Significance
Fusion of myoblasts is required for proper skeletal muscle formation during embryonic development and adult muscle regeneration. Despite the central role for plasma membrane fusion, the molecules and mechanisms that govern this process have remained elusive. Myomaker is the only identified muscle-specific protein required for myoblast fusion; however, the mechanisms by which this multipass membrane protein governs fusion are not understood. Through structure–function analysis and development of cell-based fusion assays, we begin to delineate the domains of the myomaker protein required for fusion.
Keywords: myogenesis, muscle development, cell fusion, CRISPR/Cas9
Abstract
During skeletal muscle development, myoblasts fuse to form multinucleated myofibers. Myomaker [Transmembrane protein 8c (TMEM8c)] is a muscle-specific protein that is essential for myoblast fusion and sufficient to promote fusion of fibroblasts with muscle cells; however, the structure and biochemical properties of this membrane protein have not been explored. Here, we used CRISPR/Cas9 mutagenesis to disrupt myomaker expression in the C2C12 muscle cell line, which resulted in complete blockade to fusion. To define the functional domains of myomaker required to direct fusion, we established a heterologous cell–cell fusion system, in which fibroblasts expressing mutant versions of myomaker were mixed with WT myoblasts. Our data indicate that the majority of myomaker is embedded in the plasma membrane with seven membrane-spanning regions and a required intracellular C-terminal tail. We show that myomaker function is conserved in other mammalian orthologs; however, related family members (TMEM8a and TMEM8b) do not exhibit fusogenic activity. These findings represent an important step toward deciphering the cellular components and mechanisms that control myoblast fusion and muscle formation.
Plasma membrane fusion is a fundamental cellular process required for the conception, development, and physiology of multicellular organisms. Membrane fusion occurs during viral infection of a host cell, between intracellular membranes, and between two plasma membranes to form syncytial tissues (1). Cell–cell fusion is critical for a wide array of cellular processes, including sperm–egg fertilization, macrophage function, bone and placental development, and skeletal muscle formation. This form of fusion must be precisely controlled to prevent inappropriate cellular mixing.
The fusion of myoblasts requires cell recognition, migration, adhesion, signaling, and finally, membrane coalescence (2). Much of our knowledge about myoblast fusion has originated from studies performed in Drosophila. In this system, intracellular signaling results in cytoskeletal alterations and actin polymerization, which drive the formation of cellular projections that invade neighboring cells to cause fusion (3). Recent evidence also indicates a critical function for branched actin polymerization during Drosophila indirect flight muscle fusion (4). The essential role of the cytoskeleton in fusion is conserved in mammals, in which the actin regulators Rac1, cdc42, and N-WASp are required for muscle development (5–8). In addition to actin dynamics, numerous proteins that regulate diverse cellular processes have been associated with myoblast fusion (9–13).
We recently discovered a muscle-specific membrane protein named myomaker [annotated as Transmembrane protein 8c (TMEM8c)] that is absolutely required for skeletal myocyte fusion in the mouse (14, 15). To begin to decipher the mechanisms whereby myomaker controls cell–cell fusion, we designed a heterologous cell–cell fusion assay to monitor the ability of a series of myomaker mutants to direct the fusion of labeled fibroblasts with myoblasts in vitro. Here, we present a model of myomaker topology within the membrane, in which seven amphipathic α-helical transmembrane (TM) domains and an essential C-terminal cytoplasmic domain drive the fusion process. These findings begin to enlighten the mechanistic basis of myoblast fusion.
Results
Myomaker Is Necessary for Fusion of C2C12 Cells.
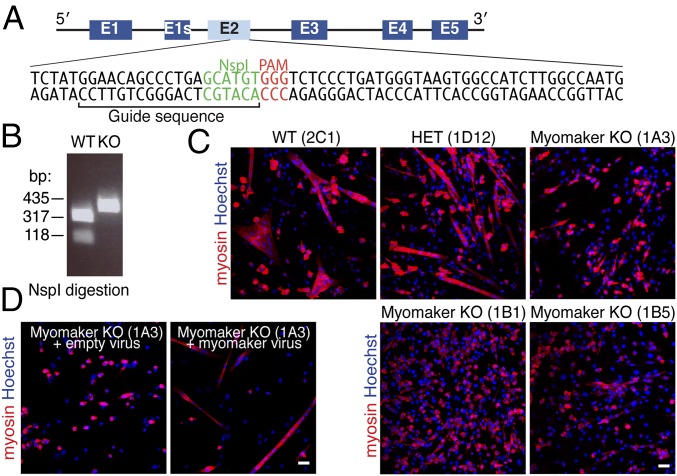
Primary embryonic myoblasts obtained from myomaker null mice can differentiate to generate myocytes but cannot fuse (14). To determine whether the immortalized C2C12 muscle cell line also required myomaker for fusion, we inactivated the gene in C2C12 cells through clustered regularly-interspaced short palindromic repeats (CRISPR)/Cas9-mediated mutagenesis (16). As a template for mutagenesis, we designed a guide RNA that recognizes exon 2 of myomaker and recruits Cas9 to induce a double-strand break (Fig. 1A). C2C12 cells were cotransfected with a vector encoding the guide RNA/Cas9 and a separate GFP plasmid, which was used to label the transfected cells. Forty-eight hours after transfection, single cells that expressed the highest levels of GFP were sorted by flow cytometry into 96-well plates. These cells were propagated and genotyped by assaying for the presence of an NspI restriction enzyme site that was disrupted after CRISPR/Cas9-mediated cleavage (Fig. 1A). Genomic DNA from a WT clone had an NspI restriction enzyme site and after NspI digestion, exhibited the expected 317- and 118-bp fragments, whereas the amplicon from genomic DNA of a homozygous targeted clone was not cut by NspI because of the lack of the restriction enzyme site (Fig. 1B). Of 46 genotyped clones, 2 were WT, 14 were heterozygous (HET), and 30 were homozygous KO, showing the robust activity of this genome editing system in C2C12 cells. We selected one WT (2C1), one HET (1D12), and three KO (1A3, 1B1, and 1B5) clones for additional analysis.
Fig. 1.
Myomaker disruption in C2C12 cells using CRISPR/Cas9 genome editing. (A) Myomaker locus with exons depicted. Exon 1s is an annotated alternative isoform; however, it is not highly conserved, and we did not investigate it in this study. Exon 2 of myomaker was mutagenized in C2C12 cells after transfection with a vector containing the indicated guide sequence and Cas9. NspI was used to assay for genome disruption at this site. (B) C2C12 clones were genotyped by PCR-amplifying a 435-bp region surrounding the protospacer adjacent motif (PAM) site followed by digestion with NspI. Genomic DNA from myomaker KO clones exhibits an uncut 435-bp fragment because of CRISPR/Cas9-mediated disruption of the NspI site. WT genomic DNA shows 317- and 118-bp bands after NspI digestion. (C) WT (2C1) and HET (1D12) clones formed multinucleated myotubes after 4 d in differentiation media. The three myomaker KO (1A3, 1B1, and 1B5) clones differentiated, shown by myosin staining (red), but failed to fuse. (D) Expression of myomaker using a retrovirus in KO clone 1A3 rescued fusion. Representative images are shown from experiments that were performed twice in duplicate. (Scale bar: 50 μm.)
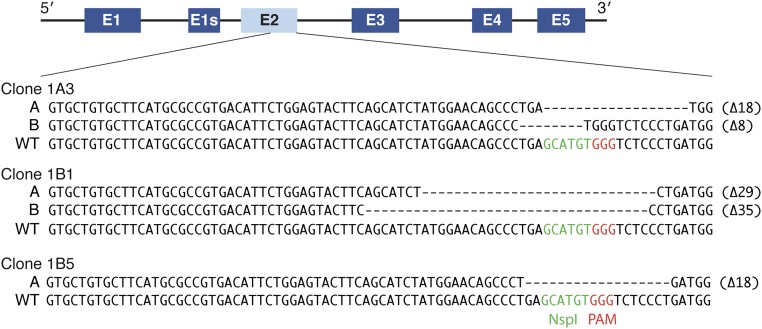
A DNA template for homology-directed insertion was not introduced during the transfection; thus, the genomic region was repaired through nonhomologous end-joining, which causes insertion/deletion mutations. To determine the deletions and/or insertions in each KO clone, the exon 2 PCR products were cloned into TOPO vector, and then, multiple TOPO clones were sequenced to identify mutations on both alleles. Clone 1A3 contained 18- and 8-bp deletions, and clone 1B1 exhibited 29- and 35-bp deletions (Fig. S1). We only detected an 18-bp deletion in 10 TOPO clones from clone 1B5, suggesting that both alleles were repaired in the same manner (Fig. S1).
Fig. S1.
Genomic deletion of myomaker exon 2 after CRISPR/Cas 9 mutagenesis. A 435-bp fragment within exon 2 was amplified from genomic DNA from each myomaker KO clonal cell line and ligated into a shuttle vector (TOPO vector). Ten TOPO clones were then sequenced to identify the insertions or deletions after nonhomologous end joining (NHEJ) repair of the double-strand break. The missing nucleotides are indicated by dashes, and the total number of nucleotides disrupted is shown at the end of each line. For clones 1A3 and 1B1, the repair differed for each allele (displayed as A and B sequences). Sequencing of 10 TOPO clones from 1B5 showed the same sequence, suggesting identical repair of both alleles.
Consistent with the requisite role of myomaker in myoblast fusion in vivo, we observed a dramatic lack of fusion in the C2C12 clonal lines lacking myomaker when maintained in differentiation medium (Fig. 1C). Myomaker KO cells differentiated, as shown by myosin staining, but remained mononucleated, whereas myotubes were clearly present in cultures from the WT and HET clones (Fig. 1C). These findings suggest that the in-frame deletions in the myomaker KO clones create a nonfunctional protein or prevent protein expression. The mutation introduced in clone 1B5 is predicted to cause an in-frame deletion of amino acids 77–82 (Fig. 2A).
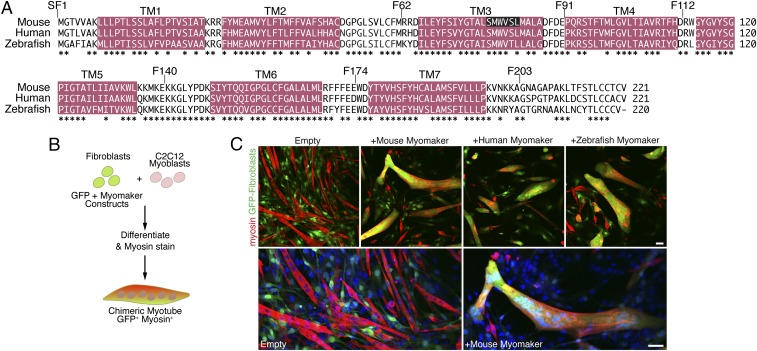
Fig. 2.
Myomaker orthologs are fusogenic. (A) Amino acid alignment of mouse, human, and zebrafish myomaker proteins. Magenta regions depict significant stretches of hydrophobic amino acids. TM refers to TM domains. Also shown are locations of FLAG epitopes preceding the indicated amino acid (SF1, F62, F91, F112, F140, F174, and F203). Residues deleted in the CRISPR/Cas9 myomaker KO clone 1B5 are displayed with white font and black background. *Sequence conservation. (B) Schematic showing the heterologous cell–cell fusion system. Fibroblasts were infected with GFP and myomaker constructs and mixed with C2C12 myoblasts. The cultures were differentiated for 4 d and then immunostained with myosin antibody as a marker for myocytes. GFP-positive (fibroblast origin), myosin-positive (myoblast origin) chimeric myotubes, in yellow/orange, indicate fusion between the two cell populations. (C) Fibroblasts infected with control (empty) and GFP viruses do not fuse to C2C12 cells. Expression of mouse, human, or zebrafish myomaker in fibroblasts promotes dramatic fusion with muscle. Lower shows enlarged images of cells infected with empty and mouse myomaker retroviruses with Hoechst-stained nuclei. Representative images are shown from experiments that were performed at least three times. (Scale bar: 50 μm.)
To confirm that the block to fusion was a specific consequence of the loss of myomaker rather than a nonspecific effect of CRISPR/Cas9 mutagenesis, we infected KO clone 1A3 with a retrovirus encoding myomaker or an empty plasmid as a control and then, allowed the cells to differentiate. Reexpression of myomaker rescued the fusion block of myomaker KO C2C12 cells (Fig. 1D). These data indicate that myomaker is absolutely essential for fusion in this cell system.
Fusogenic Activity of Myomaker Orthologs.
Myomaker is a hydrophobic protein containing 221 aa that localizes to intracellular vesicles and the plasma membrane of skeletal myocytes (14). Analysis of hydrophobic stretches of at least 20 aa, corresponding o potential TM regions, revealed seven potential TM domains (Fig. 2A). Myomaker is also highly conserved, exhibiting ∼60% direct sequence identity within vertebrate species. The human and mouse proteins share 196 identical amino acids, and the zebrafish ortholog has 163 identical residues (Fig. 2A). Morpholino knockdown of myomaker in zebrafish revealed that myomaker is essential for fast myocyte fusion (17).
We generated retroviruses expressing the human and zebrafish myomaker proteins and tested them for their ability to drive fusion of fibroblasts in a heterologous cell–cell fusion system. In this assay, fibroblasts were infected with myomaker and GFP overnight, washed extensively, mixed with C2C12 cells, and transferred to differentiation medium for 4 d (Fig. 2B). We tracked fibroblasts by GFP visualization and C2C12 cells by immunostaining for myosin heavy chain (red in Fig. 2B), a marker of a differentiated muscle cell. A myosin-positive, GFP-positive multinucleated cell indicates fusion between a fibroblast and C2C12 cell, forming a chimeric myotube that appeared yellow/orange in Fig. 2B. We used empty GFP-infected fibroblasts as control, and chimeric myotubes were rarely visualized when mixed with C2C12 cells. As shown in Fig. 2C, the human and zebrafish myomaker orthologs readily promoted the fusion of GFP-positive fibroblasts with C2C12 myoblasts that stained positively for myosin expression (red in Fig. 2C), showing that the myomaker activity to promote heterologous fusion is conserved in vertebrate species (Fig. 2C). Also shown is a magnified image with Hoechst staining to highlight the nuclei within the chimeric myotube.
Requirement of Myomaker on Both Cells for Maximal Fusion.
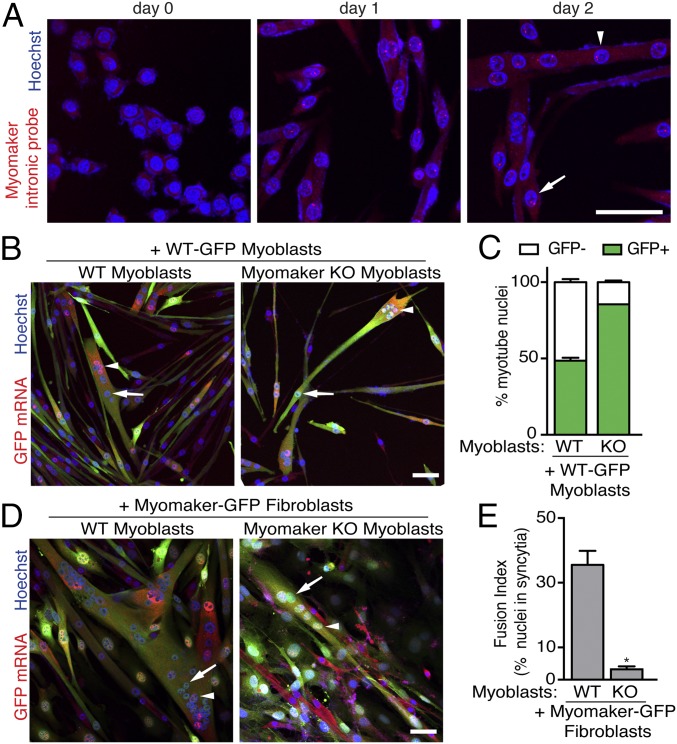
Whether the fusion machinery is similar or different on the two fusing cells is an important concept with respect to membrane fusion mechanisms. As a means of identifying nuclei that were actively expressing myomaker, we used single-molecule RNA FISH with a probe specific for the intronic myomaker sequence. Primary WT myoblast nuclei on day 0 of differentiation were not positive for myomaker transcription (Fig. 3A), consistent with our previous work showing that myomaker is induced upon differentiation (14). On day 1 of differentiation, myomaker transcription was detected in the majority of myocytes (visualized by red punctae in the nuclei in Fig. 3A). Both myocyte and myotube nuclei transcribe myomaker on day 2 of differentiation, suggesting that myomaker could contribute to fusion in each cell type (Fig. 3A).
Fig. 3.
Fusion is most efficient when both cells express myomaker. (A) RNA FISH using a probe specific for myomaker introns was performed on primary WT myoblasts during differentiation. This approach reveals nuclei that are actively transcribing myomaker (red punctae in nuclei). Arrow indicates myoblast nuclei, and arrowhead shows myotube nuclei. (B) WT myoblasts were infected with GFP retrovirus and mixed with either unlabeled WT myoblasts or unlabeled myomaker KO myoblasts. Three days after differentiation, FISH for GFP was performed to track GFP+ nuclei. Arrows depict GFP− nuclei, and arrowheads show GFP+ nuclei. (C) Quantitation of the percentage of myotube nuclei that are either GFP+ or GFP− reveals that myomaker KO cells fuse less efficiently than WT. (D) Fibroblasts were infected with myomaker and GFP and then mixed with either unlabeled WT myoblasts or unlabeled myomaker KO myoblasts. GFP FISH was performed 3 d after differentiation. Arrows depict GFP− nuclei, and arrowheads show GFP+ nuclei. (E) Quantitation of fusion index reveals that WT myoblasts fuse to fibroblasts more efficiently than KO myoblasts. Representative images are shown from experiments that were performed at least three times. (Scale bar: 50 μm.) *P < 0.05 compared with the WT.
To determine the importance of myomaker on both fusing membranes, we performed cell-mixing experiments with myomaker-modified myoblasts and fibroblasts. Although we showed previously that WT myocytes can fuse to myomaker KO myocytes, the ratio of WT and KO nuclei in the resulting myotubes was not investigated (14). In the cell-mixing experiments presented here, we infected one population of cells with GFP, mixed them with an unlabeled population of cells, and then, performed GFP FISH to track individual nuclei. This approach labels GFP mRNA in the cytoplasm and also, the nuclei that are actively transcribing GFP. Thus, the nuclei positive for GFP mRNA are from the GFP-infected population, whereas the nuclei negative for GFP mRNA correspond to the non-GFP population. This experimental strategy allows relative quantitation of fusion efficiency of myomaker KO myoblasts with either WT myoblasts or myomaker-expressing fibroblasts.
We infected primary WT myoblasts with a retrovirus encoding GFP and sorted GFP-positive cells to generate a homogenous population of myoblasts. We then mixed WT-GFP myoblasts with either unlabeled WT primary myoblasts or unlabeled myomaker KO primary myoblasts and differentiated the cultures for 3 d. GFP FISH revealed an expected 50:50 ratio of WT-GFP nuclei (GFP-positive):unlabeled WT nuclei (GFP-negative) in myotubes (Fig. 3 B and C). In contrast, WT-GFP nuclei comprised a significantly higher fraction of myotube nuclei compared with myomaker KO (GFP-negative) nuclei (Fig. 3 B and C). Thus, although myomaker KO muscle cells can fuse with WT muscle cells, they are considerably less effective in doing so than WT muscle cells.
Our previous work showed that myomaker-expressing fibroblasts cannot fuse to each other but can fuse to muscle cells, suggesting that additional factors expressed by myoblasts but not fibroblasts are required for fusion (14). To further evaluate the requirement of myomaker on both fusing cells, we tested the fusion efficiency between myomaker KO myoblasts and myomaker-expressing fibroblasts. We mixed myomaker-GFP 10T1/2 fibroblasts with either unlabeled WT primary myoblasts or unlabeled myomaker KO primary myoblasts and differentiated the cultures for 3 d. Myomaker-GFP fibroblasts readily fused to WT myoblasts, but only a low level of fusion was observed between myomaker-GFP fibroblasts and myomaker KO myoblasts (Fig. 3D). Quantification of overall fusion index in each culture revealed a significant reduction in the percentage of nuclei in syncytial cells when myomaker-GFP fibroblasts and myomaker KO myoblasts were mixed before differentiation (Fig. 3E). Taken together, these data indicate that fusion is most efficient when myomaker is present on both fusing cells, highlighting the requirement of additional myoblast-derived factors for myomaker-mediated fusion.
Analysis of Fusogenic Activity of Myomaker-FLAG Proteins.
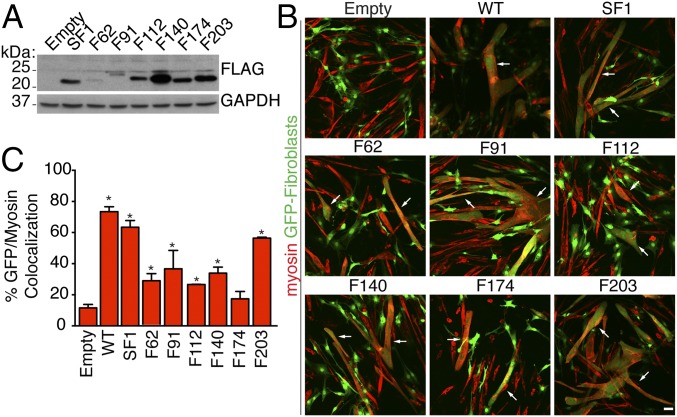
To begin to define the functional domains and topology of myomaker within the membrane, we introduced FLAG tags at specified locations throughout the protein and assayed their expression by anti-FLAG immunoblotting. We generated a construct that contained a synthetic cleavable signal sequence upstream of FLAG followed by a full-length myomaker protein referred to as signal sequence FLAG amino acid 1 of myomaker (SF1) (Fig. 2A). We also engineered FLAG epitopes in multiple hydrophilic loops of myomaker, where these constructs do not contain the synthetic signal sequence. These six versions of myomaker contained FLAG insertions before the indicated amino acids: F62, F91, F112, F140, F174, and F203 (Fig. 2A).
We infected fibroblasts with retroviruses encoding each version of myomaker and performed Western blot analysis with a FLAG antibody to detect expression. Each myomaker-FLAG protein was produced, although different mutants showed varying levels of expression (Fig. 4A), likely reflecting differing stabilities. We next determined the fusogenic activity of each FLAG construct using the heterologous cell–cell fusion system. All of the myomaker-FLAG constructs induced the fusion of fibroblasts to C2C12 cells (Fig. 4B). To quantify heterologous fusion, we used CellProfiler, an image analysis software, to assess the percentage of myosin and GFP colocalization (18). This analysis revealed clear fusogenic activity for each FLAG construct, albeit to various levels (Fig. 4C). Interestingly, the level of expression did not necessarily correlate with function. For instance, F140 was expressed at much higher levels than F62 but displayed similar fusogenic activity.
Fig. 4.
Myomaker epitope-tagged constructs are expressed and functional. (A) Protein extracts from fibroblasts expressing each FLAG-tagged version of myomaker (depicted in Fig. 2A) were analyzed by immunoblotting using an FLAG antibody. Empty-infected fibroblasts were used as a control. Each myomaker-FLAG construct was expressed at varying levels. GAPDH was a loading control. (B) Fibroblasts coinfected with myomaker-FLAG constructs and GFP can fuse with C2C12 cells. Arrows indicate GFP+ myosin+ structures. (C) Quantitation of heterologous fusion through analyzing the colocalization of GFP (fibroblasts) and myosin (myocytes) using CellProfiler. Each tagged version of myomaker exhibits different levels of expression and function. Representative images are shown from experiments that were performed at least three times. (Scale bar: 50 μm.) *P < 0.05 compared with empty.
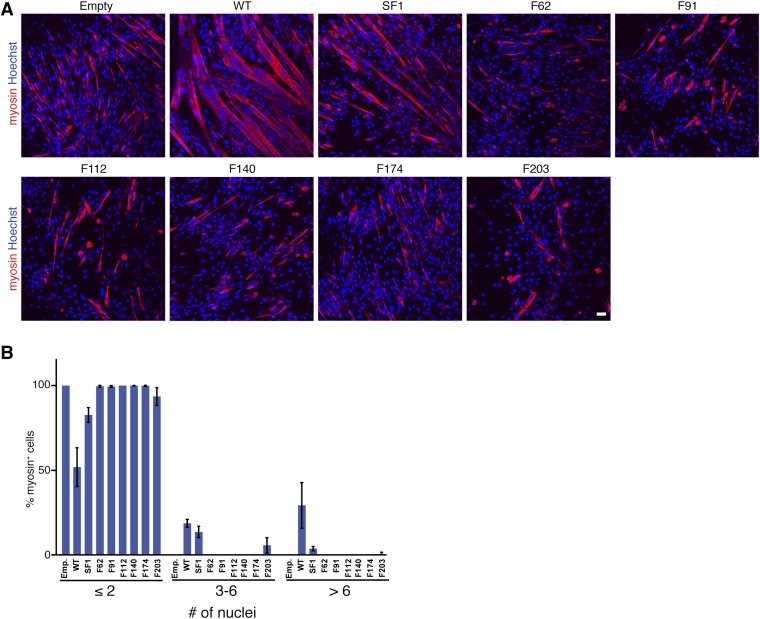
We also tested the ability of each myomaker-FLAG construct to rescue the fusion defect in myomaker KO C2C12 cells generated through Cas9 mutagenesis (Fig. 1). As expected, myomaker KO myoblasts infected with empty retrovirus expressed myosin but remained mononucleated, whereas expression of WT myomaker dramatically rescued the fusion defect (Fig. S2A). Notably, the various myomaker-FLAG constructs displayed less fusogenic activity than WT untagged myomaker (Fig. S2A). It is formally possible that the synthetic N-terminal signal peptide and FLAG epitope in the various myomaker constructs perturb normal orientation of the protein in the bilayer, including misorientation of the N terminus to the exterior of the cell. However, these constructs retain fusogenic activity.
Fig. S2.
Myomaker epitope-tagged constructs do not rescue fusion defect in myomaker KO myoblasts. (A) Myomaker KO myoblasts were infected with various myomaker-FLAG constructs and induced to differentiate. The cultures were stained with myosin antibody and Hoechst to assay fusion. (B) Quantitation of fusion induced by each construct shows a lack of fusion induction by the FLAG constructs. SF1 and F203 do exhibit fusion ability, although they decreased compared with WT. Representative images are shown from experiments that were performed at least three times. Emp., empty. (Scale bar: 50 μm.)
Fusion quantification revealed that only SF1 and F203 possessed the ability to rescue fusion in myomaker KO cells but that neither rescued to the level of WT myomaker (Fig. S2B). The disparity between fusogenic functions of the myomaker-FLAG constructs in the heterologous fusion system compared with the myomaker KO myoblast system could indicate that optimal fusion requires myomaker on both membranes (Fig. 3). In the heterologous fusion system, endogenous myomaker on myoblasts may compensate for a less functional myomaker-FLAG construct expressed on fibroblasts. However, in the myomaker KO myoblast system, where endogenous myomaker is absent, the activities of less functional myomaker-FLAG proteins are more easily revealed.
Experimental Assessment of Myomaker Topology.
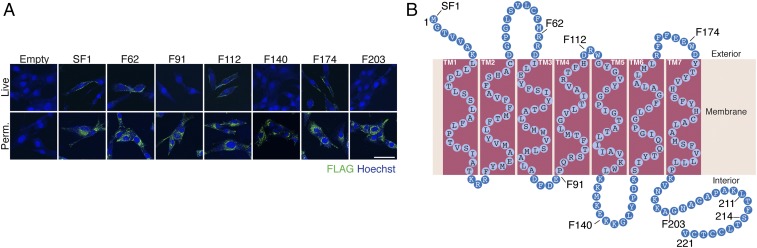
To analyze the topology of myomaker within the membrane, we tested the accessibility of FLAG epitopes by performing anti-FLAG immunocytochemistry on live C2C12 cells infected with retroviruses expressing FLAG-tagged myomaker constructs. These cells were not fixed or permeabilized before antibody incubation on ice. SF1, F62, F112, and F174 exhibited surface staining; however, no staining was detected for F91, F140, and F203 myomaker constructs (Fig. 5A). When cells were fixed and permeabilized before antibody incubation, each construct showed similar intracellular punctate staining (Fig. 5A). Positive staining of live cells expressing SF1, F62, F112, and F174 indicates that the FLAG epitopes within these myomaker mutants are exposed to the extracellular space.
Fig. 5.
Determination of myomaker membrane topology. (A) C2C12 cells were infected with myomaker-FLAG retroviral constructs and allowed to differentiate for 1 d. One set of cells was immunostained on ice with a FLAG antibody before fixation or permeabilization (Live), whereas a separate culture was stained after fixation and permeabilization (Perm.). SF1, F62, F112, and F174 exhibited surface staining in live cells, indicating the presence of the epitope on the cell surface. Each construct displayed intracellular punctate localization in permeabilized cells. Representative images are shown from experiments that were performed at least three times. (Scale bar: 50 μm.) (B) Model of myomaker topology based on the positive epitopes in live staining. According to this model, myomaker contains seven TM domains with a 25-aa intracellular C-terminal domain.
A model of myomaker topology, in which the regions positive for live staining are exposed to the exterior of the cell, is shown in Fig. 5B. Collectively, these studies support the conclusion that the N terminus of myomaker faces the exterior of the cell and that the C terminus is intracellular. Given that TM domains typically contain 18–20 hydrophobic residues (19), our results predict that myomaker spans the membrane seven times, which is in agreement with many membrane topology prediction software programs. This model does not illuminate the relative lateral membrane distances between the TM regions, which could vary the composition of the hydrophilic loops. It should also be noted that the topology model is based on the orientation within the membrane of epitope-tagged versions of the protein, which could differ in structure from the WT protein.
Other TMEM8 Proteins Lack Fusogenic Activity.
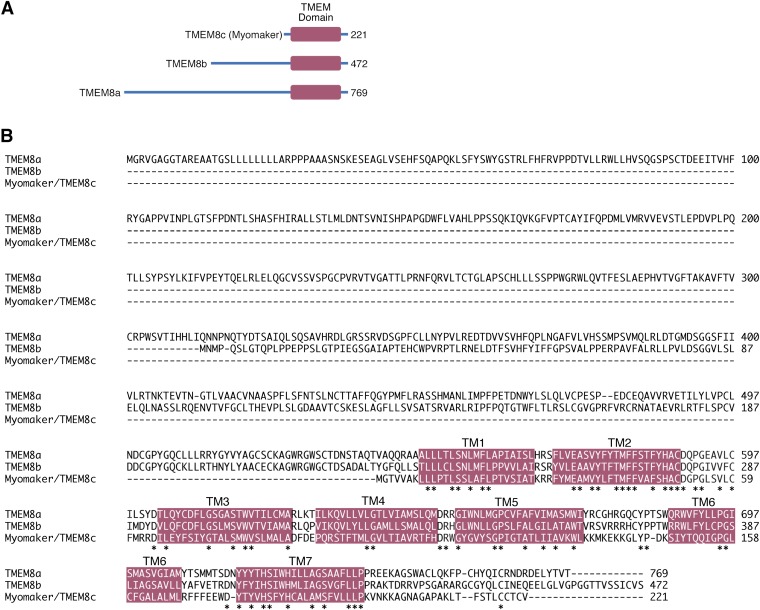
Homology searches using myomaker protein sequence resulted in identification of TMEM8a and TMEM8b, which are both orphan membrane proteins with unknown functions. TMEM8a/TMEM8b possesses much longer N-terminal regions than myomaker, and overall, these proteins are 25% identical using myomaker as a reference (Fig. S3). The majority of homology between myomaker and TMEM8a/TMEM8b occurs in the TM regions (43 of 58 amino acids), with a smaller fraction of similarity in the hydrophilic loops (15 of 58 amino acids) (Fig. S3B).
Fig. S3.
TMEM8a, TMEM8b, and TMEM8c/myomaker sequence comparison. (A) TMEM8a and TMEM8b contain longer N-terminal regions than TMEM8c/myomaker. The number of amino acid residues for each protein is shown. (B) Amino acid alignment between the three TMEM8 family members. Magenta regions depict significant stretches of hydrophobic amino acids. TM refers to TM domains. The majority of homology is localized to the TM domains between the three proteins. *Sequence conservation.
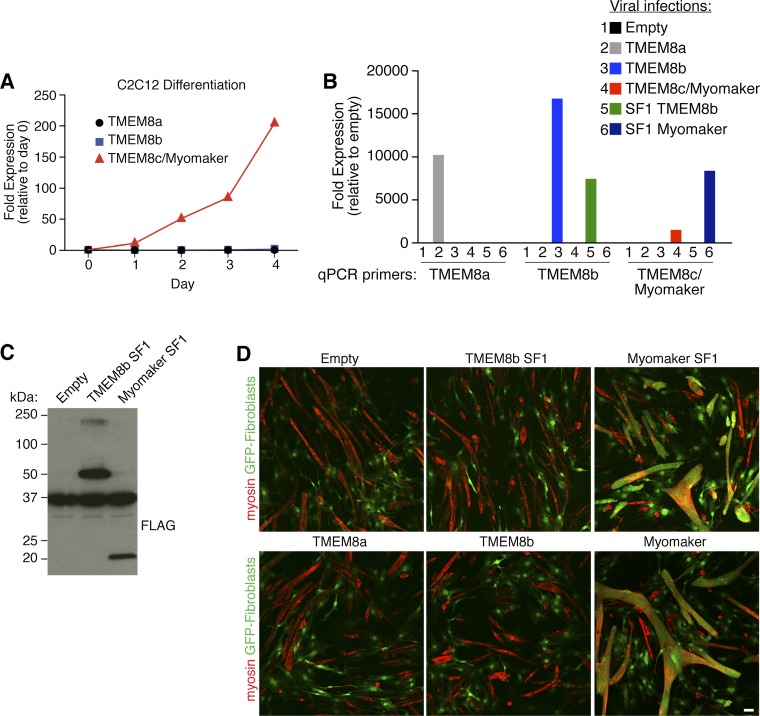
Whereas myomaker is dramatically induced at days 2 and 3 of C2C12 differentiation, TMEM8a and TMEM8b are not regulated in the same manner (Fig. S4A). We generated retroviruses expressing TMEM8a and TMEM8b to test their function in the heterologous cell fusion system. Moreover, we engineered a FLAG-tagged version of TMEM8b (TMEM8b SF1) similar to myomaker SF1, in which a signal sequence was placed upstream of FLAG on the N terminus of the protein. Infection of fibroblasts with each virus followed by quantitative PCR analysis revealed that each was appropriately expressed (Fig. S4B). TMEM8b SF1 protein was also detected by FLAG Western blot analysis, and the levels were similar to those of expression of myomaker SF1 (Fig. S4C). Despite expression of TMEM8a, TMEM8b, or TMEM8b SF1 in fibroblasts, they did not promote fusion of fibroblasts with C2C12 myoblasts (Fig. S4D).
Fig. S4.
TMEM8a and TMEM8b differ functionally from myomaker. (A) Quantitative RT-PCR analysis for TMEM8a, TMEM8b, and TMEM8c/myomaker transcripts in C2C12 cells after the indicated day of differentiation. Expression levels were normalized to 18S and expressed as fold change relative to day 0. (B) Quantitative PCR (qPCR) analysis for TMEM8a, TMEM8b, and TMEM8c/myomaker in fibroblasts after infection with empty, TMEM8a, TMEM8b, TMEM8c/myomaker, SF1 TMEM8b, and SF1 myomaker virus. Expression levels were normalized to 18S and expressed as fold change relative to empty-infected cells. (C) FLAG Western blot analysis shows expression of epitope-tagged versions of TMEM8b and myomaker in fibroblasts. (D) The heterologous cell fusion system reveals that fibroblasts infected with empty, TMEM8a, TMEM8b, or TMEM8b SF1 virus do not fuse to C2C12 cells. Fibroblasts containing myomaker or myomaker SF1 readily fuse. Representative images are shown from experiments that were performed at least three times.
Requirement of Myomaker C-Terminal Region for Function.
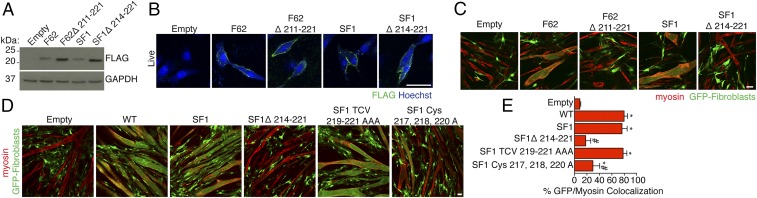
The largest domain of myomaker that is exposed to the extracellular or intracellular space is the C-terminal region. To investigate the role of this region in myomaker function, we deleted the C-terminal domain in the context of two separate myomaker-FLAG constructs. We chose two different FLAG versions because of potential disparities in expression after epitope insertion. Specifically, we generated viruses encoding a truncated myomaker F62 mutant lacking the last 11 amino acids (F62Δ211–221) and an SF1 protein deficient in the final 8 amino acids (SF1Δ214–221). Western blot analysis for FLAG showed that each C-terminal deletion mutant protein was expressed at a higher level than the control FLAG constructs (Fig. 6A). Moreover, live staining of transduced C2C12 cells revealed the presence of both of these deletion mutants on the membrane, suggesting the C-terminal region does not impact membrane localization (Fig. 6B).
Fig. 6.
The intracellular C-terminal region of myomaker is required for fusogenic activity. (A) Viruses expressing two independent C-terminal deletion myomaker-FLAG mutants were generated. The final 11 amino acids of F62 (F62Δ211–221) and the last 8 amino acids of SF1 (SF1Δ214–221) were deleted. Expression of each virus was assessed by FLAG immunoblotting. GAPDH was used as a loading control. (B) Myomaker proteins that contained a shorter C terminus were expressed on the membrane as determined by staining live cells after viral transduction. (C) Fibroblasts that express F62Δ211–221 and SF1Δ214–221 do not fuse to myoblasts, whereas F62 and SF1 exhibit robust fusogenic activity. (D) Constructs containing mutations in the final TCV, a PDZ-binding motif, of myomaker (SF1 TCV 219–221 AAA) and the C-terminal cysteines (SF1 Cys-217, 218, and 220 A) were tested for function in the heterologous fusion system. (E) Quantitation of the fusion between fibroblasts and muscle cells. Representative images are shown from experiments that were performed at least three times. (Scale bar: 50 μm.) *P < 0.05 compared with empty; #P < 0.05 compared with SF1.
We next tested if the C-terminal region was necessary for myomaker fusogenic function. Infection of fibroblasts with SF1 and F62 caused fusion with myoblasts; however, SF1Δ214–221 and F62Δ211–221 lacked similar fusogenic activity (Fig. 6C). The final 8 amino acids of myomaker harbor cysteine residues that resemble a consensus for lipid modification as well as a PDZ-binding motif (TCV). To determine if either of these domains explains the lack of function in the C-terminal mutant, we mutated these regions to alanine. Specifically, we generated SF1 constructs with either a TCV 219–221 AAA mutation or cysteines 217, 218, and 220 mutated to A and tested their ability to confer fusogenic ability to fibroblasts. Mutation of the TCV region did not alter fusion activity; however, the cysteine mutations significantly blunted fusion (Fig. 6 D and E). Our results indicate the intracellular C-terminal region of myomaker is necessary for fusion.
Discussion
The results of this study indicate that the majority of the myomaker protein is embedded in the bilayer, with the N terminus facing the extracellular space and an intracellular C terminus. We also show that human and zebrafish orthologs function similarly to mouse myomaker in a heterologous cell fusion system. In contrast, TMEM8a and TMEM8b do not function like TMEM8c/myomaker. Studies performed in Drosophila indicate that myoblast fusion is an asymmetric process, in which one cell uses actin to invade a neighboring cell, which increases tension to promote fusion (20). We show that myomaker KO myocytes can fuse to WT myocytes, but fusion is less efficient than with pairs of WT myocytes, highlighting the importance of myomaker on both fusing cells. Finally, we show that the intracellular C-terminal region of myomaker is necessary for fusogenic activity, suggesting the importance of signaling or other modes of protein–protein interactions in the control of fusion. Table S1 summarizes the expression, function, and surface staining of the various mutant myomaker constructs on which our model of myomaker topology and function is based.
Table S1.
Properties of myomaker constructs
| Construct | Expression | Fusogenic activity in KO myoblasts | Heterologous fusogenic activity | Epitope detection on surface |
| WT | + | + | + | NT |
| SF1 | + | + | + | + |
| F62 | + | − | + | + |
| F91 | + | − | + | − |
| F112 | + | − | + | + |
| F140 | ++ | − | + | − |
| F174 | + | − | + | + |
| F203 | + | + | + | − |
| CRISPR/Cas9 (Δ77–82) | NT | − | − | NT |
| F62Δ211–221 | + | − | − | + |
| SF1Δ214–221 | + | − | − | + |
| SF1TCV 214–221 AAA | + | − | − | NT |
| SF1Cys 217, 218, and 220 A | + | − | − | NT |
NT, not tested.
The hydrophilic loops that are predicted to connect the TM regions of myomaker are quite small, ranging from 3 to 14 aa. The relatively small number of amino acids that protrude from either the intracellular or extracellular space could provide a clue to the biochemical function of myomaker. For instance, it is unlikely that myomaker functions in the adhesion step of fusion, because it is not predicted to be glycosylated and contains only short extracellular segments (21). Muscle cell fusion has been shown to involve calcium and the formation of a hemifusion intermediate, in which the outer leaflets of the membranes fuse followed by fusion pore formation and cytoplasmic mixing (22–24). Additionally, fusion-competent myocytes display phosphatidylserine in the outer leaflet of the membrane (25). It is conceivable that myomaker functions during one or more of these steps.
Our finding that the C-terminal region of myomaker is oriented toward the interior of the cell and necessary for function suggests that signaling or other protein–protein interactions are important for fusion. Indeed, the C terminus (TCV) conforms to the consensus of a PDZ-binding motif (S/T-X-V/L/I) (26); however, our mutation of the TCV in myomaker does not perturb function. This C-terminal domain also contains three cysteines, which may serve as sites for lipidation, and mutation of the three cysteines was sufficient to diminish activity.
The fusion of muscle cells during the development and regeneration of skeletal muscle is a complex process that is only beginning to be understood. In this work, we have performed initial structure–function analysis of myomaker, a necessary regulator of myoblast fusion, to obtain more information regarding the precise role for this protein in the fusogenic event of myoblasts. Although these analyses revealed an initial structural model of myomaker, eventual determination of the precise structure will be required to define its 3D orientation within the membrane. Elucidation of the biochemical function of myomaker will also be an area of future interest.
Materials and Methods
Cell Culture.
Methods for myoblast isolation and cell culture are further described in SI Materials and Methods.
CRISPR-Mediated Genome Editing in C2C12 Cells.
A guide oligonucleotide corresponding to exon 2 of myomaker was ordered from Integrated DNA Technologies and cloned into the Cas9-containing pX330 vector, which was a gift from Feng Zhang, Massachusetts Institute of Technology, Cambridge, MA (plasmid 42230; Addgene) (27). Methods for myomaker targeting using Cas9 are further described in SI Materials and Methods.
Retroviral Generation and Cell Mixing.
An EcoRI restriction site was used to clone each construct into the retroviral vector pBabe-X, and Platinum E cells were used to generate virus, which is further described in SI Materials and Methods.
RNA FISH.
Primary myoblasts were seeded on fibronectin-coated eight-well Ibidi microslides at a density of 2 × 104 per well. Twenty-four hours after seeding, cultures were placed in differentiation media and assayed for myomaker transcription using probes specific for myomaker introns. Methods for myomaker RNA in situ are further described in SI Materials and Methods.
Quantitation of Heterologous Fusion Using CellProfiler.
Image analysis and quantification were performed on immunofluorescence images using the open-source software CellProfiler (www.cellprofiler.org) (18). Analysis pipelines were designed as follows. The GFP-colored region (corresponding to fibroblasts) and the myosin-stained region (myoblasts) were identified from each image and quantified on the basis of the total pixel area occupied by the GFP fluorescence or the myosin stain as determined by automatic image thresholding. The colocalized region of GFP fluorescence and myosin stain was identified and quantified in each image automatically by the MaskObjects module. The pixel area of the colocalized region was normalized by the total myosin area and defined as the heterologous fusion efficiency (percentage of colocalization).
Immunocytochemistry.
Methods for immunocytochemistry are described in SI Materials and Methods.
SI Materials and Methods
Cell Culture and Primary Myoblast Isolation.
C2C12 cells were purchased from American Type Culture Collection and propagated in DMEM (Sigma) containing 10% heat-inactivated FBS and supplemented with antibiotics; 10T1/2 fibroblasts were also purchased from American Type Culture Collection and passaged in the same media. C2C12 cells were differentiated by switching to media containing 2% heat-inactivated horse serum and antibiotics. All solutions described as a percentage are based on vol/vol. Primary myoblasts were isolated from myomakerLacZ/loxP;Pax7-CreERT2 mice after treatment with vehicle (WT) or five injections of tamoxifen (KO) at a dose of 0.075 mg/g body weight. After dosing with vehicle or tamoxifen, the gastrocnemius and tibialis anterior were injured with cardiotoxin (10 μM; Sigma). Three days after injury, muscles were harvested and placed into gentleMACS C Tubes (Miltenyi Biotec) containing 2% collagenase (Type I; Invitrogen) in low-glucose DMEM (Invitrogen). To dissociate muscle, the C tubes were placed on a Miltenyi GentleMACS Dissociator and subjected to two cycles of digestion. Dispase (4.8 units/mL; Roche) was added to the digested muscle, incubated at 37 °C for 2 h, triturated using a pipette and 20-gauge needle, passed through a 40-μm filter, and centrifuged. The mononuclear cells were preplated for 20 min to remove nonmyoblast populations and then, plated on a collagen-coated plate in growth media (20% FBS/Ham F10, 2.5 ng/mL bFGF; Invitrogen). To enrich for myoblasts, cultures were incubated in a small volume of PBS, and the myoblasts were dislodged by knocking the plate lightly. To induce myogenesis, the cultures were placed in differentiation media (2% horse serum, DMEM).
CRISPR-Mediated Genome Editing in C2C12 Cells.
Freshly plated low-passage C2C12 cells were transfected with 4 μg pX330 (with myomaker guide RNA) and 4 μg pcDNA3-EGFP using 16 μL Lipofectamine 2000. We transfected 1 × 106 C2C12 cells in a 60-mm culture dish, and the medium was changed 16 h posttransfection. Forty-eight hours after transfection, we sorted single GFP+ cells into 96-well plates using FACS. These cells were maintained in DMEM containing 20% FBS with antibiotics at subconfluent densities. The cell lines were genotyped by amplifying a 435-bp region surrounding the predicted site of Cas9 activity using the following primers: F: 5′-CAGTGCTGAGTCTGAAAGGGATA-3′ and R: 5′-TAAGCTCTGCAAAGCAGCAAG-3′. This PCR product was then digested with NspI to genotype for editing of exon 2 of myomaker. An amplicon from a WT clone (NspI site maintained) exhibited 317- and 118-bp bands after digestion, and an amplicon from a KO clone (NspI site disrupted) remained at 435 bp.
Cloning and Viral Infection.
Each myomaker-FLAG construct was generated by independently cloning the regions immediately upstream (5′ PCR product) and downstream (3′ PCR product) of the site of insertion and then, using these products in a standard PCR sewing reaction to generate full-length myomaker-FLAG plasmids. The FLAG nucleotide sequence used was GATTACAAGGATGACGACGATAAG, and for each internal FLAG construct, we engineered a glycine–glycine–serine linker on both sides of the epitope to aid in protein folding. The signal sequence used upstream of FLAG was ATGAAGACGATCATCGCCCTGAGCTACATCTTCTGCCTGGTGTTCGCC as previously described (27). TMEM8a was cloned from adult kidney cDNA, and TMEM8b was cloned from adult brain cDNA using the following primers: TMEM8a-F: 5′-ATGGGCCGGGTTGGGGCCGGG-3′ and TMEM8a-R: 5′-TCAGGTCACTGTGTACAACTC-3′; and TMEM8b-F: 5′-ATGAACATGCCCCAGTCACTA-3′ and TMEM8b-R: 5′-TCAGCTGACACAGATGCTGC-3′. Human and zebrafish myomaker cDNAs were synthesized by GenScript and cloned into pBabe-X.
RNA Analysis.
Total RNA was extracted from either mouse tissue or cultured cells with TRIZOL (Invitrogen) and cDNA-synthesized using SuperScript III Reverse Transcriptase with random hexamer primers (Invitrogen). Gene expression was assessed using standard quantitative PCR approaches with either Power Sybr Green or Taqman Master Mix (Applied Biosystems). Analysis was performed on a 7900HT Fast Real-Time PCR Machine (Applied Biosystems) with the following Sybr primers: Myomaker-F: 5′-ATCGCTACCAAGAGGCGTT-3′ and Myomaker-R: 5′-CACAGCACAGACAAACCAGG-3′; Tmem8a-F: 5′-CCAGCAACAGCAAGGAGAG-3′ and Tmem8a-R: 5′-GCTGCCATACCAGCTGTAGA-3′; and Tmem8b-F: 5′-TTAATGTCCGTGTGGGTCAC-3′ and Tmem8b-R: 5′-CAGAGCCATAGACAGCAGCA-3′. As an internal control, 18S was used.
Retroviral Generation and Cell Mixing.
Ten micrograms of retroviral plasmid DNA was transfected using FuGENE 6 (Roche) into Platinum E Cells (Cell Biolabs), which were plated on a 100-mm culture dish at a density of 3 × 106 cells per dish 24 h before transfection. Forty-eight hours after transfection, viral media were collected, filtered through a 0.45-mm cellulose filter, and mixed with Polybrene (Sigma) at a final concentration of 6 μg/mL. For live staining experiments, C2C12 cells were plated on 35-mm culture dishes at a density of 1 × 105 cells per dish 24 h before infection with 3 mL viral media. Eighteen hours after infection, virus was removed, and cells were washed with PBS, which was replaced with differentiation media; 10T1/2 fibroblasts (∼50–60% confluent on a 100-mm plate) were infected with 10 mL virus for 18 h. After infection, cells were washed three times, trypsinized, mixed with C2C12 cells at a 1:1 ratio (1.5 × 105 each cell type), and plated on a 35-mm dish in 10% FBS and DMEM. The cultures were differentiated the next day, and GFP and myosin expression were analyzed 4 d after differentiation.
RNA FISH.
To detect myomaker expression at all stages of cell differentiation, cultures were fixed (10 min at room temperature in 3.7% formaldehyde in PBS) at 24 h after seeding (day 0) and then, at 24-h intervals (days 1–3) followed by RNA FISH. After fixation, cells were washed twice with PBS and permeabilized in 70% ethanol for at least 1 h at 4 °C. Cells were subsequently incubated in Buffer A (2× SSC, 10% formamide) at room temperature for 5 min and then, hybridized with 125 nM probe in hybridization buffer (2× SSC, 10% formamide, 10% dextran sulfate) in humidified dark chambers at 37 °C for 16 h. After hybridization, cells were incubated in Buffer A for 30 min at 37 °C. Nuclei were stained with 6 ng/mL Hoechst in Buffer A for 30 min at 37 °C. Cells were subsequently incubated in 2× SSC at room temperature. Images were acquired within 5 h posthybridization with Nikon A1R+ LUNV on a Ti-E Inverted Microscope. A set of probes specific to myomaker introns with Quasar 570 Dye was designed using Stellaris Probe Designer, version 4.1 (Table S2). For GFP RNA FISH, commercially available Stellaris FISH Probe eGFP with Quasar 570 Dye was used.
Table S2.
Probes used for RNA FISH detection of myomaker introns
| Probe | Sequence |
| 1 | acaggacaatggtggtaacc |
| 2 | caaatcctgtttctgcatca |
| 3 | cagctgagcaactcatggaa |
| 4 | agtcaggaagacacagagct |
| 5 | aagagacagtgtgactcctg |
| 6 | ccagacaaactagtgaggca |
| 7 | aagtttggctctgctctaag |
| 8 | aaggagaggatgctagtctc |
| 9 | tggataccttcacatactct |
| 10 | gagcacagatgtgtgagttt |
| 11 | ctcaggatatggaacacgca |
| 12 | cactagaaagtcatgccagc |
| 13 | cacaaccaggaggtagacag |
| 14 | aagagccatagagtttccag |
| 15 | gtgacttatcattggcctaa |
| 16 | gtgatttgctggactgtatc |
| 17 | ctggtagttgtcatggagag |
| 18 | tcattcttctattcagcagc |
| 19 | gctgtgtaactgacaccaag |
| 20 | gttagaacaggagagggcag |
| 21 | ttgtgctgttcagcaagtag |
| 22 | atttgtgtgctactgagtca |
| 23 | agcaaaatgtgggtgtaggc |
| 24 | ttagctcacctctgggaaaa |
| 25 | tgtgcgttatgggcatttaa |
| 26 | attaatcaaactttccccca |
| 27 | accccacactgcaaaaatga |
| 28 | caaggggaccctaacgataa |
| 29 | catgagaacacctgttctgt |
| 30 | acctgtgtgacagtgaactt |
| 31 | ggctttctaacaagggatct |
| 32 | ttactgtgagatcctgggaa |
| 33 | aaaggaactcagctctggga |
| 34 | aagacaagctttgggttggg |
| 35 | ccaactaggtatgagctcag |
| 36 | acttagaagctcagagacct |
| 37 | cctggatgatggatgtatca |
| 38 | ctggaaaacagccttgtctg |
| 39 | gcatccttaaagaccactta |
| 40 | ggtgagaagcatgctcagaa |
| 41 | aaggagggaggatctcagag |
| 42 | gtgcatgtctctttaagcag |
| 43 | tggaaatgagagtgcaggca |
| 44 | atacacagagctgagctgac |
| 45 | attttgaatggtctgccttc |
| 46 | aactggtagactgagtgacc |
| 47 | ctattggtatgtcctctcaa |
| 48 | agaggcatagctatggtgag |
Western Blot Analysis.
Cells were washed two times with ice-cold PBS and scraped into an Eppendorf tube. The cells were pelleted and resuspended in lysis buffer (50 mM Tris⋅Cl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% Nonidet P-40, 0.1% SDS). Protein concentration was determined. and equal amounts (50 μg) were prepared with loading buffer [1× Laemmli (Bio-Rad) with 5% β-mercaptoethanol]. Samples were incubated at room temperature for 5 min (not boiled) and separated by 12% SDS/PAGE. After SDS/PAGE, the gel was transferred to a PVDF membrane Millipore), blocked in 5% milk in Tris-buffered saline (TBS)-Tween, and incubated with M2 FLAG antibody (1:5,000; Sigma). GAPDH (1:10,000; Millipore) was used as a loading control. We used a rabbit anti-mouse secondary antibody conjugated to HRP for detection.
Immunocytochemistry.
Immunocytochemistry was performed by fixing cells with 4% paraformaldehyde (PFA)/PBS, permeabilization with 0.2% Triton X-100 in PBS, blocking with 3% BSA/PBS, incubation with primary antibody for at least 2 h, and incubation with Alexa-Fluor secondary antibodies for 1 h. We used anti-mouse M2 Flag antibody (Sigma) at 1:500 and myosin antibody (MY32; Sigma) at 1:200. Nuclei were stained with Hoechst (Invitrogen). For staining of live cells, we first washed the cells with PBS and incubated in blocking buffer (3% BSA/PBS) for 15 min. Primary antibody incubation was then performed on ice followed by fixation with 4% PFA/PBS and incubation with secondary antibody. These cultures were visualized on a Zeiss LSM 780 Confocal Microscope. ImageJ was used to merge images.
Statistical Analyses.
All data are presented as means with SEMs and were performed in replicates multiple times as indicated in the figures. P values were calculated using unpaired t test using GraphPad Prism 6 software. P < 0.05 was considered significant.
Acknowledgments
We thank J. Cabrera for graphical assistance, A. Fernandez-Perez for technical assistance, and D. Rosenbaum for helpful suggestions. D.P.M. was funded by a Development Grant from the Muscular Dystrophy Association. This work was supported by grants from the Cincinnati Children’s Trustee Program (to D.P.M.); NIH Grants HL-077439, HL-111665, HL-093039, DK-099653, and U01-HL-100401; and Robert A. Welch Foundation Grant 1-0025 (to E.N.O.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600101113/-/DCSupplemental.
References
- 1.Chen EH, Olson EN. Unveiling the mechanisms of cell-cell fusion. Science. 2005;308(5720):369–373. doi: 10.1126/science.1104799. [DOI] [PubMed] [Google Scholar]
- 2.Abmayr SM, Pavlath GK. Myoblast fusion: Lessons from flies and mice. Development. 2012;139(4):641–656. doi: 10.1242/dev.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sens KL, et al. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. J Cell Biol. 2010;191(5):1013–1027. doi: 10.1083/jcb.201006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhanyasi N, et al. Surface apposition and multiple cell contacts promote myoblast fusion in Drosophila flight muscles. J Cell Biol. 2015;211(1):191–203. doi: 10.1083/jcb.201503005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laurin M, et al. The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proc Natl Acad Sci USA. 2008;105(40):15446–15451. doi: 10.1073/pnas.0805546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasyutina E, Martarelli B, Brakebusch C, Wende H, Birchmeier C. The small G-proteins Rac1 and Cdc42 are essential for myoblast fusion in the mouse. Proc Natl Acad Sci USA. 2009;106(22):8935–8940. doi: 10.1073/pnas.0902501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruenbaum-Cohen Y, et al. The actin regulator N-WASp is required for muscle-cell fusion in mice. Proc Natl Acad Sci USA. 2012;109(28):11211–11216. doi: 10.1073/pnas.1116065109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simionescu-Bankston A, et al. The N-BAR domain protein, Bin3, regulates Rac1- and Cdc42-dependent processes in myogenesis. Dev Biol. 2013;382(1):160–171. doi: 10.1016/j.ydbio.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavlath GK, Horsley V. Cell fusion in skeletal muscle--central role of NFATC2 in regulating muscle cell size. Cell Cycle. 2003;2(5):420–423. [PubMed] [Google Scholar]
- 10.Horsley V, Jansen KM, Mills ST, Pavlath GK. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 2003;113(4):483–494. doi: 10.1016/s0092-8674(03)00319-2. [DOI] [PubMed] [Google Scholar]
- 11.Quach NL, Biressi S, Reichardt LF, Keller C, Rando TA. Focal adhesion kinase signaling regulates the expression of caveolin 3 and beta1 integrin, genes essential for normal myoblast fusion. Mol Biol Cell. 2009;20(14):3422–3435. doi: 10.1091/mbc.E09-02-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwander M, et al. Beta1 integrins regulate myoblast fusion and sarcomere assembly. Dev Cell. 2003;4(5):673–685. doi: 10.1016/s1534-5807(03)00118-7. [DOI] [PubMed] [Google Scholar]
- 13.Doherty KR, et al. Normal myoblast fusion requires myoferlin. Development. 2005;132(24):5565–5575. doi: 10.1242/dev.02155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millay DP, et al. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature. 2013;499(7458):301–305. doi: 10.1038/nature12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millay DP, Sutherland LB, Bassel-Duby R, Olson EN. Myomaker is essential for muscle regeneration. Genes Dev. 2014;28(15):1641–1646. doi: 10.1101/gad.247205.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landemaine A, Rescan PY, Gabillard JC. Myomaker mediates fusion of fast myocytes in zebrafish embryos. Biochem Biophys Res Commun. 2014;451(4):480–484. doi: 10.1016/j.bbrc.2014.07.093. [DOI] [PubMed] [Google Scholar]
- 18.Kamentsky L, et al. Improved structure, function and compatibility for CellProfiler: Modular high-throughput image analysis software. Bioinformatics. 2011;27(8):1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 20.Kim JH, et al. Mechanical tension drives cell membrane fusion. Dev Cell. 2015;32(5):561–573. doi: 10.1016/j.devcel.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Przewoźniak M, et al. Adhesion proteins--an impact on skeletal myoblast differentiation. PLoS One. 2013;8(5):e61760. doi: 10.1371/journal.pone.0061760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu JH, et al. Acceleration of human myoblast fusion by depolarization: Graded Ca2+ signals involved. Development. 2003;130(15):3437–3446. doi: 10.1242/dev.00562. [DOI] [PubMed] [Google Scholar]
- 23.Leikina E, et al. Extracellular annexins and dynamin are important for sequential steps in myoblast fusion. J Cell Biol. 2013;200(1):109–123. doi: 10.1083/jcb.201207012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shainberg A, Yagil G, Yaffe D. Control of myogenesis in vitro by Ca 2 + concentration in nutritional medium. Exp Cell Res. 1969;58(1):163–167. doi: 10.1016/0014-4827(69)90127-x. [DOI] [PubMed] [Google Scholar]
- 25.van den Eijnde SM, et al. Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation. J Cell Sci. 2001;114(Pt 20):3631–3642. doi: 10.1242/jcs.114.20.3631. [DOI] [PubMed] [Google Scholar]
- 26.Songyang Z, et al. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275(5296):73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 27.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]