Significance
Epstein–Barr Virus (EBV) Nuclear Antigen 1 (EBNA1) is essential for establishing long-term EBV infection in human cells and is implicated in EBV-related malignancies. The maintenance of the EBV genome and viral gene expression from the episome are specifically driven by EBNA1 through binding to origin of plasmid replication (oriP). This study finds that EBNA1 complexes with Ribosome Protein L4 (RPL4) and Nucleolin to stabilize EBNA1 binding to oriP. Cooperation of RPL4’s N terminus with Nucleolin K429 is necessary for EBNA1 oriP binding and episome maintenance, whereas RPL4's C-terminal K380 and K393 induce H3K4me2, which promotes EBNA1 transactivation of oriP. These discoveries enhance our understanding of the complexity of EBNA1’s interactions with host proteins and EBV episomes and reveal new targets for inhibition of EBV genome persistence.
Keywords: EBNA1, RPL4, NCL, EBV, oriP
Abstract
Epstein–Barr Virus (EBV) Nuclear Antigen 1 (EBNA1)-mediated origin of plasmid replication (oriP) DNA episome maintenance is essential for EBV-mediated tumorigenesis. We have now found that EBNA1 binds to Ribosome Protein L4 (RPL4). RPL4 shRNA knockdown decreased EBNA1 activation of an oriP luciferase reporter, EBNA1 DNA binding in lymphoblastoid cell lines, and EBV genome number per lymphoblastoid cell line. EBV infection increased RPL4 expression and redistributed RPL4 to cell nuclei. RPL4 and Nucleolin (NCL) were a scaffold for an EBNA1-induced oriP complex. The RPL4 N terminus cooperated with NCL-K429 to support EBNA1 and oriP-mediated episome binding and maintenance, whereas the NCL C-terminal K380 and K393 induced oriP DNA H3K4me2 modification and promoted EBNA1 activation of oriP-dependent transcription. These observations provide new insights into the mechanisms by which EBV uses NCL and RPL4 to establish persistent B-lymphoblastoid cell infection.
Epstein–Barr virus (EBV) was recognized as an oncogenic human pathogen after the discovery of its causal associations with B-cell lymphomas (BLs), nasopharyngeal carcinomas, and gastric carcinomas. EBV infects B-lymphocytes and epithelial cells and converts resting B cells into lymphoblastoid cell lines (LCLs). LCL maintenance requires expression of EBV nuclear antigens (EBNAs), latency-associated membrane proteins (LMPs), and noncoding RNAs (1). EBNA1 is the only EBV gene expressed in all types of EBV-infected cells and has a key role in EBV genome maintenance, replication, postmitotic EBV genome segregation, and LCL growth (1, 2). EBNA1-mediated episome maintenance depends on EBNA1 binding to the EBV origin of genome replication (oriP), which has two essential components, a dyad symmetry (DS) element and a family of repeats (FR) (3). Despite a 2.4-Å resolution crystal structure of the EBNA1 DNA binding domain bound to its cognate DNA element (4), mechanistic insights into EBNA1 and oriP-mediated episome maintenance mainly come from genetic studies using EBV recombinants and biochemical studies of EBNA1’s association with cell proteins, including CTCF, Bromodomain Protein 4 (BRD4), Nucleosome Assembly Protein 1 (NAP1), the cell Origin Recognition Complex, and the Mini Chromosome Maintenance complex (5–8). Recent studies indicate that EBNA1 may use complex strategies for episome maintenance (9–16).
EBNA1-associated ribosome biogenesis factors Nucleophosmin (NPM1) and Nucleolin (NCL) have been implicated in EBNA1 and oriP-dependent functions (17, 18). Other viruses also use ribosomal proteins (RPs), such as RPL4, RPS19, and RPL40, to enhance virus protein translation (19–21). Indeed “extraribosomal functions” of RPs were discovered through RPS1 involvement in bacteriophage Qβ-mediated genome replication (22). Moreover, the noncoding EBV RNA, EBER1, causes RPL22 redistribution from the nucleolus to the nucleoplasm and stimulates cell proliferation (23, 24). We have now found complex protein interactions among EBNA1, RPL4, and NCL and have examined the role of these interactions in EBNA1 and oriP-mediated episome maintenance and transcription activation (25).
Results
EBNA1 Binds to RPL4.
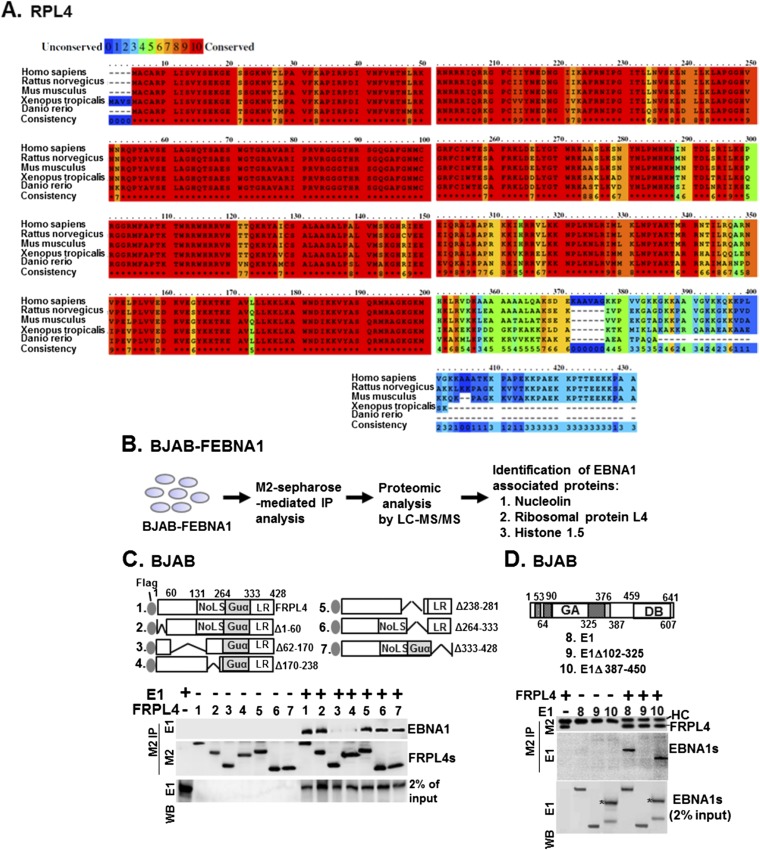
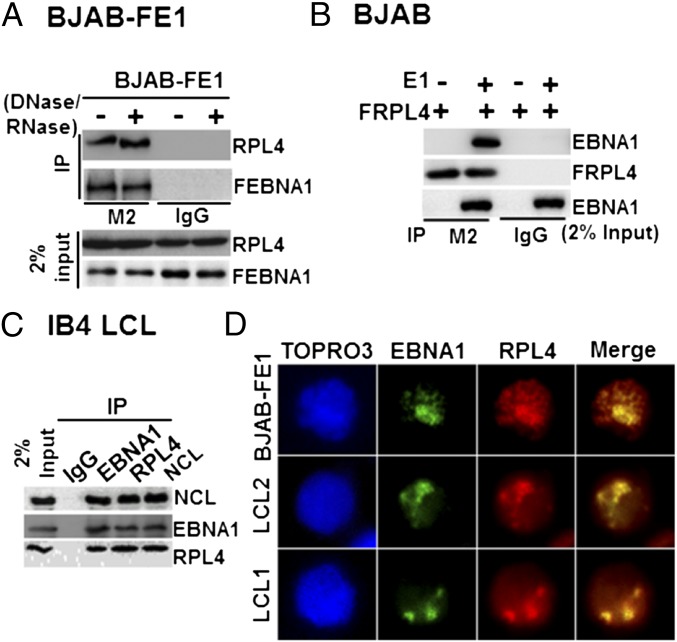
RPL4 is highly conserved in all domains of life. Eukaryotic RPL4s have an extended C terminus (26) (Fig. S1A). RPL4 was identified as a FLAG-EBNA1 (FEBNA1)-associated protein in BJAB B-lymphoma cells (BJAB-FEBNA1) by immune precipitation, liquid chromatography, and tandem mass spectrometry (LC-MS/MS) (18) (Fig. S1B). Coimmunoprecipitation (co-IP) assays were used to verify an interaction of FEBNA1 with RPL4 in BJAB-FEBNA1 cells. FEBNA1 bound RPL4, and the association was not affected by nuclease treatment (Fig. 1A). Similarly, EBNA1 was efficiently precipitated by FLAG RPL4 (FRPL4) from BJAB cells transiently transfected with FRPL4 expression plasmids (Fig. 1B). Using IB4 LCLs, EBNA1, NCL, and RPL4 could precipitate each other but not an IgG negative control (Fig. 1C). Confocal immunofluorescence microscopy visualized the nuclear colocalization of EBNA1 with RPL4 in two independent LCLs, LCL1 and LCL2, and of FEBNA1 with RPL4 in BJAB-FEBNA1 cells (Fig. 1D).
Fig. S1.
Mapping of the protein–protein interacting domain of EBNA1 and RPL4. (A) Sequence alignments of RPL4 from human, rat, mice, Xenopus, and zebra fish were shown. The degrees of related amino acid sequence conservation were indicated by numbers 0 (none) to 10 (high). (B) The flowchart of the previously described proteomic procedure for identification of EBNA1-associated proteins by LC/MS-MS (18). Both NCL and RPL4 were found to be EBNA1-associated. (C) Transfection-mediated co-IP assays were used to identify the EBNA1 binding domain of RPL4 using plasmids of FRPL4 or each of its mutant derivatives, with E1 cotransfection. We loaded 2% input of EBNA1. (D) The reciprocal experiment of C was conducted using plasmids of E1 or the indicated deletion mutants with cotransfected FRPL4. The corresponding band produced by E1 387–450 was marked with an asterisk. HC, heavy chain.
Fig. 1.
RPL4 binds to EBNA1. (A) BJAB-FEBNA1 stable cells were lyzed and treated with DNase/RNase (+) or left untreated. FEBNA1 was immune-precipitated with anti-FLAG antibody. RPL4 was detected by Western blotting. Also shown was 2% of input. (B) BJAB cells were transiently transfected with FLAG-tagged RPL4 alone or with EBNA1. FRPL4 was immune-precipitated with anti-FLAG antibody. EBNA1 was detected by Western blotting. (C) IB4 LCL lysates were immune-precipitated with antibodies against EBNA1, RPL4, and NCL, with IgG as a negative control. The precipitated proteins were blotted with the indicated antibodies, and 2% input was shown. (D) EBNA1 and RPL4 immuofluorescent staining in LCL1, LCL2, and BJAB-FEBNA1 stable cells. EBNA1 was visualized with a FITC-conjugated secondary antibody, whereas RPL4 was visualized by a rhodamine-conjugated secondary antibody.
To identify the RPL4 domains that bind to EBNA1, we transiently expressed FRPL4 deletion mutants with EBNA1 in BJAB cells. All FRPL4 deletion mutants were expressed at similar levels and precipitated with anti-FLAG antibody conjugated to agarose beads. Full-length FRPL4 and deletion mutants Δ1–60, Δ238–281, Δ264–333, and Δ333–428 precipitated ∼2–5% of input EBNA1. In contrast, FRPL4 deletion mutants Δ62–170 and Δ170–238 failed to precipitate EBNA1 (Fig. S1C). Reciprocal experiments found FRPL4 efficiently precipitated full-length EBNA1 or EBNA1 deletion mutant Δ387–450 but failed to precipitate EBNA1 Δ102–325, even though these EBNA1 proteins were expressed at similar levels (Fig. S1D). These data indicate that EBNA1 aa 102–325 and RPL4 aa 62–238 are essential for stable interaction.
RPL4 Is Essential for EBNA1 Transcription Activation, EBNA1 Binding to oriP, and EBV Genome Maintenance.
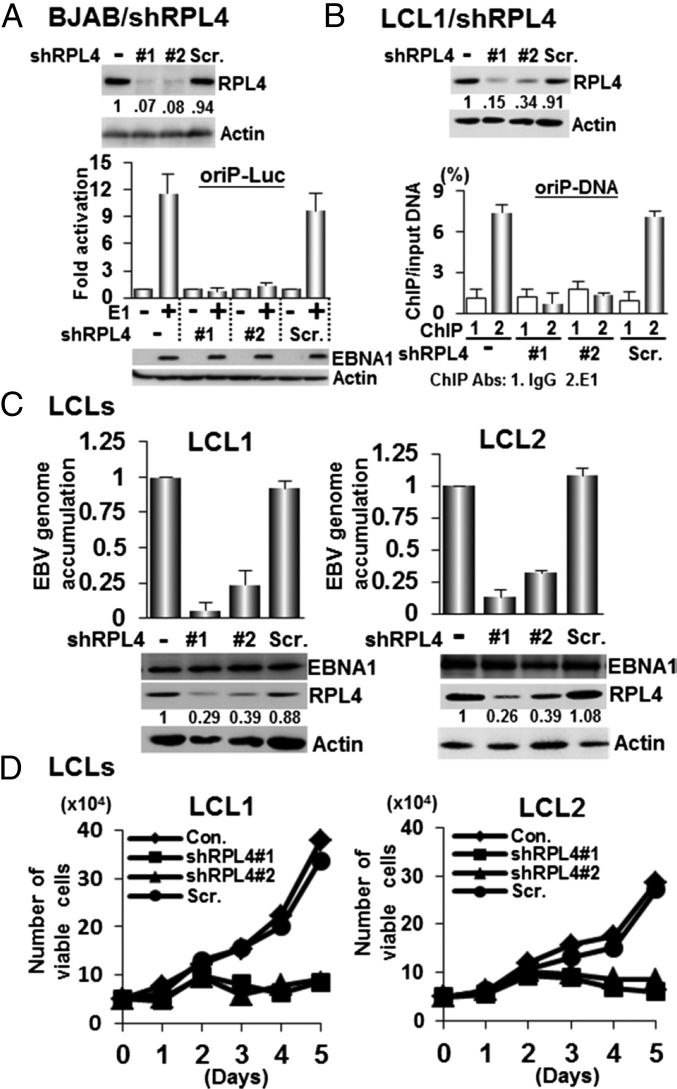
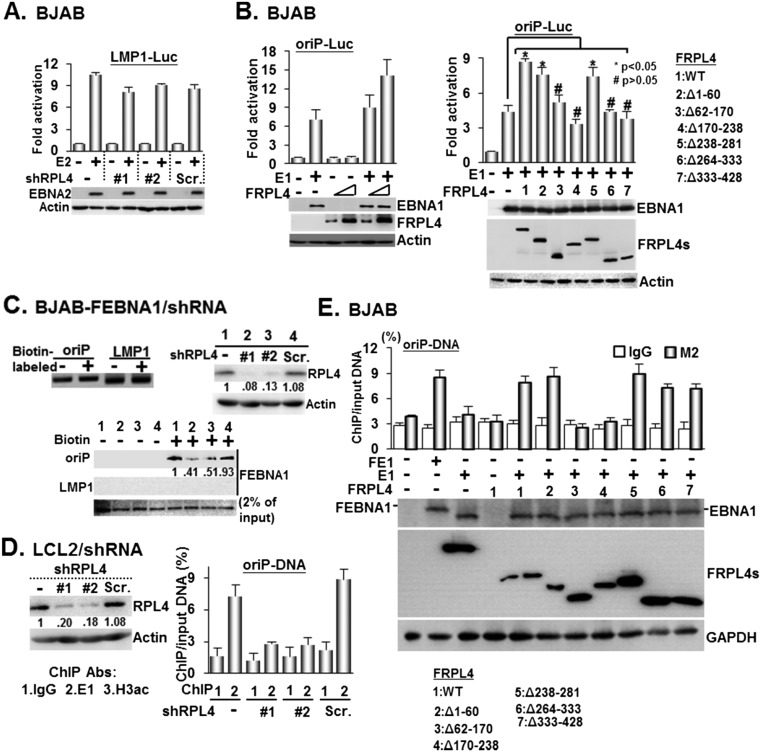
In BJAB cells, Lentivirus-expressed shRPL4 #1 or #2 targeting the RPL4 3′ untranslated region (UTR) knocked down RPL4 more than >90%, whereas a scrambled control shRNA (Scr.) did not affect RPL4 expression (Fig. 2A). These cells were transiently transfected with an EBNA1 expression plasmid together with an oriP luciferase reporter, oriP-SV40-Luc (oriP-Luc) (18). Luciferase activities were determined 24 h after transfection. EBNA1 activated oriP Luciferase ∼10-fold in control shRNA-treated cells. In contrast, EBNA1 did not activate oriP luciferase in RPL4 knockdown cells. The RPL4 knockdown did not affect EBNA2 activation of an LMP1-Luc reporter plasmid, indicative of an EBNA1-specific effect (Fig. S2A). FRPL4 overexpression increased EBNA1 activation of oriP-Luc by ∼twofold in BJAB cells (Fig. S2B). Interestingly, overexpression of FRPL4 deletion mutants Δ62–170, Δ170–238, Δ264–333, and Δ333–428 did not increase EBNA1 activation of oriP-Luc (Figs. S1C and S2B). These data indicate that RPL4 aa 62–238 and 264–428 are important for RPL4 enhancement of EBNA1 activation of oriP transcription.
Fig. 2.
RPL4 is essential for EBNA1 transcription activation and binding of oriP and LCL growth. (A) BJAB cells were transduced with lentiviruses expressing short hairpins targeting RPL4, shRPL4 #1 or shRPL4 #2, or Scr.. RPL4 depletion was shown by Western blot. These cells were then transfected with EBNA1 expression plasmid and oriP luciferase reporter plasmid and used to assay for EBNA1-dependent transcription of the oriP-Luc reporter plasmid. Luciferase activities were represented as mean ± SD from three independent experiments here and all of the following transfection luciferase reporter assays described elsewhere in this article. (B) RPL4 was depleted from LCL1 with shRNA. Antibodies against EBNA1 or control were used in ChIP assays to precipitate oriP DNA from LCLs. oriP DNA precipitation was quantitated by qPCR and presented as % of input DNA (mean ± SD) from three independent experiments and for all following ChIP experiments. (C) EBV copy number was determined by qPCR in LCL depleted for RPL4 or shRNA control, with untreated LCL set to 1. DNA was normalized to GAPDH genomic DNA. (D) Cells were counted for 5 d after shRNA knockdown. Dead cells were excluded by trypan blue staining.
Fig. S2.
Enhancement of EBNA1/oriP-Luc–dependent transcription requires RPL4/EBNA1 interactions. (A) BJAB cells transduced with shRPL4 #1 or shRPL4 #2 or Scr. were used to assay for EBNA2-dependent transcription of the LMP1-Luc reporter plasmid. The immune blots for EBNA2 and actin internal control were shown. (B) BJAB cells were cotransfected with the expression plasmids of E1, FRPL4, or FRPL4 deletion mutants (shown in Fig. S1C) and an oriP-Luc reporter plasmid and internal control CMV–β-Gal. Protein expression resulting from the indicated transfected plasmids and actin internal control were determined by Western blot. Resulting activities were presented as mean ± SD for data from three experiments. *P value < 0.05; #P > 0.05 value versus control. (C) Cell lysates derived from BJAB-FE1 expressing shRPL4 #1, shRPL4 #2, or Scr. were subjected to streptavidin agarose-mediated pulldown assays using biotin-labeled and nonlabeled oriP DNA or both types of LMP1-DNA. The DNA samples used in this assay are shown (Left). The captured proteins bound to precipitated DNA were quantified by Western blot with 2% input of FEBNA1 (Right). (D) LCL2 cells transduced with shRPL4 (#1 or #2) or Scr. were subjected to perform ChIP assays using the indicated antibodies. The amount of ChIPed oriP-EBV DNA was determined by qPCR and calculated as % of input DNA. (E) BJAB cells cotransfected with E1 or FEBNA1 plasmid (FE1) and the indicated FRPL4 expression vector and the oriP DNA-containing plasmid were subjected to perform ChIP assays using the indicated antibodies. The amount of ChIPed DNA was determined as % of input DNA for the data obtained from each experiment.
Streptavidin agarose DNA pulldown assays were used to further investigate the role of RPL4 in EBNA1 and oriP DNA binding (Fig. S2C). Biotin-labeled oriP DNA efficiently precipitated EBNA1 from BJAB-FEBNA1 stable cells and BJAB-FEBNA1 stable cells expressing scrambled control shRNA, whereas biotin-labeled LMP1 promoter control DNA did not bind EBNA1. RPL4 shRNA knockdown in BJAB-FEBNA1 stable cells reduced EBNA1 binding to oriP DNA by 50–60%.
Chromatin immunoprecipitation (ChIP) followed by quantitative PCR (ChIP-qPCR) was used to evaluate the effect of RPL4 knockdown on EBNA1 binding to oriP in LCLs. shRPL4 #1 and #2 efficiently knocked down RPL4 in LCLs, whereas control shRNA had no effect. Anti-EBNA1 antibody efficiently precipitated oriP DNA from normal or control shRNA-treated LCLs. RPL4 knockdown abolished most EBNA1 binding to oriP DNA in two different LCLs (Fig. 2B and Fig. S2D).
To evaluate if EBNA1 can recruit RPL4 to oriP DNA, FRPL4 was coexpressed with EBNA1 in BJAB cells with oriP-Luc plasmid. Anti-FLAG antibody was used to precipitate FRPL4 together with oriP DNA in ChIP-qPCR (Fig. S2E). FRPL4 only bound to oriP DNA when EBNA1 was coexpressed. FRPL4 deletion mutants were also tested for their abilities to bind to oriP in the presence of EBNA1. FRPL4 Δ1–60, Δ238–281, Δ264–333, and Δ333–428 bound oriP DNA, whereas FRPL4 EBNA1-binding null mutants Δ62–170 and Δ170–238 did not bind oriP DNA. These data indicate that EBNA1 can recruit RPL4 to oriP DNA.
RPL4 effects on EBNA1 binding to oriP DNA likely enable EBV episome maintenance and LCL survival. shRPL4 #1 and #2 depleted RPL4 60–70% in two independent LCLs (Fig. 2C). RPL4-depleted LCL1 and LCL2 had 70–90% reduced EBV genome copies and 70% reduced cell viability, compared with a control shRNA (Fig. 2D).
EBV Infection Increased RPL4 Expression and Nuclear Localization.
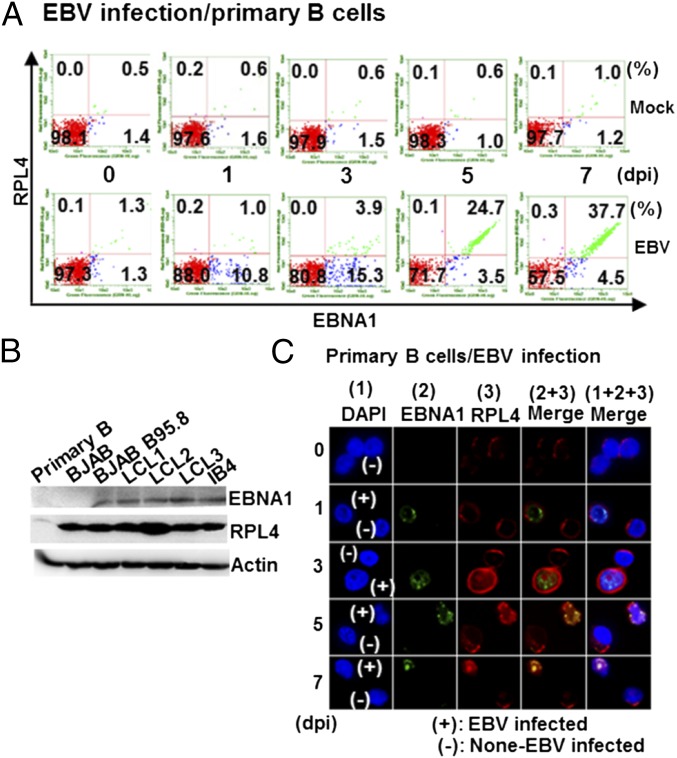
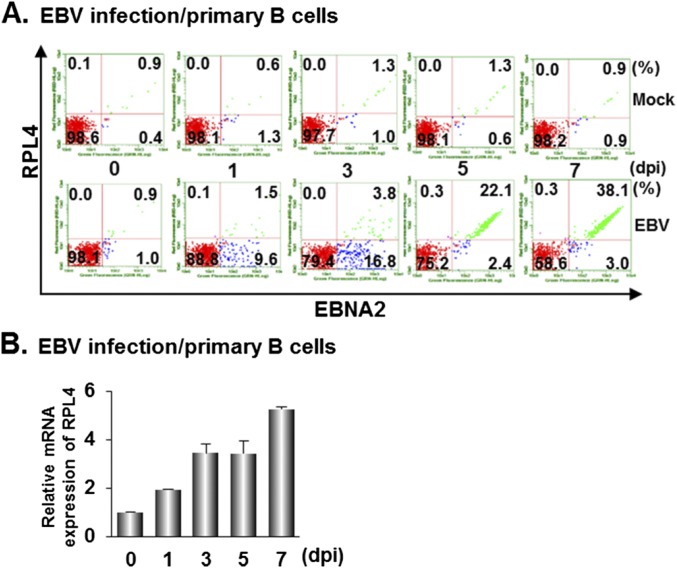
RPL4’s role in ribosome assembly links RPL4 to cell proliferation (27). Indeed, uninfected resting B lymphocytes express RPL4 at low levels. EBV infection significantly increased RPL4 expression over 7 d (Fig. 3A and Fig. S3A). EBNA1 and EBNA2 expression also increased, as did RPL4 mRNA levels, which increased by two-, 3.5-, and fivefold at 1, 3, and 7 d postinfection (dpi) (Fig. S3B). RPL4 expression levels in proliferating BJAB B-lymphoma cells, EBV-infected BJAB cells, and LCLs were remarkably higher than in resting B cells (Fig. 3B).
Fig. 3.
EBV infection induces RPL4 expression and nuclear relocalization in B cells. (A) Resting primary B lymphocytes were infected with EBV. The expression of RPL4 and EBNA1 was determined by flow cytometry at 0, 1, 3, 5, and 7 dpi. (B) The expression levels of RPL4, EBNA1, or actin internal control in EBV-associated B-linage cells were determined by Western blotting. (C) Resting primary B lymphocytes were infected with EBV. Confocal microscopy was used to evaluate the EBNA1 and RPL4 expression and localization in EBV-infected cells. The nuclei were indicated by DAPI staining (blue). The individual or merge images were shown. EBNA1 was shown in green, and RPL4 was shown in red. Cells infected with EBV were indicated as (+), whereas uninfected cells were indicated as (–).
Fig. S3.
EBV infection induced RPL4 expression at both protein and RNA levels. (A) B cells with EBV infection were used to monitor the expressing patterns of EBNA2 and RPL4 by flow cytometry at 0, 1, 3, 5, and 7 dpi after a double immunostaining procedure of RPL4/EBNA2. EBNA2-positive cells were used as the surrogate marker for EBV-infected B cells. (B) Same as in A, but cells were used for total RNA isolation followed by RT-qPCR. The abundance of RPL4 mRNA obtained for each sample was corrected by the corresponding GAPDH mRNA value.
With EBV infection, RPL4 immune fluorescence signals substantially increased in EBV-infected versus noninfected cells (Fig. 3C). EBV-infected cells expressed increasingly higher levels of EBNA1 and RPL4 from 1 to 3 dpi. RPL4 subsequently redistributed from the cytoplasm to the nucleus and partially colocalized with EBNA1 in nuclei of EBV-infected cells by 5 dpi, whereas LCL-like nuclear colocalization was apparent by 7 dpi (Figs. 1D and 3C). These data indicate that RPL4 relocalizes to the nucleus following EBV infection.
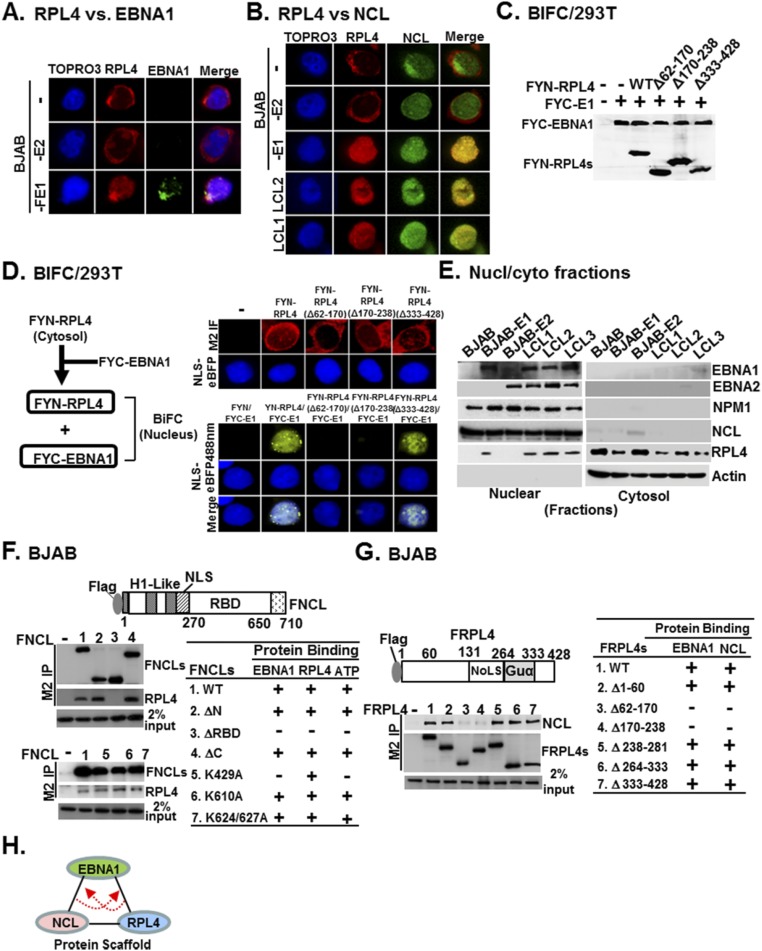
The cytosolic-dominant distribution of RPL4 in BJAB or BJAB-E2 differed from its nuclear localization in BJAB-FE1 or LCL cells (Fig. S4 A and B). RPL4 and NCL also colocalized in LCLs and BJAB-FE1 cells (Fig. S4B). Bimolecular Fluorescence Complementation (BiFC) assays were used to further evaluate the nuclear colocalization of EBNA1 and RPL4. YFP C terminus fused with a flag epitope (FYC)-RPL4 and its deletion mutants were expressed at levels similar to endogenous RPL4 (Fig. S4 C and D). When FYN (YFP N terminus fused with a flag epitope)-EBNA1 was cotransfected, FYC-RPL4 or Δ333–428 elicited nuclear BiFC signals. RPL4 EBNA1-binding null mutants FYC-RPL4 Δ62–170 and Δ170–238 did not elicit BiFC signals (Fig. S4D) (18). Nuclear and cytoplasm fractionation followed by Western blotting were also used to determine the RPL4 nuclear localization. Nearly equal RPL4 levels were detected in both nuclear and cytosolic fractions of BJAB-E1 or LCLs, whereas RPL4 was only detected in cytosolic fractions of BJAB or BJAB-E2 cells (Fig. S4E). These findings indicate that EBNA1 determines RPL4’s nuclear localization.
Fig. S4.
EBNA1 coordinates with NCL to promote RPL4 redistribution. (A) The indicated BJAB derivative cell lines were used for EBNA1/RPL4 double immunostaining. TO-PRO3 was used for nuclear counterstaining. The individual or merge confocal fluorescent images were shown. (B) Same as in A but two LCLs, LCL1 and LCL2, were additionally included and RPL4/NCL double immunostaining was performed. (C and D) 293T cells were cotransfected with the indicated BiFC plasmids, including FYC-EBNA1 (FYC-E1), FYN-RPL4, FYN-RPL4 mutant derivatives, and nuclear marker NLS-eBFP plasmid followed by immunofluorescence (D, Upper Right) or BiFC (D, Bottom Right) analysis. The cellular localization of FRPL4 or each derivative mutant was shown by M2 immunofluorescence, whereas the BiFC images were visualized by confocal fluorescent microscopy. The expression level of each corresponding protein was identified by M2 blot (C). The flowchart of BiFC is shown (D, Left). (E) Both cytosolic and nuclear fractions were extracted from EBV-associated B lineage cell lines and subjected to Western blot. The immune blots for EBNA1, EBNA2, NPM1, NCL, RPL4, and actin were shown. EBNA1, EBNA2, NPM1, and NCL are known nuclear proteins, whereas actin is a cytosolic marker. (F) BJAB cells transfected with the indicated FNCL derivative plasmids were subjected to M2-Sepharose–mediated IP, and the precipitated matrix was analyzed by Western blot using M2 or RPL4 antibody. We loaded 2% input of RPL4. The resulting phenotype of e33ach FNCL derivatives was compared with their previously identified functional profiles (18). The diagram showing that RPL4 or NCL alone could act as a scaffold protein to stabilize the association of EBNA1 with one another through their specific interactions with EBNA1. (G) The reciprocal experiment of F was conducted using seven FRPL4 derivative plasmids. In this case, M2 and NCL blots were shown with 2% input of NCL. The capability of each FRPL4 derivative to associate with EBNA1 or NCL was shown. (H) Diagram showing that RPL4 or NCL alone could act as a scaffold protein to stabilize the association of EBNA1 with one another through its specific interactions with EBNA1.
RPL4 and NCL Complexes Enhance EBNA1 oriP DNA Binding.
To further define the interactions between RPL4 and NCL, FNCL and FNCL deletion mutants were expressed in BJAB cells. Anti-FLAG antibodies immune-precipitated FNCL together with RPL4. FNCL mutants deleted for their N or C termini precipitated RPL4, whereas FNCL mutants deleted for the NCL RNA binding domain (ΔRBD) did not bind to RPL4 (Fig. S4F). Reciprocal immune precipitation revealed FRPL4 EBNA1-binding null mutants Δ62–170 and Δ170–238 failed to associate with endogenous NCL (Fig. S4G). These data indicate that RPL4, NCL, and EBNA1 are closely associated in LCLs (Fig. S4H).
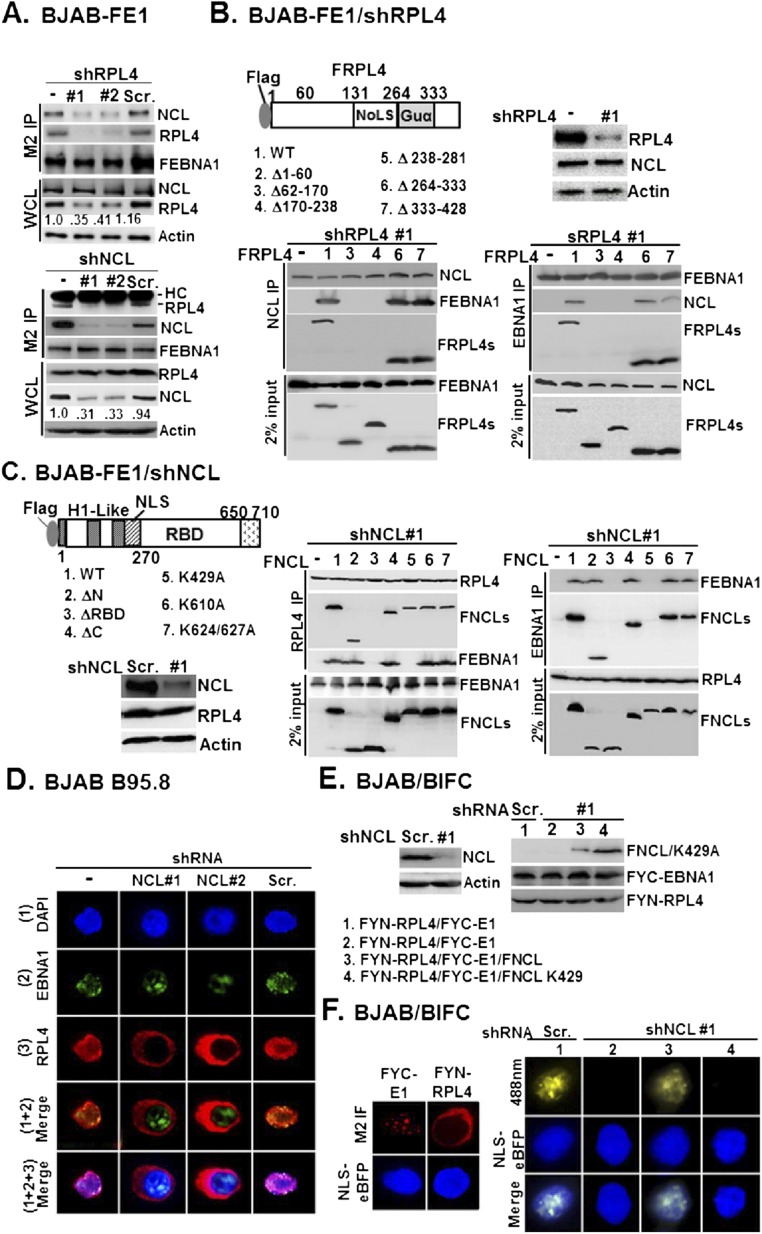
In BJAB-FEBNA1 cells, depletion of RPL4 led to decreased FEBNA1 and NCL association, and NCL knockdown also decreased RPL4 and FEBNA1 association (Fig. S5A), suggesting that the formation of a ternary complex may stabilize the protein–protein interactions. Moreover, loss of FEBNA1/NCL association in RPL4-depleted BJAB-FEBNA1 cells was readily restored by FRPL4 rescue with wild type and FRPL4Δ264–333 or Δ333–428 deletion mutants, whereas FRPL4 Δ62–170 and Δ170–238 mutants, which do not bind to EBNA1, failed to rescue RPL4 depletion (Fig. S5B). The reciprocal experiments showed that loss of FEBNA1/RPL4 association in NCL-depleted BJAB-FEBNA1 cells was readily restored by NCL rescue with wild type and FNCL ΔN, ΔC, K610A, and K624/627A mutants, whereas the EBNA1-binding mutants (18) ΔRBD and K429A failed to rescue NCL depletion (Fig. S5C). Furthermore, NCL depletion also reduced RPL4 nuclear localization (Fig. S5D). In NCL-depleted cells, EBNA1 and RPL4 did not colocalize in BiFC assays. Rescue by wild-type NCL but not the NCL K429A mutant, an ATP-binding null mutant, restored the EBNA1 and RPL4 nuclear colocalization (Fig. S5 E and F). These data indicate that EBNA1-mediated RPL4 nuclear relocalization requires NCL-K429 (Table S1).
Fig. S5.
RPL4/NCL acts as a protein scaffold for EBNA1. (A) BJAB/FE1/shRL4 #1 or #2 and BJAB/FE1/Scr. control were subjected to M2 IP analysis followed by immune blotting with antibodies for NCL, RPL4, and M2 (Upper). Similarly, BJAB/FE1/shNCL #1 or #2 and BJAB/FE1/Scr. control were used to carry out another set of experiments (Bottom). In both cases, whole-cell lysates were blotted with NCL, RPL4, and internal control actin. (B) The loss of FEBNA1/NCL association in BJAB-FE1/shRPL4 #1 was rescued by transfected FRPL4 or its mutant derivatives followed by co-IP and Western blot analyses. Immune blots for the indicated proteins from NCL and EBNA1-precipitated matrices are shown. Also shown is 2% input of FEBNA and FRPL4 or NCL and FRPL4 from each set of experiments. (C) Same as in B, except BJAB-FE1/shNCL #1- and FNCL-related plasmids were used. Immune blots for the indicated proteins from RPL4 (Middle) and EBNA1 (Right)-precipitated matrices were shown with 2% input of loading control. (D) NCL knockdown by shNCL #1 or shNCL #2 in the context of EBV-positive BJAB B95.8 reversed RPL4 redistribution, whereas Scr. had no effect. EBNA1/RPL4 double immunostaining was carried out, and nuclei were counterstained with DAPI. (E) BJAB cells were cotransfected with FYC-E1 and NLS-eBFP, or FYN-RPL4 and NLS-eBFP. (F) The cellular localization of FYC-E1 and FYN-RPL4 was represented as M2 immunofluorescence images (red), whereas the nuclear regions were marked by NLS-eBFP (blue). BJAB/shNCL #1 cells (columns 2–4) were cotransfected with FYN-RPL4 and FYC-E1, or FNCL and FNCL K429A. The resulting BiFC image in each transfectant was visualized by confocal fluorescent microscopy. The BiFC produced by transfected FYN-RPL4/FYC-E1 in Scr.-transduced BJAB (column 1) was shown.
Table S1.
Summary of the functional features of FNCL or its mutant derivatives in supporting protein–protein interactions and RPL4 redistribution
| FNCLs | Affinity binding | The ability to stabilize RPL4/EBNA1 interaction | The ability to escort RPL4/EBNA1 shuttle to the nucleus | ||
| EBNA1 | RPL4 | ATP | |||
| 1.WT | + | + | + | Y | Y |
| 2. ΔN | + | + | + | Y | ND |
| 3. ΔRBD | – | – | – | N | ND |
| 4. ΔC | + | + | + | Y | ND |
| 5. K429A | – | + | – | N | N |
| 6. K610A | + | + | + | Y | ND |
| 7. K624/627A | + | + | + | Y | ND |
N, no; ND, not done; Y, yes.
Decreased EBNA1 Activation of oriP Luciferase by RPL4 Depletion Can Be Restored by Increased RPL4 Expression, and Both RPL4 N and C Termini Are Important for EBNA1 and oriP Functionality.
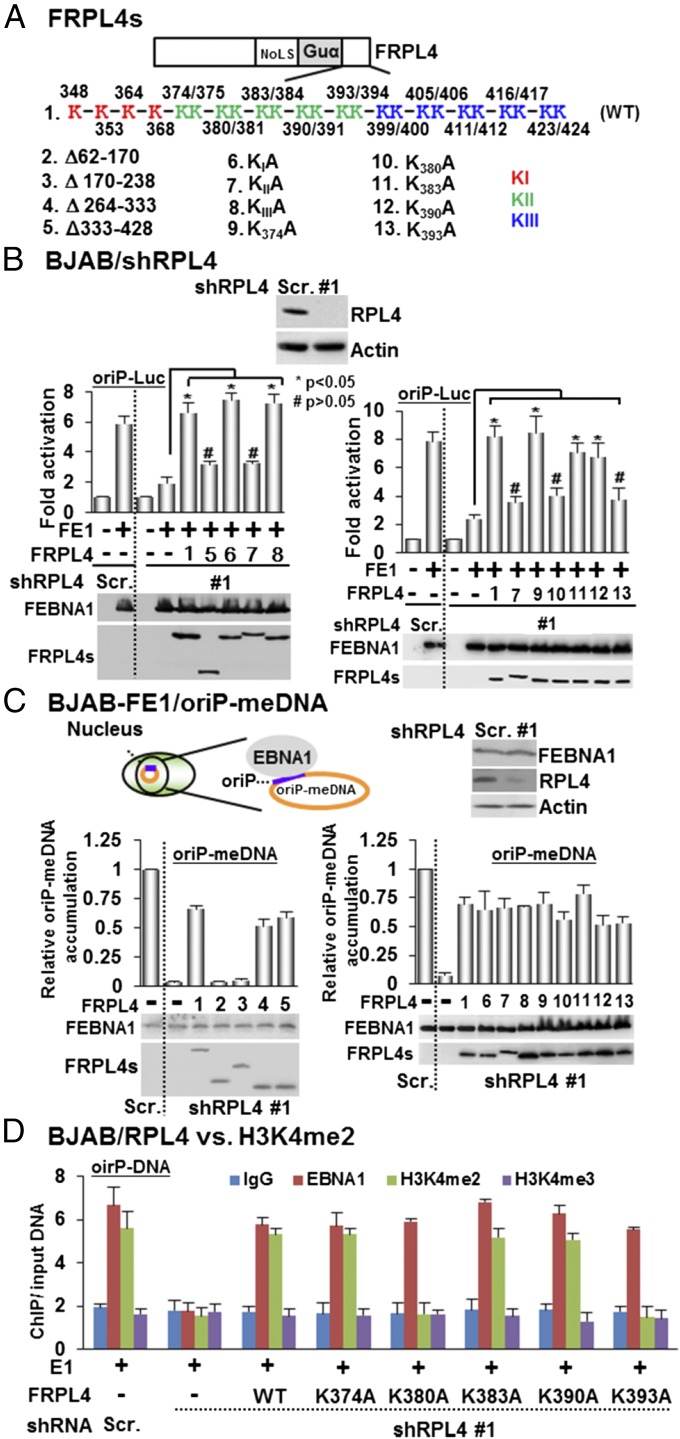
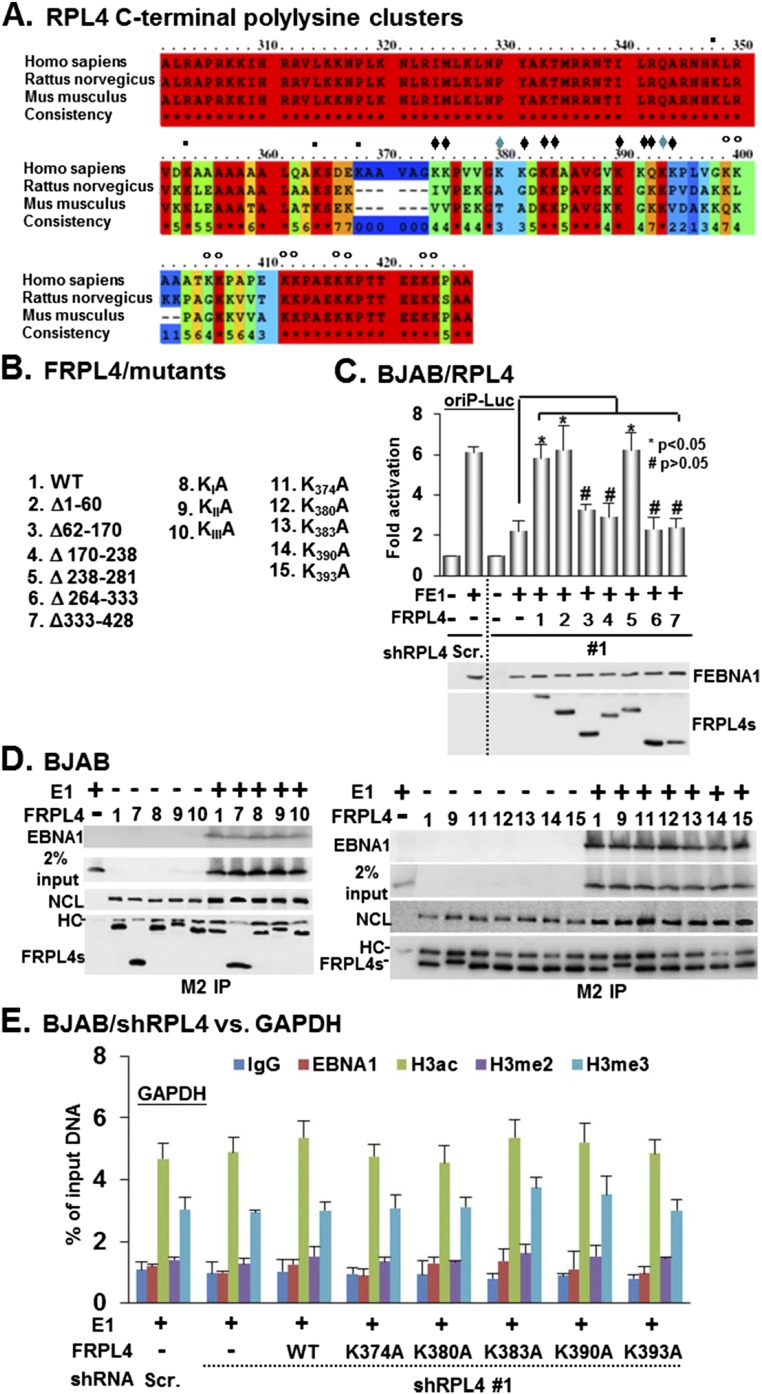
Rescue by the expression of a cDNA that is not targeted by an shRNA can establish a direct causal relationship of the knockdown and its associated phenotype. FLAG-tagged RPL4 full-length or deletion mutant expression plasmids lacking RPL4 3′ UTR were used to rescue RPL4 depletion by shRNA targeting the RPL4 3′ UTR (Fig. 4A). In RPL4-depleted cells, transfected full-length FRPL4 readily restored EBNA1 activation of oriP luciferase activity (Fig. 4B). Among FRPL4 deletion mutants, only FRPL4 Δ1–60 or Δ238–281 rescued EBNA1 activation of oriP luciferase, whereas deletion mutants Δ62–170, Δ170–238, Δ264–333, and Δ333–428 failed to rescue (Fig. 4B and Fig. S6 B and C). The RPL4 C-terminal region aa 333–428 are well conserved among Homo sapiens, Rattus norvegicus, and Mus musculus, but not in prokaryotes (26) (Figs. S1A and S6A). There are 24 lysines in this domain. The contribution of these residues to EBNA1-mediated oriP activation was evaluated. Four lysine residues between aa 333 and 369 (KIA), 10 lysine residues between aa 370 and 400 (KIIA), and 10 lysine residues between aa 401 and 428 (KIIIA) were mutated to alanine residues. Their abilities to rescue EBNA1 activation of oriP luciferase were tested. FRPL4 lysine point mutants KIA or KIIIA but not KIIA orΔ333–428 rescued EBNA1 activation (Fig. 4B). Individual lysine point mutants in KIIA were generated, and their abilities to rescue EBNA1 activation of oriP were tested. FRPL4K374A, K383A, and K390A restored full EBNA1 activity in RPL4-depleted BJAB cells, whereas K380A and K393A failed to restore EBNA1 activity. These data indicate that the RPL4 C-terminal polylysine KII cluster is essential for EBNA1 activation of oriP-dependent transcription (Fig. 4B).
Fig. 4.
The N terminus of RPL4 is involved in EBNA1 and oriP-dependent episomal DNA maintenance, whereas the C-terminal K383 and K390 have a specific role in EBNA1 and oriP-mediated transcription. (A) Schematic diagrams of FRPL4 and its deletion or point mutants. (B) RPL4 was depleted in BJAB cells by shRPL4 #1. FLAG-tagged EBNA1 and oriP luciferase reporter plasmids were cotrasfected with FLAG-tagged wild-type RPL4 or its deletion and point mutants. shRPL4 knockdown greatly reduced EBNA1 activation of oriP luciferase. Wild-type RPL4 rescued RPL4 shRNA knockdown effect and restored EBNA1 activation of oriP luciferase. Protein expression was determined by Western blotting. *P < 0.05; #P > 0.05 versus control. (C) shRPL4 was used to deplete PRL4 in BJAB cells stably expressing FEBNA1 and maintain mini-EBV episome DNA (oriP-meDNA) (28). qPCR was used to determine the mini-EBV episome copy number. RPL4 knockdown greatly reduced the mini-EBV episome copy number, whereas FLAG tagged wild-type RPL4 rescue restored the mini-EBV episome copy number. RPL4 deletion and point mutants were also tested for their effects. GAPDH genomic DNA was used to normalize the qPCR. oriP DNA in Scramble control cells was set to 1. The FEBNA1 and FRPL4 expressions were determined by Western blotting. (D) BJAB cells depleted for RBL4 expression by shRPL4 #1 or control cells were transfected with EBNA1 and oriP plasmids. Wild-type RPL4 or RPL4 lysine mutants were also cotransfected to rescue RPL4 knockdown. Antibodies against EBNA1, H3K4me2, H3K4me3, or control were used in ChIP assays to assess the oriP EBNA1 binding and histone modifications. oriP fold enrichment over input DNA was determined by qPCR.
Fig. S6.
RPL4 C terminus is not required for either EBNA1 or NCL association. (A) Sequence alignments of RPL4 C terminus from human, rat, and mice were shown. The polylysine clusters were indicated by ▪KI, ♦KII, and °KIII, respectively. (B) The schematic diagrams of FRPL4 and its mutant derivatives are shown. (C) BJAB/shRPL4 #1 cells were used to perform transfection-mediated complementation of EBNA1/oriP-Luc–dependent transcription using the indicated FRPL4 plasmids. Protein expression and actin internal control were determined by Western blot. *P < 0.05; #P > 0.05 versus control. (D) BJAB cells were cotransfected with the plasmids of EBNA1 (E1) and FRPL4 or its mutant derivatives and subjected to a M2 IP protocol. FRPL4-precipitated EBNA1 or NCL was determined by Western blot. We used 2% input of EBNA1 as a loading control. (E) RPL4 depletion did not affect the enrichment of histone modifications at the GAPDH promoter. BJAB/shRPL4 #1 or BJAB/Scr. cells were cotransfected with EBNA1 and the indicated FRPL4 plasmids. Each transfectant was subjected to ChIP assays using IgG or antibodies for EBNA1, H3ac, H3K4me2, and H3K4me3. The amount of ChIPed GAPDH promoter DNA was quantified by qPCR and determined as % of input DNA.
To further define RPL4 domains needed for episome maintenance, we evaluated the effects of RPL4 deletions on the persistence of a mini-EBV episome that depends on EBNA1 in a BJAB-FEBNA1/oriP-Luc stable cell line (28). RPL4 depletion reduced the mini-EBV episome copy number by >90% (Fig. 4C). In RPL4-depleted cells, transfected FRPL4 Δ264–333 or Δ333–428 restored 75% of the mini EBV episome copy number. In contrast, neither transfected FRPL4 Δ62–170 nor Δ170–238 rescued the mini EBV episome. The RPL4 C terminus was dispensable for EBNA1 association (Fig. S1C), and expression of each polylysine mutant rescued 75% of the mini EBV episome copy number (Fig. 4C, Fig. S6D, and Table S2). These data indicate that both the RPL4 N and C termini are important for oriP-mediated EBV episome maintenance and EBNA1 functionality.
Table S2.
Summary of the functional features of FRPL4 or its mutant derivatives in supporting protein–protein interactions and EBNA1/oriP-dependent functions
| FRPL4s | Affinity binding | Complementation for EBNA1-dependent | Recruitment of H3K4me2 | ||
| EBNA1 | NCL | oriP DNA maintenance | Transcription | ||
| WT | + | + | Y | Y | + |
| Δ1–60 | + | + | Y | Y | ND |
| Δ62–170 | – | – | N | N | ND |
| Δ170–238 | – | – | N | N | ND |
| Δ238–281 | + | + | Y | Y | ND |
| Δ264–333 | + | + | Y | Y | ND |
| Δ333–428 | + | + | Y | N | ND |
| KIA | + | + | Y | Y | ND |
| KIIA | + | + | Y | N | ND |
| KIIIA | + | + | Y | Y | ND |
| K374A | + | + | Y | Y | + |
| K380A | + | + | Y | N | – |
| K383A | + | + | Y | Y | + |
| K390A | + | + | Y | Y | + |
| K393A | + | + | Y | N | – |
N, no; ND, not done; Y, yes.
RPL4 K383 and K393 Are Required for oriP DNA H3K4me2 Modification.
H3K4me2 modifications are associated with oriP, whereas H3K4me3 and H3ac are linked to other EBNA1 binding sites (10). RPL4 depletion diminished H3K4me2 at oriP, whereas wild-type RPL4 restored H3K4me2 at oriP and RPL4 K380 and K393A failed to restore H3K4me2 at oriP (Fig. 4D). Parallel control experiments found H3K4me3 was not affected. Also, RPL4 depletion did not affect the overall abundance of these marks at the GAPDH promoter (Fig. S6E). These data indicate that RPL4 K380 and K393 are critical for oriP H3K4me2 modifications, which enable EBNA1-mediated transcription.
Discussion
Ribosome and nucleolar proteins have recently been shown to have important roles in oncogenesis through their interactions with transcription machinery (12, 29–32). RPL4 can bind to the 5′ regulatory region of MYC through cMYB (33), and NCL can bind to sites near the CCND1 gene in mantle cell lymphomas to activate CCND1 expression (34). In this study, we found that EBV exploits extra ribosomal RPL4 functions to establish latent EBV infection.
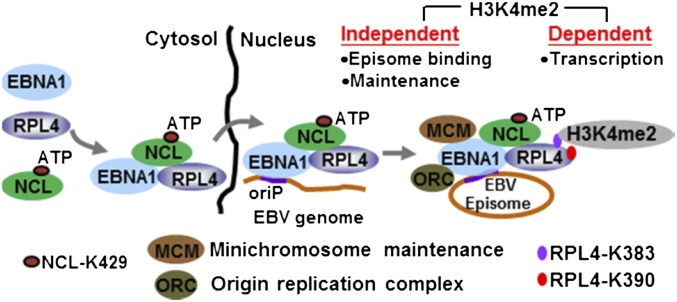
We propose a model based on our findings (Fig. S7) that (i) EBV infection induces RPL4 expression and causes RPL4 nuclear relocalization; (ii) EBNA1, NCL, and RPL4 form a complex in cells (RPL4 aa 62–238 bind to EBNA1 aa 102–325 and to the NCL RBD); (iii) EBNA1 induces RPL4 nuclear relocalization through NCL K429; (iv) RPL4’s N-terminal aa 62–238 coordinate with NCL-K429 to facilitate EBNA1 binding to oriP DNA and EBV episome maintenance; and (v) RPL4 C-terminal K380 and K393 increase oriP H3K4me2 and enhance EBNA1 activation of oriP transcription.
Fig. S7.
Model depicting that RPL4 coordinates with NCL to support EBNA1/oriP-mediated episome-dependent events.
NCL K429 is required for EBNA1 activation of oriP luciferase and for binding to oriP. A newly described NCL K429 function is to support EBNA1-mediated RPL4 nuclear relocalization. NCL K429 can bind ATP and provide ATP to enable RPL4 nuclear relocalization. Serum starvation can also induce RPL4 nuclear translocation (33).
RNAs have recently been reported to significantly affect sequence-specific transcription factor DNA binding. For example, EBV-encoded EBER RNA mediates PAX5 binding to EBV DNA (35). RNA also affects YY1 DNA binding (36). EBNA1 RG repeats bind to RNA (37), and RPL4 recruits RNA to oriP to facilitate EBNA1 DNA binding. Because EBNA1’s NCL binding domain (aa 1–100) differs from RPL4’s binding domain (aa 102–325), NCL and RPL4 may bind to EBNA1 to form a ternary complex.
Genome-wide and focused analyses have identified EBNA1 binding sites to be enriched for unique histone modifications (5, 10, 38). The EBNA1 N terminus specifically interacts with histone modifiers and nucleosome assembly proteins (2, 8). These histone modifications are likely associated with EBNA1-mediated transcription regulation of cell and virus genes. EBNA1 may recruit specific histone writers, readers, and erasers. For example, EBNA1 binds to the epigenetic reader BRD4, which binds to active H3K27ac and recruits P-TEFb to phosphorylate the RNA PolII C terminus (6). H3K4me2 is the first known histone modification at EBNA1 binding sites in the EBV genome (5). Our findings highlight the crucial role of RPL4 in H3K4me2 modification of oriP DNA. Although genomic H3K4me2-enriched regions mostly overlap with H3K4me3 near transcription start sites (39), we have found RPL4 to be able to mediate H3K4me3-independent modification of H3K4me2.
EBNA1 is the only EBV protein required for EBV persistence in host cells. Therapies targeting EBNA1 and its associated cell proteins may eradicate EBV and cure cancers caused by EBV. The findings here provide new potential targets for intervention.
Experimental Procedures
Cell Culture, EBV Infection, and Lentivirus-Mediated shRNA Knockdown.
The experimental procedures for primary B cell isolation, virus infection, cell culture, and EBV infection have been described previously (40). Lentiviral plasmids of shRNA were purchased from the National RNAi Core Facility, Academic Sinica Taiwan, and the protocol for lentivirus production was provided by the manufacturer.
Immunofluorescence Microscopy, BiFC, and ChIP.
The experimental procedures for these analytic methods have been described previously (18). Detailed experimental procedures are available in SI Experimental Procedures (along with Figs. S1–S7 and Tables S1–S3).
Table S3.
Summary of the specific set of primers used for qPCR assays
| Types of assays | Name of primers | Primer sequences |
| Genome accumulation | oriP DNA (F) | GCTAAACGAAGGAGAATGAAGAAG |
| oriP DNA (R) | CAGTGGCTGAAGATCAAGGAG | |
| GAPDH genomic DNA (F) | TGATGACATCAAGAAGGTGGTGAAG | |
| GAPDH genomic DNA (R) | TCCTTGGAGGCCATGTGGGCCAT | |
| mRNA RT-PCR | RPL4 | GCCAAGACTATGCGCAGGAAT |
| RPL4 | GTAGCTGCTGCTTCCAGCTT | |
| GAPDH (F) | AAGGTGAAGGTCGGAGTCAA | |
| GAPDH (R) | AATGAAGGGGTCATTGATGG | |
| ChIP qPCR | oriP DNA (F) | GCTAAACGAAGGAGAATGAAGAAG |
| oriP DNA (R) | CAGTGGCTGAAGATCAAGGAG | |
| GAPDH (F) | TACTAGCGGTTTTACGGGCG | |
| GAPDH (R) | TCGAACAGGAGGAGCAGAGAGCGA |
SI Experimental Procedures
Expression Plasmids and Cloning.
The expression plasmids of EBNA1, NCL, or their derivatives have been described previously (18). The expression vectors of E1 Δ102–325 and E1 Δ102–325 were gifts of John Yates, Department of Cancer Genetics, Roswell Park Cancer Institute, Buffalo, New York. The full-length cDNA of RPL4 was subcloned into XhoI and BamHI sites of the pSG5-flag vector after appropriate enzymatic digestion (41), whereas the remaining truncated depletion mutants were generated by performing a PCR-based mutagenesis protocol according to the manufacturer’s protocol (Stratagene). The synthetic RPL4 flaking sequences harboring the polylysine mutants were purchased from Genomic BioSci & Tech and subjected to a cloning procedure as described above. The RPL4 BiFC plasmid (FYN-RPL4) was created by subcloning the RPL4 cDNA into XhoI and BamHI sites of the FYN vector, whereas FYC-E1 plasmid has been previously described (18). The reporter plasmids oriP-SV40-Luc (oriP-Luc) (18) and LMP1 (–512/+72 × 2)-Luc (LMP1-Luc) (42) and the internal control CMV–β-Gal were used for transfection-mediated EBNA1 or EBNA2-dependent transcription.
Cell Culture and Transfection-Mediated EBNA1-Dependent Transcription Assay.
BJAB is an EBV-negative BL cell line, and its derivatives BJAB FEBNA1 stable clone (BJAB-FE1) and BJAB-FE1/oriP-Luc have been documented (18). In addition, BJAB B95.8 is an EBV-infected BJAB cell line. Among LCLs, IB4 is an EBV-transformed LCL that contains an integrated form of the EBV genome (43), whereas LCL1–3 are three individual classical EBV-transformed cell lines. The isolated primary B cells or B cells with EBV infection and all of the LCLs, BL cell lines, and their derivatives were cultured in RPMI-1640 supplemented with 10% (vol/vol) FCS, 2 mM l-glutamine, and penicillin/streptomycin (Life Technologies). 293T cells were cultured in DMEM (Life Technologies) with 10% (vol/vol) FCS, 2 mM l-glutamine, and penicillin/streptomycin. The experimental procedure for cell transfection was described previously (18, 40). For the EBNA1-dependent transcription assay, 107 BJAB cells and shRPL4- or Scr.-transduced BJAB cells were transfected with 10 μg of FEBNA1 expression vector, 5 μg of oriP-Luc reporter plasmid, and 1 μg of CMV-β Gal internal control reporter, or 30 μg of the FRPL4 expression vector or its mutant derivatives. The resulting luciferase activity (fold activation) produced by each transfectant was represented as the luciferase activity corrected by the β-Gal activity.
Primary B Cell Isolation and Monitoring of EBV Infection by Flow Cytometry.
Primary B cells were isolated from PBMC using Dynabeads Untouched (Life Technologies) following the protocol described by the manufacturer. EBV infectious particles were a gift of Kuei-Tai Lai, Medigen Biotechnology Corp, New Taipei City, Taiwan. We resuspended 5 × 104/100 μL primary B cells in RPMI-1640 supplemented with 15% (vol/vol) FCS, 2 mM l-glutamine, and penicillin/streptomycin and aliquoted it into a 96-well plate. We used 100 μL of EBV supernatant or PBS (mock infection) for each infection. The patterns of virus-infected cells and RPL4 expression were identified by flow cytometry after a procedure of chemical fixation and immunostaining as described previously (40).
Lentivirus Expressed RPL4, NCL, or Scrambled Control shRNAs.
Lentiviral vectors encompassing the shRNA sequences for RPL4 (shRPL4), NCL (shNCL), or Scr. were purchased from the National RNAi Core Facility, Academic Sinica Taiwan. The lentivirus particles containing the indicated shRNA were prepared according to the protocols suggested by the manufacturer. For RPL4 or NCL depletion, 1 mL of the lentiviral supernatants in the presence of 8 μg/mL polybrene were used to infect 5 × 105 BJAB, BJAB-B95.8, or LCL cells per mL in six-well plates. The transduced cells were replenished with new media and cultured for another 48 h, followed by selection with 5 ng/mL puromycin.
Immunofluorescence Microscopy, BiFC, co-IP, and Immune Blot Analyses.
For immunofluorescence analysis, cells were fixed in 2% (vol/vol) paraformaldehyde (Sigma) and subjected to an immunostaining protocol using antibodies for RPL4 (RQ-7; Santa Cruz), EBNA1 (vC-20; Santa Cruz), and NCL (MS-3 or F-18; Santa Cruz). In coimmunostainings, Rhodamin-conjugated goat anti-mouse and FITC-conjugated donkey anti-goat (Kirkegaard & Perry Laboratories, Inc.) were used as fluorochromes, and DNA was counterstained with TO-PRO3 Iodide (Life Technology) or DAPI (Sigma). For BiFC analysis, shNCL #1-transduced BJAB cells cotransfected with YN-RPL4, YC-EBNA1, nuclear marker NLS-eBFP (44), and FNCL or FNCL K429A were subjected to confocal fluorescent microscopy. Both immunofluorescence and BiFC images were visualized using CARV II Confocal Imager.
The co-IP assay was used to identify the physical interaction of EBNA1 with RPL4 in IB4 cells, FEBNA1 and RPL4 in BJAB-FE1 cells, or plasmid expressed FRPL4 and EBNA1 in BJAB cells using the following antibodies: M2 (Sigma) for flag epitope, 6F9/60 (Novas Biologicals) for EBNA1, F-18 (Santa Cruz) for NCL, and RQ-7 (Santa Cruz) for RPL4. This protocol was also used to identify the EBNA1 or NCL binding site of RPL4 and RPL4 binding site of EBNA1 or NCL using BJAB cells transfected with the necessary plasmids. Alternatively, shRPL4- or shNCL-transduced BJAB-FE1 cells were used to verify the requirement of RPL4 and NCL in stabilizing EBNA1/NCL and EBNA1/RPL4 association, respectively. Cell lysates derived from the indicated cell lines or immunoprecipitated samples were subjected to SDS/PAGE, followed by Western blotting with appropriate antibodies. The antibody for internal control actin was purchased from Santa Cruz. The proteins were detected and visualized using an ECL detection kit (Millipore). Whenever necessary, the immunoblotting images were quantified by UN-SCAN-IT.
ChIP, in Vitro DNA Pulldown, and EBV Genome Maintenance Assays.
To access the EBNA binding affinity in shRPL4- or Scr.-transduced LCLs or BJAB-FE1/oriP-Luc cells, ChIP assays were carried out followed by qPCR analysis using a set of oriP DNA primers as described previously (18, 28) (Table S3). We prepared 2 × 106 cells for each ChIP assay using antibodies for EBNA1 (6F9/60; Novas Biologicals) and IgG control (Millipore). The abundance of each assayed protein at oriP promoter DNA was determined as the percentage of input DNA (% of input). For in vitro DNA pulldown assays, the biotin-labeled oriP DNA or biotin-labeled LMP1-DNA (negative control) was amplified by PCR using a set of the biotin-labeled primers corresponding to the target DNA (28). Cell lysates from 1 × 107 shRPL4- or Scr.-transduced BJAB-FE1 were prepared and subjected to a streptavidin agarose (Life Technology)-mediated pulldown protocol (18). EBV genomic DNA or meDNA was purified using the viral DNA isolation kit following the manufacturer’s protocol (Geneaid Inc.). Genome accumulation of the full-length EBV genome and meDNA in shRPL4- or Scr.-transduced LCLs and BJAB-FE1/oriP-Luc cells were determined by qPCR using a specific pair of primers (18, 28) (Table S3).
Protein Alignment and Statistical Analyses.
The RPL4 protein sequences derived from different vertebrate groups or Tetrahymena thermophila were retrieved from the National Center for Biotechnology Information and analyzed by PRofile ALIgNEment (PRALINE) multiple sequence alignment (www.ibi.vu.nl/programs/pralinewww/), provided by the Centre for Integrative Bioinformatics VU. The data from transfection-mediated transcription reporter or ChIP assays were represented as the mean ± the SEM from three independent experiments. Whenever necessary, statistical comparisons were performed by student’s t test. A P value of less than 0.05 was considered to be statistically significant.
Nuclear and Cytosolic Fractions and Measurements of Cell Proliferation.
We subjected 5 × 106 of the indicated cell lines to cytosolic and nuclear fraction using ProteoJET Cytoplasmic and Nuclear Protein Extraction Kit (Fermentas) following the protocol provided by the manufacturer. For cell proliferation assays, a total of 5 × 104 of the indicated cell lines per 200 μL were aliquoted in triplicate in 96-well plates and subjected to perform the trypan blue exclusion method every 24 h for 5 consecutive days.
Acknowledgments
This work was supported by National Science Council Grant NSC 101-2320-B320-005-MY3, Ministry of Science and Technology Grant MOST 104-2320-B-320-013, and National Health Research Institutes Grants NHRI-EX-1025-9910BC and NHRI-EX103-10307BI (to C.-W.P.). E.K. was supported by National Institutes of Health Grants 5R01CA047006, 5R01CA085180, and 5R01CA170023.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525444113/-/DCSupplemental.
References
- 1.Longnecker R, Kieff E, Cohen J. Epstein-Barr virus. In: Knipe D, Howley P, editors. Fields Virology. 6th Ed. Vol 2. Lippincott, Williams, and WIlkins; Philadelphia: 2013. pp. 1898–1959. [Google Scholar]
- 2.Frappier L. Contributions of Epstein-Barr nuclear antigen 1 (EBNA1) to cell immortalization and survival. Viruses. 2012;4(9):1537–1547. doi: 10.3390/v4091537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5(8):1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochkarev A, et al. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell. 1996;84(5):791–800. doi: 10.1016/s0092-8674(00)81056-9. [DOI] [PubMed] [Google Scholar]
- 5.Day L, et al. Chromatin profiling of Epstein-Barr virus latency control region. J Virol. 2007;81(12):6389–6401. doi: 10.1128/JVI.02172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin A, Wang S, Nguyen T, Shire K, Frappier L. The EBNA1 protein of Epstein-Barr virus functionally interacts with Brd4. J Virol. 2008;82(24):12009–12019. doi: 10.1128/JVI.01680-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Frappier L. Nucleosome assembly proteins bind to Epstein-Barr virus nuclear antigen 1 and affect its functions in DNA replication and transcriptional activation. J Virol. 2009;83(22):11704–11714. doi: 10.1128/JVI.00931-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frappier L. EBNA1 and host factors in Epstein-Barr virus latent DNA replication. Curr Opin Virol. 2012;2(6):733–739. doi: 10.1016/j.coviro.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Ceccarelli DF, Frappier L. Functional analyses of the EBNA1 origin DNA binding protein of Epstein-Barr virus. J Virol. 2000;74(11):4939–4948. doi: 10.1128/jvi.74.11.4939-4948.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu F, et al. Genome-wide analysis of host-chromosome binding sites for Epstein-Barr Virus Nuclear Antigen 1 (EBNA1) Virol J. 2010;7:262. doi: 10.1186/1743-422X-7-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korobeinikova AV, Garber MB, Gongadze GM. Ribosomal proteins: Structure, function, and evolution. Biochemistry (Mosc) 2012;77(6):562–574. doi: 10.1134/S0006297912060028. [DOI] [PubMed] [Google Scholar]
- 12.Teng T, Thomas G, Mercer CA. Growth control and ribosomopathies. Curr Opin Genet Dev. 2013;23(1):63–71. doi: 10.1016/j.gde.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Wanzel M, et al. A ribosomal protein L23-nucleophosmin circuit coordinates Mizl function with cell growth. Nat Cell Biol. 2008;10(9):1051–1061. doi: 10.1038/ncb1764. [DOI] [PubMed] [Google Scholar]
- 14.Challagundla KB, et al. Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol Cell Biol. 2011;31(19):4007–4021. doi: 10.1128/MCB.05810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai MS, Lu H. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J Cell Biochem. 2008;105(3):670–677. doi: 10.1002/jcb.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan F, et al. Ribosomal protein S3: A KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007;131(5):927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Malik-Soni N, Frappier L. Nucleophosmin contributes to the transcriptional activation function of the Epstein-Barr virus EBNA1 protein. J Virol. 2013;88(4):2323–2326. doi: 10.1128/JVI.02521-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YL, et al. Nucleolin is important for Epstein-Barr virus nuclear antigen 1-mediated episome binding, maintenance, and transcription. Proc Natl Acad Sci USA. 2014;111(1):243–248. doi: 10.1073/pnas.1321800111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng E, et al. Characterization of the interaction between hantavirus nucleocapsid protein (N) and ribosomal protein S19 (RPS19) J Biol Chem. 2011;286(13):11814–11824. doi: 10.1074/jbc.M110.210179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee AS, Burdeinick-Kerr R, Whelan SP. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci USA. 2013;110(1):324–329. doi: 10.1073/pnas.1216454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green L, Houck-Loomis B, Yueh A, Goff SP. Large ribosomal protein 4 increases efficiency of viral recoding sequences. J Virol. 2012;86(17):8949–8958. doi: 10.1128/JVI.01053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blumenthal T, Carmichael GG. RNA replication: Function and structure of Qbeta-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 23.Houmani JL, Davis CI, Ruf IK. Growth-promoting properties of Epstein-Barr virus EBER-1 RNA correlate with ribosomal protein L22 binding. J Virol. 2009;83(19):9844–9853. doi: 10.1128/JVI.01014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toczyski DP, Matera AG, Ward DC, Steitz JA. The Epstein-Barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proc Natl Acad Sci USA. 1994;91(8):3463–3467. doi: 10.1073/pnas.91.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frappier L. Role of EBNA1 in NPC tumourigenesis. Semin Cancer Biol. 2012;22(2):154–161. doi: 10.1016/j.semcancer.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Worbs M, Huber R, Wahl MC. Crystal structure of ribosomal protein L4 shows RNA-binding sites for ribosome incorporation and feedback control of the S10 operon. EMBO J. 2000;19(5):807–818. doi: 10.1093/emboj/19.5.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334(6058):941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 28.Chen YL, Tsai HL, Peng CW. EGCG debilitates the persistence of EBV latency by reducing the DNA binding potency of nuclear antigen 1. Biochem Biophys Res Commun. 2012;417(3):1093–1099. doi: 10.1016/j.bbrc.2011.12.104. [DOI] [PubMed] [Google Scholar]
- 29.Thomson E, Ferreira-Cerca S, Hurt E. Eukaryotic ribosome biogenesis at a glance. J Cell Sci. 2013;126(Pt 21):4815–4821. doi: 10.1242/jcs.111948. [DOI] [PubMed] [Google Scholar]
- 30.de Las Heras-Rubio A, Perucho L, Paciucci R, Vilardell J, LLeonart ME. Ribosomal proteins as novel players in tumorigenesis. Cancer Metastasis Rev. 2014;33(1):115–141. doi: 10.1007/s10555-013-9460-6. [DOI] [PubMed] [Google Scholar]
- 31.Quin JE, et al. Targeting the nucleolus for cancer intervention. Biochim Biophys Acta. 2014;1842(6):802–816. doi: 10.1016/j.bbadis.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34(1):3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Egoh A, Nosuke Kanesashi S, Kanei-Ishii C, Nomura T, Ishii S. Ribosomal protein L4 positively regulates activity of a c-myb proto-oncogene product. Genes Cells. 2010;15(8):829–841. doi: 10.1111/j.1365-2443.2010.01421.x. [DOI] [PubMed] [Google Scholar]
- 34.Allinne J, et al. Perinucleolar relocalization and nucleolin as crucial events in the transcriptional activation of key genes in mantle cell lymphoma. Blood. 2014;123(13):2044–2053. doi: 10.1182/blood-2013-06-510511. [DOI] [PubMed] [Google Scholar]
- 35.Lee N, Moss WN, Yario TA, Steitz JA. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell. 2015;160(4):607–618. doi: 10.1016/j.cell.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sigova AA, et al. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350(6263):978–981. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong X, Yalamanchili R, Harada S, Kieff E. The EBNA-2 arginine-glycine domain is critical but not essential for B-lymphocyte growth transformation; the rest of region 3 lacks essential interactive domains. J Virol. 1994;68(10):6188–6197. doi: 10.1128/jvi.68.10.6188-6197.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tempera I, et al. Identification of MEF2B, EBF1, and IL6R as direct gene targets of Epstein-Barr Virus (EBV) nuclear antigen 1 critical for EBV-infected B-lymphocyte survival. J Virol. 2015;90(1):345–355. doi: 10.1128/JVI.02318-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Liu CD, et al. The nuclear chaperone nucleophosmin escorts an Epstein-Barr Virus nuclear antigen to establish transcriptional cascades for latent infection in human B cells. PLoS Pathog. 2012;8(12):e1003084. doi: 10.1371/journal.ppat.1003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng CW, et al. Hsp72 up-regulates Epstein-Barr virus EBNALP coactivation with EBNA2. Blood. 2007;109(12):5447–5454. doi: 10.1182/blood-2006-08-040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin J, Johannsen E, Robertson E, Kieff E. Epstein-Barr virus nuclear antigen 3C putative repression domain mediates coactivation of the LMP1 promoter with EBNA-2. J Virol. 2002;76(1):232–242. doi: 10.1128/JVI.76.1.232-242.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.King W, Thomas-Powell AL, Raab-Traub N, Hawke M, Kieff E. Epstein-Barr virus RNA. V. Viral RNA in a restringently infected, growth-transformed cell line. J Virol. 1980;36(2):506–518. doi: 10.1128/jvi.36.2.506-518.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ai HW, Shaner NC, Cheng Z, Tsien RY, Campbell RE. Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry. 2007;46(20):5904–5910. doi: 10.1021/bi700199g. [DOI] [PubMed] [Google Scholar]