Significance
Unlike the other three hepatitis B virus-encoded proteins, both the function and structure of HBx are poorly understood. The discovery of a BH3-like motif in HBx and the demonstration of direct association between HBx and the anti-apoptotic Bcl-2 family proteins yielded the hypothesis that HBx may rely on its BH3-like motif to antagonize the functions of Bcl-2. In this study, we show that the interaction between the HBx BH3-like motif and Bcl-2 is drastically weaker than that between a canonical BH3 motif and Bcl-2. This finding, corroborated by structural analysis, suggests that the modulation of the activity of Bcl-2 by the BH3-like motif of HBx may be different from other BH3-only proteins.
Keywords: Bcl-2, hepatitis B virus, apoptosis, crystal structure
Abstract
HBx is a hepatitis B virus protein that is required for viral infectivity and replication. Anti-apoptotic Bcl-2 family members are thought to be among the important host targets of HBx. However, the structure and function of HBx are poorly understood and the molecular mechanism of HBx-induced carcinogenesis remains unknown. In this study, we report biochemical and structural characterization of HBx. The recombinant HBx protein contains metal ions, in particular iron and zinc. A BH3-like motif in HBx (residues 110–135) binds Bcl-2 with a dissociation constant of ∼193 μM, which is drastically lower than that for a canonical BH3 motif from Bim or Bad. Structural analysis reveals that, similar to other BH3 motifs, the BH3-like motif of HBx adopts an amphipathic α-helix and binds the conserved BH3-binding groove on Bcl-2. Unlike the helical Bim or Bad BH3 motif, the C-terminal portion of the bound HBx BH3-like motif has an extended conformation and makes considerably fewer interactions with Bcl-2. These observations suggest that HBx may modulate Bcl-2 function in a way that is different from that of the classical BH3-only proteins.
The human hepatitis B virus (HBV) is a prototypical hepadnavirus that specifically infects hepatocytes (1, 2). Over 240 million people worldwide are chronically infected by HBV according to the World Health Organization. Persistent HBV infection is one of the major risk factors for the development of hepatocellular carcinoma (HCC), accounting for over 50% of all HCC cases (3, 4). The genome of HBV encodes only four proteins, among which the product of the X gene (HBx) is least understood (1).
HBx contains 154 residues and is closely associated with HCC development. The X gene is most frequently integrated and preferentially maintained in HBV-associated HCC, and HBx expression is frequently detected in HCC patients (5, 6). Several studies using transgenic mice and cell culture models yielded contrasting conclusions. Some studies suggest that the development of HCC is closely linked to HBx expression (7–10), although HBx alone may be insufficient for HCC development (11, 12). Other results suggest that HBx has the ability to suppress cell transformation through induction of apoptosis (13, 14). As such, the molecular mechanism of HBx-induced cellular transformation remains enigmatic.
Numerous cellular activities are reported to be modulated by HBx through interactions with various host targets. For example, HBx-mediated transcriptional activation is achieved through its interaction with transcription factors (15, 16) or components involved in the basal transcriptional machinery (17–19). HBx can also modulate cellular proliferation and viability through interactions with p53 and Bcl-2 family members. Reflecting its enigmatic functions, HBx has been demonstrated to either induce apoptosis (20) or prevent apoptosis (21).
HBx was recently found to contain a BH3-like motif in its C-terminal sequences (22). Experiments carried out in human hepatocytes suggest that Bcl-2 and Bcl-XL are the key cellular targets for HBx, where direct interactions between the BH3-like motif of HBx and the anti-apoptotic Bcl-2 family proteins trigger cytosolic calcium elevation, apoptosis, and viral DNA replication (22, 23). The BH3-only proteins exemplified by Bim and Bad are thought to inhibit the anti-apoptosis proteins by binding to a conserved hydrophobic groove known as the BH3-binding groove (24–26). An important question central to understanding the function of HBx is whether the noncanonical BH3-like motif of HBx also binds Bcl-2 or Bcl-xL similarly as other BH3-only proteins. In this study, we performed biochemical and structural characterization of HBx. Our results suggest that the modulation of Bcl-2 by the BH3-like motif of HBx may be different from other BH3-only proteins.
Results
Biochemical Characterization of HBx.
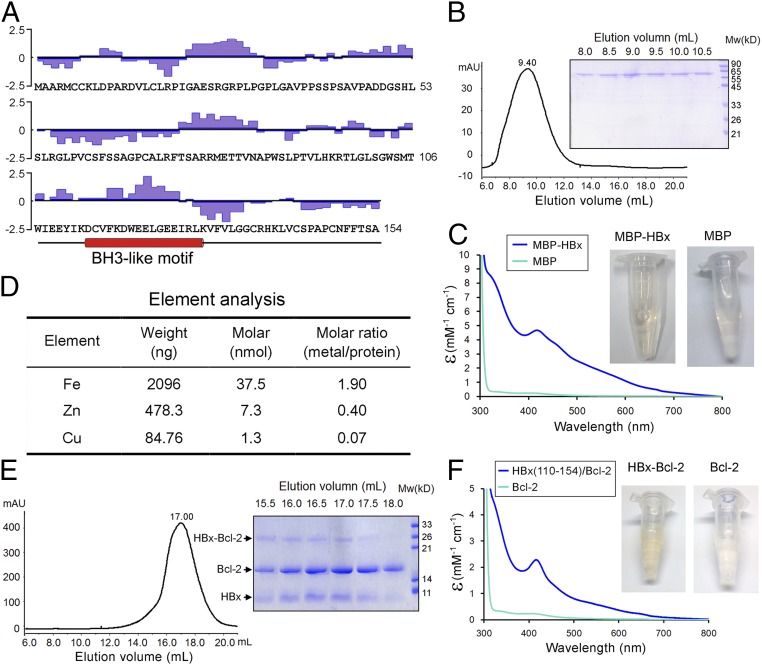
HBx comprises distinct functional domains (27) (Fig. 1A). The sequences of HBx are unusually hydrophobic for a cytosolic protein, with ∼53% of the amino acids being hydrophobic in nature. The C-terminal sequences contain a putative BH3-like motif (22). Perhaps due to the high content of hydrophobic residues, the full-length HBx protein (residues 1–154) can be expressed only in a soluble form when fused to the C terminus of maltose-binding protein (MBP). The affinity-purified full-length MBP-HBx fusion exhibited a broad elution peak on gel filtration, with the elution volume indicative of large oligomer formation (Fig. 1B).
Fig. 1.
Biochemical characterization of the hepatitis B virus protein HBx. (A) The primary sequences of HBx. The hydrophilicity plot by the Kyte–Doolittle criteria (48) is shown above the sequences. The 63 hydrophobic amino acids (Ala, Val, Leu, Ile, Phe, Cys, and Met) account for 41% of the total. In addition, there are 15 Pro and 4 Trp residues. The putative BH3-like motif is shown below the sequences in an α-helical conformation. (B) The full-length HBx appears to form large oligomers on gel filtration. The full-length MBP-tagged HBx was eluted from gel filtration with an elution volume of 9.4 mL, close to the void (Left). The peak fractions were visualized on SDS/PAGE (Right). (C) The purified full-length MBP-tagged HBx exhibits a yellow-brownish color. Wavelength scan of this protein, but not the MBP control, shows an absorption peak at 415 nm. (D) Element analysis of the full-length MBP-tagged HBx reveals the presence of metal ions. Both iron and zinc are present in significant quantities with ∼1.9 and 0.4 molar equivalence of the MBP-HBx protein. (E) The HBx fragment (residues 110–154) bound to Bcl-2 exhibits excellent behavior on gel filtration. The fusion protein between the HBx fragment and Bcl-2 (residues 1–50 and 92–207) was purified to homogeneity and separated by proteolysis. The resulting complex was subject to gel filtration analysis (Left). The peak fractions were visualized on SDS/PAGE (Right). (F) The HBx fragment (residues 110–154) bound to Bcl-2 has a yellow-brownish color. Wavelength scan of HBx-Bcl-2 shows an absorption peak at 415 nm.
The MBP-HBx fusion displays a yellow-brownish color, and a wavelength scan of 300–800 nm reveals significant absorption in the visible spectrum region with a characteristic absorption peak at 415 nm (Fig. 1C). In contrast, MBP alone has little absorption in this region (Fig. 1C). Both the color and the maximum absorption wavelength suggest the presence of metal ions. To identify the bound metal ions, we performed a preliminary element analysis, which reveals iron, zinc, and copper as the three major metals in the sample. Quantitative analysis identified iron to be the most prevalent, with an ∼1.9 molar equivalence (Fig. 1D). Unsurprisingly, the MBP-HBx fusion defied crystallization. We attempted to express the N-terminal sequences of HBx (residues 1–77). Unfortunately, despite repeated effort, the N-terminal fragment of HBx remained insoluble.
Next, we examined the C-terminal sequences of HBx. Similar to the full-length HBx protein, several constructs of the C-terminal domain yielded soluble protein only as a MBP fusion. But these proteins, similar to the full-length MBP-HBx fusion, exhibited poor solution behavior. Because the predicted BH3-like motif (residues 110–130) (22) is contained within the HBx C-terminal sequences, we fused the HBx fragment (residues 110–154) to the N terminus of human Bcl-2 (residues 1–50 and 92–207), reasoning that such a design would favor the recognition of the BH3-like motif by Bcl-2 and hence improve the behavior of the HBx C-terminal sequences. The Bcl-2 sequences 51–91 are removed to improve the behavior of the fusion protein (28). Compared with MBP fusions, both the yield and the solution behavior of the HBx-Bcl-2 fusion are markedly improved. The HBx-Bcl-2 fusion was subjected to limited proteolysis, which separates the HBx C-terminal sequences from Bcl-2. Remarkably, the free HBx C-terminal sequences were coeluted from gel filtration with Bcl-2, suggesting complex formation (Fig. 1E). The peak fractions from gel filtration, but not Bcl-2 alone, displayed the same yellow-brownish color as the MBP-HBx fusion; wavelength scan reveals the same characteristic absorption peak at 415 nm (Fig. 1F). A 12-residue stretch of the C-terminal sequences (residue 137–148) of HBx contains three cysteine residues. Replacement of these three cysteines (Cys137, Cys143, and Cys148) in the HBx fragment (residues 110–154) resulted in disappearance of the characteristic yellow-brownish color.
Quantification of HBx Interactions with Human Bcl-2.
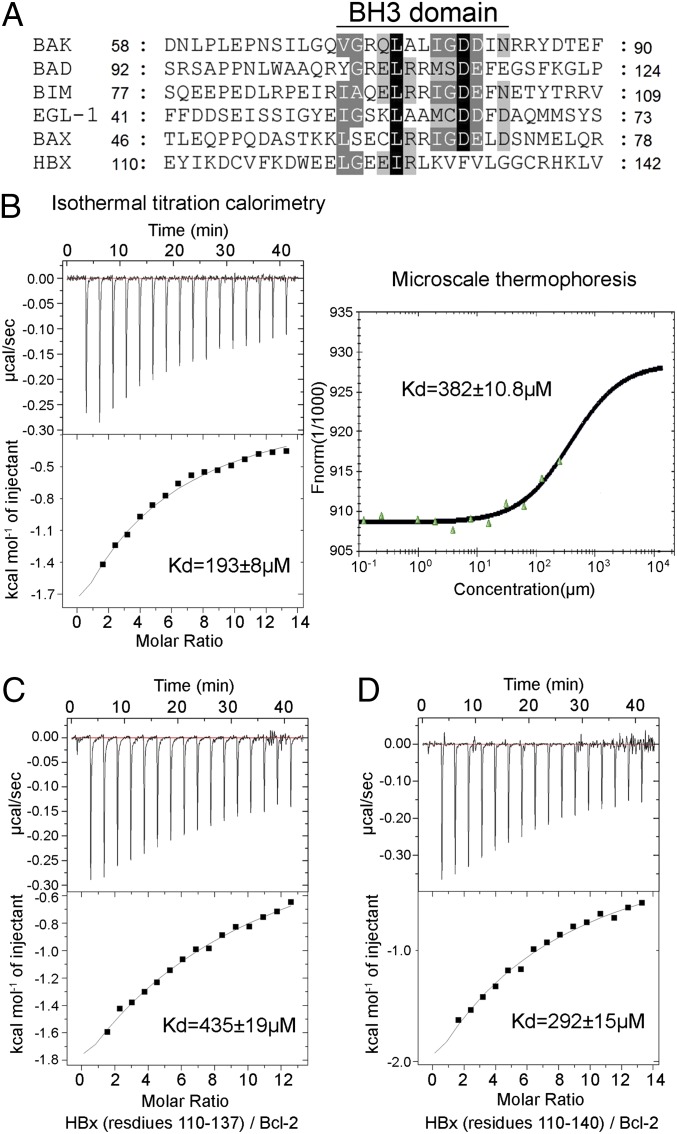
To assist structural studies, we sought to identify a minimal HBx BH3-like motif that retains binding to human Bcl-2. Sequence alignment between HBx and the canonical BH3 motifs reveals sequence conservation only at the N-terminal half, but not the C-terminal half (Fig. 2A). For example, the invariant Asp residue in the consensus BH3 motif is replaced by Phe in HBx. Because the conserved residues in a canonical BH3 motif are directly involved in interactions with the BH3-binding groove, such sequence variation in HBx predicts a diminished binding affinity to Bcl-2 in case of a conserved binding mechanism. To examine this scenario, we synthesized three HBx peptides and measured their binding affinities toward Bcl-2, using isothermal titration calorimetry (ITC).
Fig. 2.
Quantification of the interactions between the HBx BH3-like motif and Bcl-2. (A) Sequence alignment of the BH3 domains from the proapoptotic Bcl-2 proteins and HBx. Invariant and conserved residues among the canonical BH3 motifs are shaded black and gray, respectively. (B) Measurement of binding affinities between the HBx BH3-like motif (residues 110–135) and Bcl-2 (residues 1–50 and 92–207). Measurement by ITC and MST reveals dissociation constants of 193 ± 8 and 382 ± 10.8 µM, respectively. (C) Measurement of the binding affinity between the HBx fragment (residues 110–137) and Bcl-2 by ITC. Data fitting reveals a dissociation constant of 435 ± 19 µM. (D) The HBx fragment (residues 110–140) binds to Bcl-2 with a dissociation constant of ∼292 ± 15 µM by ITC.
The 26-residue HBx fragment (residues 110–135) exhibits a dissociation constant of 193 ± 8 μM toward human Bcl-2 (Fig. 2B, Left). Despite the poor sequence conservation between HBx and the canonical BH3 motifs, we were surprised by the relatively low binding affinity. To rule out potential artifacts associated with ITC, we used a different biophysical method—microscale thermophoresis (MST) (29)—which reveals a qualitatively similar dissociation constant of 382 ± 10.8 μM (Fig. 2B, Right). Use of a longer HBx fragment, such as residues 110–137 (Fig. 2C) or residues 110–140 (Fig. 2D), failed to improve the binding affinity compared with the HBx fragment 110–135. These results identify the HBx fragment 110–135 as an ideal target for cocrystallization with Bcl-2 and further indicate that the sequences immediately C-terminal to residue 135 make little contribution to Bcl-2 binding.
Notably, the Bcl-2–binding affinities by the HBx BH3-like fragments are drastically reduced compared with those by the canonical BH3 motifs. For example, the BH3-only motifs of Bad and Bim were previously reported to have dissociation constants of 17.7 and 2.44 nM, respectively, toward Bcl-2 (30).
Structure of Bcl-2 Bound to a HBx BH3-Like Motif.
To understand the recognition of the HBx BH3-like motif by Bcl-2, we sought to crystallize human Bcl-2 (residues 1–50 and 92–207) in complex with the synthetic HBx BH3-like peptide (residues 110–135). Clustered needles were obtained after screening more than 6,000 conditions. Single crystals grew to fill size at 4 °C over a period of 3 d. The structure was determined by molecular replacement at 2.1 Å resolution (Table 1).
Table 1.
Statistics of data collection and refinement
| Data | Native |
| Space group | P21 21 2 |
| Unit cell (Å) | 63.78, 82.99, 33.49 |
| Wavelength (Å) | 0.9789 |
| Resolution (outer shell, Å) | 40∼2.10 (2.18∼2.10) |
| Rmerge (outer shell, %) | 16.8 (47.5) |
| I/sigma (outer shell) | 17.4 (4.9) |
| Completeness (outer shell, %) | 99.9 (100.0) |
| No. of total reflections | 73,135 |
| No. of unique reflections | 10,943 |
| Refinement | |
| Rwork/Rfree (%) | 25.52/27.55 |
| No. of atoms | |
| Protein | 1,467 |
| Water | 93 |
| Average B value (Å2) | |
| Protein | 25.72 |
| Main chain | 25.24 |
| Side chain | 26.16 |
| Water | 35.72 |
| rmsd | |
| Bonds (Å) | 0.009 |
| Angle (°) | 1.262 |
| Ramachandran plot (%) | |
| Most favorable | 94.7 |
| Additionally allowed | 4.5 |
| Generously allowed | 0.6 |
| Disallowed | 0.0 |
Rmerge = ΣhΣi|Ih,i − Ih|/ΣhΣiIh,i, where Ih is the mean intensity of the i observations of symmetry-related reflections of h. R = Σ|Fobs − Fcalc|/ΣFobs, where Fcalc is the calculated protein structure factor from the atomic model (Rfree was calculated with 5% of the reflections selected).
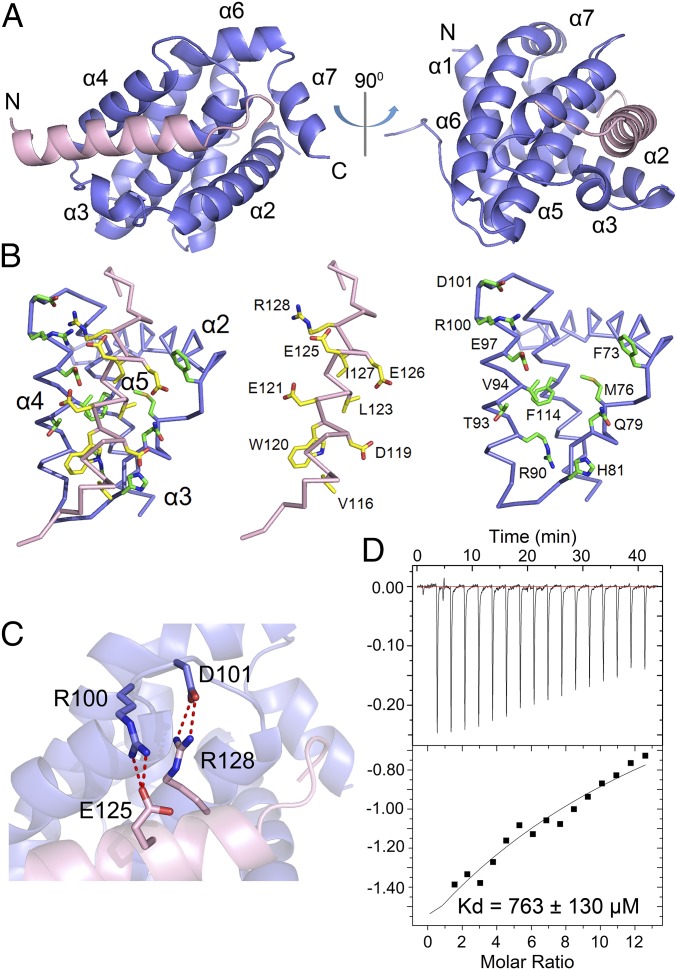
The Bcl-2-HBx complex adopts a compact globular fold, with the HBx BH3-like motif forming an amphipathic α-helix in the BH3-binding groove of Bcl-2 (Fig. 3A). The hydrophobic groove is formed by helices α2, α3, α4, α5, and α7 of Bcl-2 as previously observed (30–33). Residues 111–135 of the HBx BH3-like motif exhibit clear electron density and are modeled in the final atomic model. Of the 25 modeled amino acids of HBx, residues 114–130 form the amphipathic α-helix, followed by residues 131–135 in an extended conformation (Fig. 3A). The relative orientation of the HBx BH3-like motif with respect to Bcl-2 confirms the sequence alignment of HBx with other Bcl-2 family proteins (Fig. 2A). Notably, this alignment differs from that proposed earlier (22) by two helical turns.
Fig. 3.
Structure of Bcl-2 bound to the HBx BH3-like motif. (A) Overall structure of Bcl-2 (residues 1–50 and 92–207) in complex with the HBx BH3-like motif (residues 110–135). HBx and Bcl-2 are colored pink and slate, respectively. (B) A close-up view of the HBx/Bcl-2 interface. For clarity, the crowded interface (Left) is opened up to show its two components (Middle and Right). The backbones of HBx and Bcl-2 are colored pink and slate, respectively. (C) A close-up view of the hydrogen bonds at the HBx-Bcl-2 interface. Glu125 and Arg128 of HBx each makes a pair of charge-stabilized hydrogen bonds to residues in Bcl-2. Hydrogen bonds are represented by red dashed lines. (D) Measurement of the binding affinity between the HBx BH3-like motif (residues 110–135) and the Bcl-2 variant (R100A/D101A) by ITC. Data fitting reveals a dissociation constant of ∼763 ± 130 µM.
Analysis of the interface between the HBx BH3-like motif and the BH3-binding groove of Bcl-2 identifies van der Waals contact as a major driving force for association. However, only three hydrophobic amino acids—Trp120, Leu123, and Ile127 from the amphipathic HBx helix—interact with the hydrophobic surface groove on Bcl-2 (Fig. 3B). In addition, the guanidinium group of Arg128 from HBx donates a pair of charge-stabilized hydrogen bonds to the carboxylate side chain of Asp101 from Bcl-2, whereas the side chain of Glu125 from HBx accepts two hydrogen bonds from Arg100 of Bcl-2 (Fig. 3C). These hydrogen bonds are likely tempered by two closely spaced acidic residues Glu121 of HBx and Glu97 of Bcl-2 (Fig. 3B) and, perhaps to a lesser extent, by the basic residues Arg128 of HBx and Arg100 of Bcl-2 (Fig. 3C).
To corroborate the structural finding, we examined whether mutations on the interface residues of Bcl-2 could weaken its binding to the HBx BH3-like fragment. We generated the Bcl-2 variant R100A/D101A, with residues Arg100 and Asp101 replaced by Ala, and examined its interaction with the HBx BH3-like fragment using ITC (Fig. 3D). The Bcl-2 variant R100A/D101A exhibits a dissociation constant of 763 ± 130 μM for the HBx BH3-like fragment (Fig. 3D), which is approximately threefold lower than that by the wild-type (WT) Bcl-2. Thus, these interactions are somewhat important, but not essential, for the HBx-Bcl-2 association.
Comparison with Other Bcl-2-BH3 Structures.
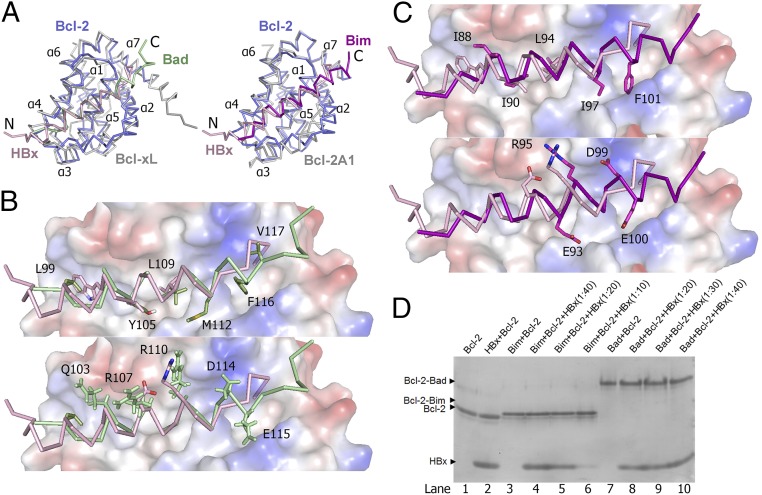
The overall structure of Bcl-2 bound to the HBx BH3-like motif is similar to those reported for the Bcl-xL-Bad and Bcl-2A1-Bim complexes (32, 34) (Fig. 4A). The structures of Bcl-2 and Bcl-2A1 (also known as Bfl-1) are nearly identical to each other, whereas the structures of Bcl-2 and Bcl-xL can be aligned to each other with a root mean squared deviation (rmsd) of about 1.83 Å for 128 Cα atoms. The HBx BH3-like motif, however, shows a distinct variation of the binding mechanism, with its amphipathic helix shifted to the N terminus by approximately two helical turns compared with that in Bad or Bim. Consequently, compared with Bad or Bim, the N terminus of the HBx fragment protrudes out of the globular complex whereas its C terminus is recessed in the BH3-binding groove (Fig. 4A). This structural feature is likely a direct consequence of the sequence variation in the C-terminal half of the HBx BH3-like motif (Fig. 2A).
Fig. 4.
Structural comparison of the Bcl-2-HBx complex with the Bcl-xL-Bad and Bcl-2A1-Bim complexes. (A) Comparison of the overall structure of the Bcl-2-HBx complex with that of Bcl-xL-Bad [PDB code 1G5J (32)] (Left) and Bcl-2A1-Bim [PDB 2VM6 (34)] (Right). The backbones are shown in ribbon representation. HBx (residues 110–135) and Bcl-2 are colored pink and slate, respectively. Bcl-xL and Bcl-2A1 are colored gray, whereas the Bad and Bim helices are colored green and orange, respectively. (B) A close-up view of the interface comparison between the Bcl-2-HBx and Bcl-xL-Bad complexes. For clarity, only the surface of Bcl-xL is shown in electrostatic potential. Bad and HBx are colored in green and pink, respectively. Hydrophobic and charged side chains of the bound BH3 motifs are shown in the Upper and Lower panels, respectively. (C) A close-up view on the interface comparison between the Bcl-2-HBx and Bcl-2A1-Bim complexes. Bim and HBx are colored in orange and pink, respectively. Hydrophobic and charged side chains of the bound BH3 motifs are shown in the Upper and Lower, respectively. (D) Analysis of Bcl-2 interactions with BH3 motifs by native PAGE. The HBx BH3-like motif and Bcl-2 failed to form a stable complex on native PAGE (lanes 1 and 2). In contrast, the BAD BH3 motif formed a stable complex with Bcl-2, and this complex remained undisturbed by excess amount of HBx BH3-like motif (lanes 7–10). Similarly, the Bim BH3 motif also formed a stable complex with Bcl-2 (lanes 3–6), which also remained undisturbed by the presence of HBx BH3-like motif.
For the Bcl-xL-Bad complex (32), six hydrophobic residues (Leu99, Tyr105, Leu109, Met112, Phe116, and Val117) from the amphipathic α-helix of Bad protrude into the hydrophobic groove of Bcl-2, making extensive van der Waals contacts (Fig. 4B, Upper). In addition, five charged residues (Gln103, Arg107, Arg110, Asp114, and Glu115) are involved in hydrogen bonds and/or salt bridge interactions with surface residues in Bcl-2 (Fig. 4B, Lower). For the Bcl-2A1-Bim complex (34), five hydrophobic residues (Ile88, Ile90, Leu94, Ile97, and Phe101) and four charged residues (Glu93, Arg95, Asp99, and Glu100) directly contribute to the binding interface (Fig. 4C). Consistent with the structural analysis, formation of the Bcl-2-HBx complex appeared to be too weak to survive native gel electrophoresis (Fig. 4D, lanes 1 and 2). In sharp contrast, the BH3 fragment derived from Bim or Bad formed a stable complex with Bcl-2, and such complex formation was unaffected by the presence of excess HBx peptide (Fig. 4D, lanes 3–10).
Discussion
How HBx expression in the liver leads to the development of HCC is poorly understood. Previous studies suggest that the C-terminal BH3-like motif of HBx is crucial for apoptosis, cytosolic calcium increase, and HBV viral replication, whereas the anti-apoptosis proteins Bcl-2 and Bcl-xL may represent the key cellular targets for such activities (22, 23). Consistent with this hypothesis, C-terminal truncation of HBx is frequently detected in HCCs (35, 36); compared with the full-length HBx, the truncated HBx (residues 1–127), which loses the C-terminal half of the BH3-like motif, could more effectively transform the liver cell line MIHA (36).
The interplay among Bcl-2 family members plays a critical role in deciding whether a cell undergoes apoptosis (37). The anti-apoptotic proteins Bcl-2 and Bcl-xL reside on the mitochondrial outer membrane and suppress apoptosis by inhibiting the release of cytochrome c from the intermembrane space into the cytoplasm (26). The proapoptotic BH3-only proteins are translocated to the mitochondrial outer membrane only upon activation, where they interact with Bcl-2 and Bcl-xL to antagonize their anti-apoptotic activity. One possibility is that, similar to the endogenous BH3-only proteins, HBx may inhibit the function of the anti-apoptotic proteins through its interactions with the conserved BH3-binding groove (22).
In this study, we characterized the interactions between Bcl-2 and the BH3-like motif of HBx, both structurally and biochemically. The BH3-like motif of HBx forms an amphipathic helix that lies in the BH3-binding groove of Bcl-2. As predicted from alignment results of the primary sequences, the binding interface diverges at the C-terminal half of the HBx BH3-like motif. The nonconserved residues 130–133 adopt an extended, rather than helical, conformation. In addition, compared with Bim or Bad, the HBx BH3-like motif has a much reduced number of hydrophobic contacts and hydrogen bonds. Together, these structural findings explain why the HBx BH3-like motif exhibits a markedly reduced binding affinity toward the BH3-binding groove of Bcl-2.
Double mutations G124L/I127A in HBx were found to abolish interactions between HBx and CED-9, the functional ortholog of Bcl-2 in Caenorhabditis elegans, and G124L was the major effector (22). Structural analysis reveals that both Gly124 and Ile127 of HBx face the BH3-binding groove, with the main chain Cα atom of Gly124 stacking against the groove and the side chain of Ile127 making direct van der Waals contacts. Substitution of Gly124 by any amino acid, let alone the bulky Leu residue, is predicted to induce steric clash with adjacent residues in the groove, destabilizing formation of the α-helix and interaction with the groove.
A surprise from the structural and biochemical characterization of HBx is the relatively low binding affinity between its BH3-like motif and Bcl-2, which is lower than that between Bim or Bad and Bcl-2 by four orders of magnitude. It is generally believed that the strength of Bcl-2 interaction by the BH3-only proteins correlates with the extent of inhibition of the anti-apoptotic function of Bcl-2. The relatively weak interaction between the HBx BH3-like motif and Bcl-2 is unlikely to be sufficient for stable association in cells, especially at low expression levels of HBx. Nonetheless, this interaction may provide a concentration-dependent modulation to the anti-apoptotic function of Bcl-2 and Bcl-xL. This analysis is consistent with the observation that only high concentrations of HBx promoted apoptosis whereas low levels inhibited apoptosis (38). We cannot rule out the possibility that full-length HBx, which was used in the in vivo experiments (22, 23), may have a higher binding affinity for Bcl-2 in cells. In addition, HBx has been reported to localize to mitochondria (39, 40). The increased local concentration of HBx may greatly favor its association with Bcl-2, which is also localized to the outer membrane of mitochondria.
There is no proven correlation between the strength of binding affinity for Bcl-2 and the functional significance of a BH3-only protein. In fact, the binding affinity for Bcl-2 varies greatly among the BH3 motifs, ranging from micromolar for Noxa and Hrk to single-digit nanomolar for Bim and Bid (30). For the BH3-like motif of HBx, the low binding affinity may serve to modulate or titrate, rather than sequester, Bcl-2. In addition, although the BH3-like motif of HBx binds Bcl-2 with low affinity, it remains to be examined how strongly it interacts with other prosurvival Bcl-2 family members such as Bcl-xL or Bcl-w. Furthermore, similar to other BH3-only motifs, HBx might also interact with the proapoptotic Bcl-2 family proteins Bax and Bak. The results of these studies may help identify additional physiologically relevant targets of HBx.
In this study, we identify HBx as a metal-binding protein, although the relationship between metal binding and the proapoptotic activity of HBx remains unknown. Because the metal-binding cysteine resides are at the C terminus of HBx, away from the BH3-like motif, the metal-binding activity by these C-terminal sequences is unlikely to have a direct impact on HBx interaction with Bcl-2. However, we cannot rule out the possibility that metal binding triggers a conformational switch in HBx that augments interactions with Bcl-2. Unfortunately, the problematic solution behavior of any recombinant fragment that contains the C-terminal sequences of HBx discourages detailed biochemical characterization. Future studies should be directed to define the role of metal binding by HBx.
We suggest that the anti-apoptosis function of HBx may involve not just its BH3-like motif but also other sequences such as the N-terminal domain and the C-terminal Cys-rich sequences. In a broader context, we further speculate that the multifunctional aspects of HBx may also rely on both the BH3-like motif and other sequence elements. The ability of HBx to interact with a large number of cellular proteins (1, 41) apparently underlies the multifaceted and sometimes seemingly conflicting functions of HBx. Research aimed at understanding HBx function in vivo is complicated by networks of protein–protein interactions. Delineating the biochemical properties of HBx in vitro requires access to soluble, well-behaved protein. Unfortunately, the full-length HBx protein exhibits poor solution behavior and is unamenable to biochemical manipulation. Nonetheless, our present study serves as a framework for understanding the proapoptotic function of HBx and for future investigations.
Materials and Methods
Protein Preparation.
Bcl-2 (residues 1–50 and 92–207) was cloned and purified as reported (28). To improve solubility, the internal loop of Bcl-2 (residues 51–91) was removed, and residues 35–50 were replaced by residues 33–48 of Bcl-xL as reported (28). The final construct was cloned into pET-15b. The full-length HBx with MBP at the N terminus was cloned into pET-15b and overexpressed in Escherichia coli BL21(DE3). The MBP-HBx fusion was purified as described (42). The C-terminal sequences of HBx (residues 110–154) were fused to the N terminus of Bcl-2 with an intervening cleavage site for the caspase drICE.
Isothermal Titration Calorimetry.
MicroCalorimetry was used for all measurements. The protein sample was prepared at a final concentration of 10 μM in a buffer containing 25 mM Tris⋅HCl (pH 8.0) and 150 mM NaCl. The peptide was dialyzed against the same buffer to a final concentration of 800 µM. All experiments were carried out at 25 °C. Data were analyzed using the software Origin (OriginLab).
Gel Filtration Analysis.
Superdex-200 (10/30; GE Healthcare) was pre-equilibrated with 150 mM NaCl, 20 mM Tris, pH 8.0, and 2 mM DTT and calibrated with molecular weight standards (GE Healthcare). After injection, the samples were eluted with a flow rate of 0.4 mL/min.
Crystallization and Structure Determination.
Crystals of Bcl-2 bound to HBx (residues 110–135) were grown at 4 °C using the hanging-drop vapor-diffusion method by mixing 1 µL of the complex protein with 1 µL of the reservoir solution containing 0.1 M sodium acetate, pH 4.6, and 0.5 M ammonium sulfate. The needle-shaped crystals grew to a full size over a period of 3 d. The crystals were directly flash-frozen in a cold nitrogen stream at 100 K. Diffraction data were collected at Shanghai Synchrotron Radiation Facility. The data were integrated and scaled using HKL2000 package (43). Further processing was carried out using the CCP4 suite (44). The crystal structure of the 1:1 complex between Bcl-2 and Bax [PDB code 2XA0 (30)] was used as the initial search model. Our structure was solved by molecular replacement using PHASER (45), manually adjusted in COOT (46), and refined with PHENIX (47).
Acknowledgments
We thank Lei Liu of Tsinghua University for synthesis of peptides. This work was supported by funds from the Ministry of Science and Technology (2014ZX09507003006) and the National Natural Science Foundation of China (31130002, 31321062, and 31430020).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5FCG).
References
- 1.Benhenda S, Cougot D, Buendia MA, Neuveut C. Hepatitis B virus X protein molecular functions and its role in virus life cycle and pathogenesis. Adv Cancer Res. 2009;103:75–109. doi: 10.1016/S0065-230X(09)03004-8. [DOI] [PubMed] [Google Scholar]
- 2.Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: Paradigms for viral-related human carcinogenesis. Oncogene. 2006;25(27):3823–3833. doi: 10.1038/sj.onc.1209559. [DOI] [PubMed] [Google Scholar]
- 3.Cougot D, Neuveut C, Buendia MA. HBV induced carcinogenesis. J Clin Virol. 2005;34(Suppl 1):S75–S78. doi: 10.1016/s1386-6532(05)80014-9. [DOI] [PubMed] [Google Scholar]
- 4.Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37(Suppl 8):S4–S66. doi: 10.1016/s0959-8049(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 5.Hwang GY, et al. Detection of the hepatitis B virus X protein (HBx) antigen and anti-HBx antibodies in cases of human hepatocellular carcinoma. J Clin Microbiol. 2003;41(12):5598–5603. doi: 10.1128/JCM.41.12.5598-5603.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng Z, et al. Integration of the hepatitis B virus X fragment in hepatocellular carcinoma and its effects on the expression of multiple molecules: A key to the cell cycle and apoptosis. Int J Oncol. 2005;26(2):467–473. [PubMed] [Google Scholar]
- 7.Kim YC, Song KS, Yoon G, Nam MJ, Ryu WS. Activated ras oncogene collaborates with HBx gene of hepatitis B virus to transform cells by suppressing HBx-mediated apoptosis. Oncogene. 2001;20(1):16–23. doi: 10.1038/sj.onc.1203840. [DOI] [PubMed] [Google Scholar]
- 8.Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351(6324):317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 9.Seifer M, Höhne M, Schaefer S, Gerlich WH. In vitro tumorigenicity of hepatitis B virus DNA and HBx protein. J Hepatol. 1991;13(Suppl 4):S61–S65. doi: 10.1016/0168-8278(91)90026-8. [DOI] [PubMed] [Google Scholar]
- 10.Koike K, et al. High-level expression of hepatitis B virus HBx gene and hepatocarcinogenesis in transgenic mice. Hepatology. 1994;19(4):810–819. [PubMed] [Google Scholar]
- 11.Billet O, et al. In vivo activity of the hepatitis B virus core promoter: tissue specificity and temporal regulation. J Virol. 1995;69(9):5912–5916. doi: 10.1128/jvi.69.9.5912-5916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perfumo S, et al. Recognition efficiency of the hepatitis B virus polyadenylation signals is tissue specific in transgenic mice. J Virol. 1992;66(11):6819–6823. doi: 10.1128/jvi.66.11.6819-6823.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H, Lee H, Yun Y. X-gene product of hepatitis B virus induces apoptosis in liver cells. J Biol Chem. 1998;273(1):381–385. doi: 10.1074/jbc.273.1.381. [DOI] [PubMed] [Google Scholar]
- 14.Schuster R, Gerlich WH, Schaefer S. Induction of apoptosis by the transactivating domains of the hepatitis B virus X gene leads to suppression of oncogenic transformation of primary rat embryo fibroblasts. Oncogene. 2000;19(9):1173–1180. doi: 10.1038/sj.onc.1203417. [DOI] [PubMed] [Google Scholar]
- 15.Barnabas S, Andrisani OM. Different regions of hepatitis B virus X protein are required for enhancement of bZip-mediated transactivation versus transrepression. J Virol. 2000;74(1):83–90. doi: 10.1128/jvi.74.1.83-90.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi BH, Park GT, Rho HM. Interaction of hepatitis B viral X protein and CCAAT/enhancer-binding protein alpha synergistically activates the hepatitis B viral enhancer II/pregenomic promoter. J Biol Chem. 1999;274(5):2858–2865. doi: 10.1074/jbc.274.5.2858. [DOI] [PubMed] [Google Scholar]
- 17.Cheong JH, Yi M, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 1995;14(1):143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haviv I, Shamay M, Doitsh G, Shaul Y. Hepatitis B virus pX targets TFIIB in transcription coactivation. Mol Cell Biol. 1998;18(3):1562–1569. doi: 10.1128/mcb.18.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haviv I, Matza Y, Shaul Y. pX, the HBV-encoded coactivator, suppresses the phenotypes of TBP and TAFII250 mutants. Genes Dev. 1998;12(8):1217–1226. doi: 10.1101/gad.12.8.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koike K, et al. Compensatory apoptosis in preneoplastic liver of a transgenic mouse model for viral hepatocarcinogenesis. Cancer Lett. 1998;134(2):181–186. doi: 10.1016/s0304-3835(98)00252-3. [DOI] [PubMed] [Google Scholar]
- 21.Shih WL, Kuo ML, Chuang SE, Cheng AL, Doong SL. Hepatitis B virus X protein inhibits transforming growth factor-beta-induced apoptosis through the activation of phosphatidylinositol 3-kinase pathway. J Biol Chem. 2000;275(33):25858–25864. doi: 10.1074/jbc.M003578200. [DOI] [PubMed] [Google Scholar]
- 22.Geng X, et al. Hepatitis B virus X protein targets the Bcl-2 protein CED-9 to induce intracellular Ca2+ increase and cell death in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2012;109(45):18465–18470. doi: 10.1073/pnas.1204652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geng X, et al. Hepatitis B virus X protein targets Bcl-2 proteins to increase intracellular calcium, required for virus replication and cell death induction. Proc Natl Acad Sci USA. 2012;109(45):18471–18476. doi: 10.1073/pnas.1204668109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petros AM, Olejniczak ET, Fesik SW. Structural biology of the Bcl-2 family of proteins. Biochim Biophys Acta. 2004;1644(2-3):83–94. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Sattler M, et al. Structure of Bcl-xL-Bak peptide complex: Recognition between regulators of apoptosis. Science. 1997;275(5302):983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 26.Cory S, Huang DC, Adams JM. The Bcl-2 family: Roles in cell survival and oncogenesis. Oncogene. 2003;22(53):8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- 27.Gong DY, et al. Role and functional domain of hepatitis B virus X protein in regulating HBV transcription and replication in vitro and in vivo. Viruses. 2013;5(5):1261–1271. doi: 10.3390/v5051261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petros AM, et al. Solution structure of the antiapoptotic protein bcl-2. Proc Natl Acad Sci USA. 2001;98(6):3012–3017. doi: 10.1073/pnas.041619798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 30.Ku B, Liang C, Jung JU, Oh BH. Evidence that inhibition of BAX activation by BCL-2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res. 2011;21(4):627–641. doi: 10.1038/cr.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Dai S, Zhu Y, Marrack P, Kappler JW. The structure of a Bcl-xL/Bim fragment complex: Implications for Bim function. Immunity. 2003;19(3):341–352. doi: 10.1016/s1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
- 32.Petros AM, et al. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 2000;9(12):2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rautureau GJ, et al. The restricted binding repertoire of Bcl-B leaves Bim as the universal BH3-only prosurvival Bcl-2 protein antagonist. Cell Death Dis. 2012;3:e443. doi: 10.1038/cddis.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herman MD, et al. Completing the family portrait of the anti-apoptotic Bcl-2 proteins: Crystal structure of human Bfl-1 in complex with Bim. FEBS Lett. 2008;582(25-26):3590–3594. doi: 10.1016/j.febslet.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 35.Tu H, et al. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 2001;61(21):7803–7810. [PubMed] [Google Scholar]
- 36.Ma NF, et al. COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin Cancer Res. 2008;14(16):5061–5068. doi: 10.1158/1078-0432.CCR-07-5082. [DOI] [PubMed] [Google Scholar]
- 37.van Delft MF, Huang DC. How the Bcl-2 family of proteins interact to regulate apoptosis. Cell Res. 2006;16(2):203–213. doi: 10.1038/sj.cr.7310028. [DOI] [PubMed] [Google Scholar]
- 38.Ye L, et al. Progressive changes in hepatoma cells stably transfected with hepatitis B virus X gene. Intervirology. 2008;51(1):50–58. doi: 10.1159/000120289. [DOI] [PubMed] [Google Scholar]
- 39.Huh KW, Siddiqui A. Characterization of the mitochondrial association of hepatitis B virus X protein, HBx. Mitochondrion. 2002;1(4):349–359. doi: 10.1016/s1567-7249(01)00040-x. [DOI] [PubMed] [Google Scholar]
- 40.Clippinger AJ, Bouchard MJ. Hepatitis B virus HBx protein localizes to mitochondria in primary rat hepatocytes and modulates mitochondrial membrane potential. J Virol. 2008;82(14):6798–6811. doi: 10.1128/JVI.00154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li T, Robert EI, van Breugel PC, Strubin M, Zheng N. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat Struct Mol Biol. 2010;17(1):105–111. doi: 10.1038/nsmb.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu D, Zou L, Li W, Wang L, Wu Y. High-level expression and large-scale preparation of soluble HBx antigen from Escherichia coli. Biotechnol Appl Biochem. 2009;54(3):141–147. doi: 10.1042/BA20090116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otwinowski ZMW. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 44.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 48.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]