with a magnesium ion occurs, but interaction with the second magnesium ion and the potassium ion is lost. Owing to the coordination through the magnesium ion, the PPNP is able to occupy the active site while the gating loop is disordered, i.e., no longer in its closed position. Furthermore, there are movements in residues that interact with the potassium ion, Glu57 and Ala259, and the PPNP moiety. Three further residues, namely His29, Lys181, and Asp291, that interact with the PPNP show movement in the absence of PPNP in the empty active site.
Catalytic Steps Captured for Human MAT.
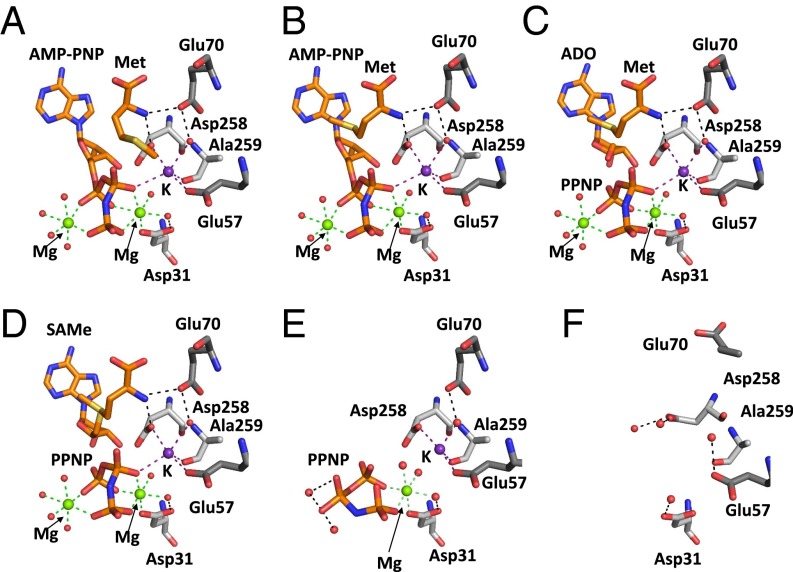
Using the structures SAMe+ADO+MET+PPNP, PPNP-bound, MATα2βV2 complex, and MAT from E. coli (eMAT; PDB ID code 1P7L), a detailed picture of SAMe synthesis can be provided from structural snapshots starting from substrate methionine. In the initial position, the functional group of methionine is too close to the phosphate group β of ATP (here AMP-PNP; PDB ID code 1P7L), suggesting that methionine will bind first, followed by ATP binding, which will change the conformation of the methionine side chain without changing its main chain position, because the nitrogen on the methionine is at a suitable distance to form two strong hydrogen bonds with OD1 of Asp258 (2.7 Å) and OE1 of Glu70 (2.8 Å) (Fig. 4A and Fig. S2).
Fig. 4.
Insights into SAMe synthesis in human MAT. (A) Methionine from SAMe+ADO+MET+PPNP and ATP analog AMP-PNP drawn as in eMAT (PDB ID code 1P7L). Methionine is in position where its nitrogen is hydrogen-bonded to Asp258 and Glu70. (B) Movement of substrate methionine as it moves toward the C5′ bond of AMP-PNP (ligands modeled from eMAT (PDB ID code 1P7L). (C) Cleavage of AMP-PNP producing ADO and PPNP from SAMe+ADO+MET+PPNP with methionine (PDB ID code 1P7L). (D) Formation of SAMe from ADO and methionine with PPNP present (SAMe+ADO+MET+PPNP). (E) Active site after SAMe has left the active site, leaving PPNP bound. (F) On breakdown of the triple phosphate, Glu70 and Asp258 change conformation, and magnesium ions no longer coordinate the PPNP. Substrates and residues are shown as sticks, and hydrogen bonds are shown as black dotted lines. Green and purple dotted lines show magnesium and potassium ion coordination, respectively. Water molecules, magnesium ions, and potassium ions are represented as red, green, and purple spheres, respectively.
The conformation of AMP-PNP is stabilized by two magnesium ions, one interacting with O1A and O2B of PPNP and the other interacting with O2A, O2G, and O3G. For the sulfur atom of methionine to bind with the C5′ atom of AMP-PNP, the methionine has to move its side chain from its initial position toward the C5′ of AMP-PNP (Fig. 4B) while the nitrogen of methionine is still interacting with the OD1 of Asp258 (2.7 Å) and OE1 of Glu70 (2.8 Å) to aid its stabilization and rotation. Once the methionine is oriented in position, its electron-rich sulfur reacts with the positively polarized C5′ of AMP-PNP in a nucleophilic substitution to produce adenosine and PPNP (Fig. 4C) before the final SAMe and PPNP (Fig. 4D) are synthesized. MATα2 (PPNP-bound) exhibits a pre-apo state in which, after the product SAMe has been formed, it is able to exit the active site, leaving the triple phosphate moiety within the active site that still possesses Mg2+ and K+ ions (Fig. 4E). The loss of one of the magnesium ions causes reorientation of the PPNP moiety, which shifts the O2B atom, leaving it too far away to coordinate with the potassium ion. For the active site to cycle and be ready to receive the substrates again, the triple phosphate must be broken down before exiting the active site. The phosphatase activity of MATα2 causes breakdown of the triple phosphate, resulting in the loss of all ion coordination. Movement of Glu57, Glu70, Asp258, and Ala259 occurs after all products have exited the active site (Fig. 4F) (PDB ID code 4NDN).
Incorporation of Nonnative Methionine Analogs.
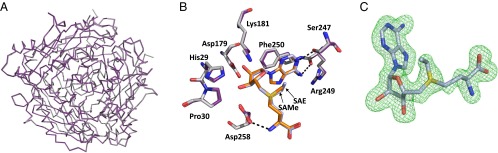
To our knowledge, the SAE structure is the first nonarchaeal MAT enzyme structure identified to contain a nonnative reaction product, SAE. Overall, there is no structural rearrangement in the main chain, and the secondary structure is maintained (rmsd 0.185 Å; Fig. 5A) compared with SAMe+ADO+MET+PPNP. SAE occupies the active site in a similar way as SAMe. There is little movement of the residues involved in the stabilization of both SAMe and SAE, including residues Ser247, Arg249, and Asp258 (Fig. 5B). The extra CH3 group can be accommodated within the active site without causing clashes with residues near the methyl/ethyl group, specifically Ile117, Ile322, Gly133, and Asp134. The omit map shows clear positions for all atoms of the SAE molecule (Fig. 5C).
Fig. 5.
Structure of SAE MATα2. (A) Superposition of MAT(α2)2 with SAE bound (in purple) and SAMe bound (in silver) displayed as lines, showing no change in folding (rmsd 0.185 Å). (B) Superposition of SAE (light blue) and SAMe within the active site of MATα2, showing no change in position of surrounding residues. Residues from the SAMe bound structure are shown as silver sticks; those from the SAE bound structure, as purple sticks. Hydrogen bonds are shown as black dotted lines. (C) Omit map (Fo-Fc) electron density map and stick representation of SAE from MATα2. The map is contoured at the three sigma level around the SAE molecule.
Discussion
Despite the plethora of MAT structures that have been published (10, 11, 21–23) or deposited in the PDB, to date only one structure of human MATα2 (12) (1.2 Å resolution) and of the MAT(α2)4(βV2)2 complex (4) (2.35–3.3 Å resolution) have been published. The mechanism of MAT enzyme function has been described for eMAT (11), and this has become the accepted mechanism for MAT enzymes. Here we present several high-resolution crystallographic structures that support the overall eMAT reaction mechanism but add to it significantly by defining the movements of methionine within the active site during catalysis. We see that methionine initially occupies the active site (Figs. 1B and 4A) in a different manner from that in eMAT (Fig. 4B). A direct attack by the sulfur of methionine on the C5′ atom of the ATP molecule was proposed to form SAMe (13), which was confirmed by kinetic isotope studies (14). With SAMe+ADO+MET+PPNP, the initial position (Fig. 1B) of methionine can be seen, which is stabilized through hydrogen bonds between the main chain nitrogen of methionine with Asp258 and Glu70, conserved in mammals and in E. coli (Fig. S4), and through the terminal oxygen and Gln113. Binding of ATP causes the side chain of MET to come too close to PPNP, causing it to rotate into a position to attack the C5′ atom of ATP without introducing a steric hindrance.
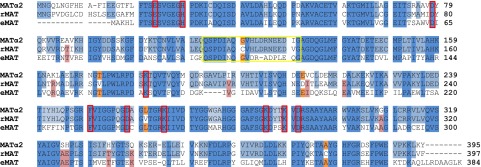
Fig. S4.
Sequence alignment of MATα2 from Homo sapiens, rMAT from Rattus norvegicus, and eMAT from E. coli. Fully conserved residues among three sequences are shown in blue, residues conserved only between human and rat are in light blue, residues conserved only between human and E. coli are in orange, and residues conserved only between E. coli and rat are in pink. Red boxes indicate residues involved in catalysis. The gating loop is highlighted by the yellow box.
PPNP is a nonhydrolyzable triple phosphate moiety that is particularly useful when studying MAT enzymes, because it cannot be cleaved to pyrophosphate or orthophosphate. The high-resolution structure SAMe+ADO+MET+PPNP shows the position not only of the PPNP, but also of Mg2+ and K+ ions within the active site (Fig. 2 and Fig. S2), which were previously difficult to model. Interestingly, the positions of the three ions within the active site are highly similar to those in E. coli (Fig. 2C). High-resolution (1.1 Å) data clearly show that all three active site ions are highly coordinated, either hexacoordinated or pentacoordinated (Fig. S2). Orientation of the PPNP is the same as that of the AMP-PNP in the E. coli structure, demonstrating that as the AMP-PNP is cleaved, the newly formed PPNP does not cause any change in orientation or in the residues that were initially anchoring the AMP-PNP in the active site. Refining the MAT(α2)4(βV2)2 complex (PDB ID code 4NDN) using the position of the PPNP from the high-resolution structure satisfies electron density. PPNP is stabilized through interactions with Mg2+ and K+ ions, as well as with a number of residues (Fig. 2A and Fig. S2). These interactions would become destabilized on the cleavage of the triple phosphate to pyrophosphate and orthophosphate, allowing them to exit the active site. Only a small structural movement within the region close to the active site is needed to aid the initial anchoring of ATP and stabilization of the cleaved triple phosphate moiety to allow breakdown of the triple phosphate into pyrophosphate and orthophosphate.
The observation that the MATα2 active site can be occupied but still have the gating loop disordered is new in human MAT enzymes and provides evidence of the exiting order of products from the active site. SAMe exits the active site on the gating loop, becoming disordered, followed by the cleaved triple phosphate products, causing residues such as Lys61, Glu70, and Lys181 to return to their unbound (apo) positions (Fig. 3). The nonhydrolyzable PPNP remains in the active site, the position of the highlighted residues remains the same as if the active site were occupied, and the gating loop remains ordered and shut.
An interesting observation is that the position of the PPNP differs between the PPNP-bound MATα2 and the SAMe+ADO+MET+PPNP form. This may represent a pre-apo structure in which the movement of the central phosphate causes a change in ion coordination and allows for the phosphatase activity of MATα2. Alternatively, the position may be due simply to a time-lapse effect of binding a nonhydrolyzable PPNP. The mechanism for the hydrolysis of the PPPi in MAT is believed to be similar to that for other metal ion-dependent ATPases and other phosphatases (24). Our data suggest that ATP (PPNP) binding will change the conformation of Glu70 from open (Fig. 3D) to substrate-bound (Fig. 3 A–C), aiding the stabilization of methionine binding. A comparison of the SAMe+ADO+MET+PPNP MATα2 and MAT(α2)4(βV2)2 complexes clearly shows that the presence of MATβ does not directly affect the catalytic site (Fig. 3 A and B). Although it was previously demonstrated that recombinant MATαβ complexes have greater activity than MATα2 alone (4), how MATα2 activity is regulated by MATβ remains unclear.
Although the ability of human MATα2 to use ethionine as a substrate (17), and the effect of this within the cell (16), are well known, no human MATα2 structure has been published with this substrate or its product, SAE. Recently, Wang et al. (15) published the structure of sMAT from Sulfolobus solfataricus containing SAE within the active site, and suggested the occurrence of moderate clashes between the ethyl group and Ile117 and Ile322 of human MATα2. In the human MATα2 SAE structure containing SAE, this is not the case, and the ethyl group is >3 Å away from the surrounding residues. SAE has the same orientation as SAMe (Fig. 5B), with very little noticeable movements.
Our results show that SAMe can leave the active site without the breakdown of the PPNP, which has been presumed to provide the energy for the opening of the gating loop. It is clear that the nature of active site in the catalytic subunit MATα2 is retained in the absence of the regulatory subunit MATβ. Both of these unexpected findings have wider implications for the formation and release of SAMe. The utilization of the same catalytic site for substrate ethionine for the production of SAE opens up therapeutic possibilities for diseases connected to SAMe and SAE utilization.
Materials and Methods
Mutant Production.
Purification of MATα2 and MATβ variants was done as described previously (4). Generation of the C-terminal MATβV2 mutant, MATβV2H323stop, was accomplished through site-directed mutagenesis with a PET28-MATβV2 vector using the QuikChange site-directed mutagenesis method (Stratagene), with the following primers: forward, 5′-GGAGACAAACGGTCTTTTAATAGCTCGAGCACCACC-3′; reverse, 5′-GGTGGTGCTCGAGCTATTAAAAGACCGTTTGTCTCC-3′. Sequencing of plasmids was performed by the external service of the Spanish National Cancer Research Center, Madrid.
Complex Formation, Crystallization, and Data Collection.
MATα2 and MATβV2H323stop were incubated for 1 h at 4 °C in 50 mM Hepes pH 7.5, 10 mM MgCl2, 50 mM KCl, 100 µM AMP-PNP (Sigma-Aldrich), and 10 mM DTT with either 10 mM methionine (Sigma-Aldrich) or 10 mM ethionine (Sigma-Aldrich). Each of these complexes was then loaded onto a Superdex 200 10/300 column (GE Healthcare) and eluted using 25 mM Hepes pH 7.5, 200 mM NaCl, 1 mM MgCl2, 5 mM KCl, and 1 mM TCEP. Before crystallization, MAT(α2)4(βV2H323stop)2 was incubated before gel filtration with AMP-PNP, and methionine was incubated with extra AMP-PNP (250 µM) or selenium-methionine (250 µM), resulting in SAMe+ADO+MET+PPNP and PPNP-bound crystals, respectively. The SAE-bound structure was obtained from MAT(α2)4(βV2H323stop)2 that was incubated before gel filtration with AMP-PNP and extra ethionine (250 µM).
For all crystallizations, the protein concentration was 5.8 mg/mL. SAMe+ADO+MET+PPNP and PPNP-bound crystals appeared at 25 °C within 2–3 d in drops containing 1 µL of 0.1 M Hepes pH 7.5 and 30% (wt/vol) PEG 600 with 1 µL of MATα2. For SAE, crystals appeared at 18 °C within 3–4 d in drops containing 800 nL of 0.1 M imidazole pH 7.0 and 50% (wt/vol) MPD with 800 nL of MATα2. Crystals were cryoprotected in the reservoir solution and flash-frozen in liquid nitrogen. For the SAE-bound crystals, up to 20% (wt/vol) ethylene glycol was added to the reservoir solution for cryoprotection. Different datasets were collected at the XALOC beamline at the ALBA synchrotron center (Barcelona) and the I04-1 beamline at the Diamond synchrotron center (Oxford) (0.92 Å wavelength). Data reduction was carried out using HKL-2000 (25) and XDS (26). Phases were calculated with Phaser (27), using MATα2 (PDB ID code 2P02) (12) as a search model for molecular replacement. The structure solution revealed that all crystals contained only MATα2 protein, irrespective of the starting material.
Refinement.
Model building and refinement were performed using REFMAC (28), COOT (29), and PHENIX (30). Participants at different stages of the enzymatic reaction were modeled in the 1.1-Å resolution SAMe+ADO+MET+PPNP structure within the active site, and their occupancies were modified to fit the electron density, taking into account the following observations: the ADO part of the molecule is always in the active site as part of ADO or SAMe; PPNP has an occupancy of 0.7, so it can be present with both SAMe (0.5) and ADO (0.5); PPNP molecules cannot be present together with the conformation observed for MET (0.3) because of steric hindrance; ADO and SAMe occupy the same space in the active site, so they cannot be present together; and SAMe and MET occupy the same space in the active site, so they cannot be present together. Data collection and refinement statistics are summarized in Table 1. Structural figures were generated using PYMOL (31).
Acknowledgments
We thank members of the Liverpool’s Biophysics group and Dr. Ramon Hurtado-Guerrero (University of Zaragoza) for the helpful discussions. We also thank the staff and management of the Diamond and ALBA synchrotron centers for the use of their facilities. Use of the Diamond synchrotron center was funded in part by the European Community’s Seventh Framework Programme (FP7/2007-2013) under BioStruct-X (Grant 283570). Support for this work was provided by the National Institutes of Health (Grant R01 DK51719, to S.C.L. and J.M.M.) and the Plan Nacional de I + D (Grant SAF 2011-29851). S.V.A. was supported by Wellcome Trust Fellowship 097826/Z/11/Z and a Santander collaborative grant. B.M. was supported in part by the Biotechnology and Biological Sciences Research Council Doctoral Training Partnership, the University of Liverpool–Center for Cooperative Research in Biosciences partnership, and Santander collaborative grants.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5A1I, 5A1G, and 5A19).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510959113/-/DCSupplemental.
References
- 1.Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev. 2012;92(4):1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landgraf BJ, Booker SJ. Biochemistry: The ylide has landed. Nature. 2013;498(7452):45–47. doi: 10.1038/nature12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeGros HL, Jr, Halim AB, Geller AM, Kotb M. Cloning, expression, and functional characterization of the beta regulatory subunit of human methionine adenosyltransferase (MAT II) J Biol Chem. 2000;275(4):2359–2366. doi: 10.1074/jbc.275.4.2359. [DOI] [PubMed] [Google Scholar]
- 4.Murray B, et al. Structure and function study of the complex that synthesizes S-adenosylmethionine. IUCrJ. 2014;1(Pt 4):240–249. doi: 10.1107/S2052252514012585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang H, et al. Expression pattern, regulation, and functions of methionine adenosyltransferase 2beta splicing variants in hepatoma cells. Gastroenterology. 2008;134(1):281–291. doi: 10.1053/j.gastro.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaull GE, Tallan HH. Methionine adenosyltransferase deficiency: New enzymatic defect associated with hypermethioninemia. Science. 1974;186(4158):59–60. doi: 10.1126/science.186.4158.59. [DOI] [PubMed] [Google Scholar]
- 7.Gaull GE, et al. Hypermethioninemia associated with methionine adenosyltransferase deficiency: Clinical, morphologic, and biochemical observations on four patients. J Pediatr. 1981;98(5):734–741. doi: 10.1016/s0022-3476(81)80833-5. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, et al. Role of methionine adenosyltransferase 2A and S-adenosylmethionine in mitogen-induced growth of human colon cancer cells. Gastroenterology. 2007;133(1):207–218. doi: 10.1053/j.gastro.2007.03.114. [DOI] [PubMed] [Google Scholar]
- 9.Attia RR, et al. Selective targeting of leukemic cell growth in vivo and in vitro using a gene silencing approach to diminish S-adenosylmethionine synthesis. J Biol Chem. 2008;283(45):30788–30795. doi: 10.1074/jbc.M804159200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.González B, et al. Crystal structures of methionine adenosyltransferase complexed with substrates and products reveal the methionine-ATP recognition and give insights into the catalytic mechanism. J Mol Biol. 2003;331(2):407–416. doi: 10.1016/s0022-2836(03)00728-9. [DOI] [PubMed] [Google Scholar]
- 11.Komoto J, Yamada T, Takata Y, Markham GD, Takusagawa F. Crystal structure of the S-adenosylmethionine synthetase ternary complex: A novel catalytic mechanism of S-adenosylmethionine synthesis from ATP and Met. Biochemistry. 2004;43(7):1821–1831. doi: 10.1021/bi035611t. [DOI] [PubMed] [Google Scholar]
- 12.Shafqat N, et al. Insight into S-adenosylmethionine biosynthesis from the crystal structures of the human methionine adenosyltransferase catalytic and regulatory subunits. Biochem J. 2013;452(1):27–36. doi: 10.1042/BJ20121580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parry RJ, et al. Studies of enzyme stereochemistry: Elucidation of the stereochemistry of S-adenosylmethionine formation by yeast methionine adenosyltransferase. J Am Chem Soc. 1982;104(3):871–872. [Google Scholar]
- 14.Markham GD, Parkin DW, Mentch F, Schramm VL. A kinetic isotope effect study and transition state analysis of the S-adenosylmethionine synthetase reaction. J Biol Chem. 1987;262(12):5609–5615. [PubMed] [Google Scholar]
- 15.Wang F, et al. Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus. FEBS J. 2014;281(18):4224–4239. doi: 10.1111/febs.12784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao KN, et al. Acute hemorrhagic pancreatic necrosis in mice: Induction in male mice treated with estradiol. Am J Pathol. 1982;109(1):8–14. [PMC free article] [PubMed] [Google Scholar]
- 17.Katoh N, Shimbayashi K, Abe K, Sakurada K. Decreased estradiol receptor concentrations in ethionine-induced fatty liver of rats. Toxicol Lett. 1991;58(3):279–285. doi: 10.1016/0378-4274(91)90039-9. [DOI] [PubMed] [Google Scholar]
- 18.Smith RC, Salmon WD. Formation of S-adenosylethionine by ethionine-treated rats. Arch Biochem Biophys. 1965;111(1):191–196. doi: 10.1016/0003-9861(65)90339-5. [DOI] [PubMed] [Google Scholar]
- 19.Skordi E, et al. Analysis of time-related metabolic fluctuations induced by ethionine in the rat research articles. J Proteomic Res. 2007;6:4572–4581. doi: 10.1021/pr070268q. [DOI] [PubMed] [Google Scholar]
- 20.Orenstein JM, Marsh WH. Incorporation in vivo of methionine and ethionine into and the methylation and ethylation of rat liver nuclear proteins. Biochem J. 1968;109(4):697–699. doi: 10.1042/bj1090697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Z, Hu Y, Markham GD, Takusagawa F. Flexible loop in the structure of S-adenosylmethionine synthetase crystallized in the tetragonal modification. J Biomol Struct Dyn. 1996;13(5):727–739. doi: 10.1080/07391102.1996.10508887. [DOI] [PubMed] [Google Scholar]
- 22.Takusagawa F, Kamitori S, Markham GD. Structure and function of S-adenosylmethionine synthetase: Crystal structures of S-adenosylmethionine synthetase with ADP, BrADP, and PPi at 28-Angstrom resolution. Biochemistry. 1996;35(8):2586–2596. doi: 10.1021/bi952604z. [DOI] [PubMed] [Google Scholar]
- 23.González B, et al. The crystal structure of tetrameric methionine adenosyltransferase from rat liver reveals the methionine-binding site. J Mol Biol. 2000;300(2):363–375. doi: 10.1006/jmbi.2000.3858. [DOI] [PubMed] [Google Scholar]
- 24.Cleland WW, Hengge AC. Enzymatic mechanisms of phosphate and sulfate transfer. Chem Rev. 2006;106(8):3252–3278. doi: 10.1021/cr050287o. [DOI] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Part A: Methods in Enzymology, eds Carter CW Jr, Sweet RM (Academic Press, New York), Vol 276, pp 307–326.
- 26.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schrödinger LLC (2010) The {PyMOL} Molecular Graphics System, Version 1.3r1.