Significance
How endocytosis regulates intracellular signaling is a major unsolved question. In this study, we labeled Ras, which plays a central role in normal and oncogenic signaling, with the fluorescent protein by gene editing, and for the first time (to our knowledge) examined the localization of endogenous Ras in living cells stimulated with epidermal growth factor (EGF). Microscopy imaging of living cells demonstrated that, although activated EGF receptors are rapidly internalized into endosomes, Ras is not present in these endosomes and mainly located in the plasma membrane. Therefore, EGF receptors signal to MAP kinases through Ras exclusively from the plasma membrane.
Keywords: EGF receptor, Ras, endocytosis
Abstract
Signaling from epidermal growth factor receptor (EGFR) to extracellular-stimuli–regulated protein kinase 1/2 (ERK1/2) is proposed to be transduced not only from the cell surface but also from endosomes, although the role of endocytosis in this signaling pathway is controversial. Ras is the only membrane-anchored component in the EGFR–ERK signaling axis, and therefore, its location determines intracellular sites of downstream signaling. Hence, we labeled endogenous H-Ras (HRas) with mVenus fluorescent protein using gene editing in HeLa cells. mVenus-HRas was primarily located at the plasma membrane, and in small amounts in tubular recycling endosomes and associated vesicles. EGF stimulation resulted in fast but transient activation of mVenus-HRas. Although EGF:EGFR complexes were rapidly accumulated in endosomes together with the Grb2 adaptor, very little, if any, mVenus-HRas was detected in these endosomes. Interestingly, the activities of MEK1/2 and ERK1/2 remained high beyond the point of the physical separation of HRas from EGF:EGFR complexes and down-regulation of Ras activity. Paradoxically, this sustained MEK1/2 and ERK1/2 activation was dependent on the active EGFR kinase. Cell surface biotinylation and selective inactivation of surface EGFRs suggested that a small fraction of active EGFRs remaining in the plasma membrane is responsible for continuous signaling to MEK1/2 and ERK1/2. We propose that, under physiological conditions of cell stimulation, EGFR endocytosis serves to spatially separate EGFR–Grb2 complexes and Ras, thus terminating Ras-mediated signaling. However, sustained minimal activation of Ras by a small pool of active EGFRs in the plasma membrane is sufficient for extending MEK1/2 and ERK1/2 activities.
Ras GTPases function as the molecular switch during signal transduction from various extracellular stimuli to intracellular signaling networks controlling cell proliferation, differentiation, motility, and apoptosis (reviewed in refs. 1 and 2). A multitude of receptors transmit signals through Ras to activate mitogen-activated protein kinases (MAPKs) and phosphotidylinositol 3-kinase. Ras-mediated activation of extracellular-stimuli–regulated protein kinase 1/2 (ERK1/2) by epidermal growth factor receptor (EGFR) is an extensively studied signaling pathway that is centrally involved in regulation of normal cell growth and EGFR-dependent tumorigenesis (reviewed in ref. 3). Ligand binding to EGFR triggers activation of its kinase and phosphorylation of tyrosine residues in the receptor carboxyl terminus that serve as docking sites for the Src homology 2 (SH2) domain of the Grb2 adaptor (4). SH3 domains of Grb2 are constitutively associated with Son-of-Sevenless 1 and 2 (SOS1/2), the primary guanine nucleotide exchange factors of Ras. Binding of the Grb2–SOS complex to EGFR positions SOS in the proximity to membrane-anchored Ras, thus allowing Ras–SOS interaction and promoting the replacement of GDP by GTP. GTP-loaded Ras recruits to the membrane and activates Raf serine-threonine kinases, which leads to consequential phosphorylation and activation of MEK1/2 and ERK1/2 (reviewed in ref. 5).
EGFR signaling is initiated at the cell surface, but ligand-induced endocytosis leads to rapid redistribution of receptors to endosomes and their subsequent sorting for degradation in lysosomes, which results in signal attenuation. Numerous studies demonstrated that EGFR remains ligand-bound and capable of signaling in early endosomes until receptors are sequestered in multivesicular endosomes (reviewed in ref. 6). The hypothesis of signaling to ERK1/2 from endosomes is under debate in the literature. Although the localization of receptor-proximal complexes containing Grb2, Shc, and SOS in endosomes is unequivocally demonstrated in various experimental models (reviewed in ref. 6), functional tests using inhibitors of endocytosis yielded contrasting conclusions about the requirement of endocytosis for EGFR-dependent activation of ERK1/2, ranging from the negative effects of endocytosis on ERK1/2 activation to an absolute requirement of endocytosis for normal duration and amplitude of ERK1/2 activity (7–11). Because methods to specifically inhibit EGFR endocytosis without affecting other receptor properties are not available, understanding the localization dynamics of the pathway constituents is critical for resolving these discrepancies.
Defining localization of Ras is the key to understanding the spatial organization of the EGFR–ERK pathway because Ras is the most downstream pathway component that is membrane-anchored but not bound to the receptor. Human cells express three main Ras isoforms, K-Ras (KRas), H-Ras (HRas), and N-Ras (NRas), which are highly homologous. Their membrane-targeting signals are located in the last 23–24 carboxyl-terminal amino acids, known as the hypervariable domain (reviewed in refs. 12 and 13). All Ras isoforms contain the “CAAX box” that is farnesylated (14). NRas and HRas are also palmitoylated in the Golgi complex, which provides an additional membrane-anchoring signal (15, 16). KRas is not palmitoylated but uses a hexalysine (polybasic) motif to interact with the negatively charged phospholipids at the plasma membrane (16). Our previous studies detected HRas and KRas in EGFR-containing endosomes in PAE and A431 cells, although HRas displayed substantially higher extent of endosomal localization compared with KRas (17). Other studies also demonstrated the localization and activation of HRas and other Ras isoforms on intracellular membranes (13, 14, 18–21). Most of the subcellular localization studies were, however, performed in cells transiently or constitutively overexpressing Ras (for example, see refs. 22 and 23). Localization of endogenous Ras isoforms has been rarely studied, owing to the lack of antibodies that efficiently detect Ras by immunofluorescence microscopy.
In the present study, we analyzed the localization of endogenous HRas in living HeLa cells in which HRas was labeled with the fluorescent protein mVenus (mV-HRas) by gene editing. The rationale for focusing on HRas was that (i) HRas exhibits prominent colocalization with EGFR in endosomes when overexpressed (17); and (ii) H-Ras is frequently mutated in head-and-neck squamous cell carcinoma, tumors that are typically EGFR driven (24). Live-cell imaging demonstrated the predominant localization of mV-HRas in the plasma membrane and detected virtually no mV-HRas in EGFR-containing endosomes, suggesting that EGFR endocytosis serves to separate Ras from EGFR-proximal activating complexes and therefore down-regulate Ras activity.
Results and Discussion
Kinetics of EGF–Ras–MAPK Pathway Activation in Gene-Edited Cells Expressing Endogenous Labeled HRas.
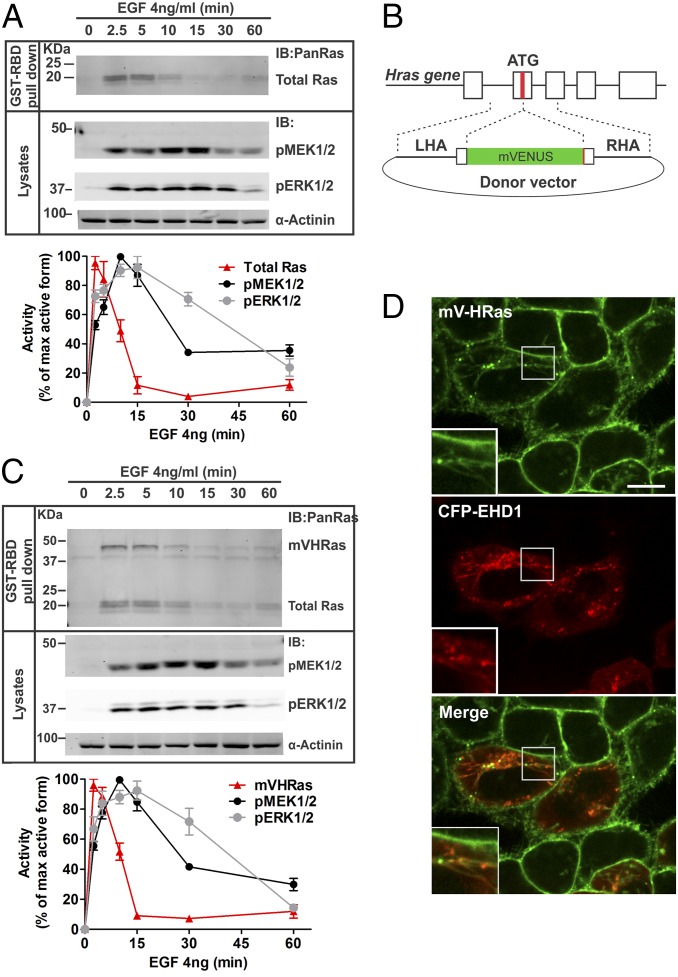
To study the spatiotemporal regulation of the EGFR–ERK1/2 signaling pathway, we first examined the time course of activation of the main components of this pathway in serum-starved HeLa cells stimulated with EGF. This variant of HeLa cells was used in our previous studies of localization of Grb2 and MEK2 (25, 26). These cells express ∼35,000 EGFRs per cell, resembling EGFR expression levels in various types of normal epithelial cells. Measurements of the amount of GTP-loaded Ras revealed that Ras·GTP peaked in the first 2.5–5 min after EGF stimulation followed by fast inactivation in the following 5–10 min down to 10–15% of the peak activity (Fig. 1). In contrast, phosphorylation of the catalytic residues of MEK1/2 and ERK1/2 reached maximum at 5–10 min and slowly decayed during the following 45 min. c-Raf was rapidly phosphorylated at Ser338 upon EGF stimulation, but the kinetics of c-Raf activity was difficult to monitor due to a complex pattern of multiple phosphorylation/dephosphorylation events controlling this activity. The kinetics of Ras, MEK, and ERK activities were similar when the cells were treated with EGF in the physiological range of concentrations (4 ng/mL; Fig. 1A) and supraphysiological concentration (20 ng/mL; Fig. S1A), suggesting that in our experimental system, concentrations of EGF up to 20 ng/mL can be used to study the regulation of the Ras–ERK activation pathway. Transient Ras activation followed by the sustained ERK activation has been previously observed (27). Prolonged Ras activity was reported in cells substantially overexpressing Ras (28).
Fig. 1.
Similar kinetics of EGF-stimulated Ras and MEK/ERK activities in parental HeLa and genome-edited HeLa cells expressing mVenus-HRas. (A) HeLa cells were incubated with 4 ng/mL at 37 °C for indicated times. GTP-loaded Ras was pulled down from cell lysates with GST-RBD. GST pulldowns and aliquots of the lysates were electrophoresed, and probed for Ras (pan-Ras antibody) in pulldowns, and phosphorylated MEK1/2 (pMEK) and ERK1/2 (pERK), and α-actinin (loading control) in lysates. Western blots of a representative experiment are shown. Graphs represent mean band intensities normalized to the amount of α-actinin (±SEM; n = 4) expressed as percentage of the maximum high-signal intensity for each protein during the time course. (B) mVenus sequence was inserted in-frame at the 5′-end of the coding sequence of the HRAS gene using a TALEN pair and a donor vector containing mVenus inserted between left and right homology arms (LHA and RHA) from the genomic HRAS sequence. (C) HeLa/mV-HRas cells were stimulated with 4 ng/mL, and the activities of mV-HRas, MEK1/2, and ERK1/2 were measured as described in A. Western blots of a representative experiment are shown. Graphs represent mean band intensities normalized to the amount of α-actinin (±SEM; n = 4) expressed as percentage of the maximum high-signal intensity for each protein during the time course. (D) HeLa/mV-HRas cells transiently expressing CFP-EHD1 were imaged through 445-nm (CFP; red) and 515-nm channels (mVenus; green). Insets represent high magnification of the region marked by the white rectangle. (Scale bar: 10 μm.)
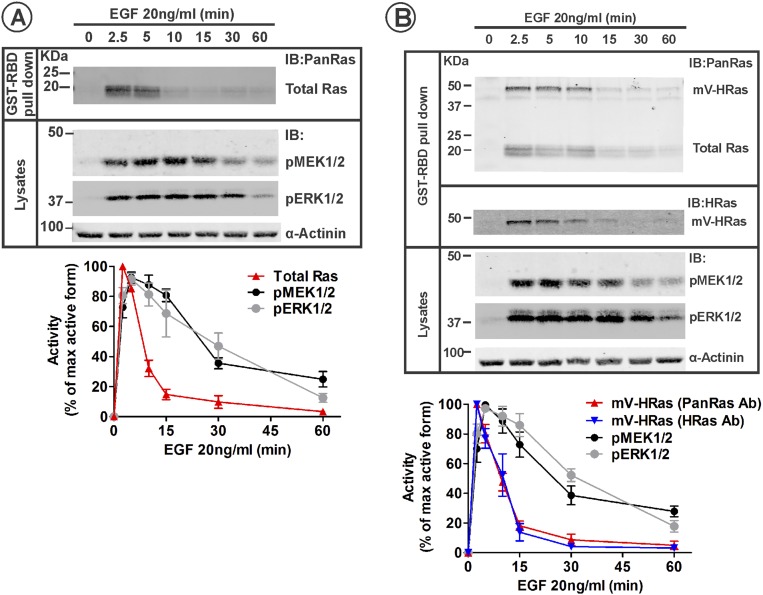
Fig. S1.
Kinetics of Ras and MEK/ERK activity in parental HeLa and HeLa/mVenus-HRas HeLa cells stimulated with 20 ng/mL EGF. (A) Serum-starved HeLa cells were incubated with 20 ng/mL EGF at 37 °C for indicated times. GTP-loaded Ras was pulled down from cell lysates with GST-RBD. GST pulldown and aliquots of the lysates were electrophoresed, and probed by Western blotting for Ras (pan-Ras antibody) in pulldowns, and phosphorylated MEK1/2 (pMEK) and ERK1/2 (pERK), and α-actinin (loading control) in lysates. Western blot images of a representative experiment are shown. Graphs represent mean band intensities normalized to the amount of α-actinin (±SEM; n = 4) expressed as percentage of the maximum high-signal intensity for each signaling protein during the time course. (B) Serum-starved HeLa/mV-HRas cells were stimulated with 20 ng/mL, and the activities of mV-HRas, MEK1/2, and ERK1/2 were measured as described in A. GST pulldown was also probed with the HRas antibody. Western blot images of a representative experiment are shown. Graphs represent mean band intensities normalized to the amount of α-actinin (±SEM; n = 4) expressed as percentage of the maximum high-signal intensity for each protein during the time course.
Different activation kinetics of Ras and its downstream effectors raises a question of how is the duration of EGFR-dependent activity of ERK controlled. In the same variant of HeLa cells, at least 90% of EGFR complexes with Grb2 were shown to be located in endosomes after 10–15 min of EGF stimulation, suggesting that EGFR is capable of signaling from endosomes (25). To examine whether these endosomal EGFRs can signal through Ras to sustain ERK activity, we generated a gene-edited HeLa cell line by inserting mVenus fluorescent protein into the endogenous locus of HRAS gene using transcription activator-like effector nuclease (TALEN)-based method (further referred to as HeLa/mV-HRas cells) (Fig. 1B and Fig. S2A). Interestingly, despite a single-allele editing (Fig. S2B), mV-HRas was expressed at ∼2.2-fold higher level than HRas in parental cells (clone 3, Fig. S2C). The mechanism underlying an increased mV-HRas expression is unknown but most probably involves an increased rate of biosynthesis. The kinetics of GTP loading of mV-HRas was essentially similar to that of total (pan) Ras in parental HeLa cells stimulated with 4 or 20 ng/mL EGF (Fig. 1C and Fig. S1B). Likewise, kinetics of MEK1/2 and ERK1/2 phosphorylation in EGF-stimulated HeLa/mV-HRas and parental HeLa cells were similar (Fig. 1 and Fig. S1). These data suggest that a moderately increased expression of HRas does not affect the signal transduction process. Given that this HRas level is within the range of HRas expression levels in cultured mammalian cells (29), HeLa/mV-HRas cells represent an appropriate model to analyze the spatiotemporal regulation of EGFR–Ras–ERK signaling pathway.
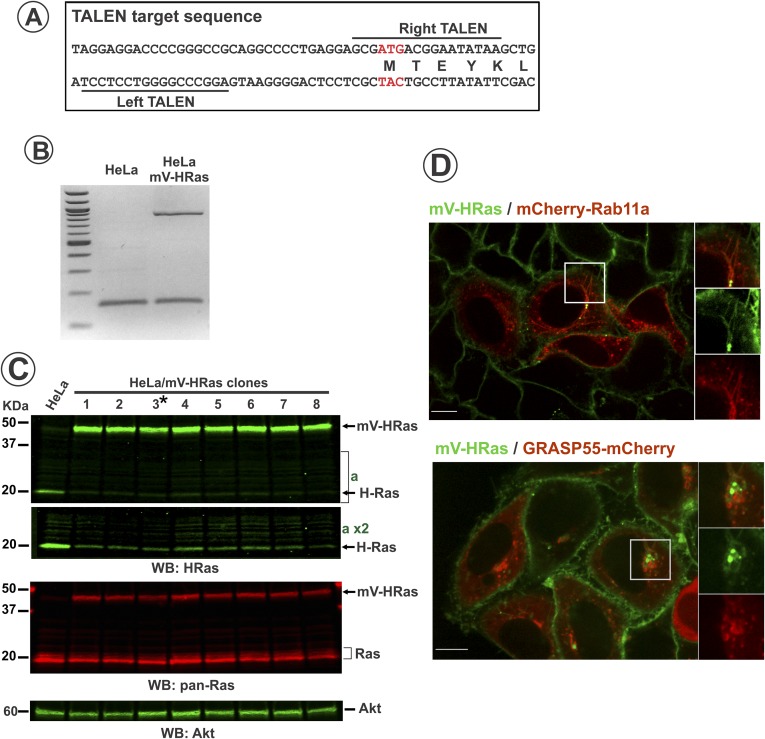
Fig. S2.
Characterization of mVenus-HRas in genome-edited HeLa cells. (A) TALEN target sequences in the HRAS gene. (B) PCR of 5′-UTR of HRAS gene from parental and edited HeLa cells. Note double band in HeLa/mVHRas sample confirms single allele edition. High–molecular-weight band corresponds to edited HRas allele. (C) Parental and several single-cell clones of HeLa/mV-HRas cells were lysed, and the lysates were probed by Western blotting with HRas, panRas, and Akt (loading control) antibodies. Asterisk shows the clone 3 used in experiments, although mVRas localization and dynamics were identical in all clones tested. Note increased (2.2-fold in clone 3 when normalized to loading control) expression of mV-Ras compared with HRas in parental cells, and reduced expression of untagged HRas in HeLa/mV-HRas cells (see part of blot “a” in high contrast), which is probably due to posttranslational control of the total expression level of HRas (mV-HRas plus untagged HRas). (D) HeLa/mVHRas cells were transfected with mCherry-Rab11 or GRASP55-mCherry. Images were acquired from living cells at 37 °C. Insets represent high-magnification images of each individual protein and a merge image of the region indicated by white rectangles. (Scale bars: 10 μm.)
Localization of mV-HRas in Cells Stimulated with EGF.
Live-cell imaging of mV-HRas by spinning disk confocal microscopy revealed highly consistent pattern of HRas distribution within the cell population. mV-HRas was mainly located in the plasma membrane (Fig. 1D). Many cells also contained mV-HRas in tubular structures that frequently span from and through the Golgi area and extend to and from the plasma membrane. These tubules were highly sensitive to chemical fixations, which precluded analysis in fixed cells. In living cells, mV-HRas tubules were positive for markers of recycling endosomes, such as EHD1 (Fig. 1D) and Rab11 (Fig. S2D). In some cells, one or two bright vesicular-shape structures of mV-HRas were seen associated with and moving along tubular recycling compartments. Some vesicular structures containing mV-HRas were colocalized with or surrounded by compartments positive for GRASP55-mCherry, a Golgi apparatus protein (Fig. S2D).
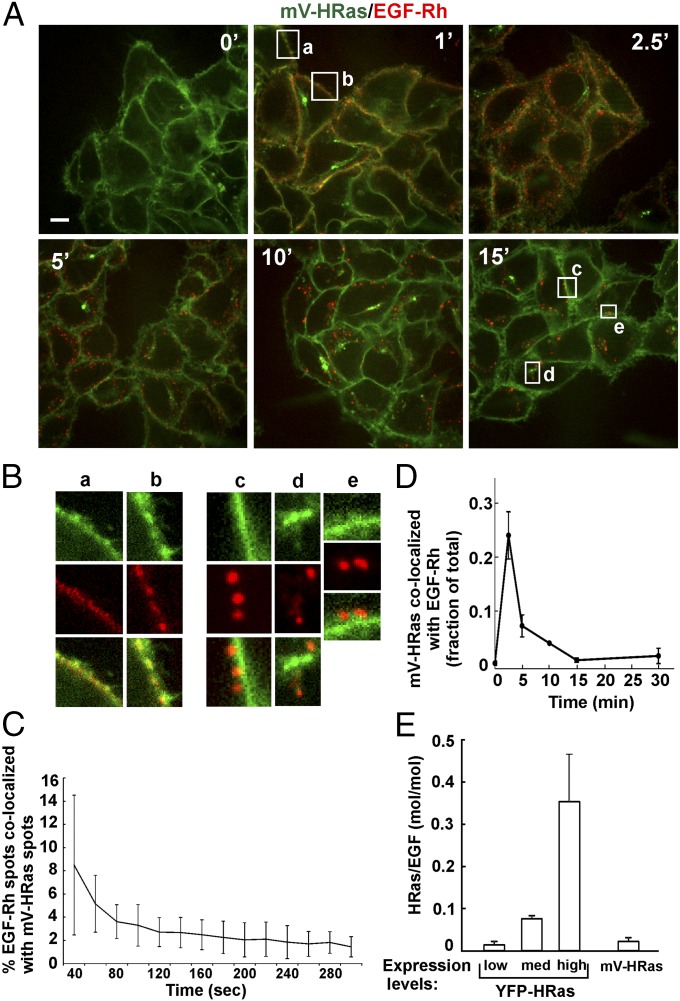
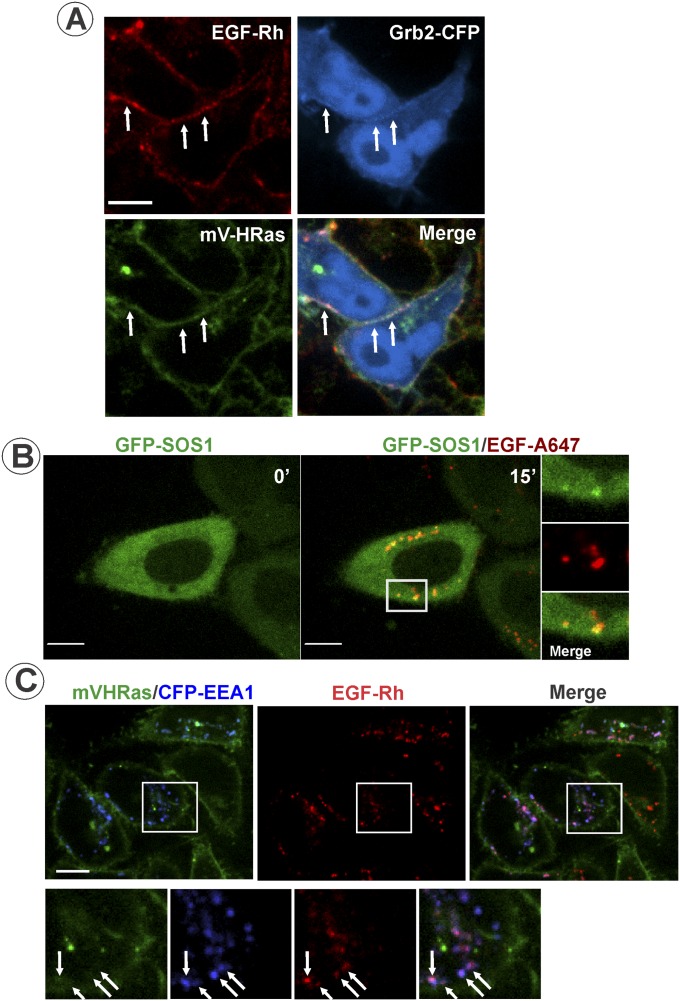
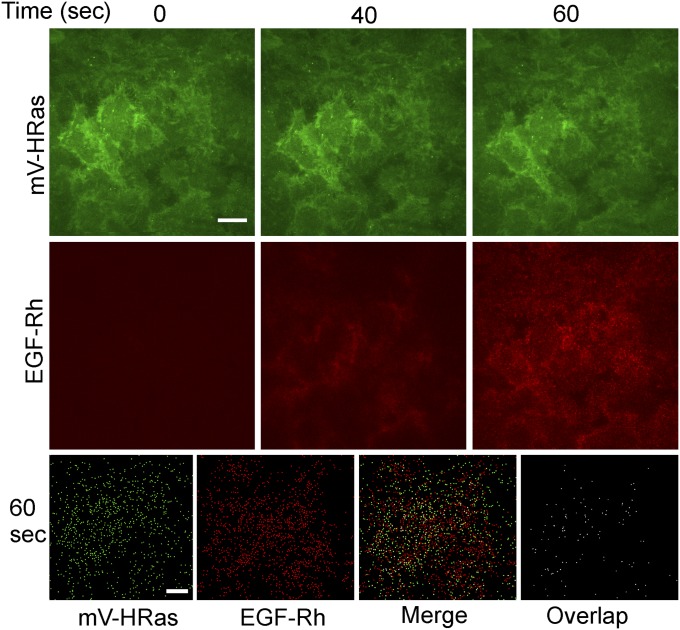
Treatment of cells with EGF or EGF conjugated to rhodamine (EGF-Rh) did not lead to detectable changes in mV-HRas localization. EGF-Rh fluorescence overlapped with mV-HRas in the plasma membrane during the first few minutes of cell stimulation (Fig. 2 A and B). Transiently expressed Grb2-CFP could also be detected together with EGF-Rh and mV-HRas in the plasma membrane at early time points (Fig. S3A). To compare dynamics of EGF-Rh and mV-HRas in the plasma membrane, the cell bottom membrane of HeLa/mV-HRas cells incubated with EGF-Rh was imaged using total internal reflection fluorescence microscopy (TIR-FM). Following rapid binding, EGF-Rh concentrated in numerous diffraction-limited spots (Fig. S4), with a pattern reminiscent of EGF:EGFR clustering in clathrin-coated pits (30). mV-HRas was diffusely distributed throughout the membrane with a few clusters representing a minor fraction of mV-HRas in the plasma membrane (Fig. S4). Only 2–3% of EGF-Rh spots overlapped with mV-HRas spots during the first 5 min of the time course (Fig. 2C and Fig. S4), suggesting that mV-HRas is not corecruited with EGF:EGFR complexes into clathrin pits and endocytic vesicles. These data are consistent with previous observations of different endocytic routes of HRas and EGFR: clathrin-independent, ARF6-dependent endocytosis of HRas (31), and clathrin-mediated endocytosis of EGFR in HeLa cells stimulated with low EGF concentrations (32).
Fig. 2.
Localization of mVenus-HRas in cells stimulated with EGF-Rh. (A) HeLa/mV-HRas cells were incubated with 4 ng/mL EGF-Rh at 37 °C for indicated times, and live-cell imaging was performed through 515-nm (mVenus; green) and 561-nm (rhodamine; red) channels. The image acquisition time was 1.2 s for mVenus throughout the time course, whereas for rhodamine it was 600 ms (0–5 min), 400 ms (10 min), and 200 ms (15 min) to avoid rhodamine signal saturation in endosomes at later time points. (Scale bar: 10 µm.) (B) Insets show high-magnification images of regions indicated by white rectangles in A to demonstrate an overlap of EGF-Rh and mV-HRas fluorescence in the plasma membrane (a and b) and no overlap of EGF-Rh and mV-HRas in endosomes (c–e). (C) The cell bottom surface of HeLa/mV-HRas cells was continuously imaged during stimulation with 4 ng/mL EGF-Rh at 37 °C using TIR-FM. EGF-Rh was added at a 10-s time point. Typically, specific cell-bound fluorescence of EGF-Rh was detected at ∼40-s time point. The percentage of EGF-Rh–positive spots colocalized with mV-HRas spots relative to the total number of EGF-Rh spots is plotted against time. Mean values (±SD; n = 4) are presented. Corresponding representative images are shown in Fig. S4. (D) HeLa/mV-HRas cells were incubated with 4 ng/mL EGF-Rh as in A, and z stacks of 18–21 confocal images were acquired through 515- and 561-nm channels using the same image acquisition parameters as in A. The percentage of mV-HRas colocalized with EGF-Rh relative to the total cellular mV-HRas fluorescence was calculated in multiple 3D images. Mean values (±SD) are presented. (E) Parental HeLa cells transiently expressing YFP-HRas or HeLa/mV-HRas cells were incubated with 4 ng/mL EGF-Rh for 15 min at 37 °C, and z stacks of confocal images were acquired through 515- and 561-nm channels. Representative images of YFP-HRas are shown in Fig. S5. Total amounts of YFP-HRas or mV-HRas per cell were calculated from 3D images using segmentation. Mean YFP-HRas expression levels were calculated as fold difference to the mean expression level of mV-HRas (× mV-HRas). Molar stoichiometry (515/561 ratio) of mV-HRas or YFP-HRas to EGF-Rh in endosomes was calculated in, respectively, HeLa/mV-HRas cells or parental cells expressing low (mean expression, 0.6× mV-HRas), intermediate (“med,” mean expression, 10.7× mV-HRas), and high (mean expression, 55.4× mV-HRas) levels of YFP-HRas. Mean 515/561 ratio values (±SD) obtained from 100 to 240 endosomes at each condition are presented.
Fig. S3.
Localization of Grb2-CFP, GFP-SOS, and mV-HRas in EGF-stimulated cells. (A) HeLa/mV-HRas cells transiently expressing Grb2-CFP were incubated with 4 ng/mL EGF-Rh for 1 min at 37 °C, and then imaged after 5-min incubation at room temperature to slow down endocytosis. Live-cell 3D images were acquired through 445-nm (CFP; blue), 515-nm (mVenus; green), and 561-nm (rhodamine; red) channels. Arrows indicate examples of the overlap of EGF-Rh, Grb2-CFP, and mV-HRas fluorescence in the plasma membrane (cell edge). (Scale bar: 10 μm.) (B) HeLa cells transiently expressing GFP-SOS1 were incubated with 10 ng/mL EGF-A647. Images were acquired from living cells at 37 °C. Insets represent high-magnification images of the region indicated by the white rectangle. (Scale bar: 10 μm.) (C) HeLa/mV-HRas cells transiently expressing CFP-EEA.1 were incubated with 4 ng/mL EGF-Rh for 15 min at 37 °C. Three-dimensional images were acquired through 445-nm (CFP; blue), 515-nm (mVenus; green), and 561-nm (rhodamine; red) channels from living cells at room temperature to reduce endosome mobility. Insets represent high-magnification images of the region indicated by the white rectangle. Arrows indicate examples of EGF-Rh and CFP-EEA.1 colocalization in endosomes. (Scale bar: 10 μm.)
Fig. S4.
TIR-FM imaging of HeLa/mV-HRas cells stimulated with EGF-Rh. Time-lapse imaging through 488-nm (mV-HRas) and 561-nm (EGF-Rh) channels was performed during stimulation of cells with 4 ng/mL EGF-Rh at 37 °C. Images of cells incubated with EGF-Rh for 0, 40, and 60 s are presented. Bottom row: binary masks corresponding to rhodamine (red) and mVenus (green) spots detected at a 60-s time point, their “Merge” mask, and the mask presenting only overlapping spots (“Overlap”). (Scale bars: 10 μm.)
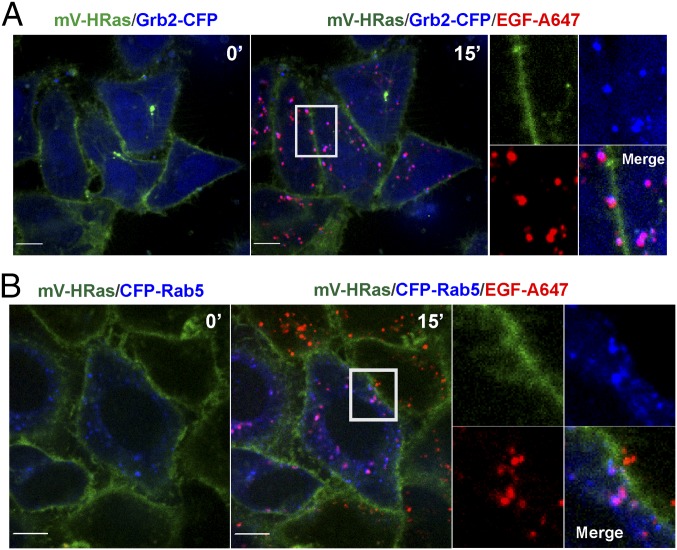
Continuous endocytosis (5–15 min) resulted in a clear separation of EGF-Rh:EGFR complexes, which were accumulated in vesicular endosomes, from mV-HRas, which was located in the plasma membrane and tubular compartments. Quantitative analysis of 3D images demonstrated that <1% of total cellular mV-HRas overlaps with EGF-Rh at the 15-min time point (Fig. 2D). Visual analysis of individual EGF-Rh–containing endosomes demonstrated no obvious mV-HRas localization in most of these endosomes, although faint mV-HRas fluorescence overlapping with some endosomes could be detected. Based on the ratio of the apparent quantum yields of YFP and rhodamine in our imaging system (25), calculations of the molar ratio of mVenus and rhodamine showed that ∼2 molecules of mV-HRas were colocalized with 100 molecules of EGF-Rh in endosomes at the 15-min time point (Fig. 2E). For comparison, the molar ratio of Grb2-YFP to EGF-Rh under the same experimental conditions was previously found to be 2:1 (25). Likewise, in gene-edited HeLa cells, transiently expressed Grb2-CFP was highly colocalized with EGF-Alexa647 (EGF-A647) but not with mV-HRas in endosomes (Fig. 3A). GFP-tagged SOS1 was also present in endosomes containing EGF-A647 in HeLa cells (Fig. S3B). Grb2-CFP and GFP-SOS1 could not be detected in the plasma membrane and tubular endosomes where mV-HRas was located after 10–15 min of EGF stimulation (Fig. 3A and Fig. S3B). Fluorescent EGF but not mV-HRas was highly colocalized with markers of early/intermediate endosomes (Rab5 and EEA.1) (Fig. 3B and Fig. S3C).
Fig. 3.
Localization of fluorescent EGF and Grb2 but not mV-HRas in early endosomes. (A) HeLa/mV-HRas cells transiently expressing Grb2-CFP were incubated with 4 ng/mL EGF-A647 for 15 min at 37 °C. Images were acquired from living cells at 37 °C. Merge images of Grb2-CFP (blue) and mV-HRas (green) in cells before stimulation, and after EGF-A647 (red) stimulation are shown. Insets represent high-magnification images of individual proteins and a merge image of the region indicated by white rectangle to show high extent of Grb2-CFP and EGF-A647 colocalization. (B) HeLa/mV-HRas cells transiently expressing CFP-Rab5 were incubated with 10 ng/mL EGF-A647. Images were acquired from living cells at 37 °C. Merge images of CFP-Rab5 (blue) and mV-HRas (green) in cells before stimulation and after EGF-A647 (red) stimulation are shown. Insets represent high-magnification images of each individual protein and a merge image of the region indicated by white rectangle to show high extent of CFP-Rab5 and EGF-A647 colocalization. (Scale bars: 10 μm.)
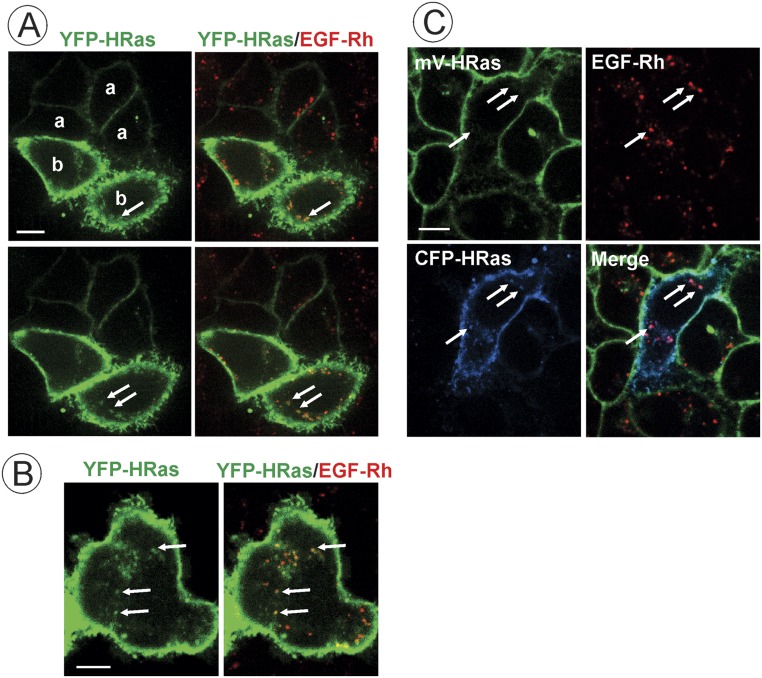
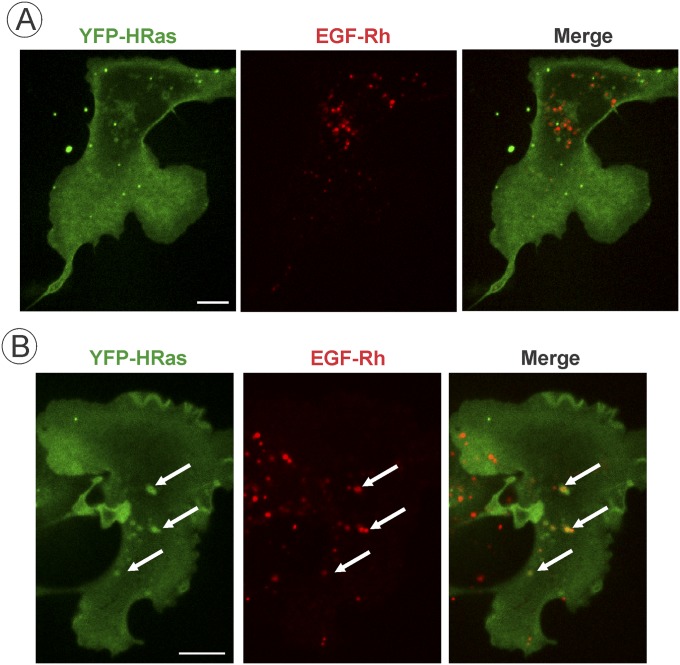
To test whether the amount of HRas present in EGFR-containing endosomes depends on the expression level of HRas, YFP-HRas or CFP-HRas were transiently expressed in parental HeLa or HeLa/mV-HRas cells, respectively. YFP-HRas and CFP-HRas were clearly detected in endosomes containing EGF-Rh when highly overexpressed (Fig. S5). Quantifications revealed that, in cells overexpressing YFP-HRas, the stoichiometry of HRas and EGF in endosomes is significantly higher (∼35:100) than in HeLa/mV-HRas cells or HeLa cells expressing low/moderate levels of YFP-HRas (Fig. 2E). Similarly, significant localization of YFP-HRas in EGF-Rh–containing endosomes could be observed in COS1 cells expressing high but not low levels of YFP-HRas (Fig. S6). Furthermore, the expression level of endogenous mV-HRas was not sufficient to detect GTP-loaded mV-Ras by imaging of various types of fluorescent-protein–tagged Ras⋅GTP sensors (22, 33). YFP-HRas expression that is >20-fold higher than that of mV-HRas was necessary for detection of EGF-induced HRas activation using the same sensors. Overall, our experiments in HeLa/mV-HRas cells demonstrate preferential targeting of the majority of endogenous HRas molecules to the plasma membrane. The relative proportion of endogenously labeled mV-HRas associated with intracellular membranes was lower than what was typically observed in cells overexpressing HRas. For example, the fluorescence intensity of tubular compartments containing overexpressed YFP-HRas was comparable to that in the plasma membrane (16, 31). Limited localization of mV-HRas in vesicular endosomal compartments in cell periphery and Golgi area was observed in contrast to what was reported in cells overexpressing HRas (17, 31, 34, 35) (Figs. S5 and S6). Certainly, differences in localization of gene-edited endogenous and exogenous HRas can be due to different cells types and levels of overexpression of the latter.
Fig. S5.
Analysis of colocalization of transiently expressed HRas with EGF-Rh. (A and B) Parental HeLa cells transiently expressing YFP-HRas were incubated with 4 ng/mL EGF-Rh for 15 min at 37 °C. Single x–y confocal sections from the image z stack acquired from cells expressing low (0.5–1.0× mV-HRas expression level; “a” in A), intermediate (5.3× mV-HRas expression; “b” in A), and high levels (38.2× mV-HRas expression; B) are presented. Different confocal sections are presented in A for better visualization of endosomes in cells expressing low and intermediate levels of YFP-HRas. Arrows indicate examples of colocalization of EGF-Rh and YFP-HRas in endosomes. (Scale bar: 10 μm.) (C) HeLa/mV-HRas cells transiently expressing intermediate level of CFP-HRas (4.3× mV-HRas expression level) were incubated with 4 ng/mL EGF-Rh for 15 min at 37 °C. Single x–y confocal section from z stack of images of mVenus, CFP, and rhodamine acquired at room temperature is presented. Arrows indicate examples of colocalization of EGF-Rh and CFP-HRas in endosomes. (Scale bar: 10 μm.)
Fig. S6.
Analysis of colocalization of HRas and EGF-Rh in COS1 cells. COS1 cells transiently expressing YFP-HRas were incubated with 4 ng/mL EGF-Rh for 15 min at 37 °C. Single x–y confocal sections from the image z stacks acquired from cells expressing low (1.6× mV-HRas expression; A) or high levels (38× mV-HRas; B) of YFP-HRas are presented. Arrows indicate examples of colocalization of EGF-Rh and YFP-HRas in endosomes. (Scale bars: 10 μm.)
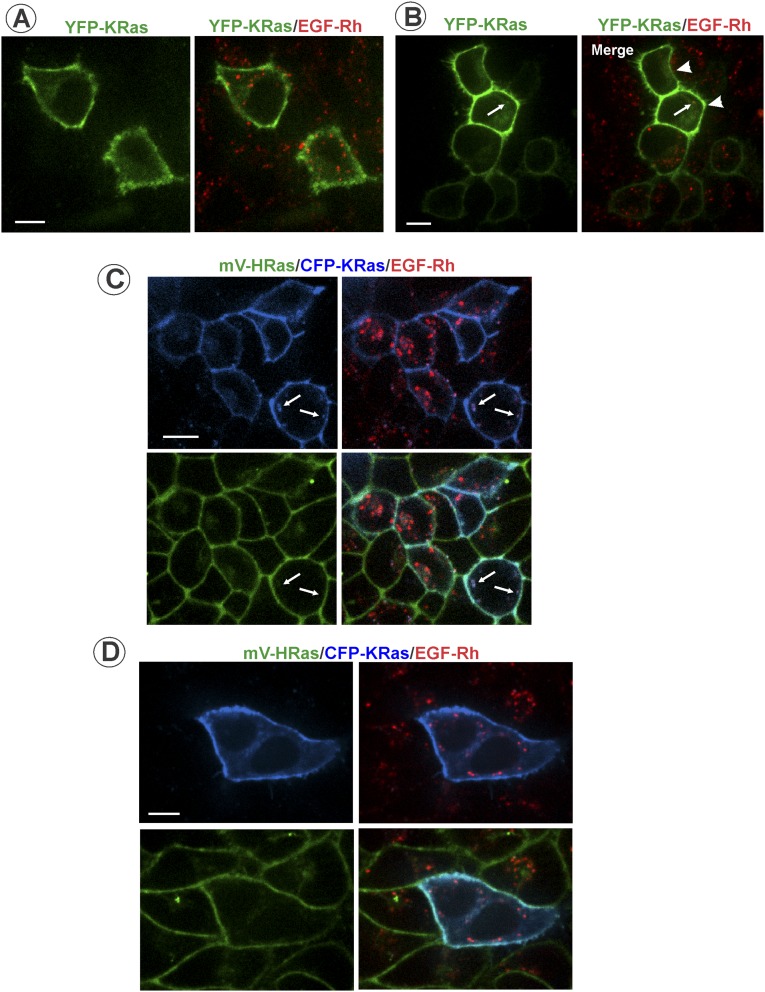
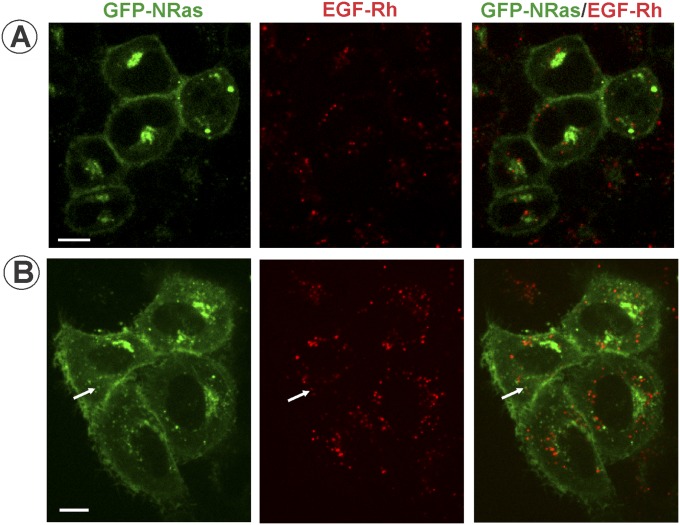
Altogether, the data in Figs. 2 and 3 and Figs. S2–S6 suggest that endocytosis of activated EGFR separates EGFR–Grb2–SOS complexes from HRas, leading to down-regulation of HRas activity. In fact, the time course of EGF-Rh/mV-HRas colocalization (Fig. 2D) was highly similar to the time course of Ras activity (Fig. 1C). The possibility that other Ras isoforms are recruited to EGFR-containing endosomes cannot be formally ruled out. To directly examine KRas (4B form) localization in our variant of HeLa cells, YFP-KRas and CFP-KRas were transiently expressed in parental HeLa and HeLa/mV-HRas cells, respectively. Regardless of the expression level, YFP-KRas and CFP-KRas were mostly located in the plasma membrane; very rarely, examples of colocalization of KRas and EGF-Rh in endosomes could be detected (Fig. S7). Similarly, transient expression of GFP-NRas revealed virtually complete separation of endosomal EGF-Rh from NRas that was located in the plasma membrane and concentrated in the pericentriolar area (Fig. S8). Given that time courses of EGF-induced activity of total Ras and mV-HRas were essentially similar (Fig. 1 and Fig. S2), indicative of a similar activation kinetics of all Ras species, the involvement of KRas and NRas in endosomal signaling by EGFR is unlikely.
Fig. S7.
Analysis of colocalization of transiently expressed KRas with EGF-Rh in HeLa cells. (A and B) Parental HeLa cells transiently expressing YFP-KRas were incubated with 4 ng/mL EGF-Rh for 15 min at 37 °C. Single x–y confocal sections from the image z stack acquired from cells expressing low (2× mV-HRas expression; Top; A) or high (19× mV-HRas; cells are indicated by arrowheads in B) levels are presented. Arrows indicate a rare example of colocalization of EGF-Rh and YFP-KRas in endosomes. (Scale bars: 10 μm.) (C and D) HeLa/mV-HRas cells transiently expressing CFP-KRas were incubated with 4 ng/mL EGF-Rh for 15 min at 37 °C. Single x–y confocal sections from the image z stack acquired from cells expressing moderate (4.5× mV-HRas expression; C) or high levels of CFP-KRas (15× mV-HRas expression; D) are presented. Arrows indicate rare examples of colocalization of EGF-Rh and CFP-KRas in endosomes. (Scale bar: 10 μm.)
Fig. S8.
Analysis of colocalization of transiently expressed GFP-NRas with EGF-Rh in HeLa cells. Parental HeLa cells transiently expressing GFP-NRas were incubated with 4 ng/mL EGF-Rh for 15 min at 37 °C. Single x–y confocal sections from the image z stack acquired through 488-nm (GFP; green) and 561-nm (rhodamine; red) channels from cells expressing low (∼1.0× mV-HRas expression; A) or high (∼10.1× mV-HRas; B) levels are presented. Arrows indicate a rare example of colocalization of EGF-Rh and GFP-NRas in endosomes. (Scale bars: 10 μm.)
Finally, it is also possible that small amounts of mV-HRas present in EGFR-containing endosomes are below the detection limit of our imaging system, although this system is capable of single-molecule imaging (36). Image acquisition times as long as 1–1.2 s were used to obtain a maximum signal-to-noise ratio in mV-HRas images, while avoiding the cross-bleed of the rhodamine fluorescence. Given that copy numbers of HRas and other Ras isoforms per cell are relatively low (37, 38), and that a very weak mV-Ras fluorescence in tubular compartments was detected (for example, see Fig. 1D), the amount of undetectable Ras in EGFR-containing endosomes is expected to be extremely small.
Sustained MEK and ERK Activities Require EGFR Kinase Activity.
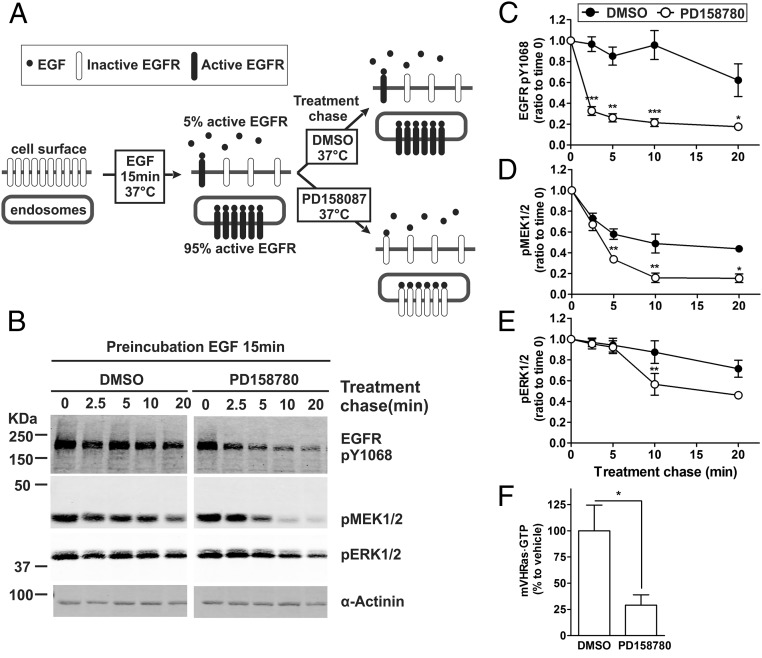
Rapid decay of Ras activity (Fig. 1) and lack of mV-HRas in EGFR–Grb2–SOS-containing endosomes (Fig. 3A) suggested that the sustained phosphorylation of MEK1/2 and ERK1/2 in the absence of upstream activating signals could be due to slow dephosphorylation of these kinases by phosphatases (39, 40). To test the hypothesis that the sustained activity of MEK1/2 and ERK1/2 is EGFR independent, EGFR kinase activity was blocked by PD158087 in cells incubated with EGF for 15 min (Fig. 4A). As expected, EGFR phosphorylation at Tyr1068 (major Grb2 binding site) was reduced by 70% following exposure to PD158087 (Fig. 4 B and C). Incomplete dephosphorylation of the receptor is likely due to a limited accessibility of pTyr1068 to phosphatases, for example, due to sequestration of a pool of active EGFRs in multivesicular endosomes. Surprisingly, inhibition of EGFR kinase also resulted in dephosphorylation of MEK1/2 and ERK1/2, demonstrating that sustained phosphorylation of these kinases does require the EGFR kinase activity. MEK1/2 dephosphorylation was significantly faster than that of ERK1/2 (Fig. 4 B–E). This delay is likely due to time- and signal-dependent expression of ERK1/2 phosphatases (40). Although mV-HRas activity in cells incubated with EGF for 15 min is only 15–20% of the peak maximum, PD158087 treatment further reduced this activity (Fig. 4F).
Fig. 4.
EGFR kinase activity is required for sustained MEK and ERK activation. (A) Schematics of the experimental protocol used in B–F. (B) HeLa/mV-HRas cells were incubated with 4 ng/mL EGF at 37 °C for 15 min. DMSO (vehicle) or PD158780 (50 nM) was then added to cells, and the cells were further incubated at 37 °C for indicated periods of time in the same medium (treatment chase). Cell lysates were probed by Western blotting with antibodies to EGFR pY1068, pMEK1/2, pERK1/2, and α-actinin (loading control). (C–E) Graphs show mean values of band intensities normalized to the amount of α-actinin (±SEM; n = 8) exemplified in B and presented as percentage of the mean value at time “0” (after the 15-min incubation with EGF but before the chase treatment). (F) HeLa/mV-HRas cells were incubated with EGF for 15 min, and then with DMSO or PD158780 for additional 5 min as described in B. The amount of mV-HRas⋅GTP was measured using the GST-RBD pull-down assay as in Fig. 1. Bar graphs show mean band intensities (±SEM; n = 5). Paired t tests were performed. *P < 0.05, **P < 0.01, and ***P < 0.001.
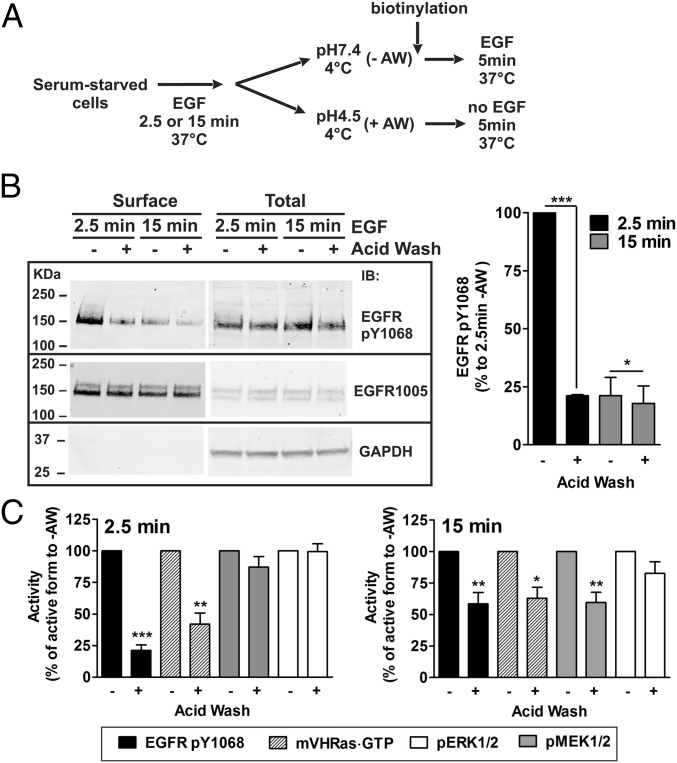
How do activated EGFRs, which are highly concentrated in endosomes, transduce signals to MEK and ERK through Ras, which is not present in these endosomes? One possibility is that Ras activity in the plasma membrane is maintained through ERK-mediated stabilization of the SOS–Ras interaction (28). However, such positive feedback would not be EGFR dependent because levels of pERK1/2 remain high and not sensitive to PD158087 for at least 5 min in our “treatment chase” experiments (Fig. 4B). Another possibility is that a small pool of active EGFRs remaining at the cell surface after 15 min of continuous endocytosis is sufficient for sustaining signaling through the Ras–ERK pathway. Technically, small pools of fluorescently tagged proteins are more difficult to detect in the plasma membrane than in endosomes because of the diffuse distribution of the fluorescence in the plasma membrane in contrast to highly concentrated fluorescence of proteins in endosomes. Biochemical measurements in the same variant of HeLa cells have previously demonstrated that about 5–10% of 125I-EGF remains at the surface after 15 min incubation of cells with 4 ng/mL 125I-EGF (25). To test whether this small pool of EGFRs remaining in the plasma membrane are active, surface biotinylation method was used. EGFR phosphorylated at Tyr1068 was detected in the biotinylated fraction recovered from cells treated with 4 ng/mL EGF for 15 min at 37 °C (Fig. 5). This surface-active EGFR pool was ∼20% of that in cells incubated with EGF for 2.5 min, conditions of the spatial overlap of EGF-Rh and mV-HRas (Fig. 2 A and B). Treatment of cells preincubated with EGF for 2.5 min with the mild acidic buffer (pH 4.5) at 4 °C, which releases surface-bound EGF without affecting cell viability (41), reduced the amount of biotinylated pTyr1068 EGFRs by 80%, confirming efficient EGF dissociation (Fig. 5 B and C). The same pH 4.5 treatment of cells incubated with EGF for 15 min inactivated only ∼20% of surface EGFRs. It is possible that receptors remaining on the cell surface after 15-min EGF stimulation form high-affinity complexes with EGF, which are resistant to low pH and require a 37 °C incubation for dephosphorylation, or are inaccessible to phosphatases due to association with clathrin lattices (42). Nevertheless, these data indicate that a small pool of active EGFRs is present in the plasma membrane after 15 min of continuous endocytosis.
Fig. 5.
A small pool of surface EGFR contributes to a sustained signaling through the Ras–ERK pathway. (A) Schematics of the experimental protocol used in B and C. (B) HeLa/mV-HRas cells were incubated with 4 ng/mL EGF for 2.5 or 15 min at 37 °C, and then incubated in either DMEM (pH 7.4) (−AW) or an acidic buffer (pH 4.5) (+AW) for 1 min at 4 °C. The cells were then biotinylated and lysed. Biotinylated proteins were pulled down by Avidin-agarose. Pulldowns and aliquots of cell lysates were electrophoresed and probed with antibodies to EGFR pY1068 and total EGFR (1005). Bar graphs on the Right represent mean band intensities (±SEM; n = 3) presented as percentage of the band intensities of pulldowns recovered from cells that were incubated with EGF for 2.5 min and not treated with the acidic buffer (lane 1). (C) Serum-starved HeLa/mV-HRas cells were incubated with EGF at 37 °C, and treated with the acidic buffer (+AW) or incubated DMEM (−AW) as described in B. Cells that were not treated with acid wash, were further incubated with 4 ng/mL EGF for 5 min at 37 °C, whereas acid wash-treated cells were incubated in DMEM containing 0.1% BSA for 5 min at 37 °C. The amounts of EGFR pY1068, pMEK1/2, and pERK1/2 in lysates were measured as described in Fig. 4B, and the amount of mV-HRas⋅GTP was measured as in Fig. 4D. Mean band intensities (±SEM; n = 6) are plotted as percentage of that intensity of the bands corresponding to cells that were not treated with the acidic buffer. Paired t test was performed. *P < 0.05, **P < 0.01, and ***P < 0.001.
The acidic wash treatment of the cells incubated with EGF for 15 min and subsequent incubation for 5 min at 37 °C reduced Ras⋅GTP level and MEK1/2 phosphorylation by 40–45% compared with cells that were continuously exposed to EGF (Fig. 5C). The extent of inactivation of Ras and MEK1/2 paralleled that of EGFR dephosphorylation (Fig. 5C). Dephosphorylation of ERK1/2 in these experiments was insignificant (Fig. 5C) because longer than 5-min “treatment chase” time was needed to dephosphorylate ERK1/2 (Fig. 4). Similar experiments in cells incubated with EGF for 2.5 min demonstrated strong dependence of Ras activity on surface EGFR signaling at this time point (Fig. 5), whereas deactivation of MEK1/2 and ERK1/2 was not observed during an onset of their activation at early times (2.5–7.5 min) after EGF stimulation. Altogether, the data in Figs. 4 and 5 support the hypothesis that a small pool of surface EGFR remaining after 15 min of continuous endocytosis may maintain low levels of plasma membrane Ras⋅GTP to sustain MEK and ERK activity.
Many studies arguing for or against the idea of the endosomal signaling to ERK were based on the detection of signaling proteins in endosomes or the use of general endocytosis inhibitors. Although it is unequivocally proven that a substantial fraction of EGFR–Grb2–SOS complexes remain intact in endosomes, our studies suggest that, at least in cells with low EGFR levels, the main function of endocytosis is to separate proximal receptor complexes from membrane-anchored Ras. Another important implication from our experiments is that signaling by a small number of active EGFRs (1,000–2,000 per cell) leading to a relatively low level of Ras activity is sufficiently amplified to maintain MEK and ERK activities for a long time. Our findings are complementary to previous observations that inhibition of endocytosis does not affect ERK1/2 activation (8–10, 43). On the other hand, the inhibitory effects of dynamin mutants on EGFR-dependent ERK activation observed in HeLa and other cells (for example, in refs. 7 and 29) could be due to the effects on the localization of the downstream components of the ERK pathway rather than on EGFR endocytosis per se. An additional scenario that may lead to EGFR–Ras endosomal signaling is possible in cells expressing high levels of EGFR and treated with high EGF concentrations, conditions causing clathrin-independent endocytosis of EGFR and massive activation of pinocytotic processes, which may lead to “mixing” of different endosomal compartments and thus substantial colocalization of active EGFR and Ras in endosomes. Future studies of the localization of endogenous Ras in an expanded panel of cells with different levels of EGFR expression will provide full understanding of the intracellular sites of Ras signaling.
Materials and Methods
The sources of EGF, antibodies, inhibitors, chemicals, and DNA plasmids are listed in Supporting Information. Parental HeLa, HeLa/mV-HRas, and COS1 cells were maintained in DMEM and 10% (vol/vol) fetal bovine serum. Before experiments, cells were serum-starved for 16 h in DMEM. Transient transfections, TALEN design and generation of HeLa/mV-HRas gene-edited cells, biochemical measurement of Ras activity, cell surface biotinylation, and Western blotting detection of phosphorylated EGFR, MEK, and ERK are described in Supporting Information. Live-cell fluorescence microscopy using spinning disk confocal microscopy, TIR-FM imaging, and methods of image analysis were performed as previously described (25, 30, 44, 45). See Supporting Information for details.
SI Materials and Methods
Reagents.
Recombinant human EGF was from BD Biosciences, Alexa Fluor 647-labeled EGF complex (EGF-A647) and EGF-Rh were from Molecular Probes. Monoclonal antibody to EGFR phosphotyrosine 1068 (pY1068), monoclonal antibody to phosphorylated ERK1/2, polyclonal rabbit antibody to phosphorylated MEK1/2, polyclonal rabbit antibody to α-actinin, and polyclonal rabbit antibody to GAPDH were from Cell Signaling Technology. HRas antibody was from Abcam. RAS10 (Pan-Ras antibody) was from EMD Millipore. Polyclonal rabbit antibody to EGFR (no. 1005) was from Santa Cruz Biotechnology. EGFR kinase inhibitor PD158780 was from Calbiochem. All chemical were purchased from Sigma-Aldrich or Thermo Fisher Scientific.
Cell Culture and Transfection.
Parental HeLa, gene-edited HeLa/mV-HRas, and COS1 cells were maintained in DMEM supplemented with 10% (vol/vol) FBS. Before experiments, cells were serum-starved for 16 h in DMEM. In transient-transfection experiments, cells at 60% confluency were transfected with DNA constructs using Lipofectamine 2000 (Invitrogen) or Effectene (Qiagen). The following constructs were used: Grb2-CFP (44); CFP-EHD1 and CFP-EEA.1 (CFP-tagged fragment 1098–1411 of EEA.1) (provided by Dr. Emilia Galperin, University of Kentucky, Lexington, KY); GFP-SOS1 (provided by Dr. Dafna Bar-Sagi, New York University, New York, NY); mCherry-Rab11a (provided by Dr. Ora Weisz, University of Pittsburgh, Pittsburgh, PA); YFP-HRas, CFP-HRas, YFP-KRas4B, and CFP-KRas4B (17); GFP-NRas (provided by Dr. J. Donaldson, NIH, Bethesda, MD).
TALEN Design and Generation of HeLa/mV-HRas Gene-Edited Cells.
Enhanced obligate heterodimeric TALENs designed to target 50 bp around the start codon of human HRAS gene were engineered by Cellectis Bioresearch. Donor plasmid was constructed in pGEM T-Easy vector (Promega) by inserting the mV sequence between 750-bp gene homology arms upstream and downstream of HRAS start codon. The 1.5-kb homology region was amplified from the BAC clone RP11-392J11 (Invitrogen) with FastStart High Fidelity PCR System (Roche). TALEN and donor plasmid were transfected into HeLa cells. The cells were grown at 30 °C, 5% CO2, for 48 h, and then maintained at 37 °C. mVenus-expressing cells were isolated by flow sorting and directly plated onto µ-plates, 96-well with optical bottom (Ibidi). Positive HeLa/mV-HRas clones were identified by microscopy screening and verified by Western blotting and genomic DNA PCR. Genomic DNA from clone 3 (Fig. S2) used in most experiments was sequenced to confirm proper insertion of mVenus.
Ras Activity Assay.
GST-fused Ras·GTP binding fragment 51–149 aa of Raf-1 (GST-RBD) in pGEX-2T was purchased from Addgene. Bacterially expressed GST-RBD was pulled down from precleared lysates using glutathione-Sepharose 4B beads (Amersham Biosciences). The beads were washed in Ca2+, Mg2+-free phosphate buffer saline and immediately used in experiments. HeLa cells were solubilized in 1% Igepal in buffer A [150 mM NaCl, 1 mM MgCl2, 10% (vol/vol) glycerol, 20 mM Tris⋅HCl, pH 8, 2 mM DTT, and phosphatase and protease inhibitors] by agitating for 10 min at 4 °C. GST-RBD beads were incubated with 2.5–3 mg of precleared cell lysates for 2.5 h at 4 °C. After extensive washes of beads with buffer A, proteins were eluted from beads in 2× sample buffer and heated for 5 min at 95 °C. GST pulldowns and cell lysates were resolved by electrophoresis, transferred to nitrocellulose membranes, and the membranes were probed by Western blotting with RAS10 and HRas antibodies. Secondary antibodies conjugated to infrared fluorescent dyes (LI-COR Biosciences) were used, and blots were imaged and analyzed using Odyssey Infrared Imaging system (LI-COR Biosciences).
EGFR, MEK, and ERK Phosphorylation.
The cells were solubilized in TGH lysis buffer [1% Triton X-100, 10% (vol/vol) glycerol, 50 mM Hepes, 50 mM NaCl, 1 mM EDTA, 10 mM NEM, 1 mM orthovanadate, and other phosphatase and protease inhibitors]. Lysates were electrophoresed and probed by Western blotting for phosphorylated EGFR Y1068, MEK1/2, and ERK1/2, as described previously (26).
Surface Biotinylation.
Cells treated or not with the acidic buffer (0.2 M acetic acid, 0.5 M NaCl, pH 4.5) were washed twice in ice-cold PBS and incubated with 0.5 mg/mL EZ-Link Sulfo-NHS-SS-Biotin (Pierce Biotechnology) at 4 °C for 30 min. The cells were then washed with PBS containing 100 mM glycine and lysed in TGH. Precleared lysates (3 mg of protein) were incubated with 40 µL of NeutrAvidin Agarose beads (Pierce Biotechnology) for 3 h at 4 °C to isolate biotinylated proteins. The beads were washed twice with TGH supplemented with 0.5 M NaCl and 1 mM sodium orthovanadate, and once with TGH alone. Proteins were eluted from beads in 2× sample buffer and heated for 5 min at 95 °C. Biotinylated proteins and aliquots of lysates were resolved by electrophoresis and transferred to nitrocellulose membranes, and the membranes were probed by Western blotting with pY1068, EGFR (1005), and GAPDH antibodies.
Live-Cell Fluorescence Microscopy Imaging.
Cells plated in 35-mm MatTek dishes (MatTek Corporation) were placed into the microscope stage adaptor, and images were acquired using a spinning disk confocal imaging system described previously (26) controlled by SlideBook 6 software (Intelligent Imaging Innovation). Two-dimensional confocal images or 3D stacks of x–y images through 445-nm (CFP), 488-nm (GFP), 515-nm (mVenus), 561-nm (rhodamine), and 640-nm (Alexa 647) channels were acquired at 37 °C or room temperature and 5% CO2. All image acquisition settings were identical for experimental variants in each experiment.
Image Analysis.
The percentage of mVenus colocalized with EGF-Rh of the total cellular mV-HRas was calculated in 3D images using a segmentation-based method as described in ref. 45.
The mole/mole stoichiometry of YFP-HRas or mV-HRas to EGF-Rh in endosomes was estimated in 3D images of HeLa cells transiently expressing YFP-HRas or HeLa/mVenus-HRas cells, respectively. The cells were incubated with 4 ng/mL EGF-Rh for 15 min at 37 °C. Z stack of confocal images was acquired through 515- and 561-nm channels at 300-nm intervals. The segment mask was generated to select EGF-Rh endosomes using identical threshold parameters in all experimental variants. Mean fluorescence intensities (MFIs) of 515- and 561-nm channels in individual endosomes were then calculated. Local backgrounds of each selected endosome were measured as MFIs in the radius of two voxels surrounding an endosome in 3D. The ratio of background-subtracted MFIs through 515-nm (mVenus) and 561-nm (rhodamine) channels (515/561) was then calculated for individual endosomes. To account for possible cross-bleed of rhodamine fluorescence and autofluorescence of endosomes, MFIs through 515-nm channel of EGF-Rh–containing endosomes were measured in parental HeLa cells not expressing YFP-HRas. The mean value of this control 515/561 ratio was 0.0153. This value was subtracted from 515/561 ratio values calculated in HeLa/mV-HRas– and YFP-HRas–expressing cells. The apparent fluorescence intensity ratio for equimolar amounts of YFP and rhodamine (when imaged using the same image acquisition time) was measured to be 0.7 as in our previous study (25). Therefore, to determine the molar stoichiometry of mV-HRas or YFP-HRas to EGF-Rh, the 515/561 ratio was corrected for the difference in image acquisition times and divided by 0.7.
Apparent expression levels of YFP or mVenus per cell were determined using segmentation of 3D images to generate YFP or mVenus masks. Total amount of fluorescence per an individual cell was then calculated after background subtraction for multiple cells in each experimental variant, and expressed in fold difference to the mean expression level of mV-HRas (1× mV-HRas). To estimate the expression level of CFP-HRas and CFP-KRas in individual cells, the ratio of apparent quantum yields of YFP to CFP (6.7:1) was determined using transiently expressed cleavable YFP-CFP fusion protein as described in ref. 44. The calculation of the total amounts of CFP per cell was performed as described above for YFP with adjustment for a 6.7-lesser quantum yield of CFP. CFP-H/KRas levels were presented as the fold difference to mV-HRas expression. The apparent expression levels of GFP-NRas relative to that of mV-HRas were determined based on theoretical calculation of the ratios of the apparent quantum yields of eGFP and YFP (515-nm channel) in our imaging system (0.95:1; GFP:YFP).
TIR-FM Imaging.
Two-color TIR-FM imaging was performed as previously described (30). Briefly, serum-starved cells in MatTek dishes were imaged on a Nikon Eclipse Ti inverted microscope (Nikon) with a 100×, 1.49 N.A. oil-immersion objective using 488-nm (mVenus) and 561-nm laser lines (rhodamine), and with 10-s intervals between time frames. EGF-Rh was added during continuous image acquisition at 37 °C. Images were collected and analyzed using Nikon Elements software (version 4.30; Nikon) and an Andor Zyla 5.5 camera; at full resolution under these conditions, the pixel size with a 1× coupler matches Nyquist sampling (120-nm xy exactly). Images were thresholded, and particle tracking function was used to define spots and quantify colocalization of mV-HRas and EGF-Rh spots. After automatic detection of mVenus and rhodamine spots, spots were filtered by quality control parameters. The total number of spots of EGF-Rh and mV-HRas, and spots containing both mV-HRas and EGF-Rh (defined as 80% of spot overlap) on the cell bottom membrane were calculated at each time point. The percentage of EGF-Rh spots containing mV-HRas relative to the total number of EGF-Rh spots was then calculated in single time frame images with multiple cells. Mean percentages from four images per a time point were calculated and plotted against time.
Acknowledgments
We are grateful to Drs. D. Bar-Sagi, J. Donaldson, E. Galperin, A. Linstedt, and O. Weisz for the gifts of reagents, to Ms. Nancy Zurowski (University of Pittsburgh) for help with cell sorting, and to Dr. A. Grassart (University of California, Berkeley) for his advice on TALEN methodology. This work was supported by National Cancer Institute Grant CA089151.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520301113/-/DCSupplemental.
References
- 1.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: Weaving a tumorigenic web. Nat Rev Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Omerovic J, Laude AJ, Prior IA. Ras proteins: Paradigms for compartmentalised and isoform-specific signalling. Cell Mol Life Sci. 2007;64(19-20):2575–2589. doi: 10.1007/s00018-007-7133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowenstein EJ, et al. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70(3):431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 5.Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79(1):143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 6.Sorkin A, Von Zastrow M. Signal transduction and endocytosis: Close encounters of many kinds. Nat Rev Mol Cell Biol. 2002;3(8):600–614. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- 7.Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274(5295):2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 8.DeGraff JL, Gagnon AW, Benovic JL, Orsini MJ. Role of arrestins in endocytosis and signaling of alpha2-adrenergic receptor subtypes. J Biol Chem. 1999;274(16):11253–11259. doi: 10.1074/jbc.274.16.11253. [DOI] [PubMed] [Google Scholar]
- 9.Johannessen LE, Ringerike T, Molnes J, Madshus IH. Epidermal growth factor receptor efficiently activates mitogen-activated protein kinase in HeLa cells and Hep2 cells conditionally defective in clathrin-dependent endocytosis. Exp Cell Res. 2000;260(1):136–145. doi: 10.1006/excr.2000.5004. [DOI] [PubMed] [Google Scholar]
- 10.Sousa LP, et al. Suppression of EGFR endocytosis by dynamin depletion reveals that EGFR signaling occurs primarily at the plasma membrane. Proc Natl Acad Sci USA. 2012;109(12):4419–4424. doi: 10.1073/pnas.1200164109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teis D, Wunderlich W, Huber LA. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev Cell. 2002;3(6):803–814. doi: 10.1016/s1534-5807(02)00364-7. [DOI] [PubMed] [Google Scholar]
- 12.Prior IA, Hancock JF. Ras trafficking, localization and compartmentalized signalling. Semin Cell Dev Biol. 2012;23(2):145–153. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: Post-translational modification of RAS. Nat Rev Mol Cell Biol. 2012;13(1):39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choy E, et al. Endomembrane trafficking of ras: The CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98(1):69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- 15.Roy S, et al. Individual palmitoyl residues serve distinct roles in H-ras trafficking, microlocalization, and signaling. Mol Cell Biol. 2005;25(15):6722–6733. doi: 10.1128/MCB.25.15.6722-6733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laude AJ, Prior IA. Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J Cell Sci. 2008;121(Pt 4):421–427. doi: 10.1242/jcs.020107. [DOI] [PubMed] [Google Scholar]
- 17.Jiang X, Sorkin A. Coordinated traffic of Grb2 and Ras during epidermal growth factor receptor endocytosis visualized in living cells. Mol Biol Cell. 2002;13(5):1522–1535. doi: 10.1091/mbc.01-11-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiu VK, et al. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4(5):343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 19.Caloca MJ, Zugaza JL, Bustelo XR. Exchange factors of the RasGRP family mediate Ras activation in the Golgi. J Biol Chem. 2003;278(35):33465–33473. doi: 10.1074/jbc.M302807200. [DOI] [PubMed] [Google Scholar]
- 20.Arozarena I, et al. Activation of H-Ras in the endoplasmic reticulum by the RasGRF family guanine nucleotide exchange factors. Mol Cell Biol. 2004;24(4):1516–1530. doi: 10.1128/MCB.24.4.1516-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmick M, et al. KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell. 2014;157(2):459–471. doi: 10.1016/j.cell.2014.02.051. [DOI] [PubMed] [Google Scholar]
- 22.Misaki R, et al. Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. J Cell Biol. 2010;191(1):23–29. doi: 10.1083/jcb.200911143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu A, et al. A clathrin-dependent pathway leads to KRas signaling on late endosomes en route to lysosomes. J Cell Biol. 2009;184(6):863–879. doi: 10.1083/jcb.200807186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anonymous Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fortian A, Sorkin A. Live-cell fluorescence imaging reveals high stoichiometry of Grb2 binding to the EGF receptor sustained during endocytosis. J Cell Sci. 2014;127(Pt 2):432–444. doi: 10.1242/jcs.137786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galperin E, Sorkin A. Endosomal targeting of MEK2 requires RAF, MEK kinase activity and clathrin-dependent endocytosis. Traffic. 2008;9(10):1776–1790. doi: 10.1111/j.1600-0854.2008.00788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamioka Y, Yasuda S, Fujita Y, Aoki K, Matsuda M. Multiple decisive phosphorylation sites for the negative feedback regulation of SOS1 via ERK. J Biol Chem. 2010;285(43):33540–33548. doi: 10.1074/jbc.M110.135517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boykevisch S, et al. Regulation of ras signaling dynamics by Sos-mediated positive feedback. Curr Biol. 2006;16(21):2173–2179. doi: 10.1016/j.cub.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 29.Omerovic J, Hammond DE, Clague MJ, Prior IA. Ras isoform abundance and signalling in human cancer cell lines. Oncogene. 2008;27(19):2754–2762. doi: 10.1038/sj.onc.1210925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fortian A, et al. Endocytosis of ubiquitylation-deficient EGFR mutants via clathrin-coated pits is mediated by ubiquitylation. Traffic. 2015;16(11):1137–1154. doi: 10.1111/tra.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porat-Shliom N, Kloog Y, Donaldson JG. A unique platform for H-Ras signaling involving clathrin-independent endocytosis. Mol Biol Cell. 2008;19(3):765–775. doi: 10.1091/mbc.E07-08-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang F, Khvorova A, Marshall W, Sorkin A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J Biol Chem. 2004;279(16):16657–16661. doi: 10.1074/jbc.C400046200. [DOI] [PubMed] [Google Scholar]
- 33.Biskup C, Rubio I. Real-time visualization and quantification of native Ras activation in single living cells. Methods Mol Biol. 2014;1120:285–305. doi: 10.1007/978-1-62703-791-4_19. [DOI] [PubMed] [Google Scholar]
- 34.Bivona TG, Philips MR. Ras pathway signaling on endomembranes. Curr Opin Cell Biol. 2003;15(2):136–142. doi: 10.1016/s0955-0674(03)00016-4. [DOI] [PubMed] [Google Scholar]
- 35.Lorentzen A, Kinkhabwala A, Rocks O, Vartak N, Bastiaens PI. Regulation of Ras localization by acylation enables a mode of intracellular signal propagation. Sci Signal. 2010;3(140):ra68. doi: 10.1126/scisignal.20001370. [DOI] [PubMed] [Google Scholar]
- 36.Kural C, et al. Dynamics of intracellular clathrin/AP1- and clathrin/AP3-containing carriers. Cell Rep. 2012;2(5):1111–1119. doi: 10.1016/j.celrep.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11(3):319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 38.Fujioka A, et al. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J Biol Chem. 2006;281(13):8917–8926. doi: 10.1074/jbc.M509344200. [DOI] [PubMed] [Google Scholar]
- 39.Bermudez O, Pagès G, Gimond C. The dual-specificity MAP kinase phosphatases: Critical roles in development and cancer. Am J Physiol Cell Physiol. 2010;299(2):C189–C202. doi: 10.1152/ajpcell.00347.2009. [DOI] [PubMed] [Google Scholar]
- 40.Ekerot M, et al. Negative-feedback regulation of FGF signalling by DUSP6/MKP-3 is driven by ERK1/2 and mediated by Ets factor binding to a conserved site within the DUSP6/MKP-3 gene promoter. Biochem J. 2008;412(2):287–298. doi: 10.1042/BJ20071512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorkin A, et al. Recycling of epidermal growth factor-receptor complexes in A431 cells: Identification of dual pathways. J Cell Biol. 1991;112(1):55–63. doi: 10.1083/jcb.112.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garay C, et al. Epidermal growth factor-stimulated Akt phosphorylation requires clathrin or ErbB2 but not receptor endocytosis. Mol Biol Cell. 2015;26(19):3504–3519. doi: 10.1091/mbc.E14-09-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sigismund S, et al. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev Cell. 2008;15(2):209–219. doi: 10.1016/j.devcel.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 44.Galperin E, Verkhusha VV, Sorkin A. Three-chromophore FRET microscopy to analyze multiprotein interactions in living cells. Nat Methods. 2004;1(3):209–217. doi: 10.1038/nmeth720. [DOI] [PubMed] [Google Scholar]
- 45.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21(6):737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]